Abstract
ICP22 is a multifunctional herpes simplex virus 1 (HSV-1) regulatory protein that regulates the accumulation of a subset of late (γ2) proteins exemplified by UL38, UL41, and US11. ICP22 binds the cyclin-dependent kinase 9 (cdk9) but not cdk7, and this complex in conjunction with viral protein kinases phosphorylates the carboxyl terminus of RNA polymerase II (Pol II) in vitro. The primary function of cdk9 and its partners, the cyclin T variants, is in the elongation of RNA transcripts, although functions related to the initiation and processing of transcripts have also been reported. We report two series of experiments designed to probe the role of cdk9 in infected cells. In the first, infected cells were treated with 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), a specific inhibitor of cdk9. In cells treated with DRB, the major effect was in the accumulation of viral RNAs and proteins regulated by ICP22. The accumulation of α, β, or γ proteins not regulated by ICP22 was not affected by the drug. The results obtained with DRB were duplicated in cells transfected with small interfering RNA (siRNA) targeting cdk9 mRNAs. Interestingly, DRB and siRNA reduced the levels of ICP22 but not those of other α gene products. In addition, cdk9 and ICP22 appeared to colocalize with RNA Pol II in wild-type-virus-infected cells but not in ΔUL13-infected cells. We conclude that cdk9 plays a critical role in the optimization of expression of genes regulated by ICP22 and that one function of cdk9 in HSV-1-infected cells may be to bring ICP22 into the RNA Pol II transcriptional complex.
The studies described in this report center on the role of infected cell protein 22 (ICP22), an α (immediate early) protein in viral replication. Mutants lacking ICP22 yield reduced levels of viral progeny in a cell-type-dependent manner (36). The protein appears to perform several functions (24). A key function expressed by the carboxyl-terminal domain (CTD) of ICP22 in conjunction with the viral UL13 protein kinase is to enhance the synthesis of a subset of late (γ2) proteins exemplified by the products of the UL38, UL41, and US11 genes (2, 24, 31, 37). In earlier studies, this laboratory reported that ICP22 and the UL13 protein kinase mediate the activation of cdc2 and degradation of its partners, cyclins A and B (3, 4). cdc2 and its new partner, the viral DNA polymerase accessory factor (UL42), bind topoisomerase IIα in an ICP22-dependent manner (1, 4). In addition, ICP22 and UL13 mediate an intermediate phosphorylation of the carboxyl terminus of RNA polymerase II (Pol II) in Vero cells (14, 18, 34, 35).
Subsequent studies designed to elucidate the interaction of ICP22 with RNA Pol II led to the discovery that ICP22 physically interacts with cyclin-dependent kinase 9 (cdk9) and that the protein complex containing ICP22 and cdk9 phosphorylated the CTD of RNA Pol II in a viral US3 protein kinase-dependent fashion in vitro (8). These studies also showed that the CTD of RNA Pol II fused to glutathione S-transferase was phosphorylated in reaction mixtures containing ICP22 or cdk9 immunoprecipitated from lysates of wild-type parent virus- or ΔUL13 mutant virus- but not from ΔUS3 mutant virus-infected cells (8). The levels of cdk9 and its partner, cyclin T, were unaltered throughout the replicative cycle. These experiments placed ICP22 and cdk9 in a complex with the CTD of RNA Pol II. At the same time, we confirmed the requirement of ICP22 and the UL13 protein kinase in the posttranslational modification of RNA Pol II that alters its electrophoretic mobility, while US3 kinase appears to play a similar role but in a cell type-dependent fashion (8). The focus of this report is on the role of cdk9 in the expression of the subset of viral genes regulated by ICP22. We found the following relevant to this report.
cdk9 is a Ser/Thr proline-directed kinase that shares 47% homology with other cdk's (22). It was originally designated PITALRE because of its similarity to the PSTAIRE-like cyclin binding motif of cdc2, a motif that has been identified in all cdk-related kinases (20, 23). cdk9 is ubiquitously expressed and exists as two isoforms, i.e., cdk942 and cdk955. cdk955 contains a 13-kDa N-terminal Pro-Gly rich extension and is expressed from a TATA-containing promoter, whereas cdk942 is not (37). The ratio of cdk942/cdk955 is cell type dependent and most likely regulated in a tissue-dependent manner (19, 37, 38). However, both cdk9 isoforms have been found to localize to the nucleus (27).
cdk9 interacts with a variety of T-type cyclins, T1, T2a, T2b, and cyclin K (10, 26). cdk9 and its cyclin T partners bind together and are referred to as positive transcription elongation factor b (P-TEFb). The various complexes that arise from binding of different cyclins are likely to be cell-type specific and differentiation and activation dependent (27). The P-TEFb complexes function to phosphorylate the CTD of RNA Pol II and positively regulate cellular transcription (21, 22, 26, 27).
P-TEFb is required for phosphorylation of the CTD of RNA Pol II and for RNA Pol II-dependent production of mature mRNA transcripts from promoters in in vitro assays—activities that are inhibited by 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (22).
The mammalian CTD of RNA Pol II consists of a 52-heptad repeat of Tyr-Ser-Pro-Thr-Ser-Pro-Ser that can be phosphorylated on the different serine residues during transcription (17, 25). For transcription, a hypophosphorylated form of RNA Pol II and other factors must bind to the promoter region of the RNA template. Once bound, RNA Pol II is phosphorylated at the Ser5 position by cdk7, which leads to clearance from the promoter and transcriptional initiation (22). Shortly after this initiation step, DRB sensitivity-inducing factor and negative elongation factor F bind to and pause RNA Pol II near the promoter. To overcome this block, P-TEFb is recruited to RNA Pol II to phosphorylate the SPT5 subunit of DRB sensitivity-inducing factor and the RNA-binding subunit of negative elongation factor F, removing the repressive block on RNA Pol II. P-TEFb then phosphorylates the CTD of RNA Pol II, primarily at the Ser2 residues, allowing for productive elongation (17, 25).
