Abstract
Integration of viral DNA into the host chromosome is an essential step in the life cycle of retroviruses and is facilitated by the viral integrase enzyme. The first generation of integrase inhibitors recently approved or currently in late-stage clinical trials shows great promise for the treatment of human immunodeficiency virus (HIV) infection, but virus is expected to develop resistance to these drugs. Therefore, we used a novel resistance selection protocol to follow the emergence of resistant HIV in the presence of the integrase inhibitor elvitegravir (GS-9137). We find the primary resistance-conferring mutations to be Q148R, E92Q, and T66I and demonstrate that they confer a reduction in susceptibility not only to elvitegravir but also to raltegravir (MK-0518) and other integrase inhibitors. The locations of the mutations are highlighted in the catalytic sites of integrase, and we correlate the mutations with expected drug-protein contacts. In addition, mutations that do not confer reduced susceptibility when present alone (H114Y, L74M, R20K, A128T, E138K, and S230R) are also discussed in relation to their position in the catalytic core domain and their proximity to known structural features of integrase. These data broaden the understanding of antiviral resistance against integrase inhibitors and may give insight facilitating the discovery of second-generation compounds.
Integration of retroviral DNA is an essential step in the life cycle of human immunodeficiency virus (HIV) (29). The integration process is facilitated by the viral integrase (IN) enzyme which catalyzes the insertion of the viral DNA into the host genome in a multistep process involving viral and host proteins. HIV IN recognizes and binds specific sequences in the long terminal repeats (LTRs) of the viral retrotranscribed DNA in the cytoplasm. After DNA binding, IN cleaves GT dinucleotides from the 3′ termini of the linear cDNA in a process called 3′ processing (2). The processed viral DNA, as part of the preintegration complex, is then translocated into the nucleus, where IN inserts the viral DNA into the host chromosome by a process called strand transfer (2, 12, 13). There are few cellular enzymes with comparable function to HIV integrase (24), apart from the V(D)J polynucleotide recombinase RAG1 (34). Therefore, the IN enzyme has been considered an attractive target for antiretroviral therapy for the last decade (27, 36). Recent progress has resulted in two IN inhibitors (5, 30), with one drug in late-stage clinical trials and one currently approved for use in treatment-experienced patients (18). For all currently targeted retroviral proteins, inhibition with antiretroviral drugs has led to emergence of resistance in treated patients, often leading to treatment failure and requiring changes in the composition of the highly active antiretroviral therapy (HAART) drug regimen (6). Nevertheless, the emergence of new classes of drugs will enable new combinations of inhibitors to be used and will offer more treatment options to HIV-infected patients at risk of failure on current HAART regimes.
Early IN inhibitors (e.g., L-731,988 and S-1360) contained β-hydroxyl diketo moieties that were purported to engage the magnesium cations coordinated by the D,D(35)E motif in the active site of the enzyme and prevented strand transfer (4, 14, 20). The early diketo-containing compounds showed poor therapeutic indices and micromolar potency in antiviral assays. A naphthyridine-based compound (L-870,810) contained a new chemical scaffold and superior potency to the early diketo-containing molecules (19). However, the development of L-870,810 was discontinued due to toxicity observed in dogs. The hydroxypyrimidinone carboxamide MK-0518 (raltegravir [RAL]) (5) and the quinolone based GS-9137 (elvitegravir [EVG]; formerly, JTK-303) were then identified as promising lead compounds, and clinical development was initiated (5, 9, 18).
The emergence of resistance mutations to EVG and RAL will presumably influence the use of these inhibitors in various HAART regimens and, hence, help to guide the requirements for a second generation of IN inhibitors (10). Currently, data concerning the emergence of resistance mutations to both EVG and RAL are very limited (26, 33). While the analysis of larger patient resistance data sets will result in better knowledge of resistance pathways, indications of likely pathways can also be achieved with in vitro selection (IVS), which often elicits similar pathways of resistance to those observed in vivo. Furthermore, the genotyping and phenotyping of viruses obtained in vitro can be achieved rapidly, and confirmation with site-directed mutants (SDMs) enables the delineation of the change in drug susceptibility due to each individual mutation. In this way, a picture of the resistance pathways, changes in susceptibility, the degree of cross-resistance, and the identification of possible compensatory mutants and other resistance-associated mutations (RAMs) to a wide range of IN inhibitors can be built. Here, we report the results of IVS of HIV type 1 (HIV-1) with the investigational IN inhibitor EVG and describe mutations that confer reduced susceptibility to EVG and other IN inhibitors, including RAL.
MATERIALS AND METHODS
Antiviral compounds.
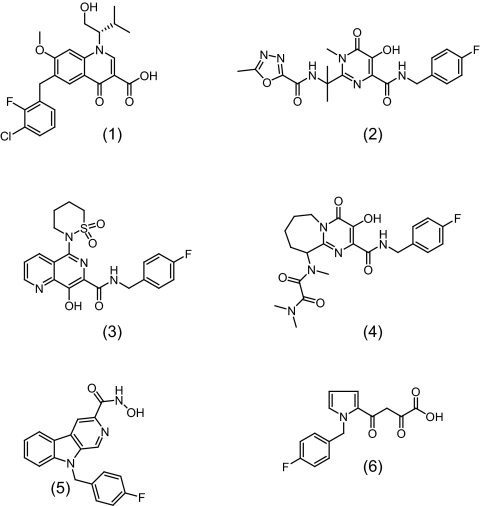
EVG (GS-9137) (30), RAL (MK-0518) (1), L-870,810 (19), PACA (a 3-hydroxy-4-oxo-4,6,7,8,9,10-hexahydro-pyrimido[1,2-a]azepine-2-carboxylamide) (3), PICA (a 9H-pyrido[3,4-b]indole-3-carboxamide) (23), L-731,988 (28), and the nucleoside reverse transcriptase inhibitor zidovudine (21) were obtained from commercial suppliers or were synthesized in-house. Chemical structures are shown in Fig. 1.
FIG. 1.
Structure of seven different IN inhibitors. (1) Elvitegravir. (2) Raltegravir. (3) L-870,810. (4) PACA. (5) PICA. (6) L-731,988.
Cells and viruses.
The human T-lymphoblastoid cell line MT4 was kindly provided by Naoki Yamamoto (National Institute of Infectious Diseases, AIDS Research Center, Tokyo, Japan). The cell line was maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, 0.1% NaHCO3, and antibiotics (0.02% gentamicin and 0.8% G418) and incubated in a humidified incubator with a 5% CO2 atmosphere at 37°C.
MT4-LTR-enhanced green fluorescent protein (EGFP) cells were obtained by transfecting MT4 cells with a selectable construct encompassing the coding sequences for the HIV LTR as a promoter for the expression of EGFP and subsequent selection of permanently transfected cells.
MT4-cytomegalovirus (CMV)-EGFP cells were obtained by selection for permanently transformed MT4 cells with a CMV-EGFP reporter gene.
HIV-1 IIIB was provided by Guido van der Groen (Institute of Tropical Medicine, Antwerp, Belgium). HIV-1 molecular clone pHXB2D was provided by the Centre for AIDS Reagents (ARP206; Brussels, Belgium).
