The development of single-cell protoplast systems is certainly one of the milestones in the history of plant virology, allowing for the analysis of viral molecular processes at the cellular level. The use of plant cell-based systems in the study of the Bromoviridae family of multipartite single-stranded plant RNA viruses facilitated the discovery and dissection of viral processes engaged in the single-cell reproduction cycle: replication, transcription, protein synthesis, movement, virion assembly, and RNA recombination. This review summarizes the application of protoplast systems to the analysis of consecutive steps of the bromovirus life cycle, emphasizing their temporal and spatial patterns during virus multiplication.
Our knowledge of viral infection at the individual cell level determines our understanding of the infection at the entire plant body level. The idea of applying single-cell systems in virology was introduced with the first attempts to transfect Escherichia coli cells with a T4 anti-E. coli bacteriophage (27). E. C. Cocking was the first (1960) to enzymatically isolate plant cells (19), which were then infected with Tobacco mosaic tobamovirus (TMV) (20). The ensuing pioneer work showed that the infection was synchronous and that the uptake of viral particles/RNAs by protoplasts was efficient enough to support virus replication (5, 118). The first viral infection of protoplasts involved the use of poly-l-ornithine (5), but the trials that followed exploited the fusogenic polymers (24), liposomes (28), or electroporation (78, 129). By 1980, these procedures had been successfully employed to transfect protoplasts from more than six species of plants, including various members of the Bromoviridae family, e.g., Brome mosaic bromovirus (BMV; 80), Cucumber mosaic cucumovirus (CMV; 69), and Cowpea chlorotic mottle virus (CCMV; 130). The first attempts were based on the transfection of plant cells with whole-virus particles (104). Later on, viral RNA and its chemical modifications were used as the inoculum (62). Further advances in nucleic acid technology, especially the accessibility of infectious transcripts, have broadened the application of protoplast systems to the study of Bromoviridae.
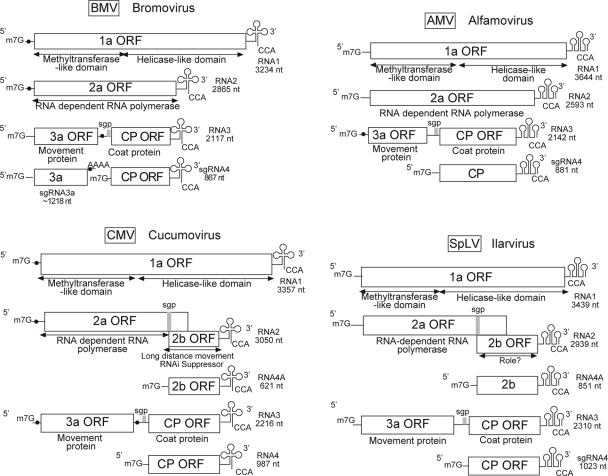
The Bromoviridae constitute one of the most important families of plant RNA viruses. They are distributed worldwide, they infect an extensive range of hosts, and some of them (e.g., CMV and Broad bean mottle bromovirus) are responsible for major crop epidemics (111, 63). The family consists of five genera named after their most representative members: Alfamovirus, Bromovirus, Cucumovirus, Ilarvirus, and Oleavirus. All these viruses possess tripartite, single-stranded, positive-sense RNA genomes (Fig. 1). RNA1 and RNA2 encode the RNA-dependent RNA polymerase (RdRp) proteins 1a and 2a, respectively. The dicistronic RNA3 encodes the movement protein (MP) and coat protein (CP). The latter is translated from subgenomic RNA 4 (sgRNA4). Many of the family members, such as CMV, BMV, and Alfalfa mosaic alfamovirus (AMV), represent excellent model systems to shed new light on viral molecular processes.
FIG. 1.
Comparison of the genome organization of viruses from the family Bromoviridae, including BMV (51, 132), AMV (9), CMV (101), and Spinach latent ilarvirus (SpLV) (9). Open boxes represent ORFs, gray rectangles represent localization of sgp's, and black dots represent B-box consensus-like sequence localization. Cloverleaf-like/pseudoknotted structures represent 3′ UTRs, while 5′ m7G stands for the 7-methylguanosine cap structure. RNA1 and -2 encode two replicase polypeptides (1a and 2a), while RNA3 encodes the 3a MP and the CP. The CP gene is translated from sgRNA4 in all Bromoviridae. RNAi, RNA interference.
PROTOPLAST VERSUS OTHER SYSTEMS
Before the introduction of the protoplast system, tissue cultures were commonly used to study plant viruses (73). Their advantages include the stability of the cells, the ease with which the cultures can be handled, and the availability of the system without season-dependent physiological variations (110). However, the heterogeneous sizes of the plant cells, the asynchrony of cell growth, and the presence of a rigid cell wall often hampered efficient viral transfections. Protoplasts permit one to solve these problems by representing a homogeneous population of mesophyll cells that assures synchronous and well-established viral infection. The most important advantage of this system is that the use of plant cells permits one-step virus growth experiments. It is only with an experiment of this type that the successive stages of virus replication can be identified, monitored, and analyzed. Nevertheless, during experiments, plant cells are maintained in an artificial milieu which differs from the natural environment of the leaf cells. Such changes in the environment may induce changes in the isolated cells and therefore influence virus accumulation. Thus, the possible discrepancies between virus behavior in the cell system and in plant tissue have to be considered. To verify these inconsistencies, it is useful to supplement cell-based studies with additional experiments using other well-developed systems.
The yeast system represents the optional cell-based approach, which was shown, for instance, to recapitulate all known features of BMV replication and gene expression in its natural plant host cells, including the formation of progeny virions (95). Yeast provides multiple advantages for studying viral replication, including the abilities to review the contributions of viral and host functions (59) and to apply strong selections to large yeast populations for detecting low-frequency events, such as RNA recombination (34). However, in working with this cell-based system, one can expect some inconsistencies. Yeast cells divide every several hours and are constantly producing new RNA replication complexes, and this dynamic expansion may amplify effects associated with competition for replication factors.
Cellular expansion of the viral infection is just one of many aspects of the virus life cycle. Late-stage processes, e.g., cell-to-cell movement and systemic spread, can be analyzed only by the application of plant-based approaches. The traditional methods for introducing virions or their genomes into plants utilize manual inoculation in the presence of abrasive substances, which damage cell walls. While these methods are in common use, only a narrow number of cells become infected, and some viruses are not amenable to manual inoculation due to tissue-specific restrictions. Agrobacterium-directed transient gene expression (agroinfiltration), involving the delivery of the desired genes into plant cells as a liquid culture through infiltration, represents the optional method suitable for virology studies in plant-based systems. Agroinfiltration has been used widely in plant virology for the identification of disease resistance genes (7), the induction and suppression of posttranscriptional gene silencing (127), and the study of various late-stage processes, e.g., viral packaging (4) or cell-to-cell movement (50). Most importantly, agroinfiltration facilitates the delivery of several transgenes to be coexpressed into the same cell from different Agrobacterium transformants. These characteristics are important for studying multicomponent viruses, such as Bromoviridae, since the high-level accumulation of genomic RNAs and their expression is replication contingent.
Cell-free systems, although remote and different from the host environment, introduce the opportunity to investigate strictly biochemical/biophysical aspects of viral processes, e.g., recombination (131), protein interactions (1), or replication (93). Although the in vitro systems allow for the dissection of the mechanism and roles of proteins, they have been found to be difficult to obtain, probably due to the membrane association of most replication complexes. Also, in vitro systems may lack important properties found only in vivo. The results obtained by the utilization of in vivo and in vitro systems might lead to differences that reflect the synergistic benefits of various experimental designs for revealing important aspects of the viral life cycle. Plant virology will continue to benefit from integrating the complementary insights from all these approaches.
VIRUS ENTRY
Plant viruses initiate infection by penetrating the cell wall, but unlike animal viruses, there are no known receptors involved in this process (100). Electron microscopy studies have suggested that pinocytosis is involved in Bromoviridae entry (12, 42). An examination of fixed protoplast sections showed that BMV and CCMV induce proliferation of the endoplasmic reticulum and formation of cytoplasmic vacuoles by nuclear membranes in a process called blebbing (12). The observations of plasmalemma invaginations at the virus attachment site and virus-containing cytoplasmic vesicles supported the pinocytosis mechanism. However, in planta studies revealed that Bromoviridae penetrate plant tissue only via mechanical or biological damage (72).