In addition to its role in transcriptional elongation, cdk9 also plays a role in RNA initiation and processing. Thus, P-TEFb was reported in the preinitiation complexes of human immunodeficiency virus and has been shown to induce transcription complex assembly by recruiting the TATA-box-binding protein (TBP) (32). In Saccharomyces cerevisiae, elongation of transcripts by RNA Pol II takes place in the absence of the cdk9 homologue Ctk1, but the recruitment of polyadenylation factors and processing of the 3′ ends are defective (5, 7, 39).
Viral gene products have been reported to modify the function of cdk9. Thus, the human immunodeficiency virus Tat protein interacts with P-TEFb and phosphorylates the CTD of RNA Pol II (12, 13, 21). Human cytomegalovirus infection has been shown to induce the hyperphosphorylation of the CTD of the large subunit of RNA Pol II. This modification is associated with changes in the abundance, activity, and localization of cdk9 and cdk7, as shown previously by S. Tamrakar et al. (40). This same group has also shown that the addition of cdk inhibitors at the start of infection alters the levels and localization of cdk9 and cdk7 (15). More recently, the Kaposi's sarcoma-associated herpesvirus K-cyclin protein has been shown to interact with cdk9 (6).
In this report, we show by two different methods, i.e., inhibition of cdk9 by DRB and reduction in the levels of cdk9 in cells transfected with small interfering RNA (siRNA) targeted to cdk9 mRNA, that cdk9 plays a role in the expression of the subset of genes regulated by ICP22. We also noted a slight but reproducible decrease in the accumulation of ICP22. In addition, we present evidence that cdk9 recruits ICP22 to the RNA Pol II complex.
MATERIALS AND METHODS
Cells and viruses.
U2OS (HTB-96), HEK 293T, and HEp-2 cells were obtained from the American Type Culture Collection (Manassas, VA) and grown in McCoy medium (Gibco-BRL) and Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, respectively. Herpes simplex virus 1 (F) [HSV-1(F)] is the prototype HSV-1 wild-type strain used in this laboratory (9). The recombinant virus R325 (Δα22) lacks 821 bp from the 3′ terminus of ICP22 of HSV-1(F) (30, 36). The R7356 mutant lacks ΔUL13 (31).
Cell infection.
Cells grown in 25- or 75-cm2 flasks were exposed to 10 PFU of virus per cell in medium consisting of mixture 199V supplemented with 1% calf serum. After 2 h of incubation at 37°C, the inoculum was replaced with growth medium alone or supplemented with different concentrations of DRB or dimethyl sulfoxide (DMSO) as described in Results. The cultures were reincubated at 37°C until harvested. Time zero is defined as the time of exposure of cells to virus.
Immunoblotting.
Cell lysates for immunoblotting were prepared as follows: the cell culture medium was removed and the cells were rinsed with phosphate-buffered saline (PBS), scraped into 5 ml of 1% NP-40-1% deoxycholate in PBS [PBS(A)], pelleted by centrifugation at 3,000 rpm, solubilized in high-salt lysis buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 0.5% NP-40, 400 mM NaCl, 0.1 mM sodium orthovanadate, 10 mM NaF, 2 mM dithiothreitol, 100 μg each of phenylmethylsulfonyl fluoride and tolylsulfonyl phenylalanyl chloromethyl ketone and leupeptin per ml), and stored on ice for 1 h. Insoluble materials were removed by centrifugation as described above. Samples were mixed with equal volumes of disruption buffer (2% sodium dodecyl sulfate, 50 mM Tris [pH 6.8], 2.75% sucrose, 5% β-mercaptoethanol, and bromophenol blue) or solubilized in 200 μl of disruption buffer and sonicated, boiled for 2 min, subjected to electrophoresis on a 10% bisacrylamide gel, transferred to nitrocellulose membranes, blocked for 2 h with 5% nonfat dry milk, and reacted with the appropriate primary antibody overnight at 4°C. The next day, the blots were reacted at room temperature for 1 h, rinsed three times in PBS, and reacted with the appropriate secondary antibody for 2 h. The blots were developed (i) by incubation in alkaline phosphatase (AP) buffer (100 mM Tris [pH 9.5], 100 mM NaCl, 5 mM MgCl2), followed by incubation in AP buffer containing BCIP (5-bromo-4-chloro-3-indolyphosphate) and nitroblue tetrazolium (the reaction was stopped by immersing the blot in a solution containing 100 mM Tris [pH 7.6] and 10 mM EDTA) or (ii) by enhanced chemiluminescence according to instructions supplied by the manufacturer (Pierce).
Antibodies.
The antibodies used in these studies were anti-actin (catalogue no. A4700; Sigma), anti-cyclin T (catalogue no. sc-10750; Santa Cruz, Inc.), anti-Cdk9 (catalogue no. sc-8338; Santa Cruz, Inc.), anti-RNA Pol II(8WG16) (catalogue no. MM5-126R-500; Covance), and anti-RNA Pol II(ARAN3) used at 1:250 dilutions. The rabbit polyclonal antibody to ICP22 (36) was used at a 1:500 dilution in PBS containing 1% bovine serum albumin and 0.05% Tween 20. Mouse monoclonal antibodies anti-ICP0, anti-ICP4, and anti-US11 and rabbit polyclonal antibody UL38 were from the Goodwin Cancer Research Institute (Plantation, FL) and used at a dilution of 1:1,000. Polyclonal antibody against UL41 was prepared by Jossman (Napa, CA) and used at a 1:1,000 dilution. Bound antibody was detected by using the following secondary antibodies: anti-mouse immunoglobulin G (IgG) peroxidase (catalogue no. A4416; Sigma), anti-rabbit IgG peroxidase (catalogue no. A0545; Sigma), anti-mouse IgG AP conjugate (catalogue no. 170-6520; Bio-Rad), and anti-rabbit IgG AP conjugate (catalogue no. 170-6518; Bio-Rad) diluted 1:3,000.
