Abstract
Human papillomavirus (HPV) types from the beta genus (beta-HPVs) have been implicated in the development of skin cancer. A potentially important aspect of their carcinogenic role is the ability of the E6 protein to degrade the proapoptotic family member Bak, which gives cells the ability to survive UV damage. However, it is unknown if the ability to degrade Bak is limited to certain beta-HPV types or whether E6 expression in keratinocytes affects other proteins important for apoptosis signaling. We tested the abilities of E6 proteins from several representative members of the beta-HPVs to degrade Bak and protect UV-treated keratinocytes from apoptosis. The E6 proteins of the beta-HPV type 5 (HPV5), -8, -20, -22, -38, -76, -92, and -96, as well as the alpha genus HPV HPV16, all degraded Bak or prevented its accumulation following UV treatment but did not degrade Bak constitutively. In addition, when tested using HPV16 E6 (16E6) and 8E6 as representative E6 proteins from the alpha and beta genera, respectively, Bak degradation was dependent on the E3 ubiquitin ligase, E6AP. Other important regulators of apoptotic signaling were examined and found to be unperturbed by the expression of the beta-HPV E6 proteins. Importantly, the expression of beta-HPV E6 proteins protected keratinocytes from apoptosis to the same extent as 16E6-expressing cells. In conclusion, several of the beta-HPV types possess the ability to protect UV-treated keratinocytes from apoptosis by reducing levels of Bak in those cells, thus blocking the intrinsic apoptotic pathway.
Human papillomaviruses (HPVs) are small DNA tumor viruses that infect cutaneous and mucosal epithelia and disseminate by replication in terminally differentiated keratinocytes. So far, over 100 different types have been identified and characterized on the basis of DNA sequence analysis (12). Only a small number of these display a strong association with cancer development. Most notably, the high-risk types (16, 18, 31, 33, etc.) of alpha genus HPVs (alpha-HPVs) play a critical role in the development of cervical cancer. These same types have also been implicated in the majority of other anogenital cancers and a subset of head and neck carcinomas (11, 43).
Recently, the beta-HPVs, which cause cutaneous lesions in humans, have been linked to the development of skin cancers (32). The association between beta-HPVs and skin cancer was first identified in patients with the rare inherited disorder epidermodysplasia verruciformis (EV) (27). These individuals have a predisposition to the early development of disseminated, persistent flat warts and macular lesions following infection with a specific group of about 20 related beta-HPV genotypes, also known as EV types. About half of EV patients develop premalignant skin lesions and squamous cell carcinomas by age 40, primarily in sun-exposed areas (30). DNA from these lesions was found to harbor HPV genomes, suggesting a cocarcinogenic role of beta-HPVs and UV radiation in the early development of EV cancers (32). By use of sensitive PCR methods, HPV DNA sequences have since been detected in nonmelanoma skin cancers (NMSC) from immunosuppressed and immunocompetent individuals (8, 33). More recently, HPV DNA loads within actinic keratoses were found to exceed those of NMSC, suggesting a role for HPVs only in the early stages of skin cancer (35, 39).
The critical role that high-risk alpha-HPVs play in the etiology of cervical cancer is now well established and supported by a wealth of epidemiological and experimental evidence (15, 42). Cervical carcinomas generally harbor integrated HPV genomes within every cell of the tumor and continue to express the E6 and E7 viral oncogenes. The high-risk E6 and E7 proteins modify the expression activity of many cellular proteins in order to promote cell proliferation. It has been shown that both the E6 and E7 proteins of high-risk HPVs are necessary for the efficient immortalization of human keratinocytes (28). Conversely, the E6 and E7 proteins of low-risk HPVs do not seem to express comparable cell-transforming activities (5, 34).
In high-risk alpha-HPVs, the E6 protein complexes with E6AP and promotes the ubiquitin-mediated degradation of p53, a cellular tumor suppressor that regulates cell cycle arrest and apoptosis in damaged cells (19, 20, 26). In addition, E6 possesses redundant mechanisms for inactivating p53 and apoptosis. E6 can bind to p300, which blocks p53 acetylation and inhibits the ability of p53 to transactivate gene expression (31). Furthermore, it has been observed that cells expressing the HPV16 E6 protein (16E6) also display reduced ability to repair DNA damage (13, 18). As the repair of UV damage is, at least in part, dependent on the p53 status of cell, this may be due to HPV16-infected cells lacking functional p53 (38).
However, the ability to degrade p53 seems to be restricted to the high-risk HPVs, as the beta-HPVs and low-risk alpha-HPVs do not retain this activity (26). Despite this inability, the E6 proteins from several cutaneous HPV types effectively inhibit apoptosis in response to UV damage (22). While UVB irradiation is known to stimulate the promoter activity of certain beta-HPV types (2), the exact mechanism for protection from apoptosis by these HPV types is not well understood. One possible explanation for this cytoprotective effect is the ability of the E6 proteins of both high- and low-risk HPVs to target the proapoptotic effector Bak for proteolytic degradation (21, 36, 37).
In healthy keratinocytes, Bak is sequestered by Mcl-1 and Bcl-xL, which keeps Bak inactive (41). Upon UV irradiation, Noxa and other BH3-only proteins interact with Mcl-1 and Bcl-xL, displacing them from Bak. Mcl-1 is also transiently targeted for degradation by the HECT ubiquitin ligase, Mule, which is necessary for the initiation of apoptosis following UV irradiation (29). The released Bak then multimerizes in the mitochondrial membrane, releasing cytochrome c and activating the caspase cascade (3, 10, 40). The ability of some E6 proteins to eliminate Bak has been demonstrated (21); however, the ability of other beta-HPV types to degrade Bak has yet to be determined, and it is unclear whether other members of the apoptotic cascade must also be inactivated by E6 to block apoptosis. We show that all beta-HPV E6 proteins tested are equivalent in the ability to degrade Bak and protect UV-damaged keratinocytes from apoptosis without altering other regulators of Bak. This suggests a universal cytoprotective mechanism of HPV infection.
MATERIALS AND METHODS
Tissue culture.
Primary human foreskin keratinocytes (HFKs) were derived from neonatal human foreskins and grown in EpiLife medium supplemented with calcium chloride (60 μM) and human keratinocyte growth supplement (Cascade Biologics, Portland, OR). 293T and HT1080 cells were grown in Dulbecco's modified Eagle's medium (Gibco-BRL) containing 10% fetal bovine serum and penicillin-streptomycin.
E6 sequence alignment.