PROTEIN SYNTHESIS
All Bromoviridae have RNA genomes with a 5′ cap and a non-poly(A) 3′ end that carries the tRNA-like structures (TLS) and/or pseudoknots (Fig. 2). Transfections of tobacco protoplasts with the reporter mRNAs showed that the poly(A) tail acts in synergy with the 5′ cap, serving as a translational enhancer (30). Similar functions are secured probably by the 3′ termini in all Bromoviridae (Fig. 2). Particularly, the aminoacylatable TLS at the 3′ termini of Bromo- and Cucumovirus RNAs were found to play a major role in translation enhancement (31, 133). The introduction of the BMV and CMV 3′ untranslated region (UTR) downstream of a reporter gene enhanced the translational efficiency of the chimeric RNAs in carrot protoplasts (31). Studies with transgenic cum1 and cum2 Arabidopsis thaliana protoplasts showed that the CMV 3′ UTR interacted with translation initiation factors eIF4E and eIF4G, which might contribute to the efficient translation via RNA circularization (133).
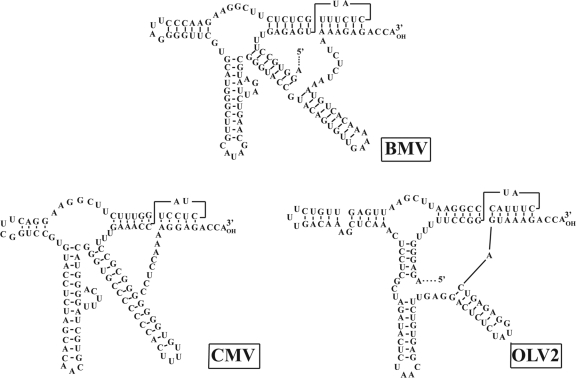
FIG. 2.
Structural similarity of the secondary conformations predicted at the 3′ UTR of BMV (98), OLV-2 (37), and CMV (113) from the family Bromoviridae.
AMV translation seems to be also enhanced by the binding of viral CP to portions of the 3′ UTR (31, 76, 77, 81). However, the CP requirement could be eliminated in carrot protoplasts by using 3′ poly(A) AMV RNAs (58), which suggested that AMV CP is also engaged in 5′-to-3′ RNA circularization (76). This idea was further supported by the discovery that AMV CP interacted with translation initiation factors in transgenic P12 tobacco protoplasts (P12 cells transformed with AMV RNA1 and RNA2, expressing replicase proteins 1a and 2a, respectively) (81). Additionally, translation of AMV RNA encoding the defective CP could be rescued by replacing the 3′ UTR with BMV 3′ UTR, suggesting that AMV translation is CP dependent, whereas BMV 3′ UTR can stimulate translation independently of CP (76, 77).
RNA REPLICATION
The outcome of the very first viral translations is the production of factors recruiting genomic RNAs into membrane-bound replication complexes (Fig. 3). Data from several recent studies showed that viral infection induced the formation of membrane patches called spherules, the sites of viral replication (6, 94, 120). A high-resolution immunofluorescence confocal microscopy study confirmed the localization of 1a and 2a replicase proteins in the endoplasmic reticula of BMV-infected barley protoplasts (95). In AMV-infected cowpea protoplasts, 1a and 2a colocalized at the membrane structures surrounding the vacuoles (120). Similarly, CMV 1a protein was shown to interact with a tonoplast intrinsic protein in A. thaliana protoplasts (54). Apparently, different Bromoviridae members are able to select different cellular membranes as their RNA replication sites.
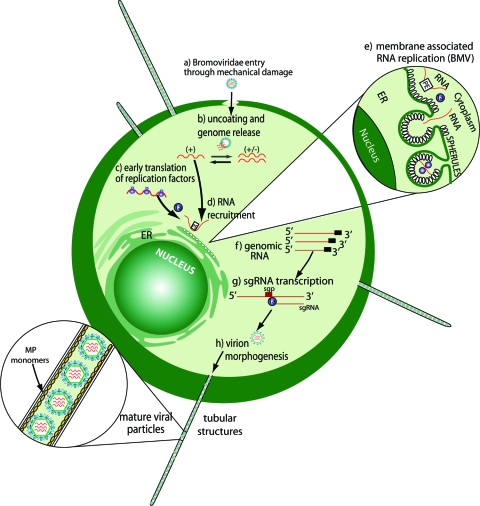
FIG. 3.
Schematic illustration of the Bromoviridae life cycle in a protoplast cell. The viral entry (a) to the protoplast cell can be supported via polyethylene glycol or electroporation-mediated changes in membrane permeability. Following uncoating (b) and early translation (c) of viral replication proteins, the induction of spherule formation (d), where viral RNA replication (e) occurs, has been observed (6). Newly synthesized mRNAs egress into the cytoplasm for sgRNA transcription (f) and translation of other viral products, such as MP and CP, that are engaged in virion maturation (g). The presence of viral MP triggers the formation of tubular structures that mediate virion cell-to-cell transfer via a tubule-guided mechanism (99, 109, 134). ER, endoplasmic reticulum.
The shift between early translation and replication must be synchronized to allow sufficient synthesis of RdRp proteins securing efficient RNA replication. Some elements of the 5′ UTR segments were shown to affect the transition between replication and transcription. The exchange of the RNA3 5′ UTR segments between different strains of AMV, tested with transgenic P12 protoplasts, proved that the B-box (a motif commonly present at the 5′ UTR of Bromoviridae RNAs, also called the ICR2-like motif due to its similarity to the internal control region of tRNA promoters [65]), might play a role in this process (122). In addition, an immunofluorescence study with yeast has suggested that 1a recruited the BMV RNAs from the translation machinery and targeted the 1a-2a viral RNA complex to the membrane replication sites (25). Particularly, the binding of 1a protein to the B-box structure recruited RNAs to the membranous replication complexes (14). These results were confirmed for barley and tobacco protoplasts by altering the B-box motif, which affected both membrane association and BMV RNA replication (16, 35, 85, 86, 116).
Replication of Bromoviridae RNA proceeds in an asymmetric manner, with 100 positive BMV RNA strands being produced for every negative [(−)] strand in barley protoplasts (66). Apparently, the maintenance of a 100:1 ratio requires accurate coordination, as was shown by using RNA3 mutants carrying nucleotide substitutions at the RNA4 initiation site (40) or bearing frameshifts/deletions in the CP gene (43, 74). The intercistronic region of RNA3 acts as the primary determinant of asymmetric BMV replication, although the CP may be an additional factor.
The CP was shown to be involved in regulation of the balance between AMV positive [(+)]- and (−)-strand synthesis in cowpea protoplasts (43, 74). Olsthoorn et al. (81) proposed the conformational switch model explaining the switch between (+)- and (−)-strand production in AMV-infected transgenic P12 protoplasts. This model stated that the 3′ ends of AMV RNAs folded into two mutually exclusive forms playing diverse functions: the CP-free pseudoknotted structure and the CP-bound extended conformation. CP binding to the 3′ end disrupts the pseudoknot, inducing an extended conformation that is no longer capable of RdRp recognition for (−)-strand synthesis. In this way, CP binding induces asymmetric (+)-sense RNA synthesis. The alternative theory, called the 3′ organization model, proposed by Guogas et al. (84) argued that CP binding to the 3′ end compacts, rather then extends, the 3′ RNA termini. The authors proposed an organized AMV RNA-CP complex as the equivalent of a TLS, which presents a uniform population of the termini to act as templates for RNA replication. Additionally, replication assays in nontransgenic tobacco protoplasts demonstrated that the nucleotide changes proposed to both disrupt and restore the pseudoknot structure are deleterious to replication and do not support the significance of the proposed pseudoknot structure for regulating replication. These opposing theories reflect differences that may due to the use of wild-type protoplasts (84) versus transgenic P12 cells that overexpress AMV replicase (81).