Isolation of total RNA.
U2OS cells were mock treated or treated for 2 h with 50 μM of DRB or DMSO alone and exposed to virus as described above. Total RNA extracted from cells harvested 10 h after infection with the aid of TRIzol reagent (Life Technologies) according to the manufacturer's instructions was digested with DNase (Life Technologies), extracted with phenol-chloroform, and precipitated with ethanol (Fisher Scientific) to remove possible DNA contamination. Total RNA was used for Northern analysis and real-time PCR analysis.
Northern blot analysis.
Total RNA (15 μg) extracted from treated or untreated U2OS cells infected with wild-type virus as described above was loaded onto a denaturing formaldehyde gel and probed with 32P-labeled probes for the viral proteins ICP4, ICP22, UL38, and UL41. The probes were PCR-amplified from viral DNA with the following set of probes: for ICP22, oligonucleotide 5 (5′TGCGCCTTGTGTAAAAGC) and oligonucleotide 6 (5′TGTCGCTGCACGGATAGG); for ICP4, oligonucleotide 7 (5′GCTCATCGTGGTCAACACC) and oligonucleotide 8 (5′CTTCCCAGTCCACAACTTCC); for UL38, oligonucleotide 9 (5′CATCATCTAACCCGCCAAGT) and oligonucleotide 10 (5′GTGAATCGTGTTGGTGATCC); and for UL41, oligonucleotide 11 (5′ATGAAGTTTGCCCACACACA) and oligonucleotide 12 (5′CACGGAGGCGTAGGTGTTAT). Prehybridization and hybridization were done with the ULTRAhyb buffer (Ambion) supplemented with 200 μg of denatured salmon sperm DNA per ml (Stratagene). The membrane was prehybridized at 42°C for 2 h and then overnight after the addition of the 32P-labeled probes. The membrane was rinsed as suggested by the ULTRAhyb manufacturer and exposed to film for signal detection.
siRNA transfections.
cdk9-directed siRNA was obtained from Ambion (catalogue no. 103567-103 and 103567-104). Cells were transfected with siRNA 24 h after seeding according to Dharmacon's Dharmafect transfection reagent protocol. The medium was replaced after 24 h, and the cells were infected 48 h after transfection and harvested 10 h later. The cell lysates were subjected to extraction for RNA or solubilized for immunoblot analyses.
Real-time PCR.
cdk9 RNA was quantified using the ABI TaqMan assays, as described by Randall et al. (33) (Applied Biosystems). Specifically, 2 μl of DNase I-treated total RNA was reverse transcribed with SuperScript II (Invitrogen) and oligo(dT) for 1 h at 42°C and then heat inactivated at 80°C for 20 min. One-twentieth of the cDNA mix was mixed with an equal volume of 2 μl SYBR Green master mix (Applied Biosystems) and the appropriate primers. PCR conditions were as follows: 50°C for 2 min and 95°C for 10 min (95°C for 15 s and 55°C for 1 min) for 40 cycles. Results were analyzed with SDS 1.7 software (Applied Biosystems). Relative cdk9 RNA levels were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) RNA levels.
Immunofluorescence.
For the infection experiments, HEp-2 cells grown on four-well confocal slides (Erie Scientific) were exposed to 10 PFU of wild-type or mutant viruses per cell for 2 h. After incubation at 37°C for a total of 10 h, the cells were fixed in 4% paraformaldehyde. After fixation, the cells were neutralized with 100 mM of glycine in PBS, permeabilized with 0.1% Triton X-100 in PBS in the presence of 2 mg/ml bovine serum albumin (PBS-TB), and reacted with primary antibodies, diluted in PBS-TB, for 2 h at room temperature. The anti-cdk9 rabbit polyclonal antibody (H-79; Santa Cruz, Inc.) was used in a dilution of 1:500; the anti-RNA Pol II mouse monoclonal antibody (8WG16) (catalogue no. MM5-126R-500; Covance) was used in a dilution of 1:1,000. The samples were then rinsed several times with PBS-TB and reacted with Alexa Fluor 594-conjugated goat anti-rabbit and Alexa Fluor 488-conjugated goat anti-mouse secondary antibodies (Molecular Probes), diluted 1:1,000 in PBS-TB, for 40 min at room temperature. After several washes, first with PBS-TB and then with PBS, the samples were mounted in Vectashield mounting medium for fluorescence (Vector Laboratories) and examined in a Zeiss confocal microscope equipped with software provided by Zeiss.
RESULTS
DRB causes a reduction in the accumulation of ICP22 and of selected late proteins in cells infected with wild-type virus.