SDMs of IN coding sequences were constructed in the pUC19-5′HXB2D vector (XbaI-SalI fragment of pHXB2D), containing the HIV-1 clone HXB2D IN coding sequence, by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and high-performance liquid chromatography-purified primers (Genset Oligos, La Jolla, CA). Altered plasmid sequences were confirmed by dideoxyribose sequencing.
Generation of the SDM virus stocks.
MT4 cells were subcultured at a density of 250,000 cells/ml on the day before transfection. Cells were pelleted and resuspended in phosphate-buffered saline at a concentration of 3.1 × 106 cells/ml. A 0.8-ml portion (2.5 × 106 cells/ml) was used for each transfection. Transfections were performed with a Bio-Rad Gene Pulser (Bio-Rad, Hercules, CA) with 0.4-cm electrode cuvettes (Bio-Rad). Cells were electroporated with 10 μg of SalI-linearized pUC19-3′HXB2D (SalI-XbaI fragment of pHXB2D) and 5 μg of SalI-digested SDM at 250 μF and 300 V, followed by a 30-min incubation at room temperature. Ten milliliters of fresh culture medium was then added to the suspension of transfected cells, and incubation was performed at 37°C in a humidified atmosphere with 5% CO2. Cell cultures were monitored for the appearance of cytopathic effect (CPE). At virus breakthrough (full CPE), culture supernatant was typically harvested by centrifugation at 8 to 10 days after transfection and was stored at −80°C for subsequent drug susceptibility determination.
Antiviral assays.
The antiviral activity of different inhibitors was determined in a cell-based HIV-1 replication assay. Briefly, MT4-LTR-EGFP cells (150,000 cells/ml) were infected with HIV-1 (IIIB, HXB2D, selected viruses, or site-directed mutant strains; multiplicity of infection [MOI] of 0.0025) in the presence or absence of inhibitor. After 3 days of incubation, the inhibition of HIV replication was quantified by measuring EGFP fluorescence, and expressed as the inhibitor concentration required for 50% inhibition of HIV-1 replication in cell culture (EC50). The cytotoxicity of inhibitors was determined in parallel on mock-infected MT4-CMV-EGFP cells (150,000 cells/ml) cultured in the presence or absence of test compound concentrations (data not shown). After 3 days of incubation, inhibition of cell proliferation was quantified by measuring the EGFP fluorescence and expressed as the compound concentration that inhibits cell growth by 50% (cytotoxicity).
Genotyping.
Viral RNA was extracted from culture supernatant or virus stock using a NucliSens easyMAG apparatus (bioMérieux, Marcy l' Etoile, France), a high-throughput automated nucleic acid extraction system. cDNA encoding reverse transcriptase and IN was generated with Expand Reverse Transcriptase (Roche Diagnostics, Basel, Switzerland), followed by amplification of the IN region by nested PCR (1,456 bp). PCR products were genotyped by automated population-based full-sequence analysis (ABI Prism BigDye Terminator cycle sequencing; Applied Biosystems, Foster City, CA). Sequencing results were reported as amino acid changes compared with the HIV-1 IIIB wild-type reference sequence. Mutations present in more than 25% of the total virus population could be detected as a mixture with the wild-type virus. HIV-1 IIIB and HIV-1 HXB2D are two distinct wild-type strains with similar phenotypes for the IN inhibitors tested in this study. These two wild-type virus strains differ genetically in three residues of the IN sequence. The HIV-1 IIIB IN sequence contains residues T124, D232, and V265, whereas the HIV-1 HXB2D IN sequence contains A124, N232, and A265. Moreover, D232N and A124T have been reported as IN polymorphisms in HIV-1 (25). Starting from HIV-1 IIIB, mutations T124A, D232N, and V265A were observed in the genotypes of viruses selected with EVG. As these mutations are consistent with the observed polymorphisms and differences between the HIV-1 IIIB and HIV-1 HXB2D IN sequences and no large EC50 difference was observed for the tested IN compounds between the strains, these mutations did not contribute to the IN resistance and consequently have been omitted from the results and discussion sections.
Classical in vitro selection experiments.
Classical in vitro selection was performed as described before (35). Briefly, in a T25 tissue culture flask, MT4-LTR-EGFP cells were propagated in a volume of 10 ml and infected with HIV-1 wild-type strain IIIB (0.01 to 0.001 the 50% cell culture infectious dose) in the presence of EVG at a concentration of 5 nM. Cultures were examined every 3 to 4 days for signs of virus replication. Upon virus breakthrough, the supernatant was collected, and an aliquot was used to infect fresh cultures in the presence of a twofold higher concentration of EVG than was used to infect the previous culture. This procedure was repeated up to an EVG concentration of 10 μM. Viral replication was assessed by microscopic scoring of the cytopathogenicity and virus-induced fluorescence, and viral breakthrough was defined as microscopic evidence of extensive viral replication in all cell clusters (full CPE). Samples for genotyping and phenotyping were obtained from the harvested supernatant.
Automated in vitro selection experiments in 96-well plates.
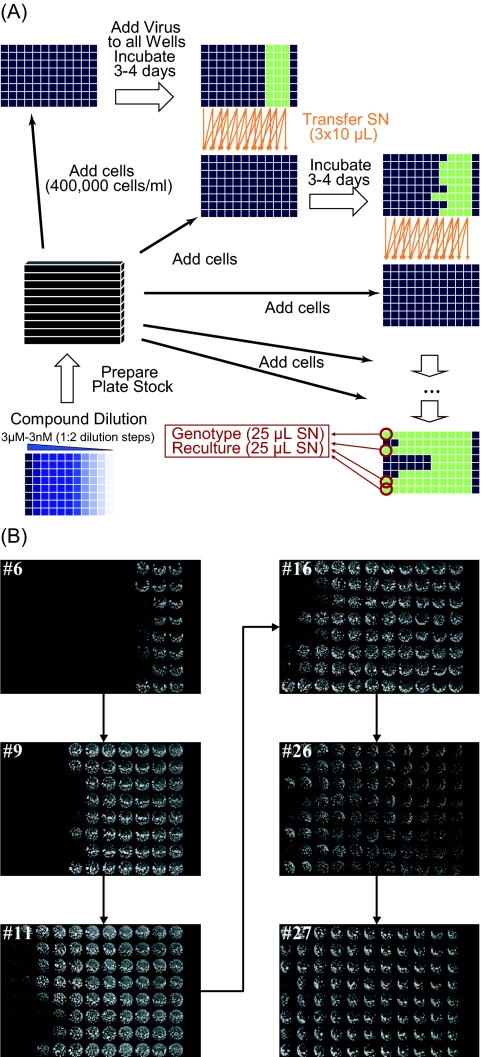
In vitro selection was performed in 96-well plates, with each row representing a separate IVS experiment (Fig. 2). MT4-LTR-EGFP cells (400,000 cells/ml) and HIV-1 IIIB (MOI of 0.01) were added to the wells of a 96-well plate containing a serial 1:2 dilution range of EVG (3 μM to 3 nM), and plates were incubated in a humidified incubator with a 5% CO2 atmosphere at 37°C. After 3 to 4 days, 30 μl of virus-containing supernatant was harvested from each well and split in three 10-μl portions. These portions were used to infect the 150 μl of fresh cells in the wells of the same row in a new compound plate with, respectively, 1× (same as originating well), 2×, and 4× the EVG concentration. This procedure was repeated every 3 to 4 days. When virus replication was evident in wells at the highest concentration of EVG, the supernatant was harvested and used for genotyping, phenotyping, and propagating new virus stock. In contrast with the classical IVS experiments, where compound concentration is only increased at viral breakthrough, this method uses a protocol where the virus is challenged with an increased compound concentration at each passage.