A cladogram was constructed from the E6 amino acid sequences of the beta-HPVs. The sequences were taken from GenBank (NCBI, http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi) and aligned with the ClustalW program (EMBL-EBI, http://www.ebi.ac.uk/clustalw/).
Plasmids.
Full-length HPV DNA from HPV5, -8, -38, and -76 (kindly provided by E-M. de Villiers), -20 and -22 (Michel Favre), and -92 and -96 (Ola Forslund) were used as the templates for subsequent PCRs in order to clone the E6 genes into the pLXSN vector. The PCR primers used for the untagged versions of E6 were as follows: 5E6 forward (5′ AAAAAGCAGGCTTGGCAATGGCTGAGGGAG 3′) and reverse (5′ AGAAAGCTGGGTGACCTCTTTACCAATCATG 3′), 8E6 forward (5′ AAAAAGCAGGCTTGGAAATGGACGGGCAGG 3′) and reverse (5′ AGAAAGCTGGGTCTCTTTACCAATCATGATAC 3′), 20E6 forward (5′ AAAAAGCAGGCTGGGACATGGCTACACCTC 3′) and reverse (5′ AGAAAGCTGGGTATCATTATTGAAAATGCTTACAC 3′), 22E6 forward (5′ AAAAAGCAGGCTTAAACATGCAACCGCTTGTG 3′) and reverse (5′ AGAAAGCTGGGTCCAATCATTCTATTGCTTTAC 3′), 38E6 forward (5′ AAAAAGCAGGCTTAATCATGGAACTACCAAAAC 3′) and reverse (5′ AGAAAGCTGGGTCCAATCATTCTATTGCTTTGC 3′), 76E6 forward (5′ AAAAAGCAGGCTGAGACATGGCTAGACCTG 3′) and reverse (5′ AGAAAGCTGGGTTTCCCAATCATTCTATTACTC 3′), 92E6 forward (5′ AAAAAGCAGGCTTCACAATGGCAAAACCTCCTTC 3′) and reverse (5′ AGAAAGCTGGGTGTTTCCCAATCATATCTCTGTAC 3′), and 96E6 forward (5′ AAAAAGCAGGCTAGGTGATGCAGTATCTGATCC 3′) and reverse (5′ AGAAAGCTGGGTTTTCCCCAATCATATCTCTCTAC 3′). The E6 genes were then inserted via the Gateway recombination-based system (Invitrogen, Carlsbad, CA) into pLXSN for retroviral transfection and into pDEST15 for glutathione S-transferase pulldowns. All constructs were verified by DNA sequencing. The pBABE-puro, pGEX2T-16E6, pLXSN, pLXSN-16E6, and pBABE-E6AP-sh constructs have been described previously (16). The full-length human Bak construct used for in vitro translations was kindly provided by D. L. George (University of Pennsylvania School of Medicine, Pittsburgh, PA).
RT-PCR.
RNA was isolated with the Trizol reagent (Invitrogen, Carlsbad, CA). Briefly, 1 ml of Trizol was added to each 10-cm plate, cells were incubated 5 min, and 200 μl of chloroform added. The aqueous phase was transferred and mixed with an equal volume of isopropanol, incubated for 10 min, and pelleted at 12,000 rpm for 10 min at 4°C. After being washed with 75% ethyl alcohol, the samples were again pelleted at 9,000 rpm for 5 min and resuspended in RNase-free H2O. cDNA copies of RNA templates were made using Superscript II reverse transcriptase (RT) (Invitrogen, Carlsbad, CA) according to the manufacturer's protocols. PCR amplification was then performed to identify 100-bp amplicons with the designated E6 primers as follows: 5E6 forward (5′ GAGGGAGCCGAACACCAA 3′) and reverse (5′ CAATCACAGGGATGCCTAAGG 3′), 8E6 forward (5′ TTAGGTGTCAAAACTGCTTGTCATT) and reverse (5′ CCTTTCCAGCCTCCTCTAACTTT 3′), 16E6 forward (5′ GCACAGAGCTGCAAACAACTATACA 3′) and reverse (5′ TCCCGAAAAGCAAAGTCATATACC 3′), 20E6 forward (5′ TTTTGCATGCTGTCGTGTTTG 3′) and reverse (5′ TGTTACTTGCTCTATGTCTCTGCCTAA 3′), 22E6 forward (5′ GCCTACGCTTCAGCCCAAT 3′) and reverse (5′ AAATTTGGCCTACAGGTCGTTGT 3′), 38E6 forward (5′ GAGGATTTTGTTTTTGCATGTTGT 3′) and reverse (5′ CAATTTCACGGCCAAAGACA 3′), 76E6 forward (5′ TGTGGAAGGACGGATTTTGC 3′) and reverse (5′ ATGCCTACCACAGTTTCCTGATG 3′), 92E6 forward (5′ TATGCTTGCTGTGGTGCTTGT 3′) and reverse (5′ AGTCCCTTTCTATAGCATCCTTTCC 3′), and 96E6 forward (5′ ACCGATCCAGTGGCTTTGC 3′) and reverse (5′ AGTTGCGAAACTTACCGTTAACG 3′). The primers for 36B4 were previously described (16). For real-time RT-PCR, cells were grown and treated with UV in six-well plates and harvested by use of 0.5 ml Trizol reagent. RNA was isolated as described above and subsequently processed using an RNeasy mini-cleanup protocol (Qiagen). For analysis of mRNA levels, TaqMan gene expression assays Hs00832876 (Bak) and 4333764F (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) were used (Applied Biosystems, Foster City, CA). For data analysis, the ΔΔCT method (where CT is for threshold cycle) was used, with GAPDH as a control to calculate the change compared to LXSN control cell data.
Retrovirus production and infection.
Retroviruses were produced either in established viral producer cell lines (PA317 or PG13) or transiently in 293T cells by a vesicular stomatitis virus G-pseudotyped virus production protocol as previously described (6). Briefly, after concentration of the virus by ultracentrifugation, HFKs were infected at ∼60% confluence in 10-cm plates with the addition of Polybrene (8 μg/ml). Four hours after infection, cells were washed with phosphate-buffered saline (PBS) and the medium was replaced. The cells were allowed to recover for 24 h before the addition of selective media. HFKs were selected in G418 (50 μg/ml) or puromycin (0.5 μg/ml) as appropriate. Selection in G418 was usually complete within 7 days. Cells expressing both E6 (LXSN) and E6APsh (pBABE-puro) were additionally selected in puromycin-containing medium for 5 days following G418 selection.
UVB irradiation.