In this series of experiments, U2OS cells grown in a 25-cm2 flask were mock infected or exposed to 10 PFU of HSV-1(F) per cell and then incubated in medium without additives or in medium containing 25 or 50 μM DRB or a concentration of DMSO corresponding to the concentration of DMSO in the medium of cells exposed to 50 μM DRB. The cells were harvested at 10 h after infection, lysed, denatured, subjected to electrophoresis in denaturing gels, and then reacted with antibodies as described in Materials and Methods. The results shown in Fig. 1A and B are as follows. We have repeatedly observed a slight reduction in the levels of ICP22 and particularly in the fast-migrating forms of the protein. We have also observed significant decreases in the accumulation of UL41, UL38, and US11 proteins in cells exposed to either 25 or 50 μM DRB. We did not observe a decrease in the levels of ICP4, ICP0, ICP27, gB, gC, gD, or thymidine kinase. The results indicate that DRB has a profound effect on the accumulation of the subset of proteins whose accumulation is dependent on ICP22. Concomitantly, we have observed a decrease in the levels of processed forms of ICP22. A similar loss was observed in infected cell cultures exposed to DRB as late as 6 h after infection (data not shown).
FIG. 1.
DRB causes a decrease in the accumulation of ICP22-regulated proteins. U2OS cells were mock infected (Mock) or exposed to HSV-(F). Two hours after infection, the inoculum was replaced with medium alone, medium plus different concentrations of DRB, or medium plus DMSO. The cells were harvested 10 h after infection, lysed, subjected to electrophoresis in denaturing gels, transferred to a nitrocellulose membrane, and reacted with an antibody to various viral proteins.
DRB affects the accumulation of UL41, UL38, and US11 viral mRNAs but not that of ICP22, gC, or ICP4.
In this series of experiments, U2OS cells grown as described above were exposed to 10 PFU of HSV-1(F) per cell. At 24 h after infection, the cultures were replenished with medium without additives or medium containing 25 or 50 μM DRB or DMSO at a concentration identical to that in medium containing 50 μM DMSO. The cells were harvested 10 h after infection. The RNA was then harvested, electrophoretically separated under denaturing conditions, and reacted with specific probes as described in Materials and Methods. Figure 2 shows the results of four experiments that may be summarized as follows. DRB had no effect on the accumulation of ICP4, ICP22, (Fig. 2A), gC (Fig. 2B), or thymidine kinase (Fig. 2D) mRNAs. On the other hand, both 25 and 50 μM DRB had a profound effect on the accumulation of UL38, UL41, (Fig. 2A), US11 (Fig. 2B), or UL13 (Fig. 2C) mRNAs. We conclude that the inhibition of accumulation of late (γ) mRNAs is selective. DRB inhibited the accumulation of mRNAs dependent on ICP22 (e.g., UL38, UL41, or US11) but not the mRNA of a late (γ2) gene (gC) whose expression is not dependent on ICP22. The effect of DRB on the accumulation of UL13 mRNA was unexpected, but since UL13 kinase is a major modifier of ICP22 and is required for the expression of UL38, UL41, or US11, the absence of UL13 could explain the absence of the slow-migrating, posttranslationally modified forms of ICP22 (Fig. 1).
FIG. 2.
DRB affects the accumulation of UL41, UL38, and US11 viral mRNAs but not the accumulation of those of ICP22, gC, or ICP4. Total RNA was harvested from U2OS cells that were exposed to HSV-1(F) alone (lane 1) or treated with DMSO (lane 2) or different concentrations of DRB as shown (lanes 3 and 4) at 10 h after infection. The RNAs (15 μg/lane) were subjected to electrophoresis in denaturing formaldehyde gels and probed with 32P-labeled probes for ICP4 (A), ICP22 (A), UL38 (A), UL41 (A), thymidine kinase (tk) (D), US11 (B), gC (B), or UL13 (C) mRNAs. Panels A to D show autoradiographic images of 32P-labeled probes hybridized to the electrophoretically separated RNA. Et Br, ethidium bromide.
The decrease in accumulation of US11 and ICP22 protein is not due to degradation by a common protease.
Replicate cultures of U2OS cells were either mock infected or exposed to 10 PFU of wild-type HSV-1(F) per cell for 2 h. The inoculum was then replaced with medium without additives (Fig. 3, lanes 1, 2, and 6), with DMSO (Fig. 3, lanes 4 and 7), with 50 mM DRB (Fig. 3, lanes 5 and 8), with DMSO plus a proteasome inhibitors (MG132 or lactacystin) (Fig. 3, lanes 9 and 10), or with DRB plus one of the proteasome inhibitors (Fig. 3, lanes 11 and 12) or single individual inhibitors (Fig. 3, lanes 13 and 14). One infected culture (Fig. 3, lane 2) was harvested at 2 h after infection. Another set (Fig. 3, lanes 3, 4, and 5) was harvested at 4 h after infection. All other cultures were harvested at 10 h after infection, solubilized, subjected to electrophoresis in denaturing gels, and probed with antibodies against either ICP22 or US11 protein.
FIG. 3.
The decrease in accumulation of US11 and ICP22 protein is not due to proteolytic digestion by common proteases. U2OS cells were either mock infected or exposed to 10 PFU of wild-type HSV-1(F) per cell for 2 h. The inoculum was then replaced with medium alone, medium plus 50 μM of DRB, or medium plus DMSO. The cells were incubated at 37°C for three additional hours before being exposed to the proteasome or protease inhibitors in the presence (lanes 4 to 8) or absence (lanes 9 to 11) of 50 μM DRB. Cells were harvested 10 h after infection, and the electrophoretically separated proteins were immunoblotted for ICP22 and US11.
The results were as follows. (i) The levels of ICP22 at 4 h after infection increased relative to those at 2 h (Fig. 3, compare lanes 3 and 2). At 4 h after infection, exposure of the infected cells for 2 h to DMSO had no effect on accumulation of ICP22 (Fig. 3, compare lanes 3 and 4). In contrast, exposure of the infected cells for 2 h to DRB resulted in a decrease in the accumulation of ICP22 (Fig. 3, compare lanes 4 and 5).
