Abstract
Given the failures of nonreplicating vaccines against chronic hepatitis C virus (HCV) infection, we hypothesized that a replicating viral vector may provide protective immunity. Four chimpanzees were immunized transdermally twice with recombinant vaccinia viruses (rVV) expressing HCV genes. After challenge with 24 50% chimpanzee infective doses of homologous HCV, the two control animals that had received only the parental VV developed chronic HCV infection. All four immunized animals resolved HCV infection. The difference in the rate of chronicity between the immunized and the control animals was close to statistical significance (P = 0.067). Immunized animals developed vigorous gamma interferon enzyme-linked immunospot responses and moderate proliferative responses. To investigate cross-genotype protection, the immunized recovered chimpanzees were challenged with a pool of six major HCV genotypes. During the acute phase after the multigenotype challenge, all animals had high-titer viremia in which genotype 4 dominated (87%), followed by genotype 5 (13%). However, after fluctuating low-level viremia, the viremia finally turned negative or persisted at very low levels. This study suggests the potential efficacy of replicating recombinant vaccinia virus-based immunization against chronic HCV infection.
Nearly half a billion people, 1 in 12 of the world's population, are infected with hepatitis B or C viruses, resulting in about 1.5 million deaths each year (3). While hepatitis C virus (HCV) infection accounts for a heavy burden of chronic liver disease, cirrhosis, and hepatocellular carcinoma, vaccines are not yet available. A candidate HCV vaccine was developed by investigators at Chiron Corporation in 1994 (13). This vaccine, composed of recombinant HCV E1E2 proteins, produced in cell culture, protected a high proportion of chimpanzees against development of acute infection after challenge with small quantities of homologous genotype HCV; however, it failed to uniformly prevent chronic infections. Furthermore, it failed to protect against a heterologous subtype challenge (26). Immunity induced by this candidate vaccine appeared to depend on induction of an antibody response, which was extremely short lived (13), suggesting that immunity would be of limited duration. Puig et al. (50) also evaluated E1E2 protein immunization and found that this induced a delay in virus replication but did not prevent chronic infection.
As it is now considered probable that cell-mediated immunity is necessary for the control of chronic HCV infection (52, 63), recent candidate HCV vaccines have been designed to induce cell-mediated immunity. These include vaccines using DNA-based immunization (21-23, 29, 30, 35-38, 68, 72), DNA priming followed by HCV protein boosting (53, 59, 70), DNA priming followed by recombinant avipoxvirus (43, 44), recombinant modified vaccinia virus (rVV) Ankara (54), or recombinant adenovirus boosting (40, 46, 69), recombinant adenovirus priming and DNA boosting (20), recombinant modified vaccinia virus Ankara (1) or adenoviruses (39, 67), recombinant baculovirus derived virus-like particles (17, 31), hepatitis B virus surface antigen (HBsAg)-HCV recombinants (41), peptides (56), and peptides incorporated in lysosomes (18). Although most of these candidate vaccines have produced humoral and cell-mediated immune responses, all except one of these studies (17) failed to completely prevent chronic infections, in the relatively few studies in which immunization and challenge of chimpanzees was carried out.
VV has many advantages as a vector for immunization, such as stimulation of long-lasting humoral and cell-mediated immunity after a single injection, low cost, heat stability, and lack of requirement for needles and syringes for administration (19). The latter is important for elimination of the risk of transmission of blood-borne viruses in the developing world. The fact that up to 25 kb of foreign DNA can be stably inserted into the VV genome without impairing its replication (58) provides a major opportunity for polyvalent immunization. The use of rVV to immunize against multiple pathogens was first reported by Perkus et al. (48), who introduced genes for HBV, herpes simplex virus, and influenza virus into a single VV. As a proof-of-concept study, we selected a highly replicating vaccinia virus vector which induces vigorous T-cell responses and investigated its immunogenicity and protective efficacy against chronic HCV infection. In the present study we used recombinant HCV-vaccinia virus (rVV-HCV) encoding HCV core, E1, E2, p7, NS2, and NS3.
After challenge with homologous HCV, all four immunized animals resolved the infection after acute-phase viremia, with a 1.3-log reduced peak viral load (PVL) compared to the control animals, both of which developed chronic infection. For evaluation of cross-protective efficacy, the protected animals were then challenged with a pool of HCV strains representing all of the six major genotypes. High-level acute-phase viral replication was seen in all animals. Heterologous genotypes 4 and 5 replicated predominantly during the early phase but were eventually replaced by genotype 1a during the late phase at low levels. Two of these animals became negative and one was on the borderline after 60 weeks, while one animal showed persisting viremia with a very low titer. We conclude that replicating rVV represents a promising vector for HCV immunization.
MATERIALS AND METHODS
Chimpanzees.
All chimpanzees were housed in groups in spacious outdoor cages at Vilab II at the Liberian Institute for Biomedical Research. Six HBV- and HCV-naïve chimpanzees were selected for the experiment, based on the absence of anti-HBsAg and anti-HCV antibodies, HCV viremia, and HCV-specific gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) responses. The experimental protocol was reviewed and approved by the IACUC committees of the New York Blood Center and the Liberian Institute for Biomedical Research, and the New York Blood Center's approved IACUC assurance is on file with the Office for Protection from Research Risks of the National Institutes of Health.
Vectors for immunization.
rVVs (WR strain L-var) used for immunizations were prepared by Marion Perkus at Virogenetics Inc., using methods which have been described elsewhere (47). All vectors were diluted in RPMI 1640, 10% fetal calf serum to 108 PFU/ml and flash-frozen. They were hand carried to Liberia on dry ice and stored in liquid N2. Controls received parental vaccinia virus WR strain. rVV-HCV contained stock vP1451, expressing HCV genotype 1b capsid, E1, E2, P7, NS2, and NS3 genes (C-N3), at 108 PFU/ml. HBV recombinant VV in WR strain L-var consisted of a pool of stock vP551, expressing HBV surface antigen plus PreS1 plus PreS2 and stock vP541, expressing HBV core antigen, each at 108 PFU/ml. The HBV genes were derived from an adw strain.
Immunization and challenge of chimpanzees.
This study was designed to evaluate the efficacy of a polyvalent immunization using rVVs expressing HBV and HCV genes, called PolyVax. PolyVax was administered to shaved ethanol-rinsed and dried skin using 10 sticks with bifurcated needles, after dilution with an equal volume of glycerol. Each chimpanzee received about 2 × 107 PFU of each vector, assuming delivery of 0.025 ml/stick. Controls were housed separately from animals receiving the recombinant vectors. Four weeks after single immunization with PolyVax all animals were challenged by intravenous inoculation of 10 50% chimpanzee infectious doses (CID50) of HBV adw. Three of the four immunized animals developed either no viremia or low-level short-lived viremia (data not shown).
After complete termination of HBV infection, 52 weeks after PolyVax immunization, the animals were boosted with rVV-HCV and 4 weeks later challenged with an estimated 10 CID50 of HCV-bk, kindly provided by Toshio Shikata (Nihon, Tokyo, Japan). However, this did not produce viremia even in the control animals. Recalculation of the dilution revealed that only an estimated 2.5 CID50 had been administered. Therefore, the animals were rechallenged with a fresh stock, diluted to 24 CID50/ml 17 weeks later.
