Abstract
Gene transfer vectors containing adenovirus (Ad) serotype 35 (Ad35) fibers have shown promise for cancer and stem cell gene therapy. In this study, we attempted to improve the in vitro and in vivo infection properties of these vectors by increasing their affinity to the Ad35 fiber receptor CD46. We constructed Ad vectors containing either the wild-type Ad35 fiber knob (Ad5/35) or Ad35 knob mutants with 4-fold- and 60-fold-higher affinity to CD46 (Ad5/35+ and Ad5/35++, respectively). In in vitro studies with cell lines, the higher affinities of Ad5/35+ and Ad5/35++ to CD46 did not translate into correspondingly higher transduction efficiencies, regardless of the CD46 receptor density present on cells. However, in vivo, in a mouse model with preestablished CD46high liver metastases, intravenous injection of Ad5/35++ resulted in more-efficient tumor cell transduction. We conclude that Ad5/35 vectors with increased affinity to CD46 have an advantage in competing with non-CD46-mediated sequestration of vector particles after intravenous injection.
Human adenoviruses (Ads) have been classified into six species (A to F) containing 51 defined serotypes. Most Ad serotypes utilize the coxsackievirus Ad receptor (CAR) as a primary attachment receptor. Recently, we suggested a new grouping of species B Ads based on their receptor usage (32). Group 1 (Ad serotype 16 [Ad16], Ad21, Ad35, and Ad50) nearly exclusively utilize CD46 as a cellular receptor; group 2 (Ad3, Ad7, and Ad14) share the same nonidentified receptor, which we refer to as receptor X; group 3 (Ad11p) preferentially interacts with CD46 but also utilizes receptor X if CD46 is blocked (32).
Recent studies performed in U.S. military training facilities indicate an emergence of diverse species B serotypes at the majority of sites. This included the group 1 serotype 21 and the group 2 serotypes 3, 7, and 14 (17, 39). Species B-derived, replication-deficient vectors have recently shown promise as vehicles for gene transfer into multiple human cell types including cancer cells and tissue stem cells (30, 35).
Because of the importance of species B Ads as pathogens and application of species B-derived vectors for gene transfer, we studied the interaction between group 2 Ad35 and CD46 in more detail. Ad35 engages CD46 via residues in the C-terminal trimeric fiber knob domain (7). Within CD46 the Ad35-interacting areas are located in the two distal extracellular domains of the receptor (6, 8). More recently, we used an expression library of the Ad35 fiber knob with random mutations to identify the amino acid residues within the Ad35 knob that mediate binding to CD46. We identified four critical residues (Phe242, Arg279, Ser282, and Glu302) which, when mutated, ablated Ad35 knob binding to CD46 without affecting knob trimerization (36).
In the present study we used the same Ad35 expression library to screen for Ad35 knob mutants with increased binding to CD46 compared to the wild-type Ad35 knob. Our goal was to construct Ad vectors with substantially increased affinity for CD46. The rationale for such vectors comes from studies with phage antibody expression libraries (33) and more recently from studies with aptamers, protein-binding oligonucleotides (23). The goal of phage and aptamer library screening is to identify variants with the highest affinity, because in in vitro and in vivo studies with single-chain variable fragment fragments and aptamers, higher affinity usually directly translates into more-efficient binding to receptor-positive cells. Along this line, attempts were undertaken to incorporate high-affinity ligands into measles virus (9) and Ad vectors (2, 3, 40) in order to increase efficacy and specificity of target cell infection in vivo or to establish new receptor-ligand systems for the propagation of vectors.
In this study we created Ad35 fiber containing vectors with affinities for CD46 that were 4- and 60-fold higher than that of a vector with wild-type Ad35 fibers. For in vitro studies higher affinity did not result in correspondingly higher transduction rates. However, we found that a vector with a 60-fold higher affinity was superior in transducing CD46high liver metastases after intravenous injection into mice.
MATERIALS AND METHODS
Cell lines.
Dulbecco's modified Eagle's medium (DMEM) and RPMI 1640 were purchased from GIBCO-BRL (Gaithersburg, MD). Fetal calf serum (FCS) was from HyClone (Logan, UT). 293 cells, HeLa cells, and CHO-CD46 cells were maintained in DMEM supplemented with 10% FCS, 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 1× MEM-nonessential-amino-acid solution (Invitrogen, Carlsbad, CA). CHO-CD46 cells (1) are described elsewhere and were grown in 1 mg/ml G418. K562, MO7e, and Ramos cells were maintained in RPMI 1640 medium containing 10% FCS, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. For MO7e cells, 0.1 ng/ml of granulocyte-macrophage colony-stimulating factor (Amgen, Seattle, WA) was added to the medium. For Ramos cells, medium was supplemented with 10 mM HEPES and 1 mM sodium pyruvate.
Screening of Ad35 knob library for CD46 binding.
Random mutagenic PCR was performed on the fiber knob sequence based on a protocol published earlier (37). The mutant PCR products (645 bp) were purified, digested with appropriate enzymes, and cloned into the pQE30 vector (Qiagen, Valencia, CA). The Ad35 knob mutant plasmid library was transformed into M15 host strains and plated on Luria-Bertani agar plates (master plate) with kanamycin and ampicillin. About 800 to ∼1,000 colonies per 15-cm petri dish were grown. After overnight growth, a 0.45-μm-pore-size Durapore filter membrane (Millipore, Billerica, MA) was placed on top of the colonies. The membrane was peeled off and placed carefully, with the colonies facing upwards, on two sheets of Whatman 3MM paper soaked in Luria-Bertani medium supplemented with antibiotics and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Protein expression of the colonies was induced for 20 min at room temperature, after which the filter with the colonies was placed on top of a nitrocellulose filter and a Whatman 3MM paper soaked in native lysis buffer (20 mM Tris-Cl, pH 8, 300 mM NaCl, 50 mM MgCl2, 0.1 mg/ml lysozyme, 0.75 mg/ml DNase I, and one-half of a complete EDTA-free protease inhibitor cocktail tablet/10 ml [Roche, Palo Alto, CA]). The “filter sandwich” was freeze-thawed four times for 10 min at −80°C and 10 min at 30°C. The nitrocellulose membrane was removed from the sandwich and blocked with 3% bovine serum albumin in TBS (10 mM Tris-Cl, pH 7.5, 150 mM NaCl) at 4°C overnight. The membranes were then incubated with soluble recombinant CD46 (sCD46) (37) in TBS and 3% milk for 1 h at room temperature (RT) and then washed three times for 10 min in TBS-0.05% Tween 20 (TBS-T) buffer. The blot was then incubated with anti-CD46 antibody (clone J4.48; Fitzgerald, Concord, MA) (1:50 dilution) in TBS and 3% milk for 1 h at RT and then washed three times for 10 min in TBS-T buffer. To visualize binding, the blot was incubated with goat anti-mouse immunoglobulin (Ig)-horseradish peroxidase (BD Pharmingen, San Jose, CA) (1:1,000 dilution) in TBS and 3% milk for 1 h at RT, washed, and subjected to ECL substrate (Pierce, Rockford, IL). Colonies that showed binding to CD46 were picked from the original master plates. DNA from these colonies was sequenced, and binding of purified knobs to CD46 was verified by Western blot analysis as described earlier (37).
Construction of chimeric Ad vectors.