(ii) At 10 h after infection, the cells exposed to DRB for 8 h accumulated significantly less ICP22 than untreated cells (Fig. 3, compare lanes 8 and 6). In contrast, DMSO had no effect on the accumulation of ICP22 over the same time interval (Fig. 3, compare lanes 8 and 7).
(iii) If the decrease in the accumulation of ICP22 in infected cells treated with DRB were due to proteasome dependent degradation, it could be expected that cells treated with MG132 or lactocystin would have accumulated more ICP22 than cells treated with DRB alone. A comparison of lanes 11 and 12 with lane 8 in Fig. 3 does not support this expectation.
(iv) In parallel assays, we examined the accumulation of US11. This protein was not detected in cells harvested at 2 or 4 h after infection (Fig. 3, lanes 2 to 5). It was absent or barely detectable in cells treated with DRB and harvested at 10 h after infection (Fig. 3, lane 8). Neither MG132 nor lactosystin had an effect on the accumulation of this protein (Fig. 3, compare lanes 11 and 12 with lane 8).
On the basis of the results presented in Fig. 2 and 3, we conclude that the major effect of DRB is on the accumulation of mRNAs encoding a subset of late proteins whose synthesis is regulated by ICP22 and the UL13 protein kinase. It is noteworthy that the ICP22 mRNA is not affected by DRB. The effect of DRB on ICP22 may well reflect a lack of posttranslational processing resulting from the reduction in the synthesis of UL13 protein kinase.
cdk9 siRNA selectively inhibits the accumulation of cdk9 mRNA but not cdk7 mRNA.
In this series of experiments, replicate cultures of HEK-293 cells were mock treated or transfected with Dharmafect transfection reagent 1 (Dharmacon) alone, 25 or 50 nM of siRNA 103, 25 or 50 nM of siRNA 104, or random siRNA. siRNAs 103 and 104 were directed against cdk9. The cells were exposed to 10 PFU of HSV-1(F) per cell at 48 h after transfection and were maintained at 37°C for 10 h. At that time, the cells were harvested and total RNA was extracted and analyzed by real-time PCR as described in Materials and Methods for the presence of cdk9 and cdk7 mRNAs. The amounts of cdk9 and cdk7 mRNAs were normalized with respect to the levels of the mRNAs in uninfected, untreated cells.
The results were as follows. (i) A 20% reduction in the levels of cdk9 mRNA was observed in untreated infected cells (Fig. 4, lane 2), in cells exposed to reagent and then infected (Fig. 4, lane 3), and in cells transfected with random siRNA and then infected (lane 8). Cells transfected with 103 or 104 siRNA and then infected exhibited a >70% reduction in the levels of cdk9 mRNA (Fig. 4, lanes 4 to 7). We note in particular that cells transfected with 50 nM of 104 siRNA exhibited a 90% reduction in the level of cdk9 mRNA.
FIG. 4.
siRNA targeted against cdk9 results in a reduction of cdk9 mRNA accumulation but not a reduction of cdk7 mRNA. HEK-293 cells were infected with HSV-1(F) 48 h after transfection with Dharmacon's Dharmafect (DHF) transfection reagent 1, either alone or mixed with a random siRNA or different concentrations of two different siRNAs targeted against cdk9, as shown. After 10 h of incubation, the cells were harvested and the total RNA extracted from the cells was analyzed by real-time PCR for cdk9 and cdk7 mRNAs as described in Materials and Methods.
(ii) In contrast, infection with HSV-1(F) alone or after transduction with 103 or 104 siRNAs had little, if any, effect on the levels of cdk7 mRNA (Fig. 4B, lanes 1 to 8).
siRNA targeted to cdk9 causes a decrease in UL38 viral mRNA but not in ICP22 or ICP4.
In this series of experiments, HEK-293 cells were transfected with 25, 50, or 100 nM of cdk9 siRNA 104, with random siRNA, or with Dharmacon's Dharmafect transfection reagent 1. After 48 h, the cells were mock infected or exposed to 10 PFU of HSV-1(F) per cell. The RNA extracted from cells 10 h after infection (15 μg/sample) was subjected to electrophoresis in a denaturing formaldehyde gel and probed with 32P-labeled probes for ICP4 (Fig. 5A), 18S (Fig. 5B), ICP22 (Fig. 5C), or UL38 (Fig. 5D) mRNAs. As shown in Fig. 5, there was a decrease in the accumulation of UL38 but not of ICP22 or ICP4 mRNAs.
FIG. 5.
In infected cells transduced with cdk9 siRNA, there is a decrease in the accumulation of UL38 mRNA but not in that of ICP22 or ICP4 mRNAs. HEK-293 cells were infected with HSV-1(F) 48 h after transfection with the siRNA targeted against cdk9 or the transfection reagent Dharmafect (DHF). Total RNA extracted 20 h after infection was subjected to electrophoresis in a denaturing formaldehyde gel (15 μg/lane) and probed with 32P-labeled fragments of ICP4 (A), 18S (B), ICP22 (C), and UL38 (D).
ICP22 accumulates in smaller amounts in cells transfected with siRNA targeted against cdk9.
In this series of experiments, HEK-293 cells were transfected with 25, 50, or 100 nM of 104 siRNA, with random siRNA, or with Dharmacon's Dharmafect transfection reagent 1. After 48 h, the cells were mock infected or exposed to 10 PFU of HSV-1(F) per cell. At 10 h after infection, the cells were harvested, lysed, subjected to electrophoresis in denaturing gels, transferred to a nitrocellulose sheet, and probed with antibodies to ICP4, ICP22, cdk9, and actin. As shown in Fig. 6, in cells transfected with cdk9 siRNA 104, there was a reduction in the accumulation of ICP22, UL38, US11, and, as expected, in that of cdk9, but not in the accumulation of ICP4 or ICP0.