Cells were allowed to reach 50 to 70% confluence and inoculated with fresh medium 24 h before irradiation. For treatment with UVB, cells were washed once with PBS and then irradiated through a thin film of PBS with either 15 mJ/cm2, 20 mJ/cm2, or 25 mJ/cm2 of UVB. Fresh medium was replaced and lysates were harvested at various time points, as indicated. The UVB source is a parallel bank of two FS20T12/UVB bulbs (Solarc Systems, Inc., Barrie, ON, Canada) with an output range of 280 to 320 nm. The UVB output is measured with an IL1400A radiometer coupled with the SEL240/UVB-1/TD UVB detector (International Light, Peabody, MA).
Western blot assay.
Whole-cell lysates were prepared by mechanically detaching cells in cold PBS and resuspending in WE16th lysis buffer (50 mM Tris-HCl at pH 7.5, 250 mM NaCl, 5 mM EDTA, 1% NP-40, 0.1% sodium dodecyl sulfate, 20% glycerol, 80 mM β-glycerophosphate, 50 mM sodium fluoride, 1 mM sodium orthovanadate, and a Complete protease inhibitor tablet [Roche, Alameda, CA]). Lysates were then sonicated and clarified by centrifugation. The DC protein assay (Bio-Rad, Hercules, CA) was used to determine protein concentrations. Equal amounts of protein lysates (15 to 30 μg) were electrophoresed on sodium dodecyl sulfate-polyacrylamide gels and transferred to Immobilon-P membranes (Millipore, Billerica, MA). Immunoblotting was performed with mouse anti-Bak (Ab-2; Calbiochem, San Diego, CA), mouse anti-p53 (Ab-6; Calbiochem, San Diego, CA), mouse anti-UBE3A (Abnova, Taipei City, Taiwan), mouse anti-Bcl-XL (BD Pharmingen, San Jose, CA), mouse anti-Bcl-2 (BD Transduction Laboratories, San Jose, CA), mouse anti-Bax (BD Transduction Laboratories, San Jose, CA), mouse anti-Mcl-1 (BD Pharmingen, San Jose, CA), mouse anti-Noxa (Calbiochem, San Diego, CA), rabbit anti-PUMA (Cell Signaling, Danvers, MA), mouse anti-GAPDH (Abcam, Cambridge, MA), and mouse antinucleolin (C-23; Santa Cruz Biotechnology, Santa Cruz, CA). For quantification of Western blot data, the membranes were scanned and bands were analyzed by densitometry using ImageJ (NIH).
Immunofluorescence microscopy.
HFKs or HT1080 cells (vector control, 5E6, 8E6, 38E6, and 16E6) were grown on coverslips to 70% confluence and then either treated with 20 mJ/cm2 (HT1080 cells) or 25 mJ/cm2 UV (HFKs) or left untreated as a control. At the indicated time points the medium was aspirated and the cells were fixed for 15 min at room temperature in 4% paraformaldehyde. Coverslips were processed using the Select FX Alexa Fluor 488 cytochrome c apoptosis detection kit, as per the manufacturer's instructions (Invitrogen, Carlsbad, CA). Samples were rinsed extensively in PBS before being mounted in FluoroGuard antifade reagent (Bio-Rad, Hercules, CA). Images were obtained with a Deltavision restoration microscope (Applied Precision, Issaquah, WA) fitted with an Olympus 20× or 40× objective and processed at a Silicon Graphics (Mountain View, CA) workstation with accompanying API software. The images were subsequently exported to Adobe Photoshop (version 7.0) and Adobe Illustrator (version 11.0) (Adobe Systems, San Jose, CA) for preparation.
For quantification of apoptotic cellular responses, five separate 20× fields were analyzed for each E6-expressing and vector control coverslip. Cells exhibiting loss or dispersal of cytochrome c were counted and divided by the total number of cells to give the percentage of apoptotic cells per experiment. Experiments were performed in triplicate.
Activated caspase-3 assay.
HT1080 cells (vector control, 5E6, 8E6, 38E6, and 16E6) were grown in 10-cm plates to 70% confluence and then either treated with 20 mJ/cm2 UV or left untreated as a control. Twelve hours after UV treatment, the cells were harvested and assayed for cleaved caspase-3 using the PathScan cleaved caspase-3 (Asp175) sandwich enzyme-linked immunosorbent assay kit (Cell Signaling Technology, Danvers, MA).
RESULTS
The E6 proteins of various beta-HPVs prevent the accumulation of Bak following UVB treatment of human keratinocytes.
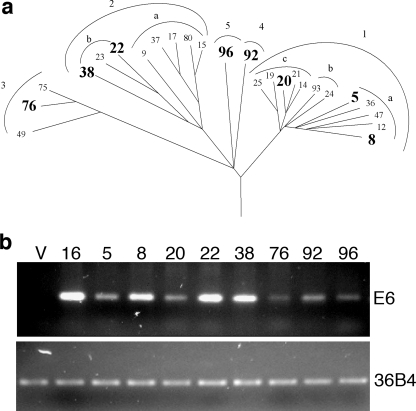
Previous studies have reported that the E6 proteins of both high- and low-risk alpha-HPVs, as well as that of HPV5 of the beta-HPVs, target Bak for proteosomal degradation (21). We investigated whether this ability is conserved among the beta-HPV types in primary HFK cultures. Currently, 25 HPV types are classified as beta-HPVs, and their phylogenetic relatedness, based on amino acid sequences from the E6 protein, is shown in Fig. 1a. Based on this alignment, representative HPV types from each of the five species were chosen for further studies. Each of these HPV E6 proteins was expressed in HFKs, and expression was confirmed using RT-PCR (Fig. 1b).
FIG. 1.
Beta-HPV cladogram and expression levels in HFKs. (a) Cladogram representation of beta-HPV species and subspecies. Types highlighted in bold, representing each of the separate species, were chosen for further study. (b) RT-PCR of E6 in the stable E6-expressing HFK lines. 36B4 levels within each cell type were used as an endogenous control. V, LXSN vector control cells.