All Ad vectors contained a 2.3-kb cytomegalovirus promoter-driven enhanced green fluorescent protein (EGFP) gene (derived from pEGFP-1) (Clontech, Palo Alto, CA) inserted into the E3 region of Ad5. The chimeric Ad5/35 vector was generated earlier and described elsewhere (29). To generate Ad5/35+ and Ad5/35++, the corresponding shuttle plasmids pAdGFP/F35+ and pAdGFP/F35++ were constructed based on pAdGFP/F35 (29). A 1,463-bp KpnI/MscI fragment of pAdGFP/F35 containing most of the knob sequence was generated by two-step PCR amplification using Pfu-Turbo DNA polymerase (Stratagene). The following primers and template were used in the first PCR step: (i) P1, 5′-GATGACACTGATGGTACCTTACAAGAAAACATACGTGC-3′, and P1-R, 5′-GTGTCTGATACACCAACTAGAGACACGTAGCC-3′, using pAdGFP/F35 as the template; (ii) P2, 5′-GGCTACGTGTCTCTAGTTGGTGTATCAGACAC-3′, and P2-R(HindIII), 5′-TGAAACATAACACAAAGCTTTCTTTATTCTTGGGCATTTTAGTTGTCGT CTTCTGTAATG-3′, using the pQE30-Ad35 plasmid encoding mutations Asp207Gly and Thr245Ala, or P2 and P2-R(XhoI), 5′-TGAAACATAACACACTCGAGTCTTTATTCTTGGGCATTTTAGTTGTCGTCTTCTGTAATG-3′, using the pQE30-Ad35 plasmid encoding mutations Asn217Asp, Thr245Pro, and Ile256Leu; (iii) P3(HindIII), 5′-CATTACAGAAGACGACAACTAAAATGCCCAAGAATAAAGAAAGCTTTGTGTTATGTTTCA-3′, or P3(XhoI), 5′-CATTACAGAAGACGACAACTAAAATGCCCAAGAATAAAGACTCGAGTG TGTTATGTTTCA-3′, and P3-R, 5′-TTGTGGTATGATGGCCACGTGGGTTCTGTGGTCCC-3′, using pAdGFP/F35 as the template. XhoI or HindIII restriction sites were introduced after the fiber stop codon for Ad5/35+ or Ad5/35++, respectively. After the first amplification, PCR products were purified by agarose gel electrophoresis, combined, and then subjected to a second PCR with primers P1 and P3-R. This new 1,463-bp KpnI/MscI fragment was cloned into pAdGFP/F35 to generate pAdGFP/F35+ and pAdGFP/F35++. To generate full-length E1/E3 vector genomes, pAdGFP/F35+ and pAdGFP/F35++ were inserted in pAdHM4 (29) by homologous recombination in Escherichia coli BJ1583 (4). The resulting plasmids pAd5GFP/F35+ and pAd5GFP/F35++ were analyzed by restriction analysis and sequencing. To produce the corresponding viruses, pAd5GFP/F35+ and pAd5GFP/F35++ were digested with PacI to release the viral genomes and transfected into 293 cells as described elsewhere (29). Recombinant viruses were propagated in 293 cells and purified by standard methods. Purified Ads were titered by optical density at 260 nm assay and plaque assay. Ad plaque titers were determined as follows. Confluent low-passage-number 293 cells plated in six-well plates were incubated for 24 h with different dilutions of virus in a total volume of 1 ml. Two weeks after infection, GFP-positive plaques on cultures overlaid with 0.5% agarose-minimal essential medium-10% FCS the day after infection were counted.
Surface plasmon resonance (SPR) analyses.
Biacore analysis of the recombinant knob-CD46 interaction was described previously (36). For analysis of virus binding, Ad5/35, Ad5/35+, and Ad5/35++ were biotinylated using an EZ-Link Sulfo-NHS-LC-Biotin kit (Pierce, Rockford, IL) according to the manufacturer's instructions. The biotinylated products were coupled to streptavidin-coated sensor chips (SA; Biacore) by manual injection until the required values for resonance units (RU) bound were obtained (Ad5/35, 206 RU; Ad5/35+, 314 RU; Ad5/35++, 307 RU). All analyses were carried out on a Biacore T100 instrument at a flow rate of 30 μl/ml in HBS-EP (Biacore). The sensor chip surface was regenerated after each cycle by injection of 10 mM glycine-HCl, pH 3. Data processing and kinetic analysis were performed using Biacore T100 evaluation software.
In vitro transduction studies.
For adherent cells, the day before infection, 3 × 105 cells per well (24-well plate) were seeded. The next day, attached cells were counted and viruses were added at the multiplicities of infection (MOI) indicated in the figure legends in 1 ml of growth medium. Cells were incubated with virus for 24 h. MO7e, K562, and Ramos cells growing in suspension were washed once with phosphate-buffered saline (PBS) before infection and resuspended in growth medium at a concentration of 3 × 105 cells per well (12-well plate). Virus at the MOI indicated in the figure legends was added to a total volume of 1 ml and incubated for 24 h. Percentages of GFP-positive cells and mean fluorescence were determined by flow cytometry.
Attachment assay.
Corresponding Ads were propagated in 293 cells, labeled with [methyl-3H]thymidine as described before (29). A total of 3 × 105 MO7e cells per tube were resuspended in 150 μl of ice-cold adhesion buffer (DMEM supplemented with 2 mM MgCl2, 1% FCS, and 20 mM HEPES) containing 3H-labeled virus at the MOI indicated in the figure legends. After 1 h of incubation on ice, cells were pelleted and washed twice with 0.5 ml of ice-cold PBS. After the last wash, the supernatant was removed, and the cell-associated radioactivity was determined with a scintillation counter. The number of virus particles (VP) bound per cell was calculated by using the virion-specific radioactivity and the number of cells. For antibody competition, 10 μg/ml anti-human CD46 antibody (clone MEM-258; Serotec Ltd., Oxford, United Kingdom) in 50 μl adhesion buffer was added and allowed to attach for 45 min at 4°C before 3H-labeled virus was added to a final volume of 150 μl.
Analysis of Ad trafficking.
To analyze intracellular trafficking of Ad vectors, MO7e cells (3 ×105 cells per tube) were washed with cold PBS, incubated on ice for 30 min, and infected with 4,000 VP per cell in a total volume of 100 μl. After incubation on ice for 45 min, cells were washed twice with cold PBS to remove virus-containing medium and incubated in growth medium at 37°C for 15 min or 4 h before fixation in a methanol-acetone mixture (1:1, vol/vol). For analysis of Ad attachment, cells were immediately fixed after removal of virus-containing medium. For analysis of Ad particle localization to late endosomal or lysosomal compartments, fixed cells were incubated with primary polyclonal rabbit anti-cathepsin B antibody (Ab-3) (1/40 dilution; Oncogene, Boston, MA) for 1 h at 37°C. The binding of primary antibody was developed with a 1/200 dilution of Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (Molecular Probes, Eugene, OR) at RT for 30 min. To analyze colocalization of Ad particles and CD46, the same procedure was used, except that the rabbit anti-cathepsin B antibody was replaced by mouse anti-human CD46 (1:100 dilution; clone J4.48; Fitzgerald, Concord, MA) followed by Cy2-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., Western Grove, PA). To visualize cell nuclei, samples were stained with DAPI (4′,6-diamidino-2-phenylindole) nuclear staining. The cells were visualized on a Leica DFC300 FX fluorescence microscope with a 100× oil lens and on a Zeiss confocal microscope. Multiple images for each experimental condition were obtained, and representative pictures are shown.
Electron microscopy studies.
For analysis of virus distribution, MO7e cells were washed and infected with 3,000 VP per cell in a total volume of 100 μl for 45 min on ice. After a washing, cells were incubated in growth medium at 37°C for 4 h. Then, cells were subsequently fixed with 2% glutaraldehyde in PBS and with 1% OsO4-phosphate buffer. Cells were then embedded in Medcast (Ted Pella, Redding, CA), and ultrathin sections were stained with uranyl acetate and lead citrate. Processed grids were evaluated and photomicrographed with a Phillips 410 electron microscope operated at 80 kV (magnification, ×21,000). For each Ad vector, the intracellular distribution was analyzed by counting at least 100 virus-containing cells.
Surface CD46 levels.
MO7e cells were washed with cold PBS and incubated on ice for 30 min; then, 3 × 105 cells/well were resuspended in a 12-well plate and infected with Ad vectors at the MOI indicated in the figure legends in a volume of 1 ml. After incubation for 2 h, 4 h, 6 h, 12 h, and 24 h at 37°C, cells were washed once with cold PBS containing 1% fetal bovine serum (FBS) and incubated with phycoerythrin (PE)-conjugated anti-human CD46 antibody (clone E4.3; Santa Cruz Biotechnology Inc., Santa Cruz, CA), diluted 1:20, in a 100-μl volume for 45 min on ice. Cells were then washed three times with cold PBS containing 1% FBS, and the PE mean fluorescence intensity and GFP fluorescence were analyzed by flow cytometry immediately. To analyze the data, PE mean fluorescence intensity of uninfected cells (MOI, 0) was set as 100%. For analysis at time zero, cells were incubated with virus on ice for 1 hour after infection.
In vivo studies.