FIG. 6.
In infected cells transfected with cdk9 siRNA, there is a decrease in the accumulation of ICP22, UL38, and US11 proteins. The procedures were the same as those described in the legend for Fig. 5, except that the cell lysates were subjected to electrophoresis in a denaturing polyacrylamide gel and reacted with antibodies against ICP4, ICP0, ICP22, UL38, US11, cdk9, or actin.
Colocalization of cdk9 and RNA Pol II in cells infected with wild-type and mutant viruses.
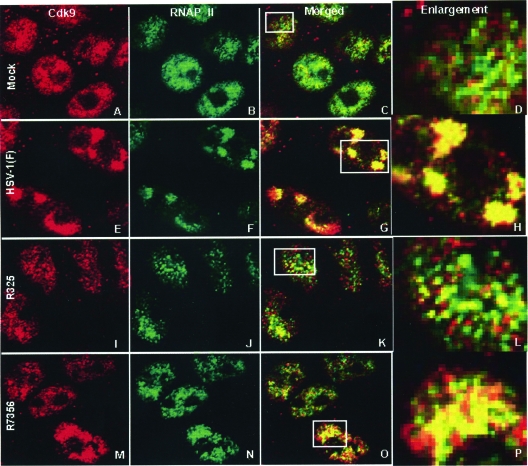
Two series of experiments were done. In the first, HEp-2 cells grown in four-well slides were mock infected or exposed to HSV-1(F). After 6 or 10 h of infection, the cells were fixed and reacted with antibodies to cdk9 and RNA Pol II. As shown in Fig. 7E to L, the images of cdk9 and RNA Pol II colocalize. This is clearly evident from the coincidence of red and green pixels in the enlargements shown in Fig. 7H and L. The foci of coincident Pol II and cdk9 evolved from punctate structures and coalesced into large structures with time after infection (compare Fig. 7G and K, containing images collected at 6 and 10 h after infection). In the second series of experiments, we examined the distribution of cdk9 and RNA Pol II in cells fixed 10 h after infection with HSV-1(F), ΔICP22 (R325), or ΔUL13 (R7356). As expected, cdk9 and RNA Pol II colocalized in cells infected with HSV-1(F) (Fig. 8E to H). In the images shown here, a portion of the nuclei was enlarged in order to observe the individual pixels. Thus, cdk9 and RNA Pol II colocalized in HSV-1(F)-infected cells (Fig. 8G and H). In contrast, the green and red pixels are separated in enlargements of nuclei of cells infected with R325, indicating that in the absence of ICP22, cdk9 and RNA Pol II did not colocalize. In the case of cells infected with ΔUL13 mutant R7356, the green and red pixels appear to be separated in most cells. Even with the nucleus enlarged (Fig. 8O and P), the colocalization is not nearly as compelling as that seen in Fig. 8G and H.
FIG. 7.
cdk9 and RNA Pol II colocalize at late times after infection. HEp-2 cells grown in four-well slides were mock infected or exposed to HSV-1(F). After 6 or 10 h of infection, the cells were fixed in 4% paraformaldehyde. After fixation, the cells were neutralized with 100 mM of glycine in PBS, permeabilized with 0.1% Triton X-100, and reacted with antibodies to cdc9 and RNA Pol II. Slides were examined in a Zeiss confocal microscope equipped with software provided by Zeiss. Panels A, E, and I show mock-infected cells and cells infected with the wild-type virus for 6 h and 10 h, respectively, stained with the anti-cdk9 antibody. Panels B, F, and J show mock infected cells and cells infected with the wild-type virus for 6 h and 10 h, respectively, stained with the anti-RNA Pol II antibody. Panels C, G, and K show the merged images of the cdk9 and RNA Pol II stains. Panels D, H, and L show an enlargement of the boxed areas in panels C, G and K, respectively.
FIG. 8.
cdk9 and RNA Pol II colocalize in cells infected with wild-type but not mutant viruses. The procedures were the same as those described in the legend for Fig. 7. Panels A, E, I, and M show mock-infected cells and cells infected with wild-type viruses R325 and R7356, respectively, stained with the anti-cdk9 antibody. Panels B, F, J, and N show mock-infected cells and cells infected with wild-type viruses R325 and R7356, respectively, stained with the anti-RNA Pol II antibody. Panels C, G, K, and O show the merged images of the cdk9 and RNA Pol II stains. Panels D, H, L, and P show an enlargement of the boxed areas in panels C, G, K, and O, respectively.
DISCUSSION
The interest of this laboratory in the function of ICP22 stems from the fact that α22, the gene encoding this protein, was the first gene deleted by genetic engineering using selectable markers (30). Numerous reports that followed this observation still left a gap in our understanding of the function of ICP22. What is known is that the deletion mutant grows moderately well in continuous human or primate cell lines but grows poorly in primary cell lines or in rodent cell lines (36). In the absence of ICP22, a subset of late protein exemplified by UL38, UL41, and US11 accumulates in smaller amounts (2, 24, 31, 37). A key viral protein required for this function of ICP22 is the UL13 protein kinase (2, 24, 31). This function maps near the carboxyl terminus of ICP22, as it is at least one of possibly several UL13 phosphorylation sites. Additional phenotypic properties of the protein have also been described previously (18, 24, 28, 29, 35). The regulation of the subset of late proteins has been linked to two series of events. First, as noted in the introduction, ICP22 mediates the degradation of cyclins A and B (1, 3). In the process, cdc2 acquires a new partner, the UL42 DNA polymerase accessory factor, and cdc2 together with UL42 binds and posttranslationally modifies topoisomerase IIα, all in an ICP22-dependent fashion (4). In a possibly related series of events, cdk9 interacted with ICP22 in a US3 protein kinase-dependent fashion, and this complex phosphorylated the CTD of RNA Pol II in vitro (8). These results raised two questions. The foremost question is the role of cdk9 in the expression of viral genes and in particular the significance of its interaction with ICP22. To address this question, we performed three series of experiments.