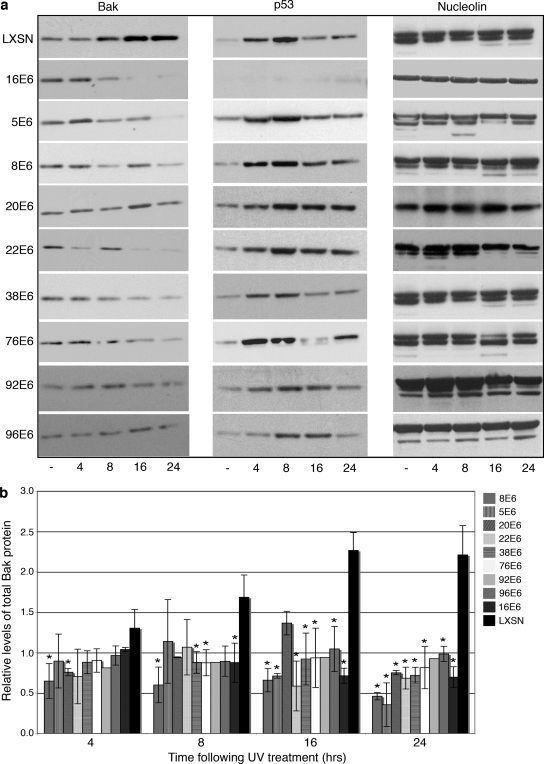
The ability of the E6 protein from each of the representative beta-HPV types to degrade Bak was then compared with high-risk 16E6 and vector LXSN control cells. The cells were treated with UVB, harvested at 4, 8, 16, and 24 h postirradiation, and subjected to immunoblot analysis to detect levels of p53 and Bak (Fig. 2a). UVB irradiation initiated an induction in p53 levels as well as an accumulation of Bak in LXSN-expressing HFKs, confirming previous reports of cellular responses to UVB irradiation (4, 24). Cells expressing 16E6 targeted p53 for degradation constitutively (14) and also reduced Bak levels by 8 h following treatment with UVB. In contrast, p53 protein was induced in all of the beta-HPV E6-expressing cells after UV exposure, with kinetics similar to those seen for the controls. Although the levels of Bak were more substantially reduced by some E6 proteins (e.g., 16E6, 8E6, and 38E6) compared to others (e.g., 20E6, 92E6, and 96E6), Bak protein did not accumulate following UV treatment in any of the E6-expressing cells (Fig. 2b). In contrast to other reports (21), the E6 proteins did not target Bak for degradation constitutively but only approximately 8 h following UV exposure.
FIG. 2.
E6 proteins degrade Bak after UVB irradiation. (a) Representative immunoblot showing the levels of Bak, p53, and nucleolin in E6-expressing and vector control HFKs. Cells were mock treated (−) or treated with 15 mJ/cm2 UVB and harvested at the indicated time points (4, 8, 16, and 24 h after UV treatment). (b) Quantitation of the levels of Bak protein following UV treatment. Values represent the mean levels of total Bak protein normalized to the levels seen for non-UV-treated cells (± standard deviation) in three independent experiments. An asterisk indicates that the statistical difference between the E6-expressing cell and the LXSN vector control at that time point has a P value of <0.05.
16E6- and 8E6-mediated Bak degradation is dependent on E6AP.
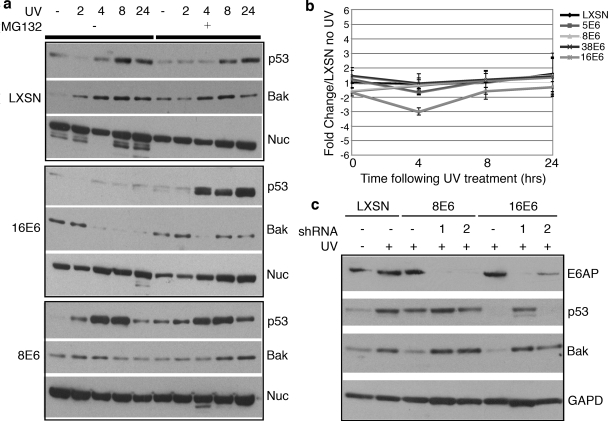
The destabilization of Bak by the E6 proteins of both alpha- and beta-HPVs has been reported to involve proteosomal degradation (21). Bak had previously been reported to be constitutively targeted for proteosomal degradation by E6 proteins (21). Because we saw degradation of Bak only following UV treatment, we tested whether the decreased levels seen for 8E6 and 16E6-expressing HFKs were in fact due to proteosome-mediated degradation (Fig. 3a). 8E6, 16E6, and LXSN vector control HFKs were exposed to 15 mJ/cm2 of UVB irradiation in the presence or absence of the proteosome inhibitor MG132 and then harvested at selected time points. The normal cellular responses to UV treatment, i.e., accumulation of Bak and p53, were seen in LXSN HFKs with or without MG132. By comparison, the decreases in the levels of Bak in both 16E6- and 8E6-expressing HFKs were reversed by the presence of MG132, thus confirming proteosomal degradation.
FIG. 3.
16E6- and 8E6-mediated proteosomal degradations of Bak require E6AP. (a) E6-expressing and vector control HFKs were mock treated (−) or treated with 15 mJ/cm2 UVB and harvested at the indicated time points (2, 4, 8, and 24 h after treatment). The proteosome inhibitor MG132 (10 μM) was added (+), or not (−), 2 h before the lysates were harvested. The levels of p53, Bak, and nucleolin were determined by immunoblot analysis. (b) Bak mRNA levels in LXSN- and E6-expressing cells following UV treatment. Cells were treated with 20 mJ/cm2 UVB and harvested at the indicated time points (0, 4, 8, and 24 h after treatment). Relative levels of Bak mRNA were calculated using the ΔΔCT method with GAPDH to normalize mRNA levels within each sample. Values shown are the mean changes in each sample compared to the untreated LXSN vector control. Error bars represent the standard deviation for each sample (n = 3). (c) E6AP protein levels were knocked down with shRNA constructs. The cells were then treated, or not (LXSN cells for comparison of basal levels), with 15 mJ/cm2 UVB irradiation. E6AP, p53, Bak, and GAPDH (GAPD) protein levels were determined by immunoblot analysis.
Bak mRNA levels were also examined following UV treatment to ensure that the lower levels of Bak protein were not due to transcriptional changes following UV. 5E6-, 8E6-, 38E6-, 16E6-, and LXSN-expressing HFKs were all exposed to UVB irradiation and harvested with Trizol after 0, 4, 8, and 24 h. Total RNA was isolated and used as a template for real-time RT-PCR with TaqMan primer/probe sets for both Bak and GAPDH. Compared to what was seen for the levels of transcript for the LXSN control cells, there was no significant change in Bak mRNA levels in any of the cells expressing E6 at any of the time points following UV treatment (Fig. 3b). These data confirm that the decreases in total Bak protein following UV treatment are due to proteosomal degradation and not to transcriptional changes.