Eighteen months after the homologous HCV challenge, when all immunized animals had clearly terminated their infections as determined by the highly sensitive PCR method (detection limit, 1.39 log10 copies/ml), the immunized animals were rechallenged intravenous with 2 ml of a pool containing 20 CID50 of each of six HCV genotypes. This pool contained genotype 1a (Hutchinson), 1b (BK), 2a (J6), 2b (J8), 3a (S52), 4a (ED43), 5a (SA13), and 6a (HK 6a). The pool was diluted in phosphate-buffered saline, 20% normal chimpanzee serum, flash-frozen, hand carried to Liberia on dry ice, and stored in liquid N2.
Blood samples were collected weekly for 8 weeks after immunization and 4 weeks post-viral challenges, then biweekly until 24 weeks, and then monthly. A 50-ml volume of blood, used for preparation of peripheral blood lymphocytes (PBLs), was collected at each bleeding.
PCR assays.
A quantitative PCR assay for HCV was carried out as previously described (34), with modifications. Primers used for a universal genotype detection (UGD) PCR assay, which detects all HCV genotypes, were 5′ GGC CTT GTG GTA CTG CCT GA 3′ and 5′ GGT CTA CGA GAC CTC CCG 3′ (Invitrogen, Carlsbad, CA). The probe was 5′-6-carboxyfluorescein CGGACC GAT AGG GTG CTT GCG AGT GC GGTCCG-DABCYL 3′ (Biosearch Technologies Inc., Novato, CA). The reverse transcription reaction was carried out in a 30-μl volume containing 20 μl purified nucleic acid, 0.2 units/μl RNase inhibitor (RNasin; Promega, Madison, WI), 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 5 mM dithiothreitol, 0.5 mM each of deoxynucleoside triphosphate, 1.5 μM HCV reverse primer, and 80 units Moloney murine leukemia virus reverse transcriptase (Gibco BRL, Rockville, MD). The reaction was run at 42°C for 45 min, followed by 95°C for 15 min. The PCR amplification reaction was carried out in a 50-μl volume containing 30 μl reverse transcription product, 2 mM MgCl2, 0.6 μM PCR primers, 0.1 μM probe, and 2.5 units AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA) in 1× Perkin-Elmer PCR buffer II and was run at 95°C for 10 min and cycled at 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s for 50 cycles. The assay was run in parallel with previously validated HCV-positive and -negative plasma samples, of which the latter remained negative even after 50 cycles of amplification.
For highly sensitive PCR detection, 2-ml plasma samples were incubated in 3% polyethylene glycol 8000 for 10 to 30 min, centrifuged at 1,500 rpm for 3 min, and concentrated to 200 μl by discarding 1.8 ml supernatant. The pellet was redissolved in the remaining fluid and used for RNA extraction. We have found this to increase sensitivity about 10-fold (unpublished data).
Genotyping.
The procedure for clone-based DNA sequencing has been described in detail previously (27, 28). Briefly, PCR products amplified by 5′-untranslated region (UTR) primers were directly cloned into a pDrive TA vector using the Qiagen PCR cloning kit (Valencia, CA). HCV sequences from positive Escherichia coli colonies were determined using M13 primers (Pharmacia Biotech, Piscataway, NJ). The amplified PCR products were sequenced using an automated DNA sequencer (Visible Genetics, Toronto, ON, Canada) and DNA sequencing kits from the same manufacturer. HCV genotypes were determined by sequence alignment using the NCBI program and by manually comparing the PCR product sequence with the HCV consensus sequences of different genotypes reported by Stuyver et al. (61).
Serologic assays.
Anti-HCV was determined with the HCV 3.0 enzyme-linked immunosorbent assay (ELISA; Ortho, Raritan, NJ). Alanine aminotransferase was determined by the Sigma calorimetric method in accordance with the manufacturer's instructions. All assays were carried out with plasma samples shipped to New York on liquid N2.
Anti-HVR1 ELISA.
A peptide-based ELISA was performed by using a synthetic peptide representing the E2 hypervariable region 1 (HVR1; amino acids [aa] 384 to 413; Peptron Inc., Daejon, South Korea) of the BK strain as described previously (69).
HCV neutralization assay.
The HCV pseudovirus particles (HCVpp) were prepared from the HCV-bk strain and used for neutralization assays as previously described (4, 5). Total immunoglobulin G from the plasma, prepared as previously described (69), was used for the assay to eliminate enhanced cellular uptake of pseudoviruses and interference with antibody-mediated neutralization due to high-density lipoproteins. As a positive control, purified and concentrated immunoglobulin from a set of over 30 chronic HCV sera of genotypes 1a, 1b, and 3 was diluted at 50 μg/ml. As a negative control, pseudovirus particles bearing glycoproteins derived from the feline endogenous retrovirus RD114 were used. HCV neutralizing activity was expressed as the percent neutralization of HCVpp infection, as previously described (6). The assays were performed three times independently and produced a similar pattern of results.
Assays for cell-mediated immunity.
PBLs were isolated from the buffy coat from 50 ml of blood anticoagulated with EDTA with Ficoll-Hypaque, washed three times with phosphate-buffered saline, and cryopreserved at 1°C/min at a cell concentration of 2 × 107 cells/ml in freezing medium containing 10% dimethyl sulfoxide and 20% fetal calf serum in RPMI 1640. Cells were stored and shipped in liquid N2.
IFN-γ ELISPOT assay.
The ELISPOT assay was performed according to the manufacturer's instructions in the IFN-γ ELISPOT kit with modifications (MABTECH, Cincinnati, OH), as previously described (69). Briefly, 3 × 105 PBLs were plated onto a 96-well plate in triplicate and were stimulated with recombinant HCV core (1b; aa 1 to 133) (69), E2 (1a; aa 383 to 745; Austral Biologicals, San Ramon, CA), or NS3 (1b; aa 1205 to 1615) (69) protein for HCV-specific responses at 3 μg/ml for 40 h. Phytohemagglutinin (Difco/Becton Dickinson, Sparks, MD) and recombinant superoxide dismutase (SOD; kindly provided by M. Houghton, Chiron) were used as positive and negative controls, respectively. The number of IFN-γ-secreting cells (ISCs) was enumerated using an ELISPOT image analyzer and KS ELISPOT 4.2 software (Axioplan 2 imaging; Zeiss, Germany). Antigen-specific ISCs were determined by subtraction from ISCs on culture with SOD. The average number of ISCs observed with SOD was 19 ± 22 (standard deviation) per well. The number of ISCs was represented as ISCs/106 PBLs.
Proliferation assay.
Briefly, 2 × 105 PBLs were plated onto 96-well plates in triplicate and stimulated with recombinant HCV protein, as described above. Phytohemagglutinin and SOD were used as positive and negative controls, respectively. After 5 days of stimulation, [3H]thymidine-containing growth medium was added and cultures were further incubated for 18 h. Antigen-specific stimulation indices were calculated as follows: (antigen-specific thymidine incorporation)/(thymidine incorporation with SOD).
Statistical analysis.