FIG. 2.
Automated IVS in 96-well plates. (A) A stock of 96-well plates was generated and stored at −20°C. Each row of a plate contained a 1:2 dilution series of EVG, with the highest concentration (3 μM) in column 1 and the lowest concentration (3 nM) in column 11. MT4-LTR-EGFP cells and HIV-1 IIIB were added to row 1-12 and row 1-11, respectively. Column 12 was used as a cell control. Every 3 to 4 days, a new growth cycle was initiated. To this end, a compound plate was thawed, and fresh cells were added. Next, virus-containing supernatant (SN) was harvested from each well of the previous plate and split in three portions that were used to infect cells in the wells on the same row in the new plate with, respectively, 1× (same as originating well), 2×, and 4× the EVG concentration. When 100% virus breakthrough was observed in the wells of column 1 (3 μM), virus was harvested for genotyping. (B) Every passage, a fluorescent scan was generated from the plates just before the recultivation cycle. Pictures for the plates of passage 6, 9, 11, 16, 26, and 27 are shown.
Replication kinetics of SDMs and selected HIV-1 strains.
MT4 cells (120,000 cells) were infected with HIV-1 (IIIB, HXB2D, selected viruses, or SDM viruses; MOI of 0.0001) in the absence of inhibitor. Every day, cells were examined for the appearance of HIV-1-induced CPE. In addition, aliquots of cell-free supernatants were taken for determination of viral p24 levels (HIV-1 p24 enzyme-linked immunosorbent assay; Perkin Elmer, Waltham, MA).
RESULTS
Classical in vitro selection of EVG-resistant viruses.
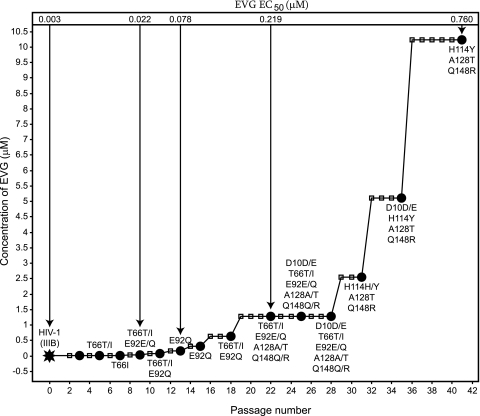
In the classical IVS experiment performed in tissue culture flasks, the virus was propagated over 41 passages (144 days) while the concentration of EVG was gradually increased from 5 nM up to 10 μM. The IN-encoding region of the pol gene of the viruses, selected at different passages, was sequenced. Multiple mutations were observed in these viruses in comparison with the IN sequence of wild-type HIV-1 IIIB (Table 1 and Fig. 3).
TABLE 1.
HIV-1 IIIB strains selected in the presence of EVG using classical IVS
| HIV-1 strain | Passage no. | Concn (μM) | Amino acid mutation(s)a
|
|||||
|---|---|---|---|---|---|---|---|---|
| D10 | T66 | E92 | H114 | A128 | Q148 | |||
| IIIB | 0 | 0.005 | ||||||
| IIIB/GS-9137(#5) | 5 | 0.01 | T66T/I | |||||
| IIIB/GS-9137(#7) | 7 | 0.02 | T66I | |||||
| IIIB/GS-9137(#9) | 9 | 0.04 | T66T/I | E92E/Q | ||||
| IIIB/GS-9137(#11) | 11 | 0.08 | T66T/I | E92Q | ||||
| IIIB/GS-9137(#13) | 13 | 0.16 | E92Q | |||||
| IIIB/GS-9137(#15) | 15 | 0.32 | E92Q | |||||
| IIIB/GS-9137(#18) | 18 | 0.64 | T66T/I | E92Q | ||||
| IIIB/GS-9137(#22) | 22 | 1.28 | T66T/I | E92E/Q | A128A/T | Q148Q/R | ||
| IIIB/GS-9137(#25) | 25 | 1.28 | D10D/E | T66T/I | E92E/Q | A128A/T | Q148Q/R | |
| IIIB/GS-9137(#28) | 28 | 1.28 | D10D/E | T66T/I | E92E/Q | A128A/T | Q148Q/R | |
| IIIB/GS-9137(#31) | 31 | 2.56 | H114H/Y | A128T | Q148R | |||
| IIIB/GS-9137(#35) | 35 | 5.12 | D10D/E | H114Y | A128T | Q148R | ||
| IIIB/GS-9137(#41) | 41 | 10.24 | H114Y | A128T | Q148R | |||
Sequencing results are reported as amino acid changes compared with the HIV-1 IIIB wild-type reference sequence. Mutations present in more than 25% of the total virus population can be detected as a mixture with the wild-type amino acids.
FIG. 3.
Classical IVS of EVG-resistant HIV-1 strains. MT4-LTR-EGFP cells were infected with HIV-1 IIIB (black star) and grown in the presence of EVG. Cells were passaged every 3 to 4 days (small squares). When full virus breakthrough was observed (black dots), viruses were harvested, and the concentration of EVG was doubled. Genotypes of harvested viruses are indicated next to the black dots; phenotypes (EVG EC50) of viruses IIIB/GS-9137(#9), IIIB/GS-9137(#13), IIIB/GS-9137(#22), and IIIB/GS-9137(#41) were determined using a cellular antiviral assay.
The first two mutations to emerge were T66I and E92Q after 5 and 9 passages, respectively. These mutations were undetectable from passage 31 and onwards. Q148R and A128T developed when 1.28 μM EVG (22 passages) was used for selection, and D10E appeared after 25 passages, but the latter mutation was not observed in the final viral population (41 passages). After 31 passages, mutation H114Y emerged. The final viral pool, emerging after 41 passages (144 days) at a concentration of 10.24 μM EVG, contained the IN sequence H114Y/A128T/Q148R.
Automated in vitro selection of EVG-resistant viruses.