All experiments involving animals were conducted in accordance with the institutional guidelines set forth by the University of Washington. All mice were housed in specific-pathogen-free facilities. CD46 transgenic C57BL/6 mouse line MCP8B (C57-CD46) mice were generously provided by Branka Horvat (INSERM, Paris, France) (16). (This line has been crossed into the C57BL/6 background for nine generations.) These mice express CD46 at levels similar to those at which it is expressed by human cells. The transgene is a CD46 C1 isoform under the control of the ubiquitously active hydroxymethyl-glutaryl coenzyme A reductase promoter. In these mice CD46 protein expression was on nonlymphoid tissues as well as on activated T and B lymphocytes, macrophages (10), and dendritic cells generated either from CD34+ bone marrow cells or from CD14+ peripheral blood monocytes (5). CD46 DNA-positive mice were identified by PCR of tail DNA using the following primers: F-CD46 (5′-GGTCAAATGTCGATTTCCAGT-3′) and R-CD46 (5′-AATCACAGCAATGACCCAAA-3′). To establish mouse models with liver metastases, animals were infused with 2 × 106 TC1-CD46 cells through a permanently placed portal vein catheter as described elsewhere (19). For intravenous application, Ad vectors in 100 μl of PBS were injected through the tail vein. For analysis of Ad transduction, 3 days after Ad injection, liver metastases were microdissected and digested with 1 mg/ml collagenase (Roche, Mannheim, Germany) plus 0.1 mM CaCl2 in RPMI 1640 for 2 h at 37°C. This was followed by incubation with versene (1:1, vol/vol) for 1 h. Protease digestion was stopped by adding FBS to a final concentration of 10%. Cells were pelleted, resuspended in 2 ml RPMI 1640-10% FBS-1 mg/ml DNase I, and incubated for 30 min at 37°C. After a washing, tumor cells were subjected to flow cytometry analysis for GFP fluorescence and CD46 expression using an anti-CD46 antibody (clone E4.3) conjugated to PE (Santa Cruz Biotechnology, Santa Cruz, CA).
RESULTS
Fiber knobs with increased avidity for CD46.
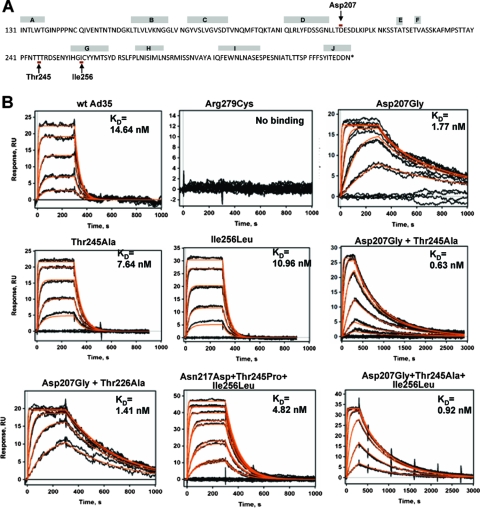
We generated a library of Ad35 knob mutants expressed in E. coli strain M15. The conditions for mutagenic PCR were optimized to achieve on average one or two amino acid mutations per knob (36). Bacteria transformed with the knob library were plated on agar plates, and knob expression was induced by IPTG. Bacterial colonies were transferred to nitrocellulose filters, and filters were incubated with sCD46, followed by anti-CD46 mouse monoclonal antibodies and anti-mouse Ig-horseradish peroxidase conjugates. Out of 10,000 colonies plated, 20 colonies with the most intense CD46 signals were picked, and plasmid DNA was sequenced. Twenty percent of colonies did not contain mutations. The knob sequences of the remaining colonies had single or combined replacements of amino acid residue Asp207, Thr245, or Ile256 (Fig. 1A). Recombinant mutant knob proteins were purified, and their affinity to sCD46 was determined by comparing SPR to those of a wild-type Ad35 knob and an Ad35 knob that contained a Arg279Cys substitution resulting in ablation of CD46 binding (36) (Fig. 1B). The KD (equilibrium dissociation constant) of the wild-type knob was 14.64 nM, while the KDs for knob mutants with the single substitutions Asp207Gly, Thr245Ala, and Ile256Leu were 1.77, 7.64, and 10.96 nM, respectively. This translates into affinities 8.3-, 1.9-, and 1.3-fold-higher than that of the wild-type knob. The majority of identified knob mutants contained two or three of the above-listed substitutions. The highest affinity (0.63 nM; 23.2-fold higher than that of the wild-type Ad35 knob) was that of a knob mutant with a double Asp207Gly, Thr245Ala substitution. Two of the identified knobs with multiple mutations had substitution Asn217Asp or Thr226Ala; however, when analyzed individually, these substitutions had no impact on knob affinity (data not shown). The association rate constants and the dissociation rate constants of the wild-type Ad35 knob and all knob mutants are shown in Table 1. The association kinetics for all knobs were comparable. However, the dissociation rate constants inversely correlated with the knob affinities. This indicates that knobs with higher affinity dissociate slower from CD46 than the wild-type Ad35 knob.
FIG. 1.
Ad35 knob mutants with increased affinity to CD46. (A) Amino acid sequence of the Ad35 fiber knob. β-sheet regions (A to J) are shown. Amino acid residues that, when mutated, can increase affinity to CD46 are marked by arrows. (B) Biacore response data for sCD46 binding to a biosensor surface containing Ad35 knob mutants. Experimental data represent the responses of duplicate injections of various concentrations of sCD46 (169 nM, 56 nM, 19 nM, 6 nM, and 2 nM). Note that the time scale for Asp207Gly plus Thr245Ala and Asp207Gly plus Thr245Ala plus Ile256Leu is 3,000 s and that one additional sCD46 concentration (1 nM) was used for these two knob mutants. wt, wild type.
TABLE 1.
SPR analysis of Ad35 knob mutantsa
| Virusb | ka (1/Ms) | kd (1/s) | KD (nM) |
|---|---|---|---|
| wt Ad35 | 1.52 × 106 | 0.0234 | 14.64 |
| Asp207Gly | 3.51 × 106 | 0.0034 | 1.77 |
| Thr245Ala | 2.25 × 106 | 0.0173 | 7.64 |
| Ile256Leu | 2.13 × 106 | 0.0236 | 10.96 |
| Asp207Gly Thr245Ala | 2.48 × 106 | 0.0017 | 0.63 |
| Asp207Gly Thr226Ala | 1.97 × 106 | 0.0026 | 1.41 |
| Asn217Asp Thr245Pro Ale256Leu | 2.25 × 106 | 0.0097 | 4.82 |
| Asp207Gly Thr245Ala Ile256Leu | 1.96 × 106 | 0.0018 | 0.92 |
ka, association rate constant (kon); kd, dissociation rate constant (koff).
Viruses other than the wild type (wt) are identified by the mutation(s) in the Ad35 knob.
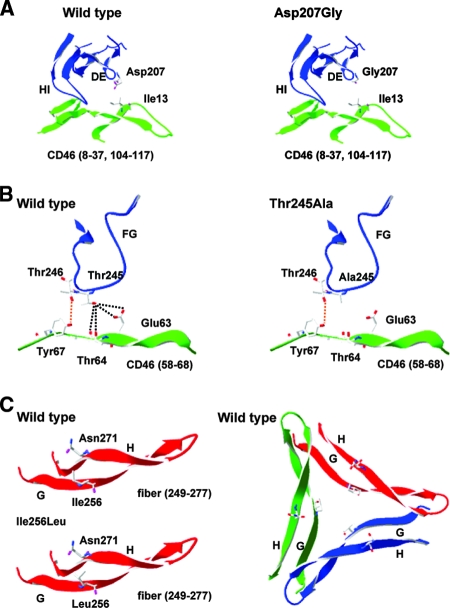
In an attempt to understand the structural basis for the increased affinity of our knob mutants to CD46, we superimposed the crystal structure of the Ad35 knob (36) with that of CD46 (20) (Fig. 2). For the Asp207Gly mutant, the hydrophobic Ile13 residue of CD46 is the closest amino acid R group to Asp207, with its hydroxyl group being 5.93 and 4.4 Å from the nearest Ile methyl groups (Fig. 2A). Unlike the polar Asp residue, glycine is hydrophobic (lacking a side chain), so an Asp207Gly substitution may enable the Ad35 DE loop to approach CD46 more closely to Ile13. Subsequently the HI loop would also get closer to CD46. For the Thr245Ala mutant, according to our original Ad35-CD46 binding model, the Ad35 Thr246 residue is important for binding and interacts with CD46 Tyr67. In this conformation the hydroxyl group of the neighboring Thr245 residue is relatively close to the backbone carbonyl and R group hydroxyl group of CD46 residue Thr64 (4.35 and 5.21 Å) and to the R group carbonyl and hydroxyl group of CD46 residue Glu63 (4.46 and 5.76 Å). Although the hydroxyl group of residue Thr245 is not close enough to form a strong H bond with any of these groups, the proximity of the groups likely influences the loop conformation in this region, so the introduction of hydrophobic alanine at this position (Thr245Ala) may make the Thr246-to-Tyr67 interaction stronger since the FG loop can move closer to CD46 (Fig. 2B). For the Ile256Leu mutant, Ad35 residue Ile256 is oriented toward the central core of the fiber trimer in the center of the G sheet (Fig. 2C) and its hydrophobic R group is closest to the polar carboxyl group in the side chain of Asn271 within the H sheet. Although the rotation of leucine in our Ile256Leu mutant is unknown, our model suggests that the methyl groups of Leu256 would be closer to Asn271 than those of Ile256, and the increased repulsion would likely push the G and H sheets further apart. How this affects Ad35-CD46 affinity is unclear, but the higher affinity of the Ad35 knob with the Ile256Leu mutation may be due to increased stability of the knob trimer.