The differences in immune responses and in protection rates between groups were determined with a nonparametric Mann-Whitney U test using the SPSS statistical package (SPSS Inc., Chicago, IL) and Fisher's exact test, respectively.
RESULTS
Results of challenge with the homologous HCV genotype.
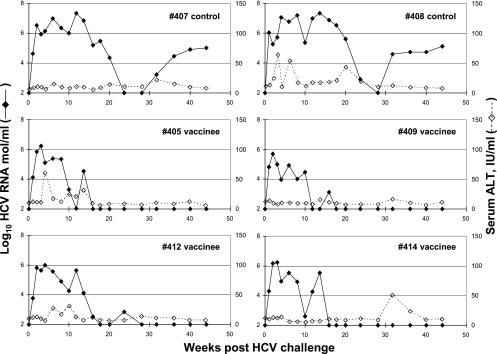
As the challenge with 2.5 CID50 at 4 weeks after rVV-HCV boost failed to induce viremia in any animals, they were rechallenged with 24 CID50 17 weeks later. Both the controls and the immunized animals then became PCR positive 1 week after challenge. The controls developed a high-titer PVL, averaging 7.32 logs 14 weeks after inoculation (Fig. 1). The titers then declined and stabilized at about 5 logs and remained in this range for the duration of the follow-up (44 weeks). By contrast, the immunized animals reached average PVLs of 6.05 logs at around 2 to 4 weeks after the challenge, after which viral loads rapidly declined, becoming nondetectable in all animals by 28 weeks after challenge. Negativity of HCV viremia at weeks 40 and 44 was confirmed by the high-sensitivity PCR method, which has a detection limit of 1.39 log10 copies/ml. The rate of self-limited infection in the immunized group was on the borderline of statistical difference from the control group (P = 0.067) and differed from our past experience with the rate of chronicity (17 chronic infections out of 38 infections) in HCV-infected chimpanzees (P = 0.11).
FIG. 1.
HCV viremia and serum alanine aminotransferase (ALT) after challenge with homologous HCV genotype 1b. HCV RNA was determined by using quantitative real-time reverse transcription-PCR with chimpanzee plasma. The detection limit of the assay was 2 log10 copies/ml. HCV RNA negativity of week 40 and 44 plasma samples from immunized animals was confirmed by the high-sensitivity PCR method (detection limit, 1.39 log10 copies/ml).
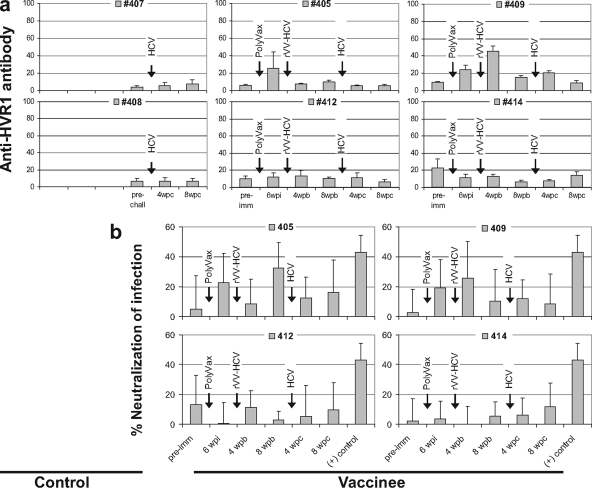
To assess the role of HCV-neutralizing antibodies in this protection we determined antibody responses against HCV-bk HVR1, which had shown a strong correlation with PVL (r = 0.969; P < 0.001) and also with neutralizing activity (r = 0.880; P < 0.01) in our previous chimpanzee study (69). As shown in Fig. 2a, chimpanzees 405 and 409 induced detectable anti-HVR1 antibody responses during the vaccination but did not maintain the responses until the time of challenge. The other two vaccinated animals and both control chimpanzees did not show significant levels of response. This result was confirmed by the HCV neutralization assays using HCV-bk pseudovirus particles (Fig. 2b). Both results showed a similar pattern and suggested that there was no significant contribution of neutralizing antibody to this protection. A small discrepancy between the two results may have been due to differences in sample materials, serum, and purified immunoglobulin, which we used in the assays.
FIG. 2.
Analysis of anti-HVR1 antibody responses and HCV-neutralizing activities. (a) Anti-HVR1 antibody responses were determined by ELISA using HCV-bk HVR1 peptide. Antibody responses were expressed as absorbance at 450 nm within the linear range, multiplied by the serum dilution factor. (b) HCV-neutralizing activity was determined by using HCVpp. The positive control (+) was immunoglobulin that was purified and concentrated from a set of over 30 chronic HCV sera of genotypes 1a, 1b, and 3 and diluted to 50 μg/ml. Pseudovirus particles bearing glycoproteins derived from the feline endogenous retrovirus RD114 were used as a negative control. HCV-neutralizing activity was expressed as the percent neutralization of HCVpp infection. Standard deviations are indicated as error bars. wpi, weeks after PolyVax immunization; wpb, weeks after rVV-HCV booster immunization; wpc, weeks after homologous HCV challenge.
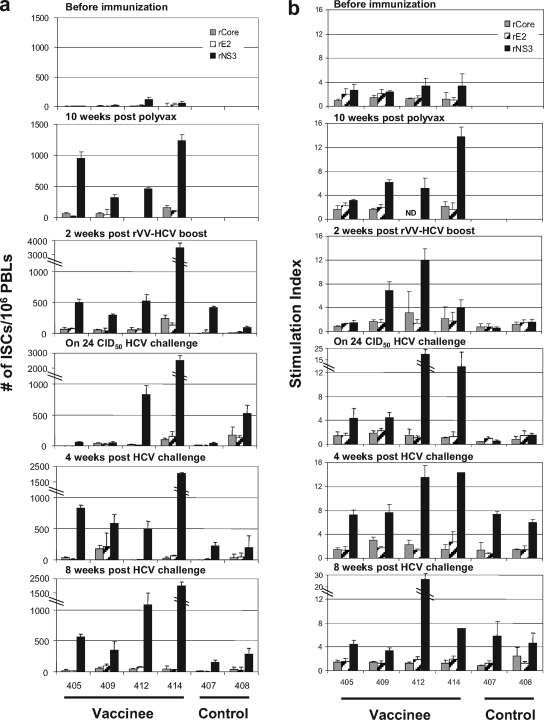
For HCV-specific T-cell responses, significant levels of IFN-γ ELISPOT responses to NS3 protein were induced in the vaccinated chimpanzees even after priming with PolyVax, and these were maintained after boosting with rVV-HCV (Fig. 3a). Two of the vaccinated chimpanzees (405 and 409) with marginal responses at the time of challenge with 24 CID50 HCV showed anamnestic recall responses after the challenge, while the other vaccinated chimpanzees with vigorous responses on challenge maintained these after challenge. By contrast, control chimpanzees showed marginal levels of responses even after challenge. Overall, vaccinated chimpanzees showed higher responses than control chimpanzees at 4 weeks after challenge (P < 0.01). Similar patterns of T-cell responses to NS3 protein were detected in proliferation assays (Fig. 3b). Modest levels of proliferative response were detected in the vaccinated chimpanzees after priming and after boosting except in chimpanzee 405. Two of them showed an anamnestic recall response after challenge, although the differences between control and vaccinated groups at 4 and 8 weeks after challenge were not statistically significant (P > 0.05).