A recently described method that uses an automated IVS approach in 96-well plates (D. Jochmans, M. Van Ginderen, I. De Baere, S. Hallenberger, and G. Kraus, presented at the 16th International HIV Drug Resistance Workshop, Barbados, West Indies, 12 to 16 June 2007) was used to select EVG-resistant strains starting from HIV-1 IIIB. Here, the virus is challenged with an increased compound concentration at each passage and not only at virus breakthrough, as is done for classical IVS. Using this method, eight parallel IVS experiments were performed simultaneously. Virus samples were taken at every passage and genotyped to identify mutations. Viral propagation continued up to a final concentration of 3 μM EVG. The genotypes observed at this concentration are shown in Table 2. All viruses were phenotyped to reveal changes in susceptibility to EVG and other inhibitors (see Fig. 5). Virus was able to grow at the final concentration of 3 μM EVG after 16 to 27 passages or 56 to 95 days (Table 2). A number of the mutations observed in the viruses selected with EVG have been observed in a phase 2b study with this IN inhibitor (e.g., Q148R, E92Q, T66I/A, and E138K) (D. McColl, S. Fransen, S. Gupta, N. Parkin, N. Margot, S. Chuck, A. Cheng, and M. Miller, presented at the 11th European Aids Conference, Madrid, Spain, 24 to 27 October 2007), supporting the relevance of this automated IVS method to clinical data.
TABLE 2.
Overview genotypes of eight viruses out of the automated IVS experiments in 96-well plates at a final concentration of 3 μM EVG
| HIV-1 strain | Passage no. | Mutation(s)a |
|---|---|---|
| Virus 1 | 16 | E138K Q148R |
| Virus 2 | 25 | T66T/A E92Q |
| Virus 3 | 25 | T66T/I A128A/T E138E/K Q148Q/R S230R |
| Virus 4 | 25 | E92Q |
| Virus 5 | 23 | E92E/Q E138E/K Q148Q/R |
| Virus 6 | 16 | T66T/A E92Q |
| Virus 7 | 27 | R20R/K T66I L74L/M S230R |
| Virus 8 | 27 | T66I |
Sequencing results are reported as amino acid changes compared with the HIV-1 IIIB wild-type reference sequence. Mutations present in more than 25% of the total virus population can be detected as a mixture with the wild-type virus.
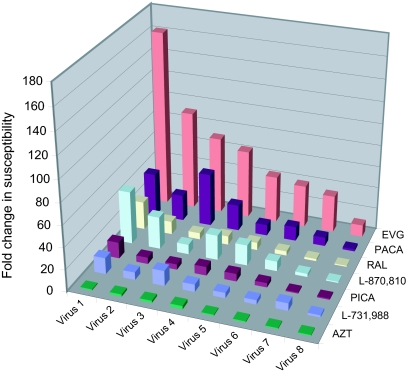
FIG. 5.
Cross-resistance of the HIV-1 selected strains (automated IVS) for different IN inhibitors. The regrown viruses from the automated IVS were tested for susceptibility to EVG and other IN inhibitors. Relative changes in susceptibility of the viruses compared with wild-type IIIB virus are displayed on the graph.
Evaluation of phenotypic (cross-) resistance of the selected HIV-1 strains.
To confirm that the selected HIV strains (Table 1) were indeed resistant to the drug, we determined the susceptibility of viruses IIIB/GS-9137(#9), IIIB/GS-9137(#13), IIIB/GS-9137(#22), and IIIB/GS-9137(#41) to EVG (Fig. 4). In parallel, the susceptibility of the resistant viruses to other HIV-1 IN inhibitors, including RAL, PACA, L-870,810, PICA, and L-731,988, were determined (Fig. 4).
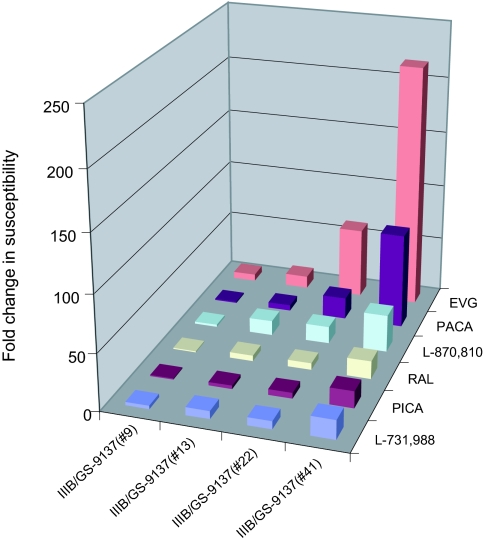
FIG. 4.
Susceptibility of the HIV-1 selected strains (classical IVS) to antiviral IN inhibitors. The susceptibilities of the HIV strains to the presence of high EVG concentrations were determined. Relative changes in the susceptibility of the viruses IIIB/GS-9137(#9), IIIB/GS-9137(#13), IIIB/GS-9137(#22), and IIIB/GS-9137(#41) compared with wild-type IIIB virus are displayed for the different IN inhibitors.
Strain IIIB/GS-9137(#9) (with the mutations T66T/I E92E/Q) showed a sixfold reduced susceptibility to EVG. This virus conferred little or no (one- to threefold) reduced susceptibility to the other IN inhibitors. The virus IIIB/GS-9137(#13) (E92Q) was 22 times less sensitive to EVG than the wild-type HIV-1 IIIB, and the susceptibility of this virus to the other IN compounds was reduced 5- to 13-fold. Virus IIIB/GS-9137(#22) (T66T/I E92E/Q A128A/T Q148Q/R) was 62 times less susceptible to EVG. A moderate reduction in susceptibility to PACA, in comparison with the previous selected virus, IIIB/GS-9137(#13), was observed. This was not the case for the other IN inhibitors, RAL, L-870,810, PICA, and L-731,988, where no or only a small reduction was seen. The finally selected strain IIIB/GS-9137(#41) (H114Y A128T Q148R) showed a 217-fold reduction in susceptibility to EVG. This virus was, respectively, 84 and 33 times less susceptible to PACA and L-870,810 while only a 15- to 17-fold reduced susceptibility to RAL, PICA, and L-731,988 was observed.
The eight resistant viruses from the automated IVS were recultured and tested for susceptibility to EVG. Susceptibility to the panel of other IN inhibitors was also determined in order to establish the degree of cross-resistance (Fig. 5). The regrown viruses were genotyped to confirm the previously observed genotypes (Table 2). The mutations E138E/K and Q148Q/R, present in virus 3 (T66T/I A128A/T E138E/K Q148Q/R S230R), were not observed after reculturing, whereas T66T/I developed to T66I. The mutation S230R present in virus 3 and virus 7 (R20R/K T66I L74L/M S230R) was observed as a mixture S230S/R after reculturing.
Virus 1 (E138K Q148R) was 162 times less susceptible to EVG and was reduced in susceptibility to the rest of the panel of IN inhibitors. Viruses 2, 3, and 4 showed large (91- to 63-fold) reductions in susceptibility to EVG. These viruses were also 5 to 49 times less susceptible to the remaining IN inhibitors. Virus 5 (E92E/Q E138E/K Q148Q/R) and virus 6 (T66T/A E92Q) were, respectively, 43- and 39-fold less sensitive to EVG. Both viruses were also less susceptible to the other IN inhibitors (4- to 19-fold). Virus 7 (R20K/R T66I L74L/M S230R/S) and virus 8 (T66I) showed, respectively, a 34- and 11-fold reduction in susceptibility to EVG and nearly wild-type susceptibility to RAL, with modest reductions in susceptibility to the other IN inhibitors (Fig. 5).
Confirmation of primary resistance mutations.