Complete replication of the Bromoviridae genomes requires the recognition of three classes of RNA promoters, directing the synthesis of genomic (−)-strand RNAs, genomic (+)-strand RNAs, and sgRNAs (described in the next section), respectively. Experiments with tobacco and barley protoplasts have demonstrated that the 3′ UTR TLS directed the initiation of (−)-strand RNA synthesis in bromo- and cucumoviruses. The process takes place when the replicase interacts with a 3-nucleotide (nt) loop (67AUA65) in the TLS-terminal stem-loop C (8, 92). Any changes in the secondary structure of stem-loop C impaired the BMV RNA replication in barley protoplasts (91). However, CMV and BMV RdRps were able to direct replication from the heterologous 3′ TLS regions in the reciprocally exchanged viral RNAs (89). Also, the sequences upstream of the tRNA-like domain, including a series of stem-loops and pseudoknots, were shown to regulate BMV RNA replication in barley cells (16, 60, 85). In the case of alfamo- and ilarviruses, the interaction between CP and the 3′ UTR secondary structures was assumed to regulate the initiation of (−)-strand RNA synthesis (75), and the process was shown to be CP concentration dependent (38). Previously proposed by Olsthoorn et al. (81), the conformational switch model stated that (−)-strand RNA synthesis occurred only on CP-free viral RNAs. However, upon viral entry, viral RNAs are surrounded by CP dimers, which would create a hostile environment for (−)-strand synthesis. Further replication assays with tobacco protoplasts have noted that the AMV replication cycle depends on the CP concentration (38). The results have demonstrated that replication was activated at low CP concentrations but was gradually repressed as the ratio of the concentration of CP dimer to RNA increased. Mutations in the CP mRNA coding sequence that blocked CP translation were found to inhibit replication, suggesting that the CP was required for stimulation. Mutations in the CP RNA 3′ UTR binding domain also reduced replication, suggesting that CP binding to the 3′ UTR was required (38).
To analyze sequences required for initiation of RNA replication, BMV RNA3 3′-terminal mutants were tested with barley protoplasts, showing that BMV (+)-strand RNA synthesis could initiate from the 3′ penultimate cytidylate on the (−)-strand RNA template and that the adenylate and uridylate residues at the +2 and +3 positions were essential (40). Furthermore, the 4-nt core (CCAA) of the cB-box sequence [motif complementary to the B-box, found in the 3′ UTR of (−)-strand RNAs in all Bromoviridae], was found to be necessary for (+)-strand RNA replication (16). In addition, the complementary 5′ UTR B-box (65), forming cloverleaf structures in CMV, CCMV, BMV, and AMV RNAs, has been predicted to contribute to efficient genomic (+)-strand synthesis (85). In the case of AMV, replication of the (+)-strand RNA was regulated by the multifunctional CP molecule (121). E. M. Jaspars and C. J. Houwing (49) demonstrated that CP was essential for the release of viral (+)-strand RNAs from the replication complexes in cowpea protoplasts. Their “messenger release” hypothesis predicts that replication complexes liberate single-stranded viral RNAs into the cytoplasm only if CP is present.
Until now, only a yeast system has been utilized to study host factors engaged in Bromoviridae replication (68). Now, the results of these high-throughput studies require verification with plant cells, which could be achieved by the application of RNA silencing technology to protoplasts, followed by viral transfection and the analysis of virus-host interactions.
TRANSCRIPTION OF sgRNA
The transcription of sgRNAs is used during the late stage of infection by Bromoviridae to express 3′-proximal genes encoding CP. SgRNAs serve as translational templates, while genomic RNAs can be recruited for replication and encapsidation (71). Three basic mechanisms for generating sgRNAs have been tested by using cell-based systems, including the premature termination during (−)-strand RNA synthesis (61), discontinuous transcription during (+)- or (−)-strand RNA synthesis (125), and internal initiation on the viral (−)-strand template (56). The last mechanism, requiring the involvement of cis-acting subgenomic promoter (sgp), is widely used by Bromoviridae.
The sgp of BMV has been characterized by using protoplast-based assays (55, 115). The promoter sequence includes an upstream AU-rich enhancer, a poly(U) tract, a 20-nt core promoter with the core hairpin, and the +1C initiation site, followed by a downstream AU-rich sequence (3, 29, 64). In order to synthesize sgRNA4, the replicase enzyme recognizes the core promoter sequence by an induced-fit mechanism (115). Recently, a novel 5′ sgRNA3a in BMV that arose by premature termination of genomic RNA3 synthesis has been discovered (132). Studies with barley and A. thaliana protoplasts have shown that both the length of the oligo(A) tract and the stability of the sgp core hairpin affected sgRNA3a synthesis (J. Sztuba-Solińska and J. J. Bujarski, unpublished data). In the case of AMV, data from transgenic P12 tobacco protoplasts have revealed a similar mechanism (121, 123). The nucleotides in the positions −26 to +1 relative to the transcription initiation site were found to control the wild-type level of sgRNA production (123). Other important elements included an enhancer (nt −136 and −94), the sgp hairpin, and the MP C terminus (123). Moreover, an RNA binding sequence on the CP could control the synthesis of AMV sgRNA (97). Two sgp's were found to be involved in the transcription of CMV sgRNAs in vitro (83, 114). The CMV sgp responsible for sgRNA4 synthesis in tobacco protoplasts has involved a 60-nt intercistronic region in RNA3 that mapped to nt −30 to nt +30 relative to the initiation cytidylate (8). However, the promoter sequence of the sgRNA4A synthesis still remains to be characterized. A potential difficulty here is that its sequences overlap in part with the coding region for replicase protein 2a (114).
GENETIC RECOMBINATION OF BROMOVIRIDAE
The multipartitism of the Bromoviridae genome facilitates genetic recombination of this family of viruses. The first recombination event was reported for BMV (in 1986), and since then, recombination processes have been studied extensively with this model bromovirus (10). The most popular model of RNA recombination is the template switching (copy choice) mechanism, which predicts that the viral replicase enzyme (RdRp) switches templates during RNA synthesis (91). Based on the pairing between the acceptor and the nascent RNAs, one can distinguish between homologous, aberrant homologous, and nonhomologous recombination events (11). Nonhomologous recombination was described for tobacco protoplasts after inoculation with BMV RNA1 and -2, and an RNA3 derivative lacking the 3′ noncoding region. It has been shown that 1 per 105 inoculated protoplasts acquired a replicating RNA3 that arose by nonhomologous recombination with RNA1 or RNA2 (45). Data obtained from protoplast systems have demonstrated that some regions of the BMV genome could support higher recombination frequencies than the others due to the presence of recombination signals, e.g., the insertion of the BMV AU-rich region into the Tomato bushy stunt virus supported frequent recombination in Nicotiana benthamiana protoplasts (112). Recently, the assistance of a novel 5′ sgRNA3a in the BMV RNA3-RNA3 homologous recombination has been tested with barley protoplasts (A. Dzianott, J. Sztuba-Solińska, and J. J. Bujarski, unpublished results), suggesting that prematurely detached sgRNA3a could prime recombination events on (−)-strand RNA3 templates within the sgp region (Fig. 4).
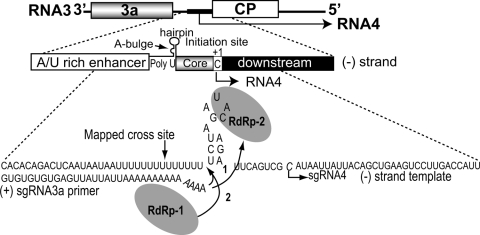
FIG. 4.
Two models of the crossing-over mechanism within the sgp region (27, 132). The top part shows the organization of sgp, while the sgp sequence is shown on the bottom. For clarity, only one recombining RNA3 molecule is shown, so the bowed arrows appear to depict crossovers toward sequences on the same template; however, more RNA3 templates might be engaged in the process. The ovals represent RdRp enzymes; one is leaving the template (RdRp-1), while another one (RdRp-2) is bound to the core hairpin of another (−)-strand template (for clarity, one template is shown). According to mechanism 1, the predetached 3′ poly(A) end “snatches” the RdRp-2 on another (−) strand, and the (+)-strand synthesis continues (represented by bow arrow 1). Mechanism 2 predicts that RdRp-1 detaches along with the sgRNA3a and rehybridizes to the poly(U) tract on another (−) strand to continue the (+)-strand synthesis (bowed arrow 2).