FIG. 3.
HCV-specific T-cell responses before and after homologous HCV challenge. (a) For the IFN-γ ELISPOT assay, PBLs were stimulated with recombinant core (gray bars), E2 (hatched bars), or NS3 (black bars) for 40 h. Responses are expressed as the number of IFN-γ-secreting cells per 106 PBLs. (b) For the proliferation assay, PBLs were stimulated with the same recombinant HCV proteins as in the ELISPOT. Standard deviations are indicated as error bars. ND, not determined.
Results of challenge of immunized chimpanzees with a multigenotype pool.
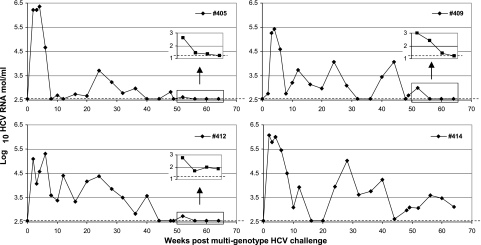
To determine whether the apparent homologous protective immunity extended to heterologous genotypes, we rechallenged the immunized animals 18 months after the homologous genotype 1b (bk) challenge with a pool containing six major HCV genotypes. HCV viremia was determined by using the UGD PCR assay, which detects all eight HCV strains used in this study with equal sensitivity (data not shown). The method was evaluated in parallel with the Roche COBAS HCM-2 genotyping kit. Both methods showed comparable results (data not shown). All the chimpanzees showed typical but low PVL (average, 5.8 ± 0.5 logs) and then fluctuating low-level viremia which eventually turned negative except in chimpanzee 414 (Fig. 4). Samples during weeks 52 to 64 from three HCV RNA-negative chimpanzees were reexamined by using high-sensitivity PCR (detection limit, 1.22 log10 copies/ml). Chimpanzees 405 and 409 were still negative, and chimpanzee 412 was slightly positive (Fig. 4, inset). In the latter animal, week 60 and 64 plasma samples were likely borderline positive, as we could not isolate PCR fragments for genotyping analysis. Only two chimpanzee plasma samples from chimps 409 and 414 were available at week 102. Chimp 409 remained negative, and chimp 414 maintained a low-level viremia (3.82 logs), respectively (data not shown).
FIG. 4.
HCV viremia after multigenotype challenge. HCV RNA was determined by quantitative real-time reverse transcription-PCR using chimpanzee plasma and the UGD assay. Dashed lines indicate the limit of detection (2.55 log10 copies/ml). The last four samples were selected to confirm low titers or negativity of HCV RNA by using high-sensitivity PCR with a detection limit of 1.22 log10 copies/ml as indicated by the dashed line, and the results are shown in the boxes on the chart.
Since these chimpanzees had previously cleared HCV-1b infection, unequal distribution of HCV genotypes was anticipated during the acute phase of multigenotype challenge. Although we observed active HCV replication in all the chimpanzees after multigenotype challenge, 1b-specific protective immunity may have provided differential immune selection pressure on different genotypes and would have resulted in preferential replication of immunologically distant genotypes from 1b. To investigate this hypothesis, we determined genotype distribution by clone-based sequencing of the 5′ untranslated region from very early (2 weeks), mid-phase (8 to 10 weeks), and late-phase (36 to 64 weeks) samples after multigenotype challenge, in which 5 to 20 clones were genotyped. As shown in Table 1, the results revealed that multiple genotypes, including 1a, 1b, 3, 4, and 5, were replicating in the very early phase according to the clonal distribution of genotypes, among which genotype 4 was dominant (70.3%). However, when actual genotype distribution, obtained by normalizing clonal genotype distribution with individual viral load, was taken into consideration, genotype 4 was still predominant (87.4%), followed by genotype 5 (12.6%), and the other genotypes were negligible. This suggested that genotypes 4 and 5 were replicating strongly during the very early phase and that cross-protection for acute infection by these genotypes was inadequate. There were shifts of major genotype from 4 to 2 during the mid-phase and to 1a during the late phase, respectively. Stable maintenance of genotype 1a was observed in chimpanzee 414 from the genotyping results of week 60 and 64 samples. Taken together, these results suggested that there may have been complicated dynamic interactions among different HCV genotypes and host immune responses over time.
TABLE 1.
Overall genotype distribution by clone-based sequencing of the 5′ UTR of HCV after multigenotype HCV challengea
| Wks postchallenge | Sample | % Genotype distributionb(no. with genotype/total clones)
|
||||||
|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 2 | 3 | 4 | 5 | 6 | ||
| 2 | #405 | 78 (14/18) | 22 (4/18) | |||||
| #409 | 17 (3/18) | 11 (2/18) | 72 (13/18) | |||||
| #412 | 100 (18/18) | |||||||
| #414 | 100 (20/20) | |||||||
| Total clones | 4.1 | 2.7 | 17.6 | 70.3 | 5.4 | |||
| Total viremia | <0.01 | <0.01 | <0.02 | 87.4 | 12.6 | |||
| 8-10 | #405 | 100 (8/8) | ||||||
| #409 | 56 (5/9) | 44 (4/9) | ||||||
| #412 | 67 (6/9) | 22 (2/9) | 11 (1/9) | |||||
| #414 | 100 (8/8) | |||||||
| Total clones | 32.4 | 11.8 | 29.4 | 2.9 | 23.5 | |||
| Total viremia | 5.3 | 0.7 | 92.3 | 0.7 | 1.0 | |||
| 36-48 | #405 | 100 (8/8) | ||||||
| #409 | 100 (9/9) | |||||||
| #412 | 100 (10/10) | |||||||
| #414 | 100 (8/8) | |||||||
| Total clones | 48.6 | 28.6 | 22.9 | |||||
| Total viremia | 89.4 | 3.6 | 6.9 | |||||
| 60, 64 | #405 | |||||||
| #409 | ||||||||
| #412 | ||||||||
| #414 | 100 (5/5) | |||||||
5′ UTRs were cloned and sequenced to determine the genotype distribution from the following samples (weeks after multigenotype HCV challenge): for animal 405 at 2, 8, 48, 60, and 64 weeks; animal 409 at 2, 8, 40, 60, and 64 weeks; animal 412 at 2, 10, 49, 60, and 64 weeks; animal 414 at 2, 8, 36, 60, and 64 weeks. The results from weeks 60 and 64 were the same.
The clonal genotype distribution in each chimpanzee was normalized with the individual viral load to represent actual genotype distributions in the animals. Briefly, viremia titers at the indicated times were multiplied by the clonal genotype distribution, and the main genotype population in the chimpanzee at the indicated times is shown in bold.
DISCUSSION
Hepatitis B and C viral infections account for a heavy burden of chronic liver disease in much of the developing world, particularly in Africa and large parts of Asia. To investigate a promising approach to low-cost immunization against these viruses, we attempted to use polyvalent recombinant vaccinia viruses, as was first suggested by Perkus et al. (48). Upon HBV challenge at 4 weeks after a single PolyVax immunization, two of the immunized animals developed no detectable HBV infection, one developed transient low-level viremia, and one reached a high peak viral load for reasons which remain unexplained (data not shown). It is likely that stronger protection would have been observed if prime boosting had been used, as was done for HCV.