E6AP is the only E3 ubiquitin ligase that is known to bind E6 proteins, forming a complex that promotes the ubiquitin-mediated degradation of target proteins, such as p53, Scribble, hDlg, and NFX1-91 (17, 20). To investigate a possible mechanism for Bak degradation in E6-expressing HFKs, E6AP expression was knocked down in 16E6-, 8E6-, and vector-expressing cells by use of short hairpin RNAs (shRNAs) (Fig. 3c). Both shRNA1 and shRNA2 knock down E6AP expression, with shRNA1 being more efficient, as seen by immunoblot analysis with an anti-E6AP antibody and the restoration of p53 levels in 16E6-expressing HFKs. In both 8E6- and 16E6-expressing HFKs, the shRNAs blocked the reduction in Bak levels following UV treatment. These data illustrate the requirement of E6AP for E6 proteins to target Bak for degradation following UVB treatment and implicate the ubiquitination pathway in this process.
Other Bcl-2 family members were not perturbed in E6-expressing HFKs following UVB exposure.
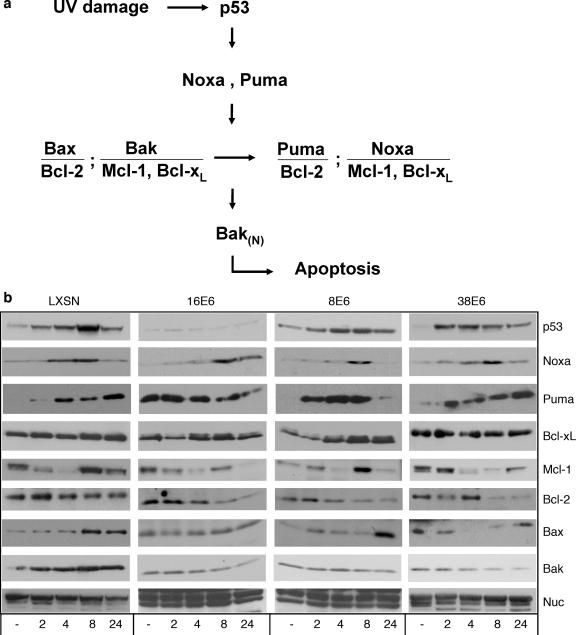
Bak levels and activation within a cell are controlled by a myriad of related Bcl-2 proteins within the intrinsic apoptosis pathway (reviewed in reference 10) (Fig. 4a). Noxa, Bcl-XL, and Mcl-1 have been shown to be crucial for the regulation of the apoptotic pathway by Bak in UVB-treated keratinocytes (41). Puma, Bax, and Bcl-2 proteins have a less-defined role in normal keratinocytes after UVB exposure but are important for the activation of apoptosis in a variety of cell types (40). We therefore wanted to investigate the effects of the E6 protein on other apoptotic regulatory proteins following UVB irradiation (Fig. 4b). UVB treatment of vector-expressing control keratinocytes resulted in the induction of p53 and Bak by 8 h, consistent with previous results (21). The proapoptotic proteins Bax, Noxa, and Puma are also induced between 4 and 8 h after UVB irradiation. Conversely, the antiapoptotic family members, Mcl-1, Bcl-XL, and Bcl-2, exhibit variability following UVB exposure. Levels of Mcl-1 are maximally reduced by 4 h and Bcl-2 levels are slowly reduced by 24 h, while Bcl-xL levels appear unchanged following UVB treatment. These data suggest that normal apoptotic signaling is occurring through the intrinsic pathway following genotoxic damage to keratinocytes (10, 41).
FIG. 4.
Normal responses of other Bcl-2 family members to UVB irradiation are not perturbed by E6 expression. (a) Proteins involved in UV-induced apoptosis signaling in keratinocytes (compiled from references 10, 29, 40, and 41). (b) E6-expressing and vector control HFKs were mock treated (−) or treated with 15 mJ/cm2 UVB and harvested at the indicated time points (2, 4, 8, and 24 h after treatment). The levels of p53, Bak, Mcl-1, Bcl-xL, Noxa, Bax, Bcl-2, Puma, and nucleolin were determined by immunoblot analysis.
In E6-expressing cells, all of the pro- and antiapoptotic proteins tested, with the exception of Bak, showed levels of induction or reduction similar to those seen for control cells. The levels of the proapoptotic members, Bax, Noxa, and Puma, were all induced by UV treatment, while the levels of the antiapoptotic proteins, Mcl-1 and Bcl-2, were reduced. The levels of Bcl-xL appeared unchanged following UVB treatment, similar to what was seen for control cells. Taken together, these results demonstrate that the normal signaling events that initiate apoptosis through the intrinsic pathway following UVB exposure remain intact for each of the E6-expressing cell lines tested, with the exception of a failure to accumulate Bak.
E6 proteins prevent apoptosis of cells following UVB exposure.
We wished to determine if the prevention of Bak accumulation in E6-expressing cells was capable of protecting these cells from UVB-mediated apoptosis. Activated Bak presumably triggers apoptosis by forming pores within the mitochondrial membrane. These pores allow the release of proapoptotic factors into the cytosol, which in turn are involved in activating the caspase cascade. Thus, the level of caspase activation is indicative of the number of cells undergoing apoptosis. Therefore, we analyzed the levels of cleavage (and thus activation) of one of these caspases, caspase-3 in E6-expressing cells following UVB treatment, as a measure of their ability to protect against apoptosis.
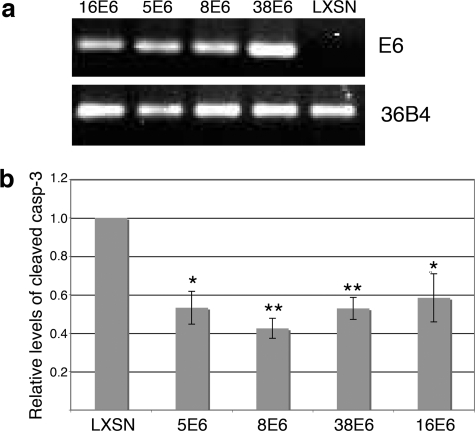
Cytoprotection has previously been demonstrated for E6 from the alpha-HPV18, -10, and -77 and beta-HPV5 by use of the HT1080 fibrosarcoma cell line (21, 22), so we first initially examined the effect of the beta-HPV E6 proteins in this established model. We expressed the E6 protein from HPV5, -8, -38, and -16 in HT1080 cells (Fig. 5a) and exposed the cells to UVB. Twelve hours after UVB treatment, cells were harvested and lysates analyzed for caspase-3 cleavage. Following UVB treatment, all of the E6-expressing cells showed levels of activated caspase-3 approximately 40 to 60% lower than those seen for vector control cells (Fig. 5b), indicating that E6 protects these cells from apoptosis.
