Abstract
Rift Valley fever virus (RVFV) is a member of the Bunyaviridae virus family (genus Phlebovirus) and is considered to be one of the most important pathogens in Africa, causing viral zoonoses in livestock and humans. Here, we report the characterization of the three-dimensional structural organization of RVFV vaccine strain MP-12 by cryoelectron tomography. Vitrified-hydrated virions were found to be spherical, with an average diameter of 100 nm. The virus glycoproteins formed cylindrical hollow spikes that clustered into distinct capsomeres. In contrast to previous assertions that RVFV is pleomorphic, the structure of RVFV MP-12 was found to be highly ordered. The three-dimensional map was resolved to a resolution of 6.1 nm, and capsomeres were observed to be arranged on the virus surface in an icosahedral lattice with clear T=12 quasisymmetry. All icosahedral symmetry axes were visible in self-rotation functions calculated using the Fourier transform of the RVFV MP-12 tomogram. To the best of our knowledge, a triangulation number of 12 had previously been reported only for Uukuniemi virus, a bunyavirus also within the Phlebovirus genus. The results presented in this study demonstrate that RVFV MP-12 possesses T=12 icosahedral symmetry and suggest that other members of the Phlebovirus genus, as well as of the Bunyaviridae family, may adopt icosahedral symmetry. Knowledge of the virus architecture may provide a structural template to develop vaccines and diagnostics, since no effective anti-RVFV treatments are available for human use.
Rift Valley fever virus (RVFV) is a member of the Bunyaviridae family, genus Phlebovirus. The Bunyaviridae family contains five genera (Orthobunyavirus, Phlebovirus, Nairovirus, Hantavirus, and Tospovirus) and, with over 300 identified viruses, is one of the largest virus families affecting mammals (12). Most viruses in the family are carried and transmitted by arthropod vectors, typically mosquitoes, ticks, sand flies, or thrips. Hantaviruses are the exception, as these viruses are rodent-borne and transmitted in aerosolized rodent excreta. The majority of bunyaviruses are significant public health concerns since they are distributed worldwide and cause human illness ranging from mild asymptomatic infection to hemorrhagic fever, fatal encephalitis, and severe adult respiratory distress syndrome (in the case of hantaviruses) (18). Exceptions are the tospoviruses, which infect plants (10, 35).
RVFV is classified as a NIAID high-priority category A biothreat agent, is an emerging public health concern, and causes serious disease in humans, with ∼1 to 2% of infections resulting in a fatal hemorrhagic fever or encephalitis. In addition, RVFV is a significant worldwide agricultural concern since infections can result in a massive loss of livestock (5, 39). Since RVFV is classified as an enhanced biosafety level 3 containment agent, our studies focused on the vaccine strain MP-12, a biosafety level 2 virus which was obtained by 12 serial passages of the virulent ZH548 isolate in the presence of 5-fluorouracil (3). A total of 25 nucleotide differences and 11 amino acid differences exist between the virulent ZH548 isolate and avirulent RVFV MP-12 (49).
Bunyaviruses are trisegmented negative-sense RNA viruses with similar morphological features (26, 39). Electron microscope (EM) images of negatively stained bunyaviruses show pleomorphic to spherical particles with diameters ranging from 80 to 120 nm (26). The three distinct viral RNA segments are apparently in a complex with the viral nucleocapsid protein (N) and the viral RNA-dependent RNA polymerase (L). These ribonucleoprotein (RNP) complexes are packaged inside a lipid bilayer during budding into the Golgi complex at internal cellular membranes (22, 30). The lipid bilayer contains the viral glycoproteins GN (also termed G1) and GC (also termed G2), which project as spikes from the viral surface. Earlier studies of negatively stained RVFV showed the RNP complexes surrounded by a membrane through which surface spikes measuring 10 to 18 nm in length and ∼5 nm in diameter were inserted (13). The virions appeared to have a distinct surface structure composed of small round capsomeres with a central cavity.
Sequence analyses of bunyavirus glycoproteins suggested that the carboxyl-terminal glycoproteins are class II viral fusion proteins (14). In contrast to other negative-sense RNA viruses, bunyaviruses do not contain a matrix protein underlying the envelope. Recently, it has been proposed that the glycoprotein cytoplasmic tails interact directly with the nucleoproteins of the RNP complex and may be important for viral genome packaging (21, 32, 42, 44).