The initial HCV challenge contained 2.5 CID50 and did not induce viremia. The animals were therefore rechallenged with 24 CID50, which resulted in acute-phase viremia in all animals. It is unlikely that the low-dose challenge was responsible for the protection against chronic infection in the immunized animals, as both controls developed chronic infection after this mild infection. Although low-dose challenge alone was not enough to provide protection, we could not rule out the possibility that combination of our vaccination regimen with low-dose challenge somehow influenced the outcome of HCV infection, possibly by enhancing HCV-specific T-cell responses. This possibility was investigated by IFN-γ ELISPOT and proliferation assays at 6 weeks after low-dose challenge, and a boosting effect was not observed (data not shown). In control chimpanzee 407, we found HCV-specific T-cell responses 2 weeks after parental VV immunization. This chimpanzee may have had a previous subclinical exposure to HCV or may have had cross-reactive T-cell responses to other viral antigens (65).
After challenge with 24 CID50 all animals rapidly developed viremia. However, peak viral loads in the immunized animals were about 1.3 logs lower than in the controls, indicating that a degree of immunity had been induced. This was further indicated by the fact that none of the four immunized animals, but both controls, developed chronic infection. This difference was statistically on the borderline of significance (P = 0.067) and has yet to be confirmed due to the relatively small number of animals used in this study. All of the immunized animals developed relatively strong cell-mediated immune responses after the booster immunization. These were predominantly directed to NS3. Neutralizing antibody responses after the vaccine booster were relatively weak and inconsistent. Thus, our findings are consistent with previous reports indicating that cell-mediated immune responses are critical for prevention of chronic HCV infections (52, 63).
We previously reported that convalescent chimpanzees resist chronic infection when rechallenged with homologous but not heterologous genotypes (49). This suggested that an HCV vaccine derived from only one genotype might not provide cross-protection against heterologous genotypes. However, a degree of cross-genotype protection has also been reported by others (32). When the immunized animals were rechallenged with a pool containing six major genotypes, PVL (5.79 logs) reached titers similar to those seen in secondary infections rather than in primary infection. Upon homologous genotype 1b (bk) challenge, we observed PVLs of 7.32 and 6.05 logs in the control and immunized animals, respectively. This suggested partial controlling immunity to acute infection by heterologous genotypes. This speculation was supported by the multiple fluctuations of viremia at low levels, which implicate a dynamic interaction between host immune surveillance and viral escape. Alternatively, it is also possible that simultaneous replication of multiple genotypes may result in lower levels of acute-phase viremia than challenge with a single genotype, due to competition among different genotypes.
During the late phase, a very low titer or undetectable viremia was observed in three of the immunized animals, although one had clearly detectable viremia of 3.82 logs up to 102 weeks after multigenotype challenge. This may correspond to the “occult HCV” infections recently reported by Carreno et al. (11, 45). It is not known whether these infections are really chronic and long-lasting or whether these will eventually resolve. It is also not clear whether such low-level infections are clinically significant. Unfortunately, the animals in our study were released to retirement islands; thus, we were not able to follow them longer.
To gain an insight regarding the correlation of T-cell immunity with protection, we were interested in the immunogenicity compared with previous chimpanzee studies that had achieved different rates of protection. Notably, the current recombinant vaccinia virus-induced immunogenicity was no higher than that of other studies (17, 20, 54, 69). Due to many variables that affect the results of cell-mediated immunological assays, our previous study was considered ideal for the comparison, because only two of six vaccinated chimpanzees were protected and results were obtained under similar experimental conditions (69). Unexpectedly, there was no significant difference in immunogenicity as determined by IFN-γ ELISPOT and proliferation assays, and the previous study induced even higher immunogenicity than the current one, which suggests that results under the current T-cell assay conditions may not completely represent protective immunity. There are at least three possibilities to explain the discrepancy, and these are not mutually exclusive. First, current immunological assays may not directly represent in vivo T-cell immunity. Stimulation with exogenous antigen to detect IFN-γ-secreting cells may far exceed the concentrations which occur in vivo, and thus T-cell responses may be overestimated (7). Second, other types of HCV-specific T cells that are multifunctional or do not exhibit readouts in conventional T-cell assays, such as cytokine production or proliferation, may affect the protection. Lastly, there may be other types of host responses than antibody and T-cell responses that better correlate with the outcome of HCV infection, as suggested from the discordance between acute-phase control of HCV replication and chronic protection (54).
The genotype shifting from 4 to eventually 1a may also result from a significant difference in the biological behavior of the different genotypes, a combined consequence of immune escape and viral fitness. As well-documented in previous reports for T-cell dysfunction during persistent lymphocytic choriomeningitis virus (66, 71) and HCV infection (2, 8, 10, 24, 60, 62), continued HCV replication might have resulted in dysfunction of preexisting HCV-specific T cells. As the degree of T-cell dysfunction increases, which results in a decrease of immune selection pressure, so does the contribution of virological factors on genotype distribution, such as “fitness cost.” Genotype 1 has been reported to replicate more efficiently (9), be more pathogenic (16, 25, 57), and be IFN resistant (15, 42) and thus is considered to have greater “fitness” than other genotypes in HCV-superinfected patients (33). This may explain why genotype 1a was eventually fixed in multigenotype-challenged chimpanzee 414. This phenomenon is likely to correspond to viral evolution of diverse quasispecies in humans, but with much broader diversity, in which both escape from host immune responses and fitness to viral replication determine dominant quasispecies (14, 51).
It has yet to be determined why genotype 4 was predominant during the very early phase. It is possible that sequence differences in critical T-cell epitopes (12, 64) or in flanking regions (55) may prevent successful recognition of genotypes by 1b-specific T cells.
We conclude that the use of replicating recombinant VV vectors provides protection against highly variable viruses, such as HCV. The WR strain of vaccinia virus is clearly too virulent for use in human immunization, particularly in immunosuppressed individuals. Thus, we are now investigating the use of highly attenuated VV strains, which we hope will retain immunogenicity and protectivity, as vectors for human use.
Acknowledgments
We are grateful to Robert Purcell and Toshio Shikata for providing HCV genotype 1a and 1b strains and to Jens Bukh, NIAID, NIH, for providing us with HCV stocks of genotypes 2 to 6 and their chimpanzee infectivity titers. George Saycoyah, Liping Li, John Zeonoway, Joseph Thomas, and Patricia McCormack provided dedicated and skillful technical assistance.
This study was supported by NIH grant no. AI 47349.
Footnotes
Published ahead of print on 27 August 2008.