RNA recombination can salvage the damaged or mutated bromoviral RNAs and/or can contribute to the genome variability (91, 92, 124). Studies of barley protoplasts have shown that RNA recombination could be a rapid and frequent phenomenon (39, 88). New viruses or strains may emerge via recombination. A study using a tobacco protoplast system with chimeric BMV and TMV RNAs that carried the exchanged 3′ UTR segments revealed their efficient replication but also showed the formation of new RNA recombinant species (45). Replication efficiency assays of viral reassortants between CMV and Tomato aspermy cucumovirus (TAV) in tobacco protoplasts (26, 105) and Cassia yellow blotch bromovirus (CYBV), BMV, Spring beauty latent bromovirus, and CCMV in N. benthamiana protoplasts, e.g., between BMV and CYBV or between CMV and TAV (48, 26), have demonstrated the replication advantage of the chimeric viruses.
Replicase errors and RNA recombination are also responsible for the formation of defective RNAs, the deleted forms of viral RNAs that contain portions of the parental virus genome (21). For instance, the ability of defective RNA to replicate and to interfere with genomic BMV RNA replication has been demonstrated for barley (21, 22) and for zucchini (52) protoplasts.
ASSEMBLY OF BROMOVIRIDAE
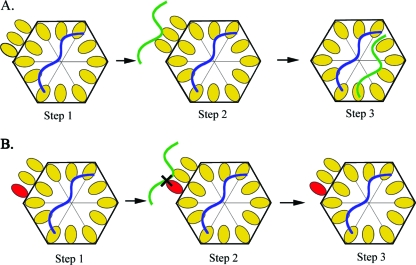
The segmented genomes of Bromoviridae are assembled within separate viral particles (87). Encapsidation studies with protoplasts allowed for the mapping of the genomic signals securing the specificity of RNA encapsidation, as well as factors stabilizing virions inside the cell (21, 23, 97). BMV RNA3 constructs carrying modifications in both the 3′ UTR and 3a gene allowed identification of two signals required for correct viral assembly in barley protoplasts: the 3′ TLS nucleating element (NE) for CP subunits and a cis-acting, position-dependent packaging element of 187 nt that was present within the MP open reading frame (ORF) (18, 22, 23). The lack of the packaging element made the sgRNA4 incompetent for autonomous assembly, whereas prepackaging of RNA3 was a prerequisite for sgRNA4 copackaging (Fig. 5) (18).
FIG. 5.
The sequential packaging model suggests that RNA3 and sgRNA4 copackage chronologically (87). Yellow ovals represent wild-type CP, the red oval symbolizes mutated CP, and blue and green ribbons indicate RNA3 and sgRNA4, respectively. (A) Assembly with wild-type coat protein subunits. Step 1, binding of wild-type CP subunits to the bipartite signal (3′TLS NE and MP NE) of RNA3 results in prior packaging of RNA3 into a virion. Step 2, the N-terminal arginine-rich motif of CP is displayed on the surface of the virion, allowing interaction of sgRNA4 with the arginine residues. Step 3 promotes copackaging of RNA3 and sgRNA4 into a virion. (B) Assembly with mutant coat protein subunits. Step 1, a mutation in the N-terminal arginine-rich motif of CP did not disrupt the assembly of RNA3. However, the sgRNA4 interaction is rigorously affected (Step 2). As a result, the virion will contain only the prepacked RNA3 (Step 3). (Adapted and reprinted from the Annual Review of Phytopathology [87] with permission of the publisher.)
The encapsidation effectiveness was shown to be dependent on the specific RNA secondary structure (23). The disruption of the stem of the 3′-proximal hairpin 1 or mutation of AUGC motif 2 in the AMV 3′ UTR inhibited CP binding to the 3′ termini, which affected both RNA replication and assembly encapsidation in barley protoplasts (97).
The selectivity of the virion assembly was proposed to be assured by the specific interaction between RNA and CP. Studies in barley protoplasts pinpointed the highly conserved N-terminal arginine-rich motif of BMV CP as being responsible for both RNA binding and RNA packaging (17, 90, 103). Likewise, the C-terminal region of the CP, especially Phe-184 and the corresponding sequence, affected both the encapsidation and the stability of the virus particles in barley and in N. benthiamaina protoplasts (79). In the case of AMV, the infection of transgenic P12 tobacco protoplasts with viral RNAs confirmed that CP molecules were required in trans for both replication and encapsidation of RNA1 and RNA2 but in cis for replication and encapsidation of RNA3 (75). Again, the role of CP C-terminal domain during interaction with viral RNA was shown to be critical for AMV encapsidation (119). The virion assembly and protection of the (+)-sense viral RNA in protoplasts seemed also to rely greatly on the CMV CP, since even a single amino acid change (Leu-129 to Phe-129) disrupted both processes (8, 117).
Furthermore, encapsidation assays of N. benthamiana protoplasts emphasized the importance of RNA-CP interaction for this process. The tested CP chimeras between BMV and CMV were unable to direct efficient RNA encapsidation (82). In contrast, the exchange of CP ORFs between AMV and Tobacco streak ilarvirus (TSV) showed that the heterologous CP sequence supported the encapsidation of TSV in P12 tobacco protoplasts (96), confirming the similarities between alfamo- and ilarvirus encapsidation.
VIRUS MOVEMENT
Although the process of cell-to-cell movement in protoplasts cannot be addressed directly by this system, some useful conclusions about the molecular mechanisms can be gained. The structural phenotype of the MPs of AMV and BMV, studied with cowpea, Hordeum vulgare, and N. benthamiana protoplasts, have shown that the MPs of these viruses did assemble into tubular structures at the surfaces of the protoplast cells (53). Electron microscopy and immunogold analyses confirmed the presence of both MP and virus particles in the tubules, suggesting that AMV and BMV can move from cell to cell as virions through tubule-like structures (53, 79, 108, 134). CMV MP also induced tubules in transfected protoplasts, but since the RNA3 tubule-defective mutants were capable of cell-to-cell spread in the infected tissue, these structures seem not to contribute to CMV movement (13). Thus, CMV is assumed to move as a ribonucleoprotein complex. Also, MPs of other members of Bromoviridae, TSV (67), and Olive latent virus 2 (OLV-2) (36), were reported to be the structural elements inside the tubules.
To identify the region of MP that is dispensable for protoplast protrusions, a set of deletion mutants within the AMV MP-GFP construct has been transfected onto tobacco protoplasts (44, 107, 134). The removal of MP amino acids 1 to 77, 84 to 142, and 226 to 300 (134), the introduction of point mutations at positions 25, 53, 123, and 138, and both N- and C-terminal deletions (107) all have affected tubule formation.
Studies in planta confirmed the role of microtubules during viral spread. These structures were shown to traverse the cell wall through modified plasmodesmata, and they mediate virion transfer via a tubule-guided mechanism (13, 41, 99, 109, 134). Recent studies performed with N. benthamiana plants and protoplasts have shown that host factors were engaged in the regulation of the BMV MP localization to the plasmodesmata (50).
Movement of the Bromoviridae sometimes requires compatibility between the MP and CP, manifested by the fact that not all chimerical MP and CP combinations are capable of cell-to-cell movement. The study of biological properties of some pseudorecombinants between CMV and TAV has shown that, even though they retained the ability to replicate in protoplasts, they were incapable of cell-to-cell movement (105, 106). An analysis of the recombinant clones suggested a requirement for compatibility between the C-terminal 29 amino acids of the MP and the C terminus of the CP (106). Following this discovery, computer analyses of the AMV CP constructs that were first tested with transgenic P12 plants and protoplasts have identified differences in the charge distribution of the CP/MP contact zone. Mutations in the N-terminal arm of AMV CP affected the cell-to-cell movement, whereas mutations at both the N and C termini affected the movement through the vascular system (119). Recent studies with barley protoplasts have confirmed interactions between BMV CP and MP that are regulated by the phosphorylation of a serine residue(s) by cellular protein kinase(s) (2).
WHAT'S NEXT?
Protoplasts will continue to serve as multipurpose systems for further lessons about the Bromoviridae single-cell reproduction cycle. We do not possess unambiguous answers concerning viral and cellular determinants that recruit genomic RNA into a membrane-bound replication complex (94, 120). The analysis of protoplast cells expressing viral proteins will help to identify key viral factors. On the other hand, the use of fluorescent tags in plant cells and small interfering RNA technology will be useful to characterize signals important for interactions between cellular and viral components. The use of the latest methods for three-dimensional analysis of cellular structures, such as electron tomography (70), will assist in our understanding of the architecture of these unique structures and in explaining such key aspects as how Bromoviridae replication and assembly get functionally connected or how viral replicase distinguishes between replication and transcription.