FIG. 2.
Structural analysis of increased binding Ad35 fiber mutants. (A) Ribbon representation of the DE and HI loops of the Ad35 fiber with Asp or Gly residues shown at position 207. CD46 residues 8 to 37 and 104 to 117 are also present, with Ile13 shown. (B) Structural ribbon representation of the FG loop of the Ad35 fiber with Thr and Ala residues shown at position 245 and Thr246 also shown. CD46 residues 58 to 68 are present, with residues Glu63, Thr64, and Tyr67 shown. The orange dashed line shows an interaction previously described as important for Ad35-CD46 binding (36), while black lines indicate a region of polarity lost upon mutation of Thr245 to Ala245. (C) Structural ribbon representation of the G and H sheets of the Ad35 fiber (residues 249 to 277), with Ile and Leu residues shown at position 256 and Asn271 also shown. Representations of the Ad35 monomer and trimer are shown.
Recombinant Ad35 knob mutant proteins were produced in E. coli and purified by Ni-nitrilotriacetic acid affinity chromatography. All mutant knobs listed in Fig. 1B formed trimers when analyzed by polyacrylamide gel electrophoresis under nondenaturing conditions (data not shown). To begin assessing the functional properties of mutants, we used Ad35 knob mutants as competitors for attachment of 3H-labeled Ad35 virus to HeLa cells at 4°C. When used in a concentration range of 0.5 to 10 ng knob per 105 cells, none of the mutants was able to block Ad35 binding more than the wild-type Ad35 knob (data not shown). This is not surprising, considering that Ad particles have 12 (trimeric) fibers per virion and therefore have a higher avidity to CD46 than recombinant (trimeric) fiber knob molecules.
Ad vectors containing Ad35 fiber knobs with increased affinity to CD46.
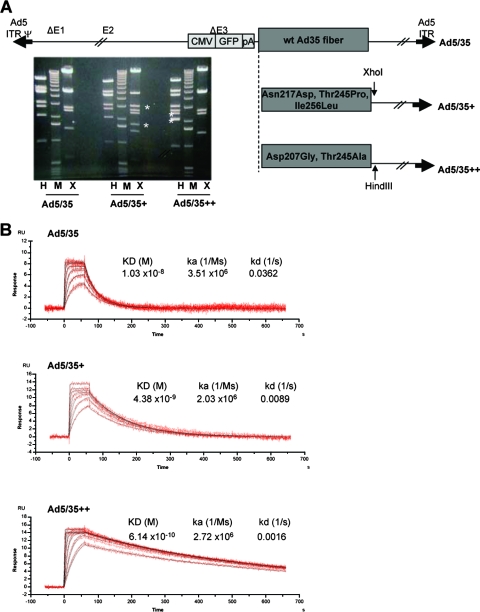
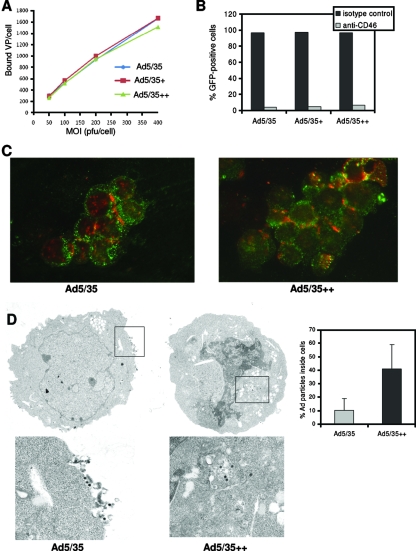
Ad particles possess 12 fibers. Since one Ad35 fiber knob binds to three CD46 molecules at the same time, one virus can potentially bind to a maximum of 36 CD46 molecules on a cell. We hypothesized that upon virus binding subsequent events such as clustering of receptor proteins and receptor-mediated signaling will be different between individual soluble Ad35 knob molecules and corresponding knobs in the context of virions. In order to investigate this, we generated viruses containing Ad35 knobs with increased affinity to CD46. The coding sequences of Ad35 fibers, which is the CD46-interacting moiety within the Ad35 capsid, were transferred into Ad5 vectors to replace the corresponding Ad5 fiber sequences. We created three types of such chimeric vectors: Ad5/35, which contained the wild-type Ad35 fiber; Ad5/35+, which contained the Asn217Asp, Thr245Pro, and Ile256Leu mutations, resulting in a threefold-higher affinity of the fiber knob to CD46; and Ad5/35++, which contained the Asp207Gly and Thr245Ala substitutions, resulting in a 23.2-fold-higher affinity (Fig. 3). All vectors were first-generation, E1/E3-deleted vectors and contained a GFP expression cassette in the deleted E3 region (Fig. 3A). The GFP expression cassette contained the cytomegalovirus promoter, the GFP gene, and the bovine growth hormone polyadenylation signal. Ad5/35+ and Ad5/35++ genomes also contained additional XhoI and HindIII restriction sites, which allowed us to confirm the identity of virus preparation after amplification and purification in CsCl gradients by restriction endonuclease analysis of viral DNA (Fig. 3A). The titers of Ad5/35, Ad5/35+, and Ad535++ which were 3.05 × 1011, 4.03 × 1011, and 5.12 × 1011 PFU/ml (based on PFU assay), respectively. The ratio of viral particles to PFU was 12:1 for all three viruses. To measure the binding between purified Ad particles and CD46 by SPR, biotinylated virions were immobilized on Bioacore sensor chips and subsequently injected with sCD46 analyte. The affinities of Ad5/35+ and Ad5/35++ were 4.2 and 60 times higher than that of the virus that contained the wild-type Ad35 knob (Fig. 3B). While the association kinetics were comparable for all three viruses, dissociation from sCD46 was slower for Ad5/35+ and Ad5/35++ viruses.
FIG. 3.
Ad vectors with increased affinity to CD46. (A) Analysis of vector genomes. (Top) Structure vector genomes. (Bottom) Restriction endonuclease analysis (X, XhoI; H, HindIII) of vectors genomes isolated from purified Add5/35, Ad5/35+, and Ad5/35++ particles. The distinguishing DNA fragments are, for Ad5/35+, the 3.14- and 1.78-kb XhoI fragments and, for Ad5/35++, the 2.53- and 2.14-kb HindIII fragments. M, 1-kb ladder marker. wt, wild type; CMV, cytomegalovirus; ITR, inverted terminal repeat. (B) Biacore analysis of Ad-sCD46 interactions. Purified Ad particles were immobilized, and binding to sCD46 was analyzed. Sensorgrams are shown for injections at seven different concentrations of sCD46 (50 to 0.781 μg/ml in a twofold dilution series). The analysis was performed in triplicate. KD, equilibrium dissociation constant; ka, association rate constant; kd, dissociation rate constant.
Transduction of cell lines.