REFERENCES
- 1.Abraham, J. D., N. Himoudi, F. Kien, J. L. Berland, A. Codran, B. Bartosch, T. Baumert, G. Paranhos-Baccala, C. Schuster, G. Inchauspe, and M. P. Kieny. 2004. Comparative immunogenicity analysis of modified vaccinia Ankara vectors expressing native or modified forms of hepatitis C virus E1 and E2 glycoproteins. Vaccine 223917-3928. [DOI] [PubMed] [Google Scholar]
- 2.Accapezzato, D., V. Francavilla, M. Paroli, M. Casciaro, L. V. Chircu, A. Cividini, S. Abrignani, M. U. Mondelli, and V. Barnaba. 2004. Hepatic expansion of a virus-specific regulatory CD8+ T cell population in chronic hepatitis C virus infection. J. Clin. Investig. 113963-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 2008. A dozen good ideas to battle hepatitis. Lancet 3711637. [DOI] [PubMed] [Google Scholar]
- 4.Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA 10014199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch, B., G. Verney, M. Dreux, P. Donot, Y. Morice, F. Penin, J. M. Pawlotsky, D. Lavillette, and F. L. Cosset. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 798217-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennett, M. S., H. L. Ng, M. Dagarag, A. Ali, and O. O. Yang. 2007. Epitope-dependent avidity thresholds for cytotoxic T-lymphocyte clearance of virus-infected cells. J. Virol. 814973-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boettler, T., H. C. Spangenberg, C. Neumann-Haefelin, E. Panther, S. Urbani, C. Ferrari, H. E. Blum, F. von Weizsacker, and R. Thimme. 2005. T cells with a CD4+ CD25+ regulatory phenotype suppress in vitro proliferation of virus-specific CD8+ T cells during chronic hepatitis C virus infection. J. Virol. 797860-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 1541-63. [DOI] [PubMed] [Google Scholar]
- 10.Cabrera, R., Z. Tu, Y. Xu, R. J. Firpi, H. R. Rosen, C. Liu, and D. R. Nelson. 2004. An immunomodulatory role for CD4+ CD25+ regulatory T lymphocytes in hepatitis C virus infection. Hepatology 401062-1071. [DOI] [PubMed] [Google Scholar]
- 11.Carreno, V. 2006. Occult hepatitis C virus infection: a new form of hepatitis C. World J. Gastroenterol. 126922-6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, K. M., B. Rehermann, J. G. McHutchison, C. Pasquinelli, S. Southwood, A. Sette, and F. V. Chisari. 1997. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J. Clin. Investig. 1002376-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choo, Q. L., G. Kuo, R. Ralston, A. Weiner, D. Chien, G. Van Nest, J. Han, K. Berger, K. Thudium, C. Kuo, et al. 1994. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc. Natl. Acad. Sci. USA 911294-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cox, A. L., T. Mosbruger, Q. Mao, Z. Liu, X. H. Wang, H. C. Yang, J. Sidney, A. Sette, D. Pardoll, D. L. Thomas, and S. C. Ray. 2005. Cellular immune selection with hepatitis C virus persistence in humans. J. Exp. Med. 2011741-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis, G. L., and J. Y. Lau. 1997. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology 26122S-127S. [DOI] [PubMed] [Google Scholar]
- 16.Donato, F., A. Tagger, R. Chiesa, M. L. Ribero, V. Tomasoni, M. Fasola, U. Gelatti, G. Portera, P. Boffetta, and G. Nardi. 1997. Hepatitis B and C virus infection, alcohol drinking, and hepatocellular carcinoma: a case-control study in Italy. Brescia HCC Study. Hepatology 26579-584. [DOI] [PubMed] [Google Scholar]
- 17.Elmowalid, G. A., M. Qiao, S. H. Jeong, B. B. Borg, T. F. Baumert, R. K. Sapp, Z. Hu, K. Murthy, and T. J. Liang. 2007. Immunization with hepatitis C virus-like particles results in control of hepatitis C virus infection in chimpanzees. Proc. Natl. Acad. Sci. USA 1048427-8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engler, O. B., R. A. Schwendener, W. J. Dai, B. Wolk, W. Pichler, D. Moradpour, T. Brunner, and A. Cerny. 2004. A liposomal peptide vaccine inducing CD8+ T cells in HLA-A2.1 transgenic mice, which recognise human cells encoding hepatitis C virus (HCV) proteins. Vaccine 2358-68. [DOI] [PubMed] [Google Scholar]
- 19.Fenner, F., D. A. Henderson, I. Arita, Z. Jezek, and I. D. Ladni. 1988. Smallpox and its eradication. World Health Organization, Geneva, Switzerland.
- 20.Folgori, A., S. Capone, L. Ruggeri, A. Meola, E. Sporeno, B. B. Ercole, M. Pezzanera, R. Tafi, M. Arcuri, E. Fattori, A. Lahm, A. Luzzago, A. Vitelli, S. Colloca, R. Cortese, and A. Nicosia. 2006. A T-cell HCV vaccine eliciting effective immunity against heterologous virus challenge in chimpanzees. Nat. Med. 12190-197. [DOI] [PubMed] [Google Scholar]
- 21.Forns, X., P. J. Payette, X. Ma, W. Satterfield, G. Eder, I. K. Mushahwar, S. Govindarajan, H. L. Davis, S. U. Emerson, R. H. Purcell, and J. Bukh. 2000. Vaccination of chimpanzees with plasmid DNA encoding the hepatitis C virus (HCV) envelope E2 protein modified the infection after challenge with homologous monoclonal HCV. Hepatology 32618-625. [DOI] [PubMed] [Google Scholar]
- 22.Gehring, S., S. H. Gregory, N. Kuzushita, and J. R. Wands. 2005. Type 1 interferon augments DNA-based vaccination against hepatitis C virus core protein. J. Med. Virol. 75249-257. [DOI] [PubMed] [Google Scholar]
- 23.Gordon, E. J., R. Bhat, Q. Liu, Y. F. Wang, C. Tackney, and A. M. Prince. 2000. Immune responses to hepatitis C virus structural and nonstructural proteins induced by plasmid DNA immunizations. J. Infect. Dis. 18142-50. [DOI] [PubMed] [Google Scholar]
- 24.Gruener, N. H., F. Lechner, M. C. Jung, H. Diepolder, T. Gerlach, G. Lauer, B. Walker, J. Sullivan, R. Phillips, G. R. Pape, and P. Klenerman. 2001. Sustained dysfunction of antiviral CD8+ T lymphocytes after infection with hepatitis C virus. J. Virol. 755550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatzakis, A., A. Katsoulidou, E. Kaklamani, G. Touloumi, Y. Koumantaki, N. C. Tassopoulos, G. Karvountzis, A. Gioustozi, S. Hadziyannis, and D. Trichopoulos. 1996. Hepatitis C virus 1b is the dominant genotype in HCV-related carcinogenesis: a case-control study. Int. J. Cancer 6851-53. [DOI] [PubMed] [Google Scholar]
- 26.Houghton, M., Q. L. Choo, D. Chien, G. Kuo, A. Weiner, S. Coates, et al. 1997. Development of an HCV vaccine, p. 656-659. In M. Rizzetto, R. H. Purcell, J. L. Gerin, and G. Verme (ed.), Viral hepatitits and liver disease. Edizioni Minerva Medica, Turin, Italy.