In order to identify the primary RAMs, recombinant viruses with single point mutations in the IN sequence were constructed. The IN mutations R20K, T66A/I, L74M, E92Q, H114Y, A128T, E138K, Q148R, N155H, and S230R were incorporated as SDMs in the backbone of HIV-1 HXB2D. The resulting viruses were phenotyped to determine the change in susceptibility to EVG and other IN inhibitors (Fig. 6).
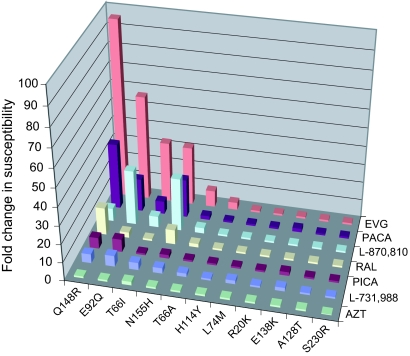
FIG. 6.
Susceptibility of HIV-1 IN recombinant clones (SDMs). The displayed IN mutations were incorporated as SDMs in the backbone of HIV-1 (HXB2D). Relative changes in susceptibility of the viruses for the different compounds are shown on the graph.
The Q148R mutation conferred large reductions in susceptibilities to EVG, RAL, PACA, L-870,810, PICA, and L-731,988, indicating that this mutation can confer cross-resistance to a range of IN inhibitors with diverse chemical scaffolds. Furthermore, the E92Q mutation, while conferring only a small (threefold) reduction in susceptibility to RAL, showed large reductions in susceptibility to EVG and the other IN inhibitors, suggesting that the E92Q mutation is, like Q148R, capable of causing reduced sensitivity to a wide range of IN inhibitors. A T66I mutation caused a 33-fold reduction in susceptibility to EVG, but this mutant retained nearly wild-type susceptibility to RAL. This mutation was also capable of reducing susceptibility to the other IN inhibitors, albeit to a lesser degree than Q148R and E92Q. The T66A mutation, while conferring ninefold reduced susceptibility to EVG, caused only approximately twofold reduction in susceptibility to other IN inhibitors. No or only a small reduction in virus susceptibility to EVG, RAL, or any other IN inhibitor used in this study was seen when the impact of a singular H114Y, L74M, A128T, E138K, S230R, or R20K mutation was tested with SDMs (Fig. 6). E92Q, Q148R, T66I, and N155H are the major IN RAMs observed in the EVG clinical trials (D. McColl, et al., presented at the 11th European Aids Conference, Madrid, Spain). In this study we selected viruses resistant to EVG, which contained among many mutations the primary RAMs T66I, E92Q, and Q148R. Only the mutation N155H was not detected in the mutant viruses obtained in our selection experiments; however, we also investigated the effect of this mutation, in the context of an SDM, on the viral susceptibility to our panel of IN inhibitors. The reduction in susceptibility to EVG and RAL conferred by the single amino acid change N155H was 32-fold and 8-fold, respectively, whereas the reduction in susceptibility to PACA and L-870,810 was 21- and 29-fold, respectively. Only a twofold reduction in viral sensitivity to PICA and L-731,988 was conferred by N155H.
Replication kinetics of mutant HIV-1 viruses.
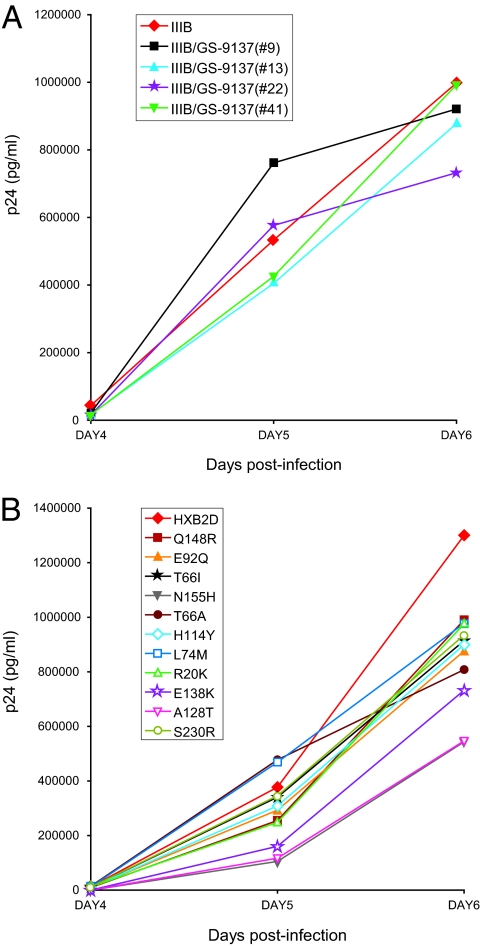
To investigate whether the drug-induced mutations affect viral replication capacity, the HIV-1 viruses selected with the classical IVS and the recombinant viruses were examined for their ability to replicate in MT4 cells in comparison with their respective parental strains. Replication kinetics of the selected viruses IIIB/GS-9137(#9), IIIB/GS-9137(#13), IIIB/GS-9137(#22), and IIIB/GS-9137(#41) were comparable to the replication of the parental HIV-1 IIIB strain (Fig. 7A). All recombinant strains showed reduced replication fitness in comparison with wild-type HXB2D (Fig. 7B). Recombinant viruses with the mutation N155H or A128T showed large reductions in p24 production relative to the replication of the wild-type HIV-1 HXB2D, while the recombinant virus with the mutation E138K had only a moderate reduction in p24 level. The recombinant viruses with the single mutation Q148R, E92Q, T66I, T66A, H114Y, L74M, R20K, or S230R showed only a small reduction in replication capacity relative to the wild type.
FIG. 7.
Replication kinetics of mutant viruses. MT4 cells were infected with the selected viruses (classical IVS) (A) or recombinant viruses (B) at an MOI of 0.0001. Cells were evaluated microscopically for HIV-induced CPE, and viral p24 levels were determined daily in cell-free supernatant. Experiments were done in duplicate.
Location of the mutations associated with resistance to EVG.
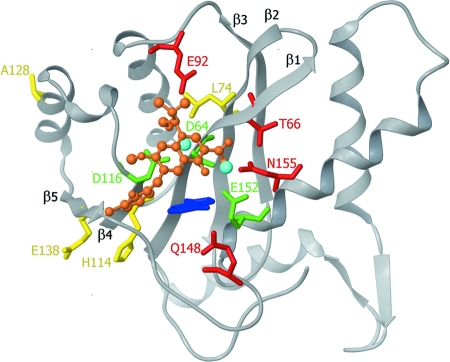
The location of the mutations associated with resistance to EVG, discussed above, were highlighted on a three-dimensional docking model of EVG with the IN catalytic core domain, based on the model of Savarino (31) (Fig. 8). Savarino generated a surrogate platform for molecular docking of EVG and other IN strand transfer inhibitors by transposing the one-metal structure of Goldgur et al. (16) onto a two-metal model of the HIV-1 IN catalytic core domain, after performing a structural alignment. Importantly, the primary mutations which cause the largest changes in susceptibility to EVG, i.e., Q148R, E92Q, T66I, and N155H, are located proximally to the catalytic triad represented by the D,D(35)E motif of IN. The position of the mutations L74M, H114Y, A128T, and E138K are also shown on the model and are more distant from the active site. These mutations do not confer large changes in susceptibility to EVG (Fig. 6), and their role in the development of resistance remains a topic of speculation. The R20K and S230R mutations lie in the terminal domains of IN and are, therefore, not shown in our model of the catalytic core domain.