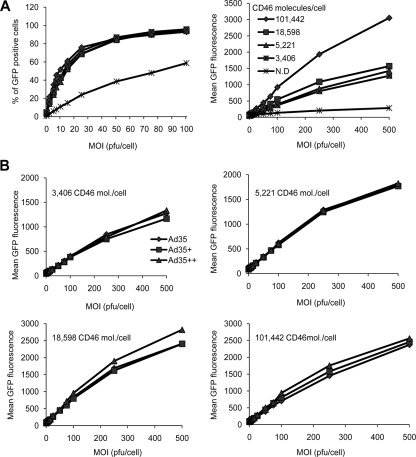
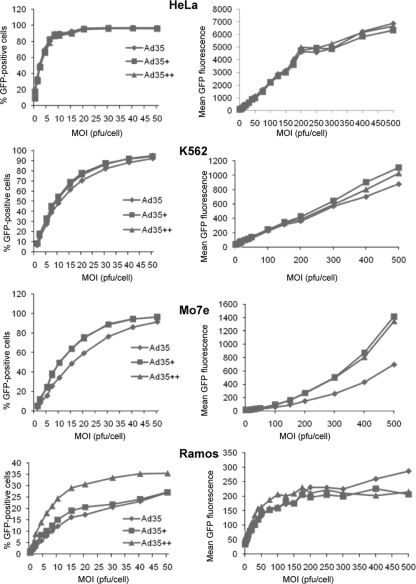
Although the SPR data showed higher affinities for Ad5/35+ and Ad5/35++ to sCD46, this had to be investigated in the cellular context, since the receptor affinities of the individual knobs, the density and accessibility on the receptors on the cell, and the resulting binding avidity of the viral particles to the cell are interrelated. We tested whether higher knob affinities might translate into higher viral particle avidities and better vector transduction of cells. To test the effect of receptor density, we used a set of CHO cell clones that expressed different densities of human CD46 (3,406, 5,221, 18,598, and 101,442 CD46 molecules per cell) (1). We first infected unmodified CHO cells and CHO-CD46 cell clones with Ad5/35 at increasing MOIs, and GFP fluorescence was measured 24 h later by flow cytometry (Fig. 4A). Unmodified CHO cells do not express CD46 and are relatively resistant to Ad5/35 infection (Fig. 4A, N.D.). Transduction rates of CHO-CD46 cell clones with the Ad5/35 vector (measured based on mean GFP fluorescence) correlated with CD46 receptor density on clones. Disappointingly, when Ad5/35+ and Ad5/35++ were included into the studies, significantly higher transduction rates were not observed, regardless of the CD46 receptor density present on CHO-CD46 cells (Fig. 4B). CHO cells do not express surface integrins, which are thought to be required for Ad5/35 infection (18, 24). Furthermore, nonprimate cells are refractory to infection with Ad5/35 (29). We therefore performed further transduction studies on human cell lines of different tissue origins, including HeLa cells (cervical epithelial cancer), A549 cells (lung epithelial cancer), 293 cells (embryonic kidney), K562 and MO7e cells (erythroleukemia), and Ramos cells (B lymphoma) (11) (Fig. 5). No differences in transduction between Ad5/35, Ad5/35+, and Ad5/35++ on the adherent, epithelium-derived cell lines HeLa (Fig. 5), A549, and 293 (data not shown) were seen. However, transduction of K562, MO7e, and Ramos suspension cultures was higher for Ad5/35+ and Ad5/35++, although the difference reached significance (P < 0.05) only for Ad5/35+ and Ad5/35++ on MO7e cells. In an attempt to understand the different outcomes of transduction studies in different cells lines, we measured the CD46 densities on our test cells. CD46 densities were 183,000 molecules per cell for HeLa cells, 43,590 molecules per cell for MO7e cells, 47,564 molecules per cell for K562 cells, and 5,730 molecules per cells for Ramos cells. Apparently, there was no correlation between CD46 density and transduction properties of our three vectors for the cell lines tested. In summary, transduction studies showed that increased affinity of Ad particles to CD46 did not correspond to higher transduction efficiency in vitro.
FIG. 4.
Transduction of CHO-CD46 cell clones. (A) CHO cells that did not express CD46 (N.D.) and CHO cell clones that expressed CD46 at different levels (1) were infected with Ad5/35 at the indicated MOIs, and the percentages of GFP-expressing cells (left) and GFP mean fluorescence (right) were measured at 24 h by flow cytometry. (B) CHO-CD46 cell clones were infected with Ad5/35, Ad5/35+, and Ad5/35++, and GFP expression was analyzed as described for panel A. Shown are the averages of three independent experiments. The standard deviations were less then 10% from the averages for all data points.
FIG. 5.
Transduction of human cell lines. Cells were infected with Ad5/35, Ad5/35+, and Ad5/35++ at the indicated MOIs, and the percentages of GFP-expressing cells (left) and GFP mean fluorescence (right) at 24 h were measured by flow cytometry. Shown are the averages of three independent experiments. The standard deviations were less then 10% from the averages for all data points.
Virus trafficking in MO7e cells.
As outlined above, higher virion avidity might affect virus entry and intracellular trafficking. We studied these events in MO7e cells. Levels of attachment of 3H-labeled Ad5/35, Ad5/35+ and Ad5/35++ particles to MO7e cells were comparable at all MOIs tested (Fig. 6A). Internalization rates, measured as described previously (24), also did not differ between the three viruses (data not shown). Transduction of cells could be blocked >94% by preincubation of cells with anti-CD46 antibody (Fig. 6B), confirming that CD46 is the primary attachment receptor for our vectors. Intracellular trafficking of Ad5/35 and Ad5/35++ virions was studied using fluorescein isothiocyanate-labeled anti-cathepsin B antibodies to visualize late endosomes. In agreement with earlier studies (27), the vast majority of Ad5/35 particles colocalized with the endosomal marker at 1 h postinfection and there was no difference between Ad5/35 and Ad5/35++ (data not shown). However, when trafficking was analyzed at 4 h postinfection, more Ad5/35++ particles were still associated with endosomes inside the cytoplasm, whereas a large number of Ad5/35 particles were found on the cell surface as free particles or associated with endosomal membranes (Fig. 6C). We previously reported that Ad5/35 particles can be recycled back to the cell surface (26, 28). Apparently, the slower dissociation of Ad5/35++ from CD46 decreases this retrograde transport and allows more Ad genomes to reach the nucleus, which might, in part, explain the higher transduction efficiencies of Ad5/35++ in MO7e cells. To support this preliminary conclusion, we performed electron microscopy studies on infected MO7e cells at 4 h postinfection. The microphotographs clearly show a higher number of Ad particles at the cell periphery for Ad5/35-infected cells (Fig. 6D). When particles in 100 random cells were counted, it was found that 89.6% of Ad5/35 particles and 59.0% of Ad5/35++ particles were associated with the peripheral cell membrane or were outside the cells. Correspondingly, 10.4% of Ad5/35 particles and 41.0% of Ad5/35++ particles were found localized inside the cells (Fig. 6D).
FIG. 6.
Analysis of intracellular trafficking in MO7e cells. (A) Attachment of 3H-labeled Ad particles to MO7e cells. MO7e cells were incubated with 3H-labeled Ads at different MOIs, and the numbers of VP bound per cell after 1 h of incubation on ice were determined. (B) Inhibition of Ad transduction by anti-CD46 antibodies. MO7e cells were incubated with 10 μg/ml anti-human CD46 antibody (clone MEM-258; Serotec Ltd., Oxford, United Kingdom) or 10 μg/ml anti-human isotype control antibody for 45 min on ice, washed once with PBS, and then infected with 40 PFU/cell of Ad5/35, Ad5/35+, or Ad5/35++. Twenty-four hours after infection, GFP expression was analyzed. Shown are the averages of three independent experiments. The standard deviations were less then 10% from the averages for all data points in panels A and B. (C) Immunofluorescence analysis of Ad particles (red) and the late endosomal marker cathepsin B (green) in MO7e cells 4 h after infection with Ad5/35 (left) or Ad5/35++ (right) at an MOI of 4,000 VP per cell. (D) Electron microscopy analysis of intracellular distribution of Ad5/35 and Ad5/35++ vector particles in MO7e cells 4 h after virus infection. Representative fields are shown. Low magnification, ×6,000; high magnification, ×20,000. (Right) Quantification of Ad particles inside cells (n = 100 cells).