In the first experiment, we blocked the function of cdk9 by exposing cells to DRB. These studies showed that in the presence of DRB, the accumulations of ICP22-regulated proteins and mRNAs were reduced, whereas the accumulation of thymidine kinase, a β gene product, and the accumulation of ICP0 and ICP4, α proteins, were unaffected. We also noted a decrease in ICP22 protein but not mRNA and a decrease in the levels of UL13 mRNAs. As shown in Fig. 3, we cannot relate the decrease in ICP22 protein levels to protease activity as tested to date.
In the second series of experiments, we blocked the synthesis of cdk9 with the aid of siRNA directed against cdk9. In these studies, we observed a 75 to 90% reduction in the levels of cdk9, but this had no significant effect on cdk7. The siRNA caused a reduction in the levels of UL38 mRNA, a representative of the genes regulated by ICP22, but not of those of ICP22. At the protein levels, we observed a reduction in the accumulation of ICP22- and ICP22-regulated proteins UL38 and US11 but not in the accumulation of those of ICP0 or ICP4. We conclude from these two series of experiments that cdk9 is required for the optimal accumulation of mRNAs and proteins regulated by ICP22. The puzzling feature of the results is the decrease in the level of ICP22 protein but not ICP22 mRNA in cells treated with DRB or cells in which the level of cdk9 was reduced by siRNA. One hypothesis that remains to be tested is that the stability of ICP22 depends on the UL13 protein kinase and that in the absence of cdk9, the levels of UL13 decrease concomitantly with the decrease in the levels of UL13 mRNA in cells treated with DRB. We should stress that the studies with the inhibitors of selected proteases failed to produce convincing evidence of turnover of ICP22 protein.
The third series of experiments focused on the localization of cdk9 and ICP22 in infected cells. Our results show that in wild-type virus-infected cells, cdk9 and RNA-Pol II colocalize. In cells infected with the ΔICP22 mutant or in cells infected with the ΔUL13 mutant, cdk9 and RNA Pol II are in close proximity but do not colocalize.
The second question raised by preceding (8) and current studies is the apparent discrepancy in the roles of the viral protein kinases US3 and UL13 in the phosphorylation of RNA Pol II. Earlier studies have shown that the expression of the subset of genes regulated by ICP22 requires, in addition, a functional UL13 protein kinase (31) and that this enzyme is also required for the shift in the electrophoretic mobility of RNA Pol II observed by Rice et al. (35) in wild-type, virus-infected Vero cells. On the other hand, we reported that RNA Pol II was phosphorylated by the complexes immunoprecipitated by ICP22 and cdk9 from wild-type, virus-infected cells, to a lesser extent from ΔUL13-infected cells, and not at all from ΔUS3-infected cells. In this study, we report that cdk9 colocalized poorly if at all with RNA Pol II in cells infected with ΔUL13 mutant virus. One explanation of the results presented here is that the viral protein kinases perform multiple functions defined in part by posttranslational modification. Thus, UL13 has been shown to phosphorylate the US3 kinase (16, 29), and very likely UL13 kinase is similarly phosphorylated by both cellular and viral kinases. Given this complexity, the roles of the protein kinases may have to be defined by direct interaction of suitably modified, purified kinases with RNA Pol II and its cellular and viral partners.
Our studies indicate that cdk9 plays a key role in the accumulation of the subset of late gene products whose abundance is regulated by ICP22. The precise mechanism by which cdk9 acts is unclear. The results suggest, however, that cdk9 is in the same complex with RNA Pol II in wild-type, virus-infected cells. One hypothesis that is consistent with all available data is that cdk9 brings posttranslationally modified ICP22 into the transcriptional complex.
Acknowledgments
These studies were aided by National Cancer Institute grant CA83939.
Footnotes
Published ahead of print on 27 August 2008.