FIG. 5.
E6 proteins abrogate caspase-3 cleavage in HT1080 cells. (a) RT-PCR of E6 in the stable E6-expressing HT1080 cell lines. 36B4 levels within each cell type were used for an endogenous RNA control. (b) Levels of activated caspase-3 in LXSN- and E6-expressing cells 12 h following a 20-mJ/cm2 UV treatment. Values represent the mean level of cleaved caspase-3 detected in four independent experiments (± standard error of the mean). A single asterisk represents a difference between the E6-expressing cells and control cells with a P value of <0.05, while a double asterisk represents the same difference with a P value of <0.01.
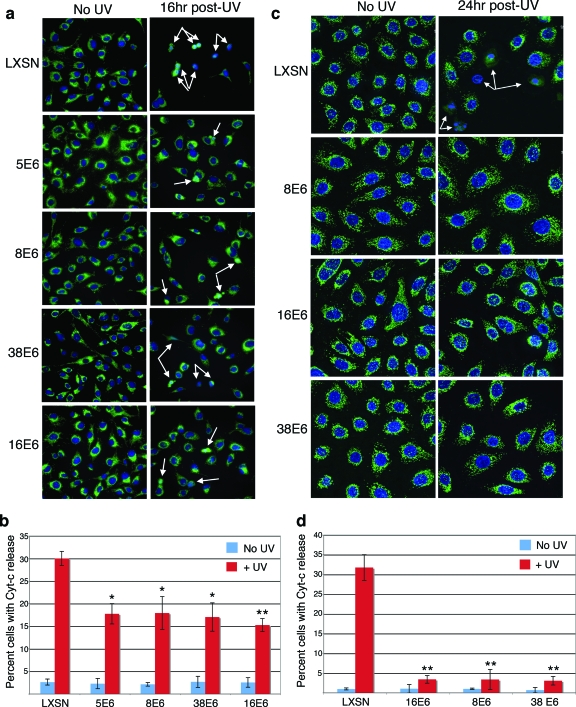
One of the apoptotic events immediately preceding caspase cleavage is the release of cytochrome c from the mitochondrial inner membrane space (3). Given that Bak pore formation is thought to mediate this event, decreased levels of total Bak protein should result in fewer pores and, in turn, a defect in the release of cytochrome c following UVB treatment. To test this, we analyzed the extent of cytochrome c release in HT1080 cells in the presence or absence of UV treatment by the immunostaining of treated cells for cytochrome c. Following UVB treatment, fewer vector control cells than non-UV-treated cells were present, indicating either that the cells had their growth arrested or that they sloughed off the coverslips as a result of treatment. Additionally, a large percentage of the remaining cells had either no cytochrome c staining or dispersed cytochrome c staining indicative of its release from mitochondria (Fig. 6a). In comparison, 5-, 8-, 38-, and 16E6-expressing cells showed only a slight decrease in the number of cells following UV treatment, and much lower numbers of cells with a dispersed cytochrome c staining pattern. When these results were quantified, approximately 30% of vector control cells appeared apoptotic, compared to approximately 15% of each of the E6-expressing cell lines tested (Fig. 6b).
FIG. 6.
E6 proteins prevent cytochrome c release in HT1080 cells and HFKs. Indirect immunofluorescence of mock- or 20-mJ/cm2 UV-treated LXSN- and E6-expressing HT1080 cells (a) or HFKs (c) demonstrating cytochrome c staining. White arrows indicate apoptotic cells with a reduced or dispersed pattern of staining. (b and d) Quantitation of levels of cytochrome c (Cyt-c) release in LXSN- and E6-expressing HT1080 cells (b) or HFKs (d). Values are the mean percentages of cells with no or dispersed cytochrome c staining patterns from three independent experiments (± standard deviations). A single asterisk indicates differences between control cells and E6 cells with a P value of <0.05, while a double asterisk indicates differences between control cells and E6 cells with a P value of <0.01.
Finally, as our Bak degradation experiments had been done using primary HFKs, we wished to determine if apoptosis signaling was abrogated in HFKs that expressed the E6 proteins. As described above, we examined the extent of cytochrome c release in HFKs in the presence or absence of UV treatment. Following UVB treatment, the vector control HFKs appeared both to be fewer in number and to have more dispersed cytochrome c staining (Fig. 6c), similar to the results seen with the HT1080 vector control cells. As with the HT1080 E6-expressing cells, the E6-expressing HFKs all had fewer cells with cytochrome c dispersion. Quantified, 30% of the vector control HFKs were apoptotic (Fig. 6d), a result that closely mimics that of the vector control HT1080 cells. However, only 5% of the E6-expressing cells appeared to be apoptotic. Taken together, these results indicate that the E6 protein protects both HT1080 cells and HFKs from apoptosis by preventing the release of cytochrome c and the downstream activation of caspase-3, thus disrupting the intrinsic pathway of apoptosis signaling.
DISCUSSION
Previous studies have determined that the E6 proteins from several HPV types confer protection from apoptosis after genotoxic stress (22). This protective effect supports a possible mechanism for the association of beta-HPVs with the development of skin cancers. A surviving pool of damaged keratinocytes may allow the accumulation of deleterious mutations, such as in p53, which are commonly found in skin cancers (25). It is unclear whether there are differences in the abilities of the various beta-HPVs to contribute to skin carcinogenesis. On one hand, several studies have identified a large number of HPV types in skin cancers and their precursors, without finding one or a few types predominating (23, 35, 39). On the other hand, some types have been found to possess activities that would contribute to carcinogenesis that other types have not shown. For example, UVB irradiation can stimulate the noncoding region of HPV5 and -8 but not other types within the beta-HPVs (2). Furthermore, the repair of UV-induced thymine dimers is compromised in cells expressing 5E6 protein but not in cells expressing E6 proteins from other cutaneous HPVs (18).
We investigated the ability of several representative HPV types from the various species within the beta genus to degrade Bak in primary keratinocyte cultures. This would reveal whether there are any inherent differences in the abilities of certain beta-HPV types to contribute to skin carcinogenesis based on their ability to protect cells from apoptosis. All of the representative beta-HPV E6 proteins tested were able to degrade Bak and/or prevent its accumulation following UVB treatment (Fig. 2).