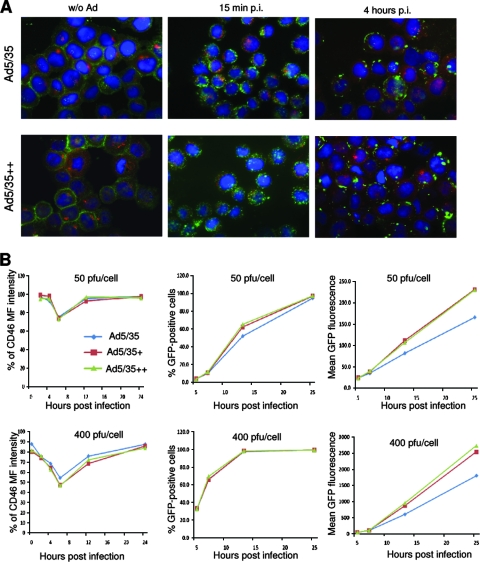
Effect of Ad infection on CD46 distribution.
Considering differences in dissociation from CD46 between Ad5/35 and Ad5/35++, we studied the colocalization of the virion hexon and CD46 at different time points after infection of MO7e cells. While uninfected MO7e cells show CD46 lining the cell membranes (Fig. 7A, left), as early as 15 min after addition of Ads, CD46 can be seen as clusters at the membrane and inside the cytoplasm, reminiscent of coated pits (Fig. 7A, middle). This is probably a result of the presence of multiple CD46 binding sites on the virion and recruitment to endocytic structures. Interestingly, clustered intracellular CD46 can still be observed at 4 h postinfection (Fig. 7A, right). To quantitate the amount of surface CD46, we used an anti-CD46-PE antibody without cell fixation on cells infected at 50 and 400 PFU/cell with Ad5/35, Ad5/35+, and Ad5/35++. After infection at an MOI of 50 PFU/cell, mean fluorescence intensity of surface CD46 decreased, reaching a minimum at 8 h postinfection, after which it slowly increased again to preinfection levels at 24 h postinfection (Fig. 7B, top). After infection at an MOI of 400 PFU/cell, this decrease was more pronounced for Ad5/35+ and Ad5/35++ than for Ad5/35. Both the percentage of GFP-expressing cells and the mean GFP fluorescence correlated with the degree of CD46 internalization at earlier time points (5 and 10 h) postinfection (Fig. 7, middle and right). This indicates that Ad35 particles are taken up together with CD46 into MO7e cells.
FIG. 7.
Analysis of CD46 distribution in infected MO7e cells. (A) Colocalization of CD46 and virus. CD46 (green) was analyzed before infection and at 15 min and 4 h after infection with Ad5/35 and Ad5/35++ at an MOI of 4,000 VP per cell. Cathepsin B-positive endosomal compartments are green. Confocal images of cells were taken in both green and red channels. Representative fields are shown. (B) Analysis of CD46 on the surfaces of MO7e cells and GFP expression at different time points after infection with Ad5/35, Ad5/35+, or Ad5/35++ at MOIs of 50 PFU/cell and 400 PFU/cell. Notably, staining for the viral hexon at 8 h postinfection did not reveal any signal at the cell surface, indicating that the decrease in CD46-PE fluorescence was not due to competition with Ad particles bound to surface CD46. Shown are the averages of three independent experiments. The standard deviations were less then 10% from the averages for all data points.
Tumor targeting after intravenous vector injection.
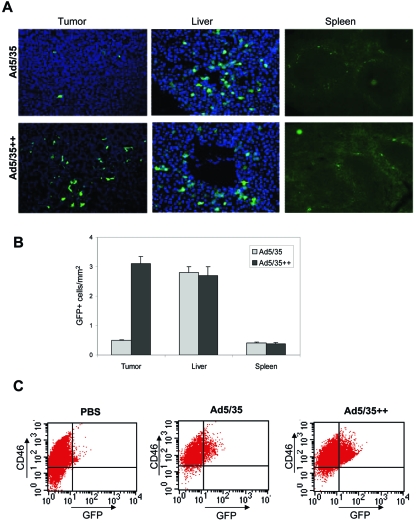
Ad5/35 vectors have shown promise for the targeting of tumors because the majority of tumor types overexpress CD46 compared to normal tissue (30). However, after intravenous injection, interaction of Ad5/35 vectors with CD46high tumors is affected by unspecific sequestration, involving high-affinity interactions with soluble blood factors (12, 25, 34), blood cells (15, 31), and tissue macrophages (38). The unspecific Ad sequestration and degradation affect the majority of intravenously injected viral particles and greatly reduce the efficiency of tumor cell transduction. We hypothesized that Ad5/35 vectors with increased affinity to CD46 would have an advantage in competing with non-CD46-mediated sequestration and demonstrate better tumor transduction after intravenous injection. To test this hypothesis, we employed C57BL/6-CD46tg mice, which express human CD46 in a pattern and at levels comparable to those for humans (16). As target cells we engineered a C57BL/6-derived tumor cell line to express human CD46 with about 20,000 molecules per cell (TC1-CD46) (5). TC-1 cells are mouse lung carcinoma cells, do not express human CD46, and are refractory to Ad5/35 infection (5). CD46 density on TC1-CD46 cells is in the range of that seen in human tumor cell lines (see above). TC1-CD46 cells were injected into the portal veins of CD46tg mice to establish liver metastases. In a previous study we reported that, in this mouse model, CD46 mRNA expression levels are >50-fold less in liver tissue than in microdissected TC1-CD46-derived liver metastases (19). Three weeks after TC1-CD46 transplantation, tumor-bearing mice were intravenously injected with Ad5/35 and Ad5/35++ at a dose of 5 × 109 PFU per mouse, and GFP expression on tumor and organ sections was analyzed 3 days later (Fig. 8). Quantitation of GFP-positive tumor cells/mm2 in consecutive liver sections (20 sections per mouse, three mice per virus) revealed a 6.1-fold (+/−1.8-fold)-higher transduction efficiency of TC1-CD46 metastases with Ad5/35++ than with Ad5/35. Other than in tumors, GFP-expressing cells were found only in liver parenchyma and the marginal zones of the spleen. There was no significant difference in the number of GFP-expressing cells between the two viruses in these tissues. This is not surprising as transduction of liver with Ad5/35 vectors is not mediated through CD46 but through interaction of the Ad5 hexon with coagulation factor X and cellular heparan sulfate proteoglycans (12, 34).
FIG. 8.
GFP expression in organs and liver metastases after intravenous injection of Ad5/35 and Ad5/35++ into CD46-transgenic mice bearing CD46high liver metastases. Mice were intraportally injected with TC1-CD46 tumor cells. Three weeks later, when liver metastases were established, mice received tail vein injection of 3 × 109 PFU of Ad5/35 and Ad5/35++ (n = 4). Organ sections were analyzed for GFP expression 3 days later. (A) Representative sections. (B) Quantification of GFP-positive cells/mm2 in liver metastases (tumor), liver, and spleen (20 sections per mouse, four mice per virus). (C) Quantification of GFP-positive cells in cell suspensions of microdissected liver metastases. Tumor tissue was digested with collagenase and versene, and cell suspensions were analyzed by flow cytometry for GFP and CD46 expression. Representative samples are shown. Transduced TC1-CD46 cells appear as GFP+ CD46high cells (upper right quadrant). Cells in the upper and lower left quadrants represent cell debris that stained positive with propidium iodide (data not shown).
To better quantitate the number of GFP-expressing cells in TC1-CD46-derived liver metastases after intravenous injection of Ad5/35 and Ad5/35++ vectors, microdissected tumors were digested by collagenase and versene. Cell suspensions were analyzed for CD46 and GFP using flow cytometry (Fig. 8C). In cell suspensions, transduced TC1-CD46 tumor cells appear as CD46high GFP+ cells. In mice injected with Ad5/35 and Ad5/35++, the percentages of GFP+ cells in the CD46high cell fractions were 6.3% (+/−2.9%) and 23.9% (+/−9.5%), respectively. Notably, we were not able to generate viable hepatocyte suspensions by direct digestion of liver tissue with collagenase/versene.
Taken together, our in vivo data demonstrated that Ad5/35++ is superior in transduction of CD46high liver metastases after intravenous vector administration.
DISCUSSION
The goal of this study was to produce Ad vectors containing fibers with affinities to CD46 that were higher than that of the naturally occurring Ad35 serotype and to test whether these vectors have a higher efficiency in target cell transduction in vitro and in vivo. We used an E. coli library expressing Ad35 fiber knob mutants with random mutations to select for knob variants with stronger binding to CD46. The affinities of mutants that were selected were up to 23.2-fold higher than that of the wild-type Ad35 knob. Replacements of amino acid residues Asp207, Thr245, and Ile256 contributed most to the increase in CD46 binding.