The use of protoplast systems for the analysis of host mutants will expand progress in identifying host factors that are crucial to the viral life cycle. A well-known illustration of this approach from Arabidopsis protoplasts has been the identification of the tom1 and tom2A host factors that did not support TMV replication (46, 47). Enhanced knowledge in this area would shape the groundwork for plant breeding efforts to develop virus resistance in crops. For instance, with the discovery of the eIF(iso)4E interaction with Turnip mosaic potyvirus VPg protein, the eIF4E was confirmed as a resistance gene and is now widely used by breeders to protect crops against potyviruses (33, 102). The cellular resistance characterized by the usage of protoplast systems along with RNA silencing technology will address questions about the limits of the involvement of host gene products in virus replication before the cell-to-cell movement occurs. By switching off the host genes followed by viral transfection, one will be able to recognize the pathogen-associated molecular patterns of innate immune responses, which will enhance our understanding of virus-host interactions. One well-known example of such a response includes the resistance controlled by the Rx locus against Potato virus Y (Potyvirus) in potato protoplasts (57).
The use of cell-based systems for studying molecular and biological properties of viruses that are considered new Bromoviridae family members might provide reliable answers concerning their membership and etiology. Differences of opinion exist concerning the taxonomy of OLV-2 within the family, whether Raspberry bushy dwarf virus and Pelargonium zonate spot virus should be considered members of the Bromoviridae, and even the accurate clustering of viruses within the Ilarvirus genus (32). Therefore, plant cell systems might help us to set up proper evolutionary relationships between Bromoviridae and to define the mechanisms of their life cycle or of the emergence of new viral strains.
Protoplast systems will open up new possibilities toward establishing cell culture assays, similar to bacterial, yeast, or animal monolayer cultures. Besides determining the molecular activities of RNA viruses in new hosts, they would help to define exact host-range determinants or to predict the role of molecular factors in virus evolutionary adaptation. For example, Chen et al. (15) have reported that sequences of lily strains of CMV were highly similar to one another, in spite of their origins, and that they became adapted to lily plants in their evolutionary history. It would be of utmost interest to employ protoplast system for tracing the host factors that increase the fitness of CMV isolates. Since the frequency of recombination is an obvious factor that determines viral fitness, the study of the new RNA arrangements in protoplasts will be essential to enhance our understanding of the Bromoviridae evolutionary dynamics and to determine what, if anything, limits their diversification.
Finally, the molecular strategies of viral gene expression used by plant viruses are also used by animal RNA viruses (126, 128). Thus, progress in our understanding of the Bromoviridae single-cell replication cycle might lead to the discovery and dissection of factors controlling the major agents of worldwide viral epidemics.
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB-0317039) and the National Institutes of Health (G1A62203) and by the Plant Molecular Biology Center at Northern Illinois University.
Footnotes
Published ahead of print on 6 August 2008.
REFERENCES
- 1.Ahlquist, P., S. X. Wu, P. Kaesberg, C. C. Kao, R. Quadt., W. DeJong, and R. Hershberger. 1994. Protein-protein interactions and glycerophospholipids in bromovirus and nodavirus RNA replication. Arch. Virol. Suppl. 9135-145. [DOI] [PubMed] [Google Scholar]
- 2.Akamatsu, N., A. Takeda, M. Kishimoto, M. Kaido, T. Okuno, and K. Mise. 2007. Phosphorylation and interaction of the movement and coat protein of brome mosaic virus in infected barley protoplasts. Arch. Virol. 1522087-2093. [DOI] [PubMed] [Google Scholar]
- 3.Allison, R. F., M. Janda, and P. Ahlquist. 1988. Infectious in vitro transcripts from cowpea chlorotic mottle virus cDNA clones and exchange of individual components with brome mosaic virus. J. Virol. 623581-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annamalai, P., and A. L. Rao. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 33896-111. [DOI] [PubMed] [Google Scholar]
- 5.Aoki, S., and I. Takebe. 1969. Infection of tobacco mesophyll protoplasts by tobacco mosaic virus ribonucleic acid. Virology 39439-448. [DOI] [PubMed] [Google Scholar]
- 6.Beckham, C. J., H. R. Light, A. Nissan, P. Ahlquist, R. Parker, and A. Noueiry. 2007. Interactions between brome mosaic virus RNAs and cytoplasmic processing bodies. J. Virol. 819759-9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendahmane, A., M. Querci, K. Kanyuka, and D. C. Baulcombe. 2000. Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 2173-81. [DOI] [PubMed] [Google Scholar]
- 8.Boccard, F., and D. Baulcombe. 1993. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology 193563-578. [DOI] [PubMed] [Google Scholar]
- 9.Bol, J. F. 1999. Alfalfa mosaic virus and ilarviruses: involvement of coat protein in multiple steps of the replication cycle. J. Gen. Virol. 801089-1102. [DOI] [PubMed] [Google Scholar]
- 10.Bujarski, J. J., and P. Kaesberg. 1986. Genetic recombination between RNA components of a multipartite plant virus. Nature 321528-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bujarski, J. J., and P. D. Nagy. 1996. Different mechanisms of homologous and nonhomologous recombination in brome mosaic virus: role of RNA sequence and replicase proteins. Semin. Virol. 7363-372. [Google Scholar]
- 12.Burgess, J., F. Motoyoshi, and E. N. Fleming. 1973. The mechanism of infection of plant protoplasts by viruses. Planta 112323-332. [DOI] [PubMed] [Google Scholar]
- 13.Canto, T., and P. Palukaitis. 1999. Are tubules generated by the 3a protein necessary for cucumber mosaic virus movement? Mol. Plant-Microbe Interact. 12985-993. [Google Scholar]
- 14.Chen, J., A. Noueir, and P. Ahlquist. 2003. An alternate pathway for recruiting template RNA to the brome mosaic virus RNA replication complex. J. Virol. 772568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, Y. K., A. F. L. M. Derks, S. Langeveld, R. Goldbach, and M. Prins. 2001. High sequence conservation among cucumber mosaic virus isolates from lily. Arch. Virol. 1461631-1636. [DOI] [PubMed] [Google Scholar]
- 16.Choi, S.-K., M. Hema, K. Gopinath, J. Santos, and C. C. Kao. 2004. Replicase-binding sites on plus- and minus-strand brome mosaic virus RNAs and their roles in RNA replication in plant cells. J. Virol. 7813420-13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi, Y. G., G. L. Grantham, and A. L. N. Rao. 2000. Molecular studies on bromovirus capsid protein. VII. Selective packaging of BMV RNA4 by specific N-terminal arginine residues. Virology 275207-217. [DOI] [PubMed] [Google Scholar]
- 18.Choi, Y. G., and A. L. N. Rao. 2003. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 779750-9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cocking, E. C. 1960. A method for the isolation of plant protoplasts and vacuoles. Nature 187962-963. [Google Scholar]
- 20.Cocking, E. C. 1966. An electron microscopic study of the initial stages of infection of isolated tomato fruit protoplasts by tobacco mosaic virus. Planta 68206-214. [DOI] [PubMed] [Google Scholar]
- 21.Damayanti, T. A., H. Nagano, K. Mise, I. Furusawa, and T. Okuno. 1999. Brome mosaic virus defective RNAs generated during infection of barley plants. J. Gen. Virol. 802511-2518. [DOI] [PubMed] [Google Scholar]
- 22.Damayanti, T. A., H. Nagano, K. Mise, I. Furusawa, and T. Okuno. 2002. Positional effect of deletions on viability, especially on encapsidation, of Brome mosaic virus D-RNA in barley protoplasts. Virology 293314-319. [DOI] [PubMed] [Google Scholar]
- 23.Damayanti, T. A., S. Tsukaguchi, K. Mise, and T. Okuno. 2003. cis-Acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts. J. Virol. 779979-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dawson, J. R. O., P. E. Dickerson, J. M. King, F. Sakai, A. R. H. Trim, and J. W. Watts. 1978. Improved methods for infection of plant protoplasts with viral ribonucleic acid. Z. Naturforsch. 33548-551. [Google Scholar]
- 25.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 7512370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding, S.-W., B.-J. Shi, W.-X. Li, and R. H. Symons. 1996. An interspecies hybrid RNA virus is significantly more virulent than either parental virus. Proc. Natl. Acad. Sci. USA 937470-7474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis, E. L., and M. Delbrück. 1939. The growth of bacteriophage. J. Gen. Physiol. 22365-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fraley, R., S. Subramani, P. Berg, and D. Papahadjopoulos. 1980. Introduction of liposome-encapsulated SV40 DNA into cells. J. Biol. Chem. 25510431-10435. [PubMed] [Google Scholar]