- 27.Hu, Y. W., E. Balaskas, M. Furione, P. H. Yen, G. Kessler, V. Scalia, L. Chui, and G. Sher. 2000. Comparison and application of a novel genotyping method, semiautomated primer-specific and mispair extension analysis, and four other genotyping assays for detection of hepatitis C virus mixed-genotype infections. J. Clin. Microbiol. 382807-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, Y. W., L. Rocheleau, B. Larke, L. Chui, B. Lee, M. Ma, S. Liu, T. Omlin, M. Pelchat, and E. G. Brown. 2005. Immunoglobulin mimicry by hepatitis C virus envelope protein E2. Virology 332538-549. [DOI] [PubMed] [Google Scholar]
- 29.Inchauspe, G., M. E. Major, I. Nakano, L. Vitvitski, and C. Trepo. 1997. DNA vaccination for the induction of immune responses against hepatitis C virus proteins. Vaccine 15853-856. [DOI] [PubMed] [Google Scholar]
- 30.Inchauspe, G., M. E. Major, I. Nakano, L. Vivitski, M. Maisonnas, and C. Trepo. 1998. Immune responses against hepatitis C virus structural proteins following genetic immunisation. Dev. Biol. Stand. 92163-168. [PubMed] [Google Scholar]
- 31.Jeong, S. H., M. Qiao, M. Nascimbeni, Z. Hu, B. Rehermann, K. Murthy, and T. J. Liang. 2004. Immunization with hepatitis C virus-like particles induces humoral and cellular immune responses in nonhuman primates. J. Virol. 786995-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanford, R. E., B. Guerra, D. Chavez, C. Bigger, K. M. Brasky, X. H. Wang, S. C. Ray, and D. L. Thomas. 2004. Cross-genotype immunity to hepatitis C virus. J. Virol. 781575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskus, T., L. F. Wang, J. Rakela, H. Vargas, A. D. Pinna, A. C. Tsamandas, A. J. Demetris, and J. Fung. 1996. Dynamic behavior of hepatitis C virus in chronically infected patients receiving liver graft from infected donors. Virology 220171-176. [DOI] [PubMed] [Google Scholar]
- 34.Lee, D. H., and A. M. Prince. 2001. Automation of nucleic acid extraction for NAT screening of individual blood units. Transfusion 41483-487. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S. W., J. H. Cho, K. J. Lee, and Y. C. Sung. 1998. Hepatitis C virus envelope DNA-based immunization elicits humoral and cellular immune responses. Mol. Cell 8444-451. [PubMed] [Google Scholar]
- 36.Lee, S. W., J. H. Cho, and Y. C. Sung. 1998. Optimal induction of hepatitis C virus envelope-specific immunity by bicistronic plasmid DNA inoculation with the granulocyte-macrophage colony-stimulating factor gene. J. Virol. 728430-8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, X., X. Forns, R. Gutierrez, I. K. Mushahwar, T. Wu, P. J. Payette, J. Bukh, R. H. Purcell, and H. L. Davis. 2002. DNA-based vaccination against hepatitis C virus (HCV): effect of expressing different forms of HCV E2 protein and use of CpG-optimized vectors in mice. Vaccine 203263-3271. [DOI] [PubMed] [Google Scholar]
- 38.Major, M. E., L. Vitvitski, M. A. Mink, M. Schleef, R. G. Whalen, C. Trepo, and G. Inchauspe. 1995. DNA-based immunization with chimeric vectors for the induction of immune responses against the hepatitis C virus nucleocapsid. J. Virol. 695798-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Makimura, M., S. Miyake, N. Akino, K. Takamori, Y. Matsuura, T. Miyamura, and I. Saito. 1996. Induction of antibodies against structural proteins of hepatitis C virus in mice using recombinant adenovirus. Vaccine 1428-36. [DOI] [PubMed] [Google Scholar]
- 40.Matsui, M., O. Moriya, and T. Akatsuka. 2003. Enhanced induction of hepatitis C virus-specific cytotoxic T lymphocytes and protective efficacy in mice by DNA vaccination followed by adenovirus boosting in combination with the interleukin-12 expression plasmid. Vaccine 211629-1639. [DOI] [PubMed] [Google Scholar]
- 41.Netter, H. J., T. B. Macnaughton, W. P. Woo, R. Tindle, and E. J. Gowans. 2001. Antigenicity and immunogenicity of novel chimeric hepatitis B surface antigen particles with exposed hepatitis C virus epitopes. J. Virol. 752130-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neumann, A. U., N. P. Lam, H. Dahari, M. Davidian, T. E. Wiley, B. P. Mika, A. S. Perelson, and T. J. Layden. 2000. Differences in viral dynamics between genotypes 1 and 2 of hepatitis C virus. J. Infect. Dis. 18228-35. [DOI] [PubMed] [Google Scholar]
- 43.Pancholi, P., Q. Liu, N. Tricoche, P. Zhang, M. E. Perkus, and A. M. Prince. 2000. DNA prime-canarypox boost with polycistronic hepatitis C virus (HCV) genes generates potent immune responses to HCV structural and nonstructural proteins. J. Infect. Dis. 18218-27. [DOI] [PubMed] [Google Scholar]
- 44.Pancholi, P., M. Perkus, N. Tricoche, Q. Liu, and A. M. Prince. 2003. DNA immunization with hepatitis C virus (HCV) polycistronic genes or immunization by HCV DNA priming-recombinant canarypox virus boosting induces immune responses and protection from recombinant HCV-vaccinia virus infection in HLA-A2.1-transgenic mice. J. Virol. 77382-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pardo, M., J. M. Lopez-Alcorocho, E. Rodriguez-Inigo, I. Castillo, and V. Carreno. 2007. Comparative study between occult hepatitis C virus infection and chronic hepatitis C. J. Viral Hepat. 1436-40. [DOI] [PubMed] [Google Scholar]
- 46.Park, S. H., S. H. Yang, C. G. Lee, J. W. Youn, J. Chang, and Y. C. Sung. 2003. Efficient induction of T helper 1 CD4+ T-cell responses to hepatitis C virus core and E2 by a DNA prime-adenovirus boost. Vaccine 214555-4564. [DOI] [PubMed] [Google Scholar]
- 47.Perkus, M. E., K. Limbach, and E. Paoletti. 1989. Cloning and expression of foreign genes in vaccinia virus, using a host range selection system. J. Virol. 633829-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perkus, M. E., A. Piccini, B. R. Lipinskas, and E. Paoletti. 1985. Recombinant vaccinia virus: immunization against multiple pathogens. Science 229981-984. [DOI] [PubMed] [Google Scholar]
- 49.Prince, A. M., B. Brotman, D. H. Lee, W. Pfahler, N. Tricoche, L. Andrus, and M. T. Shata. 2005. Protection against chronic hepatitis C virus infection after rechallenge with homologous, but not heterologous, genotypes in a chimpanzee model. J. Infect. Dis. 1921701-1709. [DOI] [PubMed] [Google Scholar]
- 50.Puig, M., M. E. Major, K. Mihalik, and S. M. Feinstone. 2004. Immunization of chimpanzees with an envelope protein-based vaccine enhances specific humoral and cellular immune responses that delay hepatitis C virus infection. Vaccine 22991-1000. [DOI] [PubMed] [Google Scholar]