We constructed Ad vectors containing either the wild-type Ad35 knob (Ad5/35), an Ad35 knob mutant with 3-fold-higher affinity to CD46 (Ad5/35+), or an Ad35 mutant with 23.2-fold-higher affinity (Ad5/35++). The KDs of Ad5/35+ and Ad5/35++ for sCD46 were 4.2-fold and 60-fold higher than that of Ad5/35. The discrepancies in avidities of recombinant mutant knobs and mutant knobs in the context of a virion can be explained by the higher number of CD46 binding sites on virions. While the association kinetics of all three viruses were comparable, dissociation from CD46 was 4.4-fold slower for Ad5/35+ and 22.6-fold slower for Ad5/35++ (Fig. 3B).
For in vitro studies, the higher affinities of Ad5/35+ and Ad5/35++ to CD46 did not translate to correspondingly higher transduction efficiencies, regardless of the CD46 receptor density present on cells. An advantage of these vectors in transduction of suspension cells, particularly MO7e erythroleukemia and Ramos B-lymphoma cells, was observed. Overall our transduction studies with cell lines indicate that the natural evolution of Ads has selected for variants with an optimal affinity for infection of their natural target cells. This conclusion is supported by recent studies with measles virus, which is, in contrast to Ad, an enveloped virus. Hasegawa et al. (9) generated a panel of measles viruses displaying HER2/neu-specific single-chain variable fragments with affinities ranging from 1.6 × 10−6 M to 1.5 × 10−11 M and characterized them on a panel of cells expressing various numbers of HER2 receptors. They demonstrated that receptor affinity has little impact on viral attachment. However, higher receptor affinity directly correlated with higher efficiency of viral cell entry and infection, whereby there was an affinity threshold level above which infection and intercellular fusion proceed with equal efficiencies, even over a 1,000-fold affinity range.
Our in vitro findings have implications for retargeting of adenovirus vectors using high-affinity ligand-receptor pairs for vector propagation. Examples of such systems include de novo-designed peptides (E-Coil and K-Coil) that interact with each other with high affinity (40) and biotin-avidin interaction pairs (3). Our data indicate that picomolar and higher affinities to a cell receptor might not necessarily improve in vitro infection properties.
Previously it has been shown that, while the CAR-interacting species C Ad5 is rapidly released from endosomes upon infection, CD46-interacting Ad vectors containing species B fibers, such as Ad5/35, remain longer in late endosomes/lysosomes and use these cellular compartments to traffic to the nucleus (14). While the CAR-interacting residues within the Ad5 fiber knob cluster in one area of the knob monomer (13), CD46 binding residues within the Ad35 knob are localized on two opposite ends of the knob. This implies that one CD46 monomer binds between two Ad35 knob monomers (37), which creates a more rigid structure and, theoretically, results in a slower dissociation of Ad35 fiber from CD46, hence slower release from endosomes for Ad5/35 viruses than for Ad5. In this study, we show that infection of MO7e cells by Ad5/35 vectors results in internalization of CD46 and accumulation in endosomes/lysosomes, as colabeling with the late endosomal marker cathepsin B shows (Fig. 7A). Disappearance of CD46 from the cell surface through internalization is MOI dependent and reached a peak at about 8 h postinfection. Our observation is in agreement with a study by Sakurai et al., who reported downregulation of CD46 after Ad35 infection (21). Furthermore, we found that, in MO7e cells, Ad5/35 particles with higher affinity to CD46 remain longer in endosomes (Fig. 6C) and are expelled from cells less as a result of retrograde transport than virions containing wild-type Ad35 fibers (Fig. 6D). Considering that the dissociation rate constants of these viruses differ by 22-fold (Fig. 3B), we hypothesize that differences in trafficking can, in part, be explained by a slower release of Ad5/35++ fiber from CD46, which is trapped together with Ad virions in endosomes/lysosomes.
Factors that determine in vivo Ad tropism are complex, particularly after intravenous vector administration, a route that is often used to target metastatic cancer. For example, vectors containing Ad5 hexons bind with high affinity to coagulation factor X, and this complex is taken up by hepatocytes through cellular heparan sulfate proteoglycans (12, 25, 34). Furthermore, intravenously injected Ad is sequestered by blood cells (15, 31) and tissue macrophages (38), which leads to Ad degradation and greatly reduces the amount of virus available for tumor cell transduction. We hypothesized that Ad5/35 vectors with increased affinity to CD46 would have an advantage in competing with non-CD46-mediated sequestration. In support of this hypothesis, we demonstrate in a mouse model with preestablished CD46high liver metastases better tumor transduction with Ad5/35++ after intravenous vector injection than with Ad5/35. Transduction of other organs in transgenic mice such as the liver was not increased with Ad5/35++. This is not surprising because hepatocyte transduction is primarily mediated by factor X. Considering that Ad35-based vectors do not bind factor X (12) and do not unspecifically transduce hepatocytes in relevant animal models (22), our current focus is to incorporate the Ad35++ fiber into an Ad35-based oncolytic vector. Our in vivo studies are encouraging for investigators who attempt to construct Ad vectors with high affinity to tumor cell receptors such as HER2/neu (2).
Overall, our studies contribute to a better understanding of the Ad35 fiber-CD46 interaction and the role of receptor affinity in Ad infection. Furthermore, they give a rationale for generating capsid-modified vectors containing high-affinity ligands for specific receptors on in vivo target cells.
Acknowledgments
This work was supported by NIH grants HLA078836 (A.L.) and AI065429 (D.S.) and grants from the Deutsche Forschungsgemeinschaft (SFB685 and STE1163) (T.S.).
We thank Roland Strong (FHCRC, Seattle) for providing access to Biacore analysis.
Footnotes
Published ahead of print on 27 August 2008.
REFERENCES
- 1.Anderson, B. D., T. Nakamura, S. J. Russell, and K. W. Peng. 2004. High CD46 receptor density determines preferential killing of tumor cells by oncolytic measles virus. Cancer Res. 644919-4926. [DOI] [PubMed] [Google Scholar]
- 2.Belousova, N., G. Mikheeva, J. Gelovani, and V. Krasnykh. 2008. Modification of adenovirus capsid with a designed protein ligand yields a gene vector targeted to a major molecular marker of cancer. J. Virol. 82630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos, S. K., M. B. Parrott, and M. A. Barry. 2004. Avidin-based targeting and purification of a protein IX-modified, metabolically biotinylated adenoviral vector. Mol. Ther. 9942-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 704805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DiPaolo, N., S. Ni, A. Gaggar, R. Strauss, S. Tuve, Z. Y. Li, D. Stone, D. Shayakhmetov, N. Kiviat, P. Toure, S. Sow, B. Horvat, and A. Lieber. 2006. Evaluation of adenovirus vectors containing serotype 35 fibers for vaccination. Mol. Ther. 13756-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleischli, C., S. Verhaagh, M. Havenga, D. Sirena, W. Schaffner, R. Cattaneo, U. F. Greber, and S. Hemmi. 2005. The distal short consensus repeats 1 and 2 of the membrane cofactor protein CD46 and their distance from the cell membrane determine productive entry of species B adenovirus serotype 35. J. Virol. 7910013-10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaggar, A., D. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 91408-1412. [DOI] [PubMed] [Google Scholar]