FIG. 8.
Mapping of the IN mutations within the HIV-1 IN catalytic site. A three-dimensional docking model of the catalytic core domain of HIV-1 IN with EVG (orange), based on the two-metal model of Savarino (31) is shown. The catalytic triad residues (D64, D116, and E152) are shown in green, and the magnesium ions are in blue. The indole ring of 5CITEP (cyan) is used as a surrogate for the adenine of the terminal residue of the 3′ processed viral DNA. Amino acid residues conferring resistance to EVG as primary mutations are shown in red while the remaining mutations are highlighted in yellow.
DISCUSSION
HIV IN is a recently validated viral target for treating HIV infection and preventing AIDS. Currently, the IN inhibitor EVG is in the advanced stages of human clinical trials, and RAL has been approved recently. These two IN inhibitors show excellent antiretroviral efficacy in the clinic in both naive and treatment-experienced HIV-1-infected patients (8). However, viral strains resistant to the IN inhibitors will undoubtedly emerge, reducing the effectiveness of these drugs in patients. As with other classes of antiretrovirals, it is likely that the first generation of IN inhibitors will be complemented by a second generation. One of the attributes of any second-generation IN inhibitor will be the activity of the new drug against mutant viruses arising from the first-generation IN inhibitors. Thus, the identification of primary resistance mutations and the degree to which they confer resistance and of secondary or compensatory mutations is of critical importance to future generations of IN inhibitors.
Two different methods for the selection of viruses resistant to EVG were employed here. In addition to a classical IVS experiment, we report a resistance selection protocol in 96-well plates, automated IVS (Fig. 2), where each row of the plate contains an independent selection experiment. This protocol enabled us to perform up to eight IVS experiments in parallel, which gives a greater throughput than the classical selection experiments and results in larger data sets. The eight viruses obtained using automated IVS with EVG contained distinct genotypes, demonstrating that automated IVS can serve to increase diversity of results obtained by enabling the emergence of complementary or alternative resistance pathways. Furthermore, the novel method uses a protocol whereby the virus is challenged with an increased compound concentration at each passage and not only at virus breakthrough, as is done for classical IVS.
To gain further understanding of the antiviral (cross-) resistance patterns and mechanisms of EVG, the HIV-1 IIIB strains selected in the presence of increasing concentrations of this IN inhibitor were analyzed genotypically and phenotypically at different stages of the selection process. In total, nine selection experiments with EVG were performed, one by classical IVS and eight by automated IVS. At least one of the four mutations, T66A, T66I, E92Q, and Q148R, was present in the nine resulting viruses (Tables 1 and 2 and Fig. 3). These four mutations conferred resistance to EVG, which was confirmed by recombinant viruses with SDMs. The changes in susceptibility due to the Q148R and E92Q mutations were 96-fold and 57-fold, respectively, whereas the changes conferred by the T66I and T66A mutations were 33-fold and 9-fold, respectively. Hence, these mutations could also be considered primary mutations associated with EVG resistance. The SDMs were also assessed for their susceptibility to other IN inhibitors, including RAL, PACA, L-870,810, PICA, and L-731,988. Q148R yielded a 15-fold reduction in susceptibility to RAL. Whereas E92Q and T66A displayed, respectively, a threefold and twofold reduction in susceptibility to RAL, T66I did not affect sensitivity to RAL. Taking into account all IN inhibitors (Fig. 6), the Q148R mutation is proposed as the most important RAM for a wide range of IN inhibitors with diverse chemical scaffolds. The E92Q mutation is identified as a mutation capable of conferring reduced susceptibility to diverse IN inhibitors, albeit with a small reduction in susceptibility to RAL. In addition, T66I confers reduced susceptibility to the IN inhibitors but to a lesser degree than both Q148R and E92Q. Mutation T66A confers reduced susceptibility to EVG but not to the other IN inhibitors.
The mutations H114Y, L74M, R20K, A128T, E138K, and S230R were observed accompanying one or more of the four primary mutations. In the context of single mutations in SDMs, however, they resulted in little or no change in EVG susceptibility (Fig. 6). Moreover, these mutations did not cause large changes to RAL, PACA, L-870,810, PICA, or L-731,988.
The mutation T66I has been previously reported as a resistance mutation conferring reduced susceptibility to the diketo acid inhibitors S-1360 (14) and L-731,988 (20). Shimura (33) also reported T66I and E92Q as RAMs of EVG. However, in those studies the Q148R mutation was not detected after selection with EVG. Other studies already identified Q148R to be associated with in vivo resistance to RAL (D. J. Hazuda, M. Miller, B. Y. Nguyen, and J. Zhao, presented at the XVI International HIV Drug Resistance Workshop, Barbados, West Indies, 12 to 16 June 2007) and to EVG (D. McColl, et al., presented at the 11th European Aids Conference, Madrid, Spain). Also N155H has been reported as a primary IN resistance mutation among patients with virologic failure for RAL (26) or EVG. We did not select the latter mutation, but our N155H-containing SDM showed a 32-fold reduction in EVG susceptibility, underscoring its importance. Shimura et al. (33) previously selected HIV-1 strains in the presence of EVG. The selected strains showed two distinct resistance pathways: one pathway consisting of T66I, Q95K, E138K, Q146P, and S147G and a second pathway comprising the mutations H51Y, E92Q, S147G, and E157Q in the IN sequence. Our experimental data confirm the emergence of E138K and T66I but not Q95K, Q146P, H51Y, S147G, and E157Q, highlighting the need for further research into the resistance pathways employed by HIV to evade drug pressure from IN inhibitors. It is possible that new mutations will emerge from other resistance selection experiments, and data from clinical trials will likely indicate which mutations are the most significant.
The selected (classical IVS) and recombinant strains were examined for their ability to replicate in MT4 cells in comparison with their respective wild-type strains (Fig. 7). The four selected viruses, IIIB/GS-9137(#9), IIIB/GS-9137(#13), IIIB/GS-9137(#22), and IIIB/GS-9137(#41), were similar to the wild type in their replication capacity. The recombinant viruses with the single mutation Q148R, E92Q, T66I, T66A, H114Y, L74M, R20K, or S230R showed a moderate or small reduction in replication capacity relative to the wild type. A large reduction in replication compared with the wild type was observed for the viruses with the single mutation N155H or A128T. As N155H is a primary IN resistance mutation among patients with virologic failure for IN inhibitors, it is possible that additional mutations might have developed after the N155H mutation was generated to compensate for the reduced replication. Clearly, more investigation will be necessary to identify possible compensatory mutations and their role on the behavior of the IN enzyme.