- 51.Ray, S. C., L. Fanning, X. H. Wang, D. M. Netski, E. Kenny-Walsh, and D. L. Thomas. 2005. Divergent and convergent evolution after a common-source outbreak of hepatitis C virus. J. Exp. Med. 2011753-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rehermann, B., and M. Nascimbeni. 2005. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 5215-229. [DOI] [PubMed] [Google Scholar]
- 53.Rollier, C., E. Depla, J. A. R. Drexhage, E. J. Verschoor, B. E. Verstrepen, A. Fatmi, C. Brinster, A. Fournillier, J. A. Whelan, M. Whelan, D. Jacobs, G. Maertens, G. Inchauspe, and J. L. Heeney. 2004. Control of heterologous hepatitis C virus infection in chimpanzees is associated with the quality of vaccine-induced peripheral T-helper immune response. J. Virol. 78187-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rollier, C. S., G. Paranhos-Baccala, E. J. Verschoor, B. E. Verstrepen, J. A. Drexhage, Z. Fagrouch, J. L. Berland, F. Komurian-Pradel, B. Duverger, N. Himoudi, C. Staib, M. Meyr, M. Whelan, J. A. Whelan, V. C. Adams, E. Larrea, J. I. Riezu, J. J. Lasarte, B. Bartosch, F. L. Cosset, W. J. Spaan, H. M. Diepolder, G. R. Pape, G. Sutter, G. Inchauspe, and J. L. Heeney. 2007. Vaccine-induced early control of hepatitis C virus infection in chimpanzees fails to impact on hepatic PD-1 and chronicity. Hepatology 45602-613. [DOI] [PubMed] [Google Scholar]
- 55.Seifert, U., H. Liermann, V. Racanelli, A. Halenius, M. Wiese, H. Wedemeyer, T. Ruppert, K. Rispeter, P. Henklein, A. Sijts, H. Hengel, P. M. Kloetzel, and B. Rehermann. 2004. Hepatitis C virus mutation affects proteasomal epitope processing. J. Clin. Investig. 114250-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shirai, M., M. Chen, T. Arichi, T. Masaki, M. Nishioka, M. Newman, T. Nakazawa, S. M. Feinstone, and J. A. Berzofsky. 1996. Use of intrinsic and extrinsic helper epitopes for in vivo induction of anti-hepatitis C virus cytotoxic T lymphocytes (CTL) with CTL epitope peptide vaccines. J. Infect. Dis. 17324-31. [DOI] [PubMed] [Google Scholar]
- 57.Silini, E., R. Bottelli, M. Asti, S. Bruno, M. E. Candusso, S. Brambilla, F. Bono, G. Iamoni, C. Tinelli, M. U. Mondelli, and G. Ideo. 1996. Hepatitis C virus genotypes and risk of hepatocellular carcinoma in cirrhosis: a case-control study. Gastroenterology 111199-205. [DOI] [PubMed] [Google Scholar]
- 58.Smith, G. L., and B. Moss. 1983. Infectious poxvirus vectors have capacity for at least 25 000 base pairs of foreign DNA. Gene 2521-28. [DOI] [PubMed] [Google Scholar]
- 59.Song, M. K., S. W. Lee, Y. S. Suh, K. J. Lee, and Y. C. Sung. 2000. Enhancement of immunoglobulin G2a and cytotoxic T-lymphocyte responses by a booster immunization with recombinant hepatitis C virus E2 protein in E2 DNA-primed mice. J. Virol. 742920-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spangenberg, H. C., S. Viazov, N. Kersting, C. Neumann-Haefelin, D. McKinney, M. Roggendorf, F. von Weizsacker, H. E. Blum, and R. Thimme. 2005. Intrahepatic CD8+ T-cell failure during chronic hepatitis C virus infection. Hepatology 42828-837. [DOI] [PubMed] [Google Scholar]
- 61.Stuyver, L., A. Wyseur, W. van Arnhem, F. Hernandez, and G. Maertens. 1996. Second-generation line probe assay for hepatitis C virus genotyping. J. Clin. Microbiol. 342259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sugimoto, K., F. Ikeda, J. Stadanlick, F. A. Nunes, H. J. Alter, and K. M. Chang. 2003. Suppression of HCV-specific T cells without differential hierarchy demonstrated ex vivo in persistent HCV infection. Hepatology 381437-1448. [DOI] [PubMed] [Google Scholar]
- 63.Thimme, R., D. Oldach, K. M. Chang, C. Steiger, S. C. Ray, and F. V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 1941395-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Timm, J., G. M. Lauer, D. G. Kavanagh, I. Sheridan, A. Y. Kim, M. Lucas, T. Pillay, K. Ouchi, L. L. Reyor, J. Schulze zur Wiesch, R. T. Gandhi, R. T. Chung, N. Bhardwaj, P. Klenerman, B. D. Walker, and T. M. Allen. 2004. CD8 epitope escape and reversion in acute HCV infection. J. Exp. Med. 2001593-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wedemeyer, H., E. Mizukoshi, A. R. Davis, J. R. Bennink, and B. Rehermann. 2001. Cross-reactivity between hepatitis C virus and Influenza A virus determinant-specific cytotoxic T cells. J. Virol. 7511392-11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wherry, E. J., J. N. Blattman, K. Murali-Krishna, R. van der Most, and R. Ahmed. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 774911-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wuest, T., G. W. Both, A. M. Prince, C. Hofmann, and P. Loser. 2004. Recombinant ovine atadenovirus induces a strong and sustained T cell response against the hepatitis C virus NS3 antigen in mice. Vaccine 222717-2721. [DOI] [PubMed] [Google Scholar]
- 68.Youn, J. W., S. H. Park, J. H. Cho, and Y. C. Sung. 2003. Optimal induction of T-cell responses against hepatitis C virus E2 by antigen engineering in DNA immunization. J. Virol. 7711596-11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Youn, J. W., S. H. Park, D. Lavillette, F. L. Cosset, S. H. Yang, C. G. Lee, H. T. Jin, C. M. Kim, M. T. Shata, D. H. Lee, W. Pfahler, A. M. Prince, and Y. C. Sung. 2005. Sustained E2 antibody response correlates with reduced peak viremia after hepatitis C virus infection in the chimpanzee. Hepatology 421429-1436. [DOI] [PubMed] [Google Scholar]
- 70.Yu, H., L. A. Babiuk, and S. van Drunen Littel-van den Hurk. 2004. Priming with CpG-enriched plasmid and boosting with protein formulated with CpG oligodeoxynucleotides and Quil A induces strong cellular and humoral immune responses to hepatitis C virus NS3. J. Gen. Virol. 851533-1543. [DOI] [PubMed] [Google Scholar]
- 71.Zhou, S., R. Ou, L. Huang, G. E. Price, and D. Moskophidis. 2004. Differential tissue-specific regulation of antiviral CD8+ T-cell immune responses during chronic viral infection. J. Virol. 783578-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, L. X., J. Liu, Y. Ye, Y. H. Xie, Y. Y. Kong, G. D. Li, and Y. Wang. 2004. A candidate DNA vaccine elicits HCV specific humoral and cellular immune responses. World J. Gastroenterol. 102488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]