- 29.French, R., and P. Ahlquist. 1988. Characterization and engineering of sequences controlling in vivo synthesis of brome mosaic virus subgenomic RNA. J. Virol. 622411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallie, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 52108-2116. [DOI] [PubMed] [Google Scholar]
- 31.Gallie, D. R., and M. Kobayashi. 1994. The role of the 3′-untranslated region of non-polyadenylated plant viral mRNAs in regulating translational efficiency. Gene 142159-165. [DOI] [PubMed] [Google Scholar]
- 32.Gallitelli, D., M. Finetti-Sialer, and G. P. Martelli. 2005. Anulavirus, a proposed new genus of plant viruses in the family Bromoviride. Arch. Virol. 150407-411. [DOI] [PubMed] [Google Scholar]
- 33.Gao, Z., E. Johansen, S. Eyers, C. L. Thomas, T. H. Noel Ellis, and A. J. Maule. 2004. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 40376-385. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Ruiz, H., and P. Ahlquist. 2006. Inducible yeast system for viral RNA recombination reveals requirement for an RNA replication signal on both parental RNAs. J. Virol. 808316-8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gopinath, K., B. Dragnea, and C. C. Kao. 2005. Interaction between brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability. J. Virol. 7914222-14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grieco, F., M. A. Castellano, G. P. Di Sansebastiano, G. Maggipinto, J.-M. Neuhaus, and G. P. Martelli. 1999. Subcellular localization and in vivo identification of the putative movement protein of olive latent virus 2. J. Gen. Virol. 801103-1109. [DOI] [PubMed] [Google Scholar]
- 37.Grieco, F., M. Dell'Orco, and G. P. Martelli. 1996. The nucleotide sequence of RNA1 and RNA2 of olive latent virus 2 and its relationships in the family Bromoviridae. J. Gen. Virol. 772637-2644. [DOI] [PubMed] [Google Scholar]
- 38.Guogas, L. M., S. M. Laforest, and L. Gehrke. 2005. Coat protein activation of alfalfa mosaic virus replication is concentration dependent. J. Virol. 795752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hema, M., K. Gopinath, and C. C. Kao. 2005. Repair of the tRNA-like CCA sequence in a multipartite positive-strand RNA virus. J. Virol. 791417-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hema, M., and C. C. Kao. 2004. Template sequence near initiation nucleotide can modulate brome mosaic virus RNA accumulation in plant protoplasts. J. Virol. 781169-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herranz, M. C., J.-A. Sanchez-Navarro, A. Saurí, I. Mingarrob, and V. Pallás. 2005. Mutational analysis of the RNA-binding domain of the Prunus necrotic ringspot virus (PNRSV) movement protein reveals its requirement for cell-to-cell movement. Virology 33931-41. [DOI] [PubMed] [Google Scholar]
- 42.Honda, Y., C. Matsui, Y. Otsuki, and I. Takebe. 1974. Ultrastructure of tobacco mesophyll protoplasts inoculated with cucumber mosaic virus. Phytopathology 6430-34. [Google Scholar]
- 43.Houwing, C. J., P. van de Putte, and E. M. J. Jaspars. 1998. Regulation of single strand RNA synthesis of alfalfa mosaic virus in non-transgenic cowpea protoplasts by the viral coat protein. Arch. Virol. 143489-500. [DOI] [PubMed] [Google Scholar]
- 44.Huang, M., L. Jongejan, H. Zheng, L. Zhang, and J. F. Bol. 2001. Intracellular localization and movement phenotypes of Alfalfa mosaic virus movement protein mutants. Mol. Plant-Microbe Interact. 141063-1074. [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa, M., P. Kroner, P. Ahlquist, and T. Meshi. 1991. Biological activities of hybrid RNAs generated by 3′-end exchanges between tobacco mosaic and brome mosaic viruses. J. Virol. 653451-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishikawa, M., S. Naito, and T. Ohno. 1993. Effects of the tom1 mutation of Arabidopsis thaliana on the multiplication of tobacco mosaic virus RNA in protoplasts. J. Virol. 675328-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishikawa, M., F. Obata, T. Kumagai, and T. Ohno. 1991. Isolation of mutants of Arabidopsis thaliana in which accumulation of tobacco mosaic virus coat protein is reduced to low levels. Mol. Gen. Genet. 23033-38 94. [DOI] [PubMed] [Google Scholar]
- 48.Iwahashi, F., K. Fujisaki, M. Kaido, T. Okuno, and K. Mise. 2005. Synthesis of infectious in vitro transcripts from Cassia yellow blotch bromovirus cDNA clones and a reassortment analysis with other bromoviruses in protoplasts. Arch. Virol. 1501301-1314. [DOI] [PubMed] [Google Scholar]
- 49.Jaspars, E. M., and C. J. Houwing. 2002. A genome-activating N-terminal coat protein peptide of alfalfa mosaic virus is able to activate infection by RNAs 1, 2 and 3 but not by RNAs 1 and 2: further support for the messenger release hypothesis. Arch. Virol. 147857-863. [DOI] [PubMed] [Google Scholar]
- 50.Kaido, M., Y. Inoue, Y. Takeda, K. Sugiyama, A. Takeda, M. Mori, A. Tamai, T. Meshi, T. Okuno, and K. Mise. 2007. Downregulation of the NbNACa1 gene encoding a movement-protein-interacting protein reduces cell-to-cell movement of Brome mosaic virus in Nicotiana benthamiana. Mol. Plant-Microbe Interact. 20671-681. [DOI] [PubMed] [Google Scholar]
- 51.Kao, C. C., and K. Sivakumaran. 2000. Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 191-97. [DOI] [PubMed] [Google Scholar]
- 52.Kaplan, I. B., K.-C. Lee, T. Canto, S.-M. Wong, and P. Palukaitis. 2004. Host-specific encapsidation of a defective RNA 3 of Cucumber mosaic virus. J. Gen. Virol. 853757-3763. [DOI] [PubMed] [Google Scholar]
- 53.Kasteel, D. T. J., N. N. van der Wel, K. A. J. Jansen, R. W. Goldbach, and J. W. M. van Lent. 1997. Tubule-forming capacity of the movement proteins of alfalfa mosaic virus and brome mosaic virus. J. Gen. Virol. 782089-2093. [DOI] [PubMed] [Google Scholar]
- 54.Kim, M. J., H. R. Kim, and K.-H. Paek. 2006. Arabidopsis tonoplast proteins TIP1 and TIP2 interact with the cucumber mosaic virus 1a replication protein. J. Gen. Virol. 873425-3431. [DOI] [PubMed] [Google Scholar]
- 55.Koev, G., and W. A. Miller. 2000. A positive-strand RNA virus with three very different subgenomic RNA promoters. J. Virol. 745988-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koev, G., B. R. Mohan, and W. A. Miller. 1999. Primary and secondary structural elements required for synthesis of barley yellow dwarf virus subgenomic RNA1. J. Virol. 732876-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kohm, B. A., M. G. Goulden, J. E. Gilbert, T. A. Kavanagh, and D. C. Baulcombe. 1993. A Potato virus X resistance gene mediates an induced, nonspecific resistance in protoplasts. Plant Cell 5913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krab, I. M., C. Caldwell, D. R. Gallie, and J. F. Bol. 2005. Coat protein enhances translational efficiency of Alfalfa mosaic virus RNAs and interacts with the eIF4G component of initiation factor eIF4F. J. Gen. Virol. 861841-1849. [DOI] [PubMed] [Google Scholar]
- 59.Kushner, D. B., B. D. Lindenbach, V. Z. Grdzelishvili, A. O. Noueiry, S. M. Paul, and P. Ahlquist. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. USA 10015764-15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lahser, F. C., L. E. Marsh, and T. C. Hall. 1993. Contributions of the brome mosaic virus RNA-3 3′-nontranslated region to replication and translation. J. Virol. 673295-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin, H-X., W. Xu, and K. A. White. 2007. A Multicomponent RNA-based control system regulates subgenomic mRNA transcription in a tombusvirus. J. Virol. 812429-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loesch-Fries, L. S., and T. C. Hall. 1980. Synthesis, accumulation and encapsidation of individual brome mosaic virus RNA components in barley protoplasts. J. Gen. Virol. 47323-332. [Google Scholar]
- 63.Makkouk, K. M., L. Rizhallah, M. Madkour, M. El-Sherbeeny, S. G. Kumari, A. W. Amriti, and M. B. Solh. 1994. Survey of faba bean (Vicia faba L.) for viruses in Egypt. Phytopathol. Mediterr. 33207-211. [Google Scholar]
- 64.Marsh, L. E., T. W. Dreher, and T. C. Hall. 1988. Mutational analysis of the core and modulator sequences of BMV RNA3 subgenomic promoter. Nucleic Acids Res. 16981-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marsh, L. E., and T. C. Hall. 1987. Evidence implicating a tRNA heritage for the promoters of positive-strand RNA synthesis in brome mosaic virus and related viruses. Cold Spring Harbor Symp. Quant. Biol. 52331-341. [DOI] [PubMed] [Google Scholar]
- 66.Marsh, L. E., C. C. Huntley, G. P. Pogue, J. P. Connell, and T. C. Hall. 1991. Regulation of (+):(−)-strand asymmetry in replication of brome mosaic virus RNA. Virology 18276-83. [DOI] [PubMed] [Google Scholar]
- 67.Martelli, G. P., and M. Russo. 1985. Virus-host relationships: symptomatological and ultrastructural aspects, p.163-205. In R. I. B. Francki (ed.), The plant viruses: polyhedral virions with tripartite genomes. Plenum Press, New York, NY.