Recently, Savarino (31) reported an in silico docking simulation of HIV-1 IN with several IN inhibitors, including EVG. The model was derived from an X-ray structure of 5CITEP [1-(5-chloroindol-3-yl)-3-(tetrazoyl)-1,3-propandione-ene] bound to the catalytic core domain of IN (16), with two magnesium ions into the active site. The indole ring of 5CITEP was used as a surrogate for the adenine of the terminal residue of the 3′-processed viral DNA. As a best docking pose for EVG, Savarino suggested that the β-hydroxyl carboxylate chelates the magnesium ion between D64 and E152 and that the hydroxylic oxygen in the isobutyl substituent coordinates the second magnesium ion. Figure 8 shows the in silico docking model of Savarino, with the mutations acquired by selection with EVG highlighted. The primary resistance mutations Q148R, E92Q, T66I/A, and N155H are located adjacent to the catalytic triad of IN and are surrounded by the more distant potentially secondary mutations H114Y, L74M, A128T, and E138K. In this model the isobutyl substituent of EVG is oriented toward E92, and the hydroxyl in the isobutyl substituent replaces one of the water molecules through which E92 coordinates the magnesium ion between D64 and D116. Therefore, it is not surprising to see that the primary mutation E92Q confers a substantial decrease in susceptibility to EVG. According to the model, there is no direct interaction between T66 and the compound, but given the short distance between T66 and EVG, a water-mediated interaction, through which a T66I/A mutation might have an effect, is plausible.
The effect of the Q148R mutation on catalytic activity could be explained in two ways. In a first hypothesis, Q148R reduces the affinity of the enzyme for magnesium (11). Q148 is located in a flexible loop (residues 141 to 148). By X-ray crystallography, Greenwald and colleagues (17) showed that mutations in the flexible loop could have an effect on the catalytic activity. Such mutations would also be expected to modulate the activity of active-site inhibitors. In addition, a recent IN inhibitor binding model concluded that strand transfer inhibitors bind close to the flexible loop and that inhibitor binding impedes loop mobility and hence catalytic activity (7). Therefore, the flexible loop and thus Q148 are important for strand transfer inhibitor binding (16). According to Dicker and coworkers (11), the Q148R mutation shifts the flexible loop equilibrium to a state that hampers magnesium binding by E152. Consequently, the susceptibility to inhibitors that are believed to chelate the two magnesium ions is reduced, but viral replication and fitness are also affected. Secondly, Q148R has an influence on the binding of the viral LTR (15, 22). In the model of Savarino, Q148 is in close contact with the indole ring of 5CITEP (Fig. 8), which shows an interaction between Q148 and the terminal adenine of the 3′ processed viral DNA. A plausible explanation for the effect of the Q148R mutation speculates on this direct interaction (15, 22). Mutations that shift the equilibrium of bound viral DNA can interfere with the effect of inhibitors (32). Previous hypotheses about Q148R all share the common features that both catalytic activity and affinity for the inhibitor are reduced. This leads to a reduction in viral susceptibility to EVG and RAL and also results in a penalty on fitness and replication efficiency of the virus that nevertheless does not preclude its selection in the clinic (D. McColl, et al., presented at the 11th European Aids Conference, Madrid, Spain).
The amino acids at positions L74, H114, E138, and A128 are distal to the catalytic site (Fig. 8) and are not expected to make direct contact with the inhibitor. No large susceptibility changes to EVG or RAL were conferred by L74M, H114Y, E138K, and A128T when the mutations were included singularly as SDMs (Fig. 6). These are possibly compensatory mutations, where the fitness or replicative capacity penalty resulting from the primary resistance mutation may be partially compensated by changes in the backbone of the protein and in the geometry of the catalytic triad. It is possible that the L74M mutation (located on β-sheet β2) could influence the positioning of D64 (located on β-sheet β1 and via H bonds interacting with V75), thereby interfering with the coordination of magnesium by the catalytic triad (Fig. 8). Additionally, H114Y may impart a subtle influence on D116, another important residue in the catalytic triad. The residue E138 is distal from the catalytic site located on β-strand β5 that is parallel to β-strand β4, on which H114 and D116 are situated (Fig. 8) and is unlikely to contact the inhibitor.
A better knowledge of the enzymatic activity, replication kinetics, and fitness of IN inhibitor-resistant mutant viruses and enzymes would improve the understanding of the resistance mechanism of the RAMs and allow the identification of compensatory mutations. New mutations will likely arise from other IVS experiments and from clinical trials. Together, these data will enhance our understanding of the nature of the binding of IN inhibitors and therefore contribute to a low-resolution structural model that may resemble the IN-DNA-inhibitor complex and offer a tool for the development of second-generation IN inhibitors.
Acknowledgments
We thank Luc Geeraert for his help in the preparation of the manuscript.
Footnotes
Published ahead of print on 20 August 2008.
REFERENCES
- 1.Anker, M., and R. B. Corales. 2008. Raltegravir (MK-0518): a novel integrase inhibitor for the treatment of HIV infection. Expert Opin. Investig. Drugs. 1797-103. [DOI] [PubMed] [Google Scholar]
- 2.Asante-Appiah, E., and A. M. Skalka. 1997. Molecular mechanisms in retrovirus DNA integration. Antiviral Res. 36139-156. [DOI] [PubMed] [Google Scholar]
- 3.Belyk, K. M., H. G. Morisson, P. Jones, and V. Summa. June 2006. Potassium salt of an HIV integrase inhibitor. International patent WO2006060712.