To ensure that the protective effect seen for cells expressing the E6 protein is limited to the ability to degrade Bak, we examined other Bcl-2 family members important for apoptosis signaling through the intrinsic pathway. There were no discernible differences between 8E6-, 38E6-, or 16E6-expressing and vector control HFKs following UVB treatment, aside from slight differences in the kinetics of the apoptotic mediators tested (Fig. 4). In order to confirm the cytoprotective effects of Bak degradation, we examined E6-expressing cells for the ability to blunt the apoptotic response. The cytochrome c and activated caspase-3 assays revealed significant differences between the effects of UVB treatment on vector control-expressing versus E6-expressing HT1080 and HFKs. However, with regard to the ability to decrease the apoptotic response to UVB, there were no significant differences between the different E6 proteins expressed in either cell type (Fig. 5 and 6). Our findings indicate that if there is any disparity regarding the ability of specific beta-HPVs to contribute to the risk of developing NMSC, this is most likely independent of their ability to degrade Bak and protect cells from apoptosis following UVB exposure.
Certainly, there are many possible explanations for these findings. As stated above, certain cutaneous HPVs may exhibit other exclusive activities that promote tumorigenesis, such as the ability of HPV5 to prevent the repair of thymine dimers (18). The E6 and E7 proteins of HPV38 display transforming activity in primary human cells (9); however, the ability of other beta-HPV E6/E7 proteins to mediate this effect is uncertain. In addition, while none of the beta-HPVs degrade p53, 38E6 and -E7 expression has been reported to inhibit the ability of p53 to induce the transcription of downstream target genes involved in growth suppression and apoptosis via the accumulation of ΔNp73 (1). Recently it has been shown that a subset of beta-HPV E6 proteins can activate telomerase through an E6AP-dependent mechanism and prolong the life span of keratinocytes in culture (7). There may also be as-yet-undetermined protein binding partners specific to certain E6 and/or E7 proteins from the various beta-HPVs that may be important with regard to the initiation of tumorigenesis.
Our studies also reveal a possible mechanism for the targeting of Bak for degradation that is conserved in both 16E6- and 8E6-expressing cells. It is well known that the high-risk alpha-HPV E6 protein complexes with E6AP, a cellular E3 ubiquitin ligase homologous to the E6AP carboxyl terminus, and targets several substrates for degradation (17, 20). In addition, it has been previously shown that both E6 proteins and E6AP are able to bind Bak and that Bak levels may be normally regulated by E6AP (37). From our knockdown experiments with shRNAs to the E6AP protein, we show that the E6 proteins of both HPV16 and HPV8 are unable to degrade Bak following UVB treatment in the absence of E6AP. We are currently investigating other possible E3 ligases that may also be important for the regulation of Bak degradation by E6.
The E6-mediated proteosomal degradation of Bak reveals another interesting feature about the interaction of E6 proteins with Bak. In our study, the degradation of Bak seen for E6-expressing HFKs occurred at least 8 h after UVB treatment and only after apoptosis was initiated. The basal, or constitutive, levels of Bak in E6-expressing cells, compared to those for control keratinocytes, remained unaffected. Previous binding experiments and functional cell death assays revealed that Mcl-1 and Bcl-xL keep Bak sequestered until cytotoxic signals activate BH3-only proteins that allow the release and activation of Bak, presumably by its oligomerization (41). The inability of E6 to degrade the constitutive levels of Bak lends support to the idea that Bak is sequestered until its activation by UVB irradiation, after which E6 is able to interact with and target Bak for degradation. Additional experiments are under way in our lab to further examine this hypothesis.
In summary, we have compared the respective abilities of several beta-HPV E6 proteins to degrade Bak independent of p53 function. Each of the E6 proteins we tested from representative subspecies within the beta-HPVs, as well as 16E6, was able to degrade Bak and/or prevent its accumulation. This ability gave cells a distinct survival advantage by evading the intrinsic apoptotic pathway normally triggered by UVB irradiation. Our findings support a role for many of the beta-HPV types in the initiation of skin carcinogenesis. Furthermore, the ability to blunt apoptosis after UVB exposure does not appear to be limited to a select few beta-HPV types. Future studies focusing on additional molecular effects of the E6 proteins from beta-HPVs are being examined in order to discover other factors which may identify any beta-HPV types that may predispose patients to the development of skin cancer.
Acknowledgments
We thank E.-M. de Villiers, M. Favre, and O. Forslund for the gifts of HPV plasmids. We thank D. L. George for the human Bak construct. We also thank K. Robinson for help with cell culture and members of the Galloway lab for helpful discussions.
This work was supported by P01 CA042792 and R01 CA064795 to D.A.G. and by the following NIH research training grants: T32 DC000018 (M.P.U.), T32 AI07140 (H.L.H.), and T32 CA09229 (K.M.B.)
Footnotes
Published ahead of print on 20 August 2008.
REFERENCES
- 1.Accardi, R., W. Dong, A. Smet, R. Cui, A. Hautefeuille, A. S. Gabet, B. S. Sylla, L. Gissmann, P. Hainaut, and M. Tommasino. 2006. Skin human papillomavirus type 38 alters p53 functions by accumulation of deltaNp73. EMBO Rep. 7334-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akgul, B., W. Lemme, R. Garcia-Escudero, A. Storey, and H. J. Pfister. 2005. UV-B irradiation stimulates the promoter activity of the high-risk, cutaneous human papillomavirus 5 and 8 in primary keratinocytes. Arch. Virol. 150145-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antignani, A., and R. J. Youle. 2006. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr. Opin. Cell Biol. 18685-689. [DOI] [PubMed] [Google Scholar]
- 4.Assefa, Z., A. Van Laethem, M. Garmyn, and P. Agostinis. 2005. Ultraviolet radiation-induced apoptosis in keratinocytes: on the role of cytosolic factors. Biochim. Biophys. Acta 175590-106. [DOI] [PubMed] [Google Scholar]
- 5.Barbosa, M. S., W. C. Vass, D. R. Lowy, and J. T. Schiller. 1991. In vitro biological activities of the E6 and E7 genes vary among human papillomaviruses of different oncogenic potential. J. Virol. 65292-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12337-342. [DOI] [PubMed] [Google Scholar]