- 68.Mas, A., I. Alves-Rodrigues, A. Noueiry, P. Ahlquist, and J. Diez. 2006. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J. Virol. 801246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maule, A. J., M. I. Boulton, C. Edmunds, and K. R. Wood. 1980. Polyethylene glycol-mediated infection of cucumber protoplasts by cucumber mosaic virus and virus RNA. J. Gen. Virol. 47199-204. [Google Scholar]
- 70.Medalia, O., I. Weber, A. S. Frangakis, D. Nicastro, G. Gerish, and W. Baumeister. 2002. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 2981209-1213. [DOI] [PubMed] [Google Scholar]
- 71.Miller, W. A., and G. Koev. 2000. Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 2731-8. [DOI] [PubMed] [Google Scholar]
- 72.Milne, J. R., and G. H. Walter. 2003. The coincidence of thrips and dispersed pollen in PNRSV-infected stonefruit orchards: a precondition for thrips-mediated transmission via infected pollen. Annals Appl. Biol. 142291-298. [Google Scholar]
- 73.Mühlbach, H. P. 1982. Plant cell cultures and protoplasts in plant virus research. Curr. Top. Microbiol. Immunol. 9981-129. [DOI] [PubMed] [Google Scholar]
- 74.Nassuth, A., and J. F. Bol. 1983. Altered balance of the synthesis of plus and minus strand RNAs induced by RNA1 and 2 of alfalfa mosaic virus in absence of RNA3. Virology 12475-85. [DOI] [PubMed] [Google Scholar]
- 75.Neeleman, L., and J. F. Bol. 1999. Cis-acting functions of alfalfa mosaic virus proteins involved in replication and encapsidation of viral RNA. Virology 254324-333. [DOI] [PubMed] [Google Scholar]
- 76.Neeleman, L., H. J. M. Linthorst, and J. F. Bol. 2004. Efficient translation of alfamovirus RNAs requires the binding of coat protein dimers to the 3′ termini of the viral RNAs. J. Gen. Virol. 85231-240. [DOI] [PubMed] [Google Scholar]
- 77.Neeleman, L., R. C. Olsthoorn, H. J. Linthorst, and J. F. Bol. 2001. Translation of a nonpolyadenylated viral RNA is enhanced by binding of viral coat protein or polyadenylation of the RNA. Proc. Natl. Acad. Sci. USA 9814286-14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nishiguchi, M., W. H. R. Langridge, A. A. Szalay, and N. Zaitlin. 1986. Electroporation-mediated infection of tobacco leaf protoplasts with tobacco mosaic virus RNA and cucumber mosaic virus RNA. Plant Cell Rep. 557-60. [DOI] [PubMed] [Google Scholar]
- 79.Okinaka, Y., K. Mise, E. Suzuki, T. Okuno, and I. Furusawa. 2001. The C terminus of brome mosaic virus coat protein controls viral cell-to-cell and long-distance movement. J. Virol. 755385-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okuno, T., and I. Furusawa. 1979. RNA polymerase activity and protein synthesis in brome mosaic virus-infected protoplasts. Virology 99218-225. [DOI] [PubMed] [Google Scholar]
- 81.Olsthoorn, R. C., S. Mertens, F. T. Brederode, and J. F. Bol. 1999. A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J. 184856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Osman, F., Y. G. Choi, G. L. Grantham, and A. L. N. Rao. 1998. Molecular studies on bromovirus capsid protein. V. Evidence for the specificity of Brome mosaic virus encapsidation using RNA3 chimera of brome mosaic and cucumber mosaic viruses expressing heterologous coat proteins. Virology 251438-448. [DOI] [PubMed] [Google Scholar]
- 83.Palukaitis, P., M. J. Roossinck, R. G. Dietzgen, and R. I. B. Francki. 1992. Cucumber mosaic virus. Adv. Virus Res. 41281-348. [DOI] [PubMed] [Google Scholar]
- 84.Petrillo, J. E., G. Rocheleau, B. Kelley-Clarke, and L. Gehrke. 2005. Evaluation of the conformational switch model for alfalfa mosaic virus RNA replication. J. Virol. 795743-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pogue, G. P., and T. C. Hall. 1992. The requirement for a 5′ stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J. Virol. 66674-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pogue, G. P., L. E. March, J. P. Connell, and T. C. Hall. 1992. The requirement for ICR-like sequences in the replication of brome mosaic virus genomic RNA. Virology 188742-753. [DOI] [PubMed] [Google Scholar]
- 87.Rao, A. L. N. 2006. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 4461-87. [DOI] [PubMed] [Google Scholar]
- 88.Rao, A. L. N., T. W. Dreher, L. E. Marsh, and T. C. Hall. 1989. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. USA 865335-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rao, A. L. N., and G. L. Grantham. 1994. Amplification in vivo of brome mosaic virus RNAs bearing 3′ non-coding region from cucumber mosaic virus. Virology 204478-481. [DOI] [PubMed] [Google Scholar]
- 90.Rao, A. L. N., and G. L. Grantham. 1996. Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology 226294-305. [DOI] [PubMed] [Google Scholar]
- 91.Rao, A. L. N., and T. C. Hall. 1990. Requirement of a viral trans-acting factor encoded by brome mosaic virus RNA-2 provides strong selection in vivo for functional recombinants. J. Virol. 642437-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rao, A. L. N., and T. C. Hall. 1993. Recombination and polymerase error facilitate restoration of infectivity in brome mosaic virus. J. Virol. 67969-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reichert, V. L., M. Choi, J. E. Petrillo, and L. Gehrke. 2007. Alfalfa mosaic virus coat protein bridges RNA and RNA-dependent RNA polymerase in vitro. Virology 364214-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Restrepo-Hartwig, M., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 7310303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 708908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reusken, C. B. E. M., L. Neeleman, and J. F. Bol. 1995. Ability of tobacco streak virus coat protein to substitute for late functions of alfalfa mosaic virus coat protein. J. Virol. 694552-4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Reusken, C. B. E. M., L. Neeleman, F. T. Brederode, and J. F. Bol. 1997. Mutations in coat protein binding sites of alfalfa mosaic virus RNA 3 affect subgenomic RNA 4 accumulation and encapsidation of viral RNAs. J. Virol. 718385-8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rietveld, K., C. W. A. Pleij, and L. Bosch. 1983. Three-dimensional models of the tRNA-like 3′ termini of some plant viral RNAs. EMBO J. 21079-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ritzenthaler, C., and C. Hofmann. 2007. Tubule-guided movement of plant viruses, p. 63-84. In E. Waigmann and M. Heinlein (ed.), Plant cell monographs, vol. 7. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 100.Roenhorst, J. W., J. W. M. van Lent, and B. J. M. Verduin. 1988. Binding of cowpea chlorotic mottle virus to cowpea protoplasts and relation of binding to virus entry and infection. Virology 16491-98. [DOI] [PubMed] [Google Scholar]
- 101.Roossinck, M. J. 2001. Cucumber mosaic virus, a model for RNA virus evolution. Mol. Plant Pathol. 259-63. [DOI] [PubMed] [Google Scholar]
- 102.Ruffel, S., M.-H. Dussault, A. Palloix, B. Moury, A. Bendahmane, C. Robaglia, and C. Caranta. 2002. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 321067-1075. [DOI] [PubMed] [Google Scholar]