- 4.Billich, A. 2003. S-1360 Shionogi-GlaxoSmithKline. Curr. Opin. Investig. Drugs 4206-209. [PubMed] [Google Scholar]
- 5.Cahn, P., and O. Sued. 2007. Raltegravir: a new antiretroviral class for salvage therapy. Lancet 3691235-1236. [DOI] [PubMed] [Google Scholar]
- 6.Clavel, F., and A. J. Hance. 2004. HIV drug resistance. N. Engl. J. Med. 3501023-1035. [DOI] [PubMed] [Google Scholar]
- 7.Cox, A. G., and V. Nair. 2006. Novel HIV integrase inhibitors with anti-HIV activity: insights into integrase inhibition from docking studies. Antivir. Chem. Chemother. 17343-353. [DOI] [PubMed] [Google Scholar]
- 8.Dayam, R., R. Gundla, L. Q. Al-Mawsawi, and N. Neamati. 2008. HIV-1 integrase inhibitors: 2005-2006 update. Med. Res. Rev. 28118-154. [DOI] [PubMed] [Google Scholar]
- 9.DeJesus, E., D. Berger, M. Markowitz, C. Cohen, T. Hawkins, P. Ruane, R. Elion, C. Farthing, L. Zhong, A. K. Cheng, D. McColl, and B. P. Kearney. 2006. Antiviral activity, pharmacokinetics, and dose response of the HIV-1 integrase inhibitor GS-9137 (JTK-303) in treatment-naive and treatment-experienced patients. J. Acquir. Immune Defic. Syndr. 431-5. [DOI] [PubMed] [Google Scholar]
- 10.Deng, J., R. Dayam, L. Q. Al-Mawsawi, and N. Neamati. 2007. Design of second generation HIV-1 integrase inhibitors. Curr. Pharm. Des. 13129-141. [DOI] [PubMed] [Google Scholar]
- 11.Dicker, I. B., H. K. Samanta, Z. Li, Y. Hong, Y. Tian, J. Banville, R. R. Remillard, M. A. Walker, D. R. Langley, and M. Krystal. 2007. Changes to the HIV long terminal repeat and to HIV integrase differentially impact HIV integrase assembly, activity, and the binding of strand transfer inhibitors. J. Biol. Chem. 28231186-31196. [DOI] [PubMed] [Google Scholar]
- 12.Engelman, A., K. Mizuuchi, and R. Craigie. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 671211-1221. [DOI] [PubMed] [Google Scholar]
- 13.Esposito, D., and R. Craigie. 1999. HIV integrase structure and function. Adv. Virus Res. 52319-333. [DOI] [PubMed] [Google Scholar]
- 14.Fikkert, V., A. Hombrouck, B. Van Remoortel, M. De Maeyer, C. Pannecouque, E. De Clercq, Z. Debyser, and M. Witvrouw. 2004. Multiple mutations in human immunodeficiency virus-1 integrase confer resistance to the clinical trial drug S-1360. AIDS 182019-2028. [DOI] [PubMed] [Google Scholar]
- 15.Gerton, J. L., S. Ohgi, M. Olsen, J. DeRisi, and P. O. Brown. 1998. Effects of mutations in residues near the active site of human immunodeficiency virus type 1 integrase on specific enzyme-substrate interactions. J. Virol. 7 25046-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldgur, Y., R. Craigie, G. H. Cohen, T. Fujiwara, T. Yoshinaga, T. Fujishita, H. Sugimoto, T. Endo, H. Murai, and D. R. Davies. 1999. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl. Acad. Sci. USA 9613040-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenwald, J., V. Le, S. L. Butler, F. D. Bushman, and S. Choe. 1999. The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity. Biochemistry 388892-8898. [DOI] [PubMed] [Google Scholar]
- 18.Grinsztejn, B., B. Y. Nguyen, C. Katlama, J. M. Gatell, A. Lazzarin, D. Vittecoq, C. J. Gonzalez, J. Chen, C. M. Harvey, and R. D. Isaacs. 2007. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet 3691261-1269. [DOI] [PubMed] [Google Scholar]
- 19.Hazuda, D. J., N. J. Anthony, R. P. Gomez, S. M. Jolly, J. S. Wai, L. Zhuang, T. E. Fisher, M. Embrey, J. P. Guare, Jr., M. S. Egbertson, J. P. Vacca, J. R. Huff, P. J. Felock, M. V. Witmer, K. A. Stillmock, R. Danovich, J. Grobler, M. D. Miller, A. S. Espeseth, L. Jin, I. W. Chen, J. H. Lin, K. Kassahun, J. D. Ellis, B. K. Wong, W. Xu, P. G. Pearson, W. A. Schleif, R. Cortese, E. Emini, V. Summa, M. K. Holloway, and S. D. Young. 2004. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc. Natl. Acad. Sci. USA 10111233-11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287646-650. [DOI] [PubMed] [Google Scholar]
- 21.Horwitz, J., J. Chua, and M. Noel. 1964. Nucleosides. V. The monomesylates of 1-(2′-deoxy-β-d-lyxofuranosyl)thymidine. J. Org. Chem. 292076-2078. [Google Scholar]
- 22.Johnson, A. A., W. Santos, G. C. Pais, C. Marchand, R. Amin, T. R. Burke, Jr., G. Verdine, and Y. Pommier. 2006. Integration requires a specific interaction of the donor DNA terminal 5′-cytosine with glutamine 148 of the HIV-1 integrase flexible loop. J. Biol. Chem. 281461-467. [DOI] [PubMed] [Google Scholar]
- 23.Kuki, A., X. Li, M. B. Plewe, H. Wang, and J. Zhang. July 2005. HIV-integrase inhibitors, pharmaceutical compositions, and methods for their use. U.S. patent 7,138,408.
- 24.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 667414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lataillade, M., J. Chiarella, and M. J. Kozal. 2007. Natural polymorphism of the HIV-1 integrase gene and mutations associated with integrase inhibitor resistance. Antivir. Ther. 12563-570. [PubMed] [Google Scholar]
- 26.Malet, I., O. Delelis, M. A. Valantin, B. Montes, C. Soulie, M. Wirden, L. Tchertanov, G. Peytavin, J. Reynes, J. F. Mouscadet, C. Katlama, V. Calvez, and A. G. Marcelin. 2008. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob. Agents Chemother. 521351-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pommier, Y., A. A. Johnson, and C. Marchand. 2005. Integrase inhibitors to treat HIV/AIDS. Nat. Rev. Drug Discov. 4236-248. [DOI] [PubMed] [Google Scholar]
- 28.Reinke, R., D. J. Lee, and W. E. Robinson, Jr. 2002. Inhibition of human immunodeficiency virus type 1 isolates by the integrase inhibitor L-731,988, a diketo acid. Antimicrob. Agents Chemother. 463301-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakai, H., M. Kawamura, J. Sakuragi, S. Sakuragi, R. Shibata, A. Ishimoto, N. Ono, S. Ueda, and A. Adachi. 1993. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J. Virol. 671169-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato, M., T. Motomura, H. Aramaki, T. Matsuda, M. Yamashita, Y. Ito, H. Kawakami, Y. Matsuzaki, W. Watanabe, K. Yamataka, S. Ikeda, E. Kodama, M. Matsuoka, and H. Shinkai. 2006. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J. Med. Chem. 491506-1508. [DOI] [PubMed] [Google Scholar]
- 31.Savarino, A. 2007. In-silico docking of HIV-1 integrase inhibitors reveals a novel drug type acting on an enzyme/DNA reaction intermediate. Retrovirology 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scarth, B., and M. Götte. 2007. The A62V mutation in HIV-1 reverse transcriptase increases the inhibitory effects of the PPi analogue Foscarnet, abstr. 586. Abstr. 14th Conf. Retrovir. Opportunistic Infect., Los Angeles, CA, 25 to 28 February 2007.
- 33.Shimura, K., E. Kodama, Y. Sakagami, Y. Matsuzaki, W. Watanabe, K. Yamataka, Y. Watanabe, Y. Ohata, S. Doi, M. Sato, M. Kano, S. Ikeda, and M. Matsuoka. 2008. Broad anti-retroviral activity and resistance profile of a novel human immunodeficiency virus integrase inhibitor, elvitegravir (JTK-303/GS-9137). J. Virol. 82764-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Gent, D. C., K. Mizuuchi, and M. Gellert. 1996. Similarities between initiation of V(D)J. recombination and retroviral integration. Science 2711592-1594. [DOI] [PubMed] [Google Scholar]
- 35.Vingerhoets, J., H. Azijn, E. Fransen, I. De Baere, L. Smeulders, D. Jochmans, K. Andries, R. Pauwels, and M. P. de Bethune. 2005. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J. Virol. 7912773-12782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Witvrouw, M., B. Van Maele, J. Vercammen, A. Hantson, Y. Engelborghs, E. De Clercq, C. Pannecouque, and Z. Debyser. 2004. Novel inhibitors of HIV-1 integration. Curr. Drug Metab. 5291-304. [DOI] [PubMed] [Google Scholar]