- 8.Gaggar, A., D. M. Shayakhmetov, M. K. Liszewski, J. P. Atkinson, and A. Lieber. 2005. Localization of regions in CD46 that interact with adenovirus. J. Virol. 797503-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasegawa, K., C. Hu, T. Nakamura, J. D. Marks, S. J. Russell, and K. W. Peng. 2007. Affinity thresholds for membrane fusion triggering by viral glycoproteins. J. Virol. 8113149-13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horvat, B., P. Rivailler, G. Varior-Krishnan, A. Cardoso, D. Gerlier, and C. Rabourdin-Combe. 1996. Transgenic mice expressing human measles virus (MV) receptor CD46 provide cells exhibiting different permissivities to MV infections. J. Virol. 706673-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, S., R. I. Endo, and G. R. Nemerow. 1995. Upregulation of integrins αvβ3 and αvβ5 on human monocytes and T lymphocytes facilitates adenovirus-mediated gene delivery. J. Virol. 692257-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalyuzhniy, O., N. C. Di Paolo, M. Silvestry, S. E. Hofherr, M. A. Barry, P. L. Stewart, and D. M. Shayakhmetov. 2008. Adenovirus serotype 5 hexon is critical for virus infection of hepatocytes in vivo. Proc. Natl. Acad. Sci. USA 1055483-5488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirby, I., E. Davison, A. J. Beavil, C. P. Soh, T. J. Wickham, P. W. Roelvink, I. Kovesdi, B. J. Sutton, and G. Santis. 2000. Identification of contact residues and definition of the CAR-binding site of adenovirus type 5 fiber protein. J. Virol. 742804-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leopold, P. L., and R. G. Crystal. 2007. Intracellular trafficking of adenovirus: many means to many ends. Adv. Drug Deliv. Rev. 59810-821. [DOI] [PubMed] [Google Scholar]
- 15.Lyons, M., D. Onion, N. K. Green, K. Aslan, R. Rajaratnam, M. Bazan-Peregrino, S. Phipps, S. Hale, V. Mautner, L. W. Seymour, and K. D. Fisher. 2006. Adenovirus type 5 interactions with human blood cells may compromise systemic delivery. Mol. Ther. 14118-128. [DOI] [PubMed] [Google Scholar]
- 16.Marie, J. C., A. L. Astier, P. Rivailler, C. Rabourdin-Combe, T. F. Wild, and B. Horvat. 2002. Linking innate and acquired immunity: divergent role of CD46 cytoplasmic domains in T cell induced inflammation. Nat. Immunol. 3659-666. [DOI] [PubMed] [Google Scholar]
- 17.Metzgar, D., M. Osuna, A. E. Kajon, A. W. Hawksworth, M. Irvine, and K. L. Russell. 2007. Abrupt emergence of diverse species B adenoviruses at US military recruit training centers. J. Infect. Dis. 1961465-1473. [DOI] [PubMed] [Google Scholar]
- 18.Murakami, S., F. Sakurai, K. Kawabata, N. Okada, T. Fujita, A. Yamamoto, T. Hayakawa, and H. Mizuguchi. 2007. Interaction of penton base Arg-Gly-Asp motifs with integrins is crucial for adenovirus serotype 35 vector transduction in human hematopoietic cells. Gene Ther. 141525-1533. [DOI] [PubMed] [Google Scholar]
- 19.Ni, S., A. Gaggar, N. Di Paolo, Z. Y. Li, Y. Liu, R. Strauss, P. Sova, J. Morihara, Q. Feng, N. Kiviat, P. Toure, P. S. Sow, and A. Lieber. 2006. Evaluation of adenovirus vectors containing serotype 35 fibers for tumor targeting. Cancer Gene Ther. 131072-1081. [DOI] [PubMed] [Google Scholar]
- 20.Persson, B. D., D. M. Reiter, M. Marttila, Y. F. Mei, J. M. Casasnovas, N. Arnberg, and T. Stehle. 2007. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat. Struct. Mol. Biol. 14164-166. [DOI] [PubMed] [Google Scholar]
- 21.Sakurai, F., K. Akitomo, K. Kawabata, T. Hayakawa, and H. Mizuguchi. 2007. Downregulation of human CD46 by adenovirus serotype 35 vectors. Gene Ther. 14912-919. [DOI] [PubMed] [Google Scholar]
- 22.Sakurai, F., S. Nakamura, K. Akitomo, H. Shibata, K. Terao, K. Kawabata, T. Hayakawa, and H. Mizuguchi. 2008. Transduction properties of adenovirus serotype 35 vectors after intravenous administration into nonhuman primates. Mol. Ther. 16726-733. [DOI] [PubMed] [Google Scholar]
- 23.Shamah, S. M., J. M. Healy, and S. T. Cload. 2008. Complex target SELEX. Acc. Chem. Res. 41130-138. [DOI] [PubMed] [Google Scholar]
- 24.Shayakhmetov, D. M., A. M. Eberly, Z.-Y. Li, and A. Lieber. 2005. Deletion of penton RGD motifs affects the efficiency of both the internalization and the endosome escape of viral particles containing adenovirus serotype 5 or 35 fiber knobs. J. Virol. 791053-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shayakhmetov, D. M., A. Gaggar, S. Ni, Z. Y. Li, and A. Lieber. 2005. Adenovirus binding to blood factors results in liver cell infection and hepatotoxicity. J. Virol. 797478-7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shayakhmetov, D. M., Z. Y. Li, A. Gaggar, H. Gharwan, V. Ternovoi, V. Sandig, and A. Lieber. 2004. Genome size and structure determine efficiency of postinternalization steps and gene transfer of capsid-modified adenovirus vectors in a cell-type-specific manner. J. Virol. 7810009-10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shayakhmetov, D. M., Z. Y. Li, V. Ternovoi, A. Gaggar, H. Gharwan, and A. Lieber. 2003. The interaction between the fiber knob domain and the cellular attachment receptor determines the intracellular trafficking route of adenoviruses. J. Virol. 773712-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shayakhmetov, D. M., and A. Lieber. 2000. Dependence of adenovirus infectivity on length of the fiber shaft domain. J. Virol. 7410274-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shayakhmetov, D. M., T. Papayannopoulou, G. Stamatoyannopoulos, and A. Lieber. 2000. Efficient gene transfer into human CD34+ cells by a retargeted adenovirus vector. J. Virol. 742567-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stone, D., and A. Lieber. 2006. New serotypes of adenoviral vectors. Curr. Opin. Mol. Ther. 8423-431. [PubMed] [Google Scholar]
- 31.Stone, D., Y. Liu, D. Shayakhmetov, Z. Y. Li, S. Ni, and A. Lieber. 2007. Adenovirus-platelet interaction in blood causes virus sequestration to the reticuloendothelial system of the liver. J. Virol. 814866-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuve, S., H. Wang, C. Ware, Y. Liu, A. Gaggar, K. Bernt, D. Shayakhmetov, Z. Li, R. Strauss, D. Stone, and A. Lieber. 2006. A new group B adenovirus receptor is expressed at high levels on human stem and tumor cells. J. Virol. 8012109-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaughan, T. J., A. J. Williams, K. Pritchard, J. K. Osbourn, A. R. Pope, J. C. Earnshaw, J. McCafferty, R. A. Hodits, J. Wilton, and K. S. Johnson. 1996. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat. Biotechnol. 14309-314. [DOI] [PubMed] [Google Scholar]
- 34.Waddington, S. N., J. H. McVey, D. Bhella, A. L. Parker, K. Barker, H. Atoda, R. Pink, S. M. K. Buckley, J. Custers, T. Morita, M. J. E. Hanvenga, S. A. Nicklin, and A. H. Baker. 2008. A critical role for the adenovirus serotype 5 hexon in liver gene transfer. Cell 132397-409. [DOI] [PubMed] [Google Scholar]
- 35.Waehler, R., S. J. Russell, and D. T. Curiel. 2007. Engineering targeted viral vectors for gene therapy. Nat. Rev. Genet. 8573-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, H., Y. C. Liaw, D. Stone, O. Kalyuzhniy, I. Amiraslanov, S. Tuve, C. L. Verlinde, D. Shayakhmetov, T. Stehle, S. Roffler, and A. Lieber. 2007. Identification of CD46 binding sites within the adenovirus serotype 35 fiber knob. J. Virol. 8112785-12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Z., C. S. Kuhr, J. M. Allen, M. Blankinship, P. Gregorevic, J. S. Chamberlain, S. J. Tapscott, and R. Storb. 2007. Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression. Mol. Ther. 151160-1166. [DOI] [PubMed] [Google Scholar]
- 38.Worgall, S., G. Wolff, E. Falck-Pedersen, and R. G. Crystal. 1997. Innate immune mechanisms dominate elimination of adenoviral vectors following in vivo administration. Hum. Gene Ther. 837-44. [DOI] [PubMed] [Google Scholar]
- 39.Zabner, J., P. Freimuth, A. Puga, A. Fabrega, and M. J. Welsh. 1997. Lack of high affinity fiber receptor activity explains the resistance of ciliated airway epithelia to adenovirus infection. J. Clin. Investig. 1001144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng, Y., M. Pinard, J. Jaime, L. Bourget, P. Uyen Le, M. D. O'Connor-McCourt, R. Gilbert, and B. Massie. 2008. A ligand-pseudoreceptor system based on de novo designed peptides for the generation of adenoviral vectors with altered tropism. J. Gene Med. 10355-367. [DOI] [PubMed] [Google Scholar]