- 103.Sacher, R., and P. Ahlquist. 1989. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J. Virol. 634545-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sakai, F., J. W. Watts, J. R. O. Dawson, and J. B. Bancroft. 1977. Synthesis of proteins in tobacco protoplasts infected with cowpea chlorotic mottle virus. J. Gen. Virol. 34285-293. [Google Scholar]
- 105.Salánki, K., I. Carrère, M. Jacquemond, E. Balázs, and M. Tepfer. 1997. Biological properties of pseudorecombinant and recombinant strains created with cucumber mosaic virus and tomato aspermy virus. J. Virol. 713597-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Salánki, K., Á. Gellért, E. Huppert, G. Náray-Szabó, and E. Balázs. 2004. Compatibility of the movement protein and the coat protein of cucumoviruses is required for cell-to-cell movement. J. Gen. Virol. 851039-1048. [DOI] [PubMed] [Google Scholar]
- 107.Sánchez-Navarro, J. A., and J. F. Bol. 2001. Role of Alfalfa mosaic virus movement protein and coat protein in virus transport. Mol. Plant-Microbe Interact. 141051-1062. [DOI] [PubMed] [Google Scholar]
- 108.Sánchez-Navarro, J. A., M. C. Herranz, and V. Pallás. 2006. Cell-to-cell movement of Alfalfa mosaic virus can be mediated by the movement proteins of Ilar-, bromo-, cucumo-, tobamo- and comoviruses, and does not require virion formation. Virology 34666-73. [DOI] [PubMed] [Google Scholar]
- 109.Sánchez-Navarro, J. A., R. Miglino, A. Ragozzino, and J. F. Bol. 2001. Engineering of Alfalfa mosaic virus RNA 3 into an expression vector. Arch. Virol. 146923-939. [DOI] [PubMed] [Google Scholar]
- 110.Sander, E., and G. Mertes. 1984. Use of protoplasts and separate cells in plant virus research. Adv. Virus Res. 29215-262. [DOI] [PubMed] [Google Scholar]
- 111.Scott, S. W. 2006. Bromoviridae and allies. Encyclopedia of Life Sciences. doi: 10.1038/npg.els.0004246. [DOI]
- 112.Shapka, N., and P. D. Nagy. 2004. The AU-rich RNA recombination hot spot sequence of Brome mosaic virus is functional in tombusviruses: implications for the mechanism of RNA recombination. J. Virol. 782288-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sivakumaran, K., Y. Bao, M. J. Rossinck, and C. C. Kao. 2000. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicases of Brome mosaic virus and Cucumber mosaic virus. J. Virol. 7410323-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sivakumaran, K., M.-H. Chen, M. J. Rosssinck, and C. C. Kao. 2002. Core promoter for initiation of Cucumber mosaic virus subgenomic RNA4A. Mol. Plant Pathol. 343-52. [DOI] [PubMed] [Google Scholar]
- 115.Sivakumaran, K., S.-K. Choi, M. Hema, and C. C. Kao. 2004. Requirements for brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro. J. Virol. 786091-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smirnyagina, E., Y. H. Hsu, N. Chua, and P. Ahlquist. 1994. Second-site mutations in the brome mosaic virus RNA3 intercistronic region partially suppress a defect in coat protein mRNA transcription. Virology 198427-436. [DOI] [PubMed] [Google Scholar]
- 117.Suzuki, M., S. Kuwata, C. Masuta, and Y. Takanami. 1995. Point mutations in the coat protein of cucumber mosaic virus affect symptom expression and virion accumulation in tobacco. J. Virol. 761791-1799. [DOI] [PubMed] [Google Scholar]
- 118.Takebe, I., and Y. Otsuki. 1969. Infection of tobacco mesophyll protoplasts by tobacco mosaic virus. Proc. Natl. Acad. Sci. USA 64843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tenllado, F., and J. F. Bol. 2000. Genetic dissection of the multiple functions of alfalfa mosaic virus coat protein in viral RNA replication, encapsidation, and movement. Virology 26829-40. [DOI] [PubMed] [Google Scholar]
- 120.Van Der Heijden, M., W. Carette, P. J. Reinhoud, A. Haegi, and J. F. Bol. 2001. Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J. Virol. 751879-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.van der Kuyl, A. C., L. Neeleman, and J. F. Bol. 1991. Deletion analysis of cis- and trans-acting elements involved in replication of alfalfa mosaic virus RNA 3 in vivo. Virology 183687-694. [DOI] [PubMed] [Google Scholar]
- 122.van der Vossen, E. A. G., L. Neeleman, and J. F. Bol. 1993. Role of the 5′ leader sequence of alfalfa mosaic virus RNA 3 in replication and translation of the viral RNA. Nucleic Acids Res. 211361-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.van der Vossen, E. A. G., T. Notenboom, and J. F. Bol. 1995. Characterization of sequences controlling the synthesis of alfalfa mosaic virus subgenomic RNA in vivo. Virology 212663-672. [DOI] [PubMed] [Google Scholar]
- 124.van der Vossen, E. A. G., C. B. E. M. Reusken, and J. F. Bol. 1996. cis-Preferential stimulation of alfalfa mosaic virus RNA 3 accumulation by the viral coat protein. Virology 220163-170. [DOI] [PubMed] [Google Scholar]
- 125.van Vliet, A. L. W., S. L. Smits, P. J. M. Rottier, and R. J. de Groot. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 216571-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.van Vugt, J. J. F. A., T. Storgaard, M. B. Oleksiewicz, and A. Bøtner. 2001. High frequency RNA recombination in porcine reproductive and respiratory syndrome virus occurs preferentially between parental sequences with high similarity. J. Gen. Virol. 822615-2620. [DOI] [PubMed] [Google Scholar]
- 127.Voinnet, O., S. Rivas, P. Mestre, and D. C. Baulcombe. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33949-956. [DOI] [PubMed] [Google Scholar]
- 128.Wang, T.-C., and M. Chao. 2005. RNA Recombination of hepatitis delta virus in natural mixed-genotype infection and transfected cultured cells. J. Virol. 792221-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Watanabe, Y., I. Ooshika, T. Meshi, and Y. Okada. 1986. Subcellular localization of the 30K protein in TMV-inoculated tobacco protoplasts. Virology 152414-420. [DOI] [PubMed] [Google Scholar]
- 130.Watts, J. W., and J. R. O. Dawson. 1980. Double infection of tobacco protoplasts with brome mosaic virus and cowpea chlorotic mottle virus. Virology 105501-507. [DOI] [PubMed] [Google Scholar]
- 131.Wierzchoslawski, R., and J. J. Bujarski. 2006. Efficient in vitro system of homologous recombination in brome mosaic bromovirus. J. Virol. 806182-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wierzchoslawski, R., A. Urbanowicz, A. Dzianott, M. Figlerowicz, and J. J. Bujarski. 2006. Characterization of a novel 5′ subgenomic RNA3a derived from RNA3 of Brome mosaic bromovirus. J. Virol. 8012357-12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yoshii, M., M. Nishikiori, K. Tomita, N. Yoshioka, R. Kozuka, S. Naito, and M. Ishikawa. 2004. The Arabidopsis Cucumovirus Multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 786102-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zheng, H., G. Wang, and L. Zhang. 1997. Alfalfa mosaic virus movement protein induces tubules in plant protoplasts. Mol. Plant-Microbe Interact. 101010-1014. [Google Scholar]