REFERENCES
- 1.Advani, S. J., R. Brandimarti, R. R. Weichselbaum, and B. Roizman. 2000. The disappearance of cyclins A and B and the increase in activity of the G2/M-phase cellular kinase cdc2 in herpes simplex virus 1-infected cells require expression of the α22/US1.5 and UL13 viral genes. J. Virol. 748-15. [PMC free article] [PubMed] [Google Scholar]
- 2.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2000. The role of cdc2 in the expression of herpes simplex virus genes. Proc. Natl. Acad. Sci. USA 9710996-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2001. cdc2 cyclin-dependent kinase binds and phosphorylates herpes simplex virus 1 UL42 DNA synthesis processivity factor. J. Virol. 7510326-10333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Advani, S. J., R. R. Weichselbaum, and B. Roizman. 2003. Herpes simplex virus 1 activates cdc2 to recruit topoisomerase IIα for post-DNA synthesis expression of late genes. Proc. Natl. Acad. Sci. USA 1004825-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 1367-76. [DOI] [PubMed] [Google Scholar]
- 6.Chang, P. C., and M. Li. 2008. Kaposi's sarcoma-associated herpesvirus K-cyclin interacts with Cdk9 and stimulates Cdk9-mediated phosphorylation of p53 tumor suppressor. J. Virol. 82278-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, E.-J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 153319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Durand, L. O., S. J. Advani, and B. Roizman. 2005. The carboxyl-terminal domain of RNA polymerase II is phosphorylated by a complex containing cdk9 and infected-cell protein 22 of herpes simplex virus 1. J. Virol. 796757-6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ejercito, P., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2357-364. [DOI] [PubMed] [Google Scholar]
- 10.Fu, T., J. Peng, G. Lee, D. H. Price, and O. Flores. 1999. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J. Biol. Chem. 27434527-34530. [DOI] [PubMed] [Google Scholar]
- 11.Grana, X., A. Luca, N. Sang, Y. Fu, P. Claudio, and J. Rosenblatt. 1994. PITALRE, a nuclear CDC2-related protein kinase that phosphorylates the retinoblastoma protein in vitro. Proc. Natl. Acad. Sci. USA 913834-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrmann, C., and A. Rice. 1995. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J. Virol. 691612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrmann, C. H., and A. P. Rice. 1993. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology 197601-608. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins, H. L., and C. A. Spencer. 2001. RNA polymerase II holoenzyme modifications accompany transcription reprogramming in herpes simplex virus type 1-infected cells. J. Virol. 759872-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapasi, A. J., and D. H. Spector. 2008. Inhibition of the cyclin-dependent kinases at the beginning of human cytomegalovirus infection specifically alters the levels and localization of the RNA polymerase II carboxyl-terminal domain kinases cdk9 and cdk7 at the viral transcriptosome. J. Virol. 82394-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato, A., M. Yamamoto, T. Ohno, M. Tanaka, T. Sata, Y. Nishyama, and Y. Kawaguchi. 2006. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 801476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, D. K., Y. Yamaguchi, T. Wada, and H. Handa. 2001. The regulation of elongation by eukaryotic RNA polymerase II: a recent view. Mol. Cells 11267-274. [PubMed] [Google Scholar]
- 18.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 735593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, H., and C. H. Herrmann. 2005. Differential localization and expression of the Cdk9 42k and 55k isoforms. J. Cell. Physiol. 203251-260. [DOI] [PubMed] [Google Scholar]
- 20.Malumbres, M., and M. Barbacid. 2005. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 30630-641. [DOI] [PubMed] [Google Scholar]
- 21.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 27127176-27183. [DOI] [PubMed] [Google Scholar]
- 22.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 27012335-12338. [DOI] [PubMed] [Google Scholar]
- 23.Marshall, R. M., and X. Grana. 2006. Mechanisms controlling CDK9 activity. Front. Biosci. 112598-2613. [DOI] [PubMed] [Google Scholar]
- 24.Ogle, W. O., and B. Roizman. 1999. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J. Virol. 734305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palancade, B., and O. Bensaude. 2003. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 2703859-3870. [DOI] [PubMed] [Google Scholar]
- 26.Peng, J., N. F. Marshall, and D. H. Price. 1998. Identification of a cyclin subunit required for the function of drosophila P-TEFb. J. Biol. Chem. 27313855-13860. [DOI] [PubMed] [Google Scholar]
- 27.Peng, J., Y. Zhu, J. T. Milton, and D. H. Price. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon, A. P. W., W. O. Ogle, and B. Roizman. 2000. The posttranslational processing of infected cell protein 22 mediated by viral protein kinases is sensitive to amino acid substitutions at distant sites and can be cell-type specific. J. Virol. 7411210-11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poon, A. P. W., and B. Roizman. 2005. The herpes simplex virus 1 ICP22 regulates the accumulation of a shorter mRNA and of a truncated US3 protein kinase that exhibits altered functions. J. Virol. 798470-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Post, L. E., and B. Roizman. 1981. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell 25227-232. [DOI] [PubMed] [Google Scholar]
- 31.Purves, F., W. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 906701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raha, T., S. W. Cheng, and M. R. Green. 2005. HIV-1 tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 3e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randall, G., L. Chen, M. Panis, A. K. Fischer, B. D. Lindenbach, and J. Sun. 2006. Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology 1311584-1591. [DOI] [PubMed] [Google Scholar]
- 34.Rice, S. A., M. C. Long, V. Lam, and C. A. Spencer. 1994. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J. Virol. 68988-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rice, S. A., M. C. V. Lam, P. A. Schaffer, and C. A. Spencer. 1995. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 695550-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sears, A. E., I. W. Halliburton, B. Meignier, S. Silver, and B. Roizman. 1985. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J. Virol. 55338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shore, S. M., S. A. Byers, P. Dent, and D. H. Price. 2005. Characterization of Cdk955 and differential regulation of two Cdk9 isoforms. Gene 35051-58. [DOI] [PubMed] [Google Scholar]
- 38.Shore, S. M., S. A. Byers, W. Maury, and D. H. Price. 2003. Identification of a novel isoform of Cdk9. Gene 307175-182. [DOI] [PubMed] [Google Scholar]
- 39.Skaar, D. A., and A. L. Greenleaf. 2002. The RNA polymerase II CTD kinase CTDK-I affects pre-mRNA 3′ cleavage/polyadenylation through the processing component Pti1p. Mol. Cell 101429-1439. [DOI] [PubMed] [Google Scholar]
- 40.Tamrakar, S., A. J. Kapasi, and D. H. Spector. 2005. Human cytomegalovirus infection induces specific hyperphosphorylation of the carboxyl-terminal domain of the large subunit of RNA polymerase II that is associated with changes in the abundance, activity, and localization of cdk9 and cdk7. J. Virol. 7915477-15493. [DOI] [PMC free article] [PubMed] [Google Scholar]