- 7.Bedard, K. M., M. P. Underbrink, H. L. Howie, and D. A. Galloway. 2008. The E6 oncoproteins from human betapapillomaviruses differentially activate telomerase through an E6AP-dependent mechanism and prolong the lifespan of primary keratinocytes. J. Virol. 823894-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouwes Bavinck, J. N., M. Feltkamp, L. Struijk, and J. ter Schegget. 2001. Human papillomavirus infection and skin cancer risk in organ transplant recipients. J. Investig. Dermatol. Symp. Proc. 6207-211. [DOI] [PubMed] [Google Scholar]
- 9.Caldeira, S., I. Zehbe, R. Accardi, I. Malanchi, W. Dong, M. Giarre, E. M. de Villiers, R. Filotico, P. Boukamp, and M. Tommasino. 2003. The E6 and E7 proteins of the cutaneous human papillomavirus type 38 display transforming properties. J. Virol. 772195-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow, J., and V. A. Tron. 2005. Molecular aspects of ultraviolet radiation-induced apoptosis in the skin. J. Cutan. Med. Surg. 9289-295. [DOI] [PubMed] [Google Scholar]
- 11.Cogliano, V., R. Baan, K. Straif, Y. Grosse, B. Secretan, and F. El Ghissassi. 2005. Carcinogenicity of human papillomaviruses. Lancet Oncol. 6204. [DOI] [PubMed] [Google Scholar]
- 12.de Villiers, E. M., C. Fauquet, T. R. Broker, H. U. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 32417-27. [DOI] [PubMed] [Google Scholar]
- 13.Finzer, P., A. Aguilar-Lemarroy, and F. Rosl. 2002. The role of human papillomavirus oncoproteins E6 and E7 in apoptosis. Cancer Lett. 18815-24. [DOI] [PubMed] [Google Scholar]
- 14.Foster, S. A., G. W. Demers, B. G. Etscheid, and D. A. Galloway. 1994. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J. Virol. 685698-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furumoto, H., and M. Irahara. 2002. Human papilloma virus (HPV) and cervical cancer. J. Med. Investig. 49124-133. [PubMed] [Google Scholar]
- 16.Gewin, L., and D. A. Galloway. 2001. E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 757198-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gewin, L., H. Myers, T. Kiyono, and D. A. Galloway. 2004. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 182269-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giampieri, S., and A. Storey. 2004. Repair of UV-induced thymine dimers is compromised in cells expressing the E6 protein from human papillomaviruses types 5 and 18. Br. J. Cancer 902203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. USA 925249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell. Biol. 134918-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, S., C. Harwood, M. Thomas, L. Banks, and A. Storey. 2000. Role of Bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev. 143065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson, S., and A. Storey. 2000. E6 proteins from diverse cutaneous HPV types inhibit apoptosis in response to UV damage. Oncogene 19592-598. [DOI] [PubMed] [Google Scholar]
- 23.Karagas, M. R., H. H. Nelson, P. Sehr, T. Waterboer, T. A. Stukel, A. Andrew, A. C. Green, J. N. Bavinck, A. Perry, S. Spencer, J. R. Rees, L. A. Mott, and M. Pawlita. 2006. Human papillomavirus infection and incidence of squamous cell and basal cell carcinomas of the skin. J. Natl. Cancer Inst. 98389-395. [DOI] [PubMed] [Google Scholar]
- 24.Latonen, L., and M. Laiho. 2005. Cellular UV damage responses—functions of tumor suppressor p53. Biochim. Biophys. Acta 175571-89. [DOI] [PubMed] [Google Scholar]
- 25.Leffell, D. J. 2000. The scientific basis of skin cancer. J. Am. Acad. Dermatol. 4218-22. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., and P. Coffino. 1996. High-risk human papillomavirus E6 protein has two distinct binding sites within p53, of which only one determines degradation. J. Virol. 704509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutzner, M. A., C. Blanchet-Bardon, and G. Orth. 1984. Clinical observations, virologic studies, and treatment trials in patients with epidermodysplasia verruciformis, a disease induced by specific human papillomaviruses. J. Investig. Dermatol. 8318s-25s. [DOI] [PubMed] [Google Scholar]
- 28.Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 634417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nijhawan, D., M. Fang, E. Traer, Q. Zhong, W. Gao, F. Du, and X. Wang. 2003. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 171475-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orth, G., S. Jablonska, M. Jarzabek-Chorzelska, S. Obalek, G. Rzesa, M. Favre, and O. Croissant. 1979. Characteristics of the lesions and risk of malignant conversion associated with the type of human papillomavirus involved in epidermodysplasia verruciformis. Cancer Res. 391074-1082. [PubMed] [Google Scholar]
- 31.Patel, D., S. M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 185061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfister, H. 1992. Human papillomaviruses and skin cancer. Semin. Cancer Biol. 3263-271. [PubMed] [Google Scholar]
- 33.Pfister, H., P. G. Fuchs, S. Majewski, S. Jablonska, I. Pniewska, and M. Malejczyk. 2003. High prevalence of epidermodysplasia verruciformis-associated human papillomavirus DNA in actinic keratoses of the immunocompetent population. Arch. Dermatol. Res. 295273-279. [DOI] [PubMed] [Google Scholar]
- 34.Storey, A., K. Osborn, and L. Crawford. 1990. Co-transformation by human papillomavirus types 6 and 11. J. Gen. Virol. 71165-171. [DOI] [PubMed] [Google Scholar]
- 35.Struijk, L., L. Hall, E. van der Meijden, P. Wanningen, J. N. Bavinck, R. Neale, A. C. Green, J. Ter Schegget, and M. C. Feltkamp. 2006. Markers of cutaneous human papillomavirus infection in individuals with tumor-free skin, actinic keratoses, and squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 15529-535. [DOI] [PubMed] [Google Scholar]
- 36.Thomas, M., and L. Banks. 1999. Human papillomavirus (HPV) E6 interactions with Bak are conserved amongst E6 proteins from high and low risk HPV types. J. Gen. Virol. 801513-1517. [DOI] [PubMed] [Google Scholar]
- 37.Thomas, M., and L. Banks. 1998. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene 172943-2954. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, M., D. Pim, and L. Banks. 1999. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 187690-7700. [DOI] [PubMed] [Google Scholar]
- 39.Weissenborn, S. J., I. Nindl, K. Purdie, C. Harwood, C. Proby, J. Breuer, S. Majewski, H. Pfister, and U. Wieland. 2005. Human papillomavirus-DNA loads in actinic keratoses exceed those in non-melanoma skin cancers. J. Investig. Dermatol. 12593-97. [DOI] [PubMed] [Google Scholar]
- 40.Willis, S. N., and J. M. Adams. 2005. Life in the balance: how BH3-only proteins induce apoptosis. Curr. Opin. Cell Biol. 17617-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Willis, S. N., L. Chen, G. Dewson, A. Wei, E. Naik, J. I. Fletcher, J. M. Adams, and D. C. Huang. 2005. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 191294-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2342-350. [DOI] [PubMed] [Google Scholar]
- 43.zur Hausen, H. 1999. Papillomaviruses in human cancers. Proc. Assoc. Am. Physicians 111581-587. [DOI] [PubMed] [Google Scholar]