EM studies of negatively stained bunyaviruses revealed variations in size, structure, surface units, and morphogenesis among different bunyavirus genera, providing a preliminary structural criterion for the classification of members within these five genera (26). However, the apparent pleomorphic appearance and size heterogeneity of particles prevented high-resolution three-dimensional (3D) structure determination and the identification of specific structural components. Occasional subunit patterns observed in some virions were suggested to be consistent with icosahedral symmetry, and different T numbers were assigned to specific viruses within the bunyavirus family (26, 50). The first ultrastructure of a bunyavirus, Uukuniemi virus (UUKV; genus Phlebovirus), was recently resolved by using cryoelectron tomography (cryo-ET) (31). This cryo-ET study described the viral glycoproteins to be arranged on a T=12 icosahedral lattice, an organization not previously shown for a virus. Further, the observed nonhomogenous size distributions of the isolated particles result in this virus being described as an intermediate between pleomorphic and icosahedral viruses.
In this study, we used cryo-ET to characterize the overall 3D organization of RVFV MP-12. Because of the heterogeneity described for many bunyaviruses and to preserve structural details of RVFV particles, flash freezing of unfixed virus suspensions in a cryogen was performed (8, 11). Cryo-ET allows for 3D reconstruction of asymmetric and/or structurally heterogeneous macromolecules and their complexes, as well as cellular organelles, at a moderate resolution (3 to 6 nm) (1, 16, 24, 29, 46). RVFV MP-12 particles in our tomograms were similar in diameter and overall organization, and their different orientations allowed particle averaging to improve resolution (40) and to restore missing information due to the limited tilt range of the sample in EM. The viral glycoproteins were arranged on a T=12 icosahedral lattice. RVFV MP-12 is the second virus found to have such an arrangement. All virus particles were homogenous, implying that RVFV MP-12 is an icosahedral virus. Our reconstruction suggests that members of the Phlebovirus genus have an icosahedral structure, contrary to the earlier view that they are pleomorphic.
MATERIALS AND METHODS
Cell culture and RVFV MP-12 purification.
VeroE6 cells (American Type Culture Collection) were cultivated in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA), 100 IU of penicillin/ml, and 100 μg of streptomycin (Cellgro). Cells were seeded into T150 flasks and infected with RVFV MP-12 (a gift from J. Morrill, University of Texas Medical Branch [UTMB]) at a multiplicity of infection of 0.02. After 1 h of adsorption, 20 ml of Dulbecco's modified Eagle medium with 2% fetal bovine serum and antibiotics was added. Six to eight hours postinfection, the cell culture supernatant was replaced with fresh serum-free medium, and virus was harvested 36 to 48 h postinfection. Virus-containing supernatant was clarified by centrifugation at 2,000 rpm for 15 min with a model 5810R Eppendorf centrifuge equipped with an A-4-62 rotor. Secreted virus was concentrated by ultrafiltration with an Ultra-15 centrifugal filter device (nominal molecular weight limit, 100,000) according to the protocol of the manufacturer (Amicon) and further purified by iodixanol (OptiPrep; Sigma) density gradient centrifugation (10 to 30% in a mixture of 10 mM Tris-HCl, 100 mM NaCl, and 1 mM EDTA, pH 7.2) in a Beckmann SW41 rotor at 210,000 × g for 1.5 h at 4°C. Fractions were collected and tested for the presence of RVFV MP-12 by Western blotting using a polyclonal mouse hyperimmune ascitic fluid against RVFV (a gift from R. Tesh, World Reference Collection for Emerging Viruses and Arboviruses, UTMB) and by plaque assays of VeroE6 cells. Throughout the growth and purification procedures, the pHs of solutions were monitored and maintained above 7.
Virus particle integrity was examined by transmission electron microscopy (TEM) using negative staining with 2% aqueous uranyl acetate (UA). Formvar carbon-coated copper grids were floated on droplets of a virus suspension for 10 min, blotted with filter paper, and stained with 2% UA for 30 s. Negatively stained samples were examined in a Philips 201 EM at 60 keV. Fractions containing the highest titer of the virus were further concentrated by ultrafiltration with an Amicon Ultra-4 centrifugal filter device (nominal molecular weight limit, 100,000). The final virus concentration was 3 × 108 PFU/ml.
Cryo-ET.
Plunge freezing was performed as described previously (41). Briefly, the purified virus was mixed with a suspension of 15-nm gold beads to provide fiducial markers for the alignment of images in a tilt series. Samples of 3.5 μl of virus suspensions were applied to 200-mesh holey EM grids (R2/2 Quantifoil; Micro Tools GmbH, Jena, Germany), and the grids were blotted with filter paper and plunged into liquid ethane. Quantifoil grids with RVFV MP-12 embedded in a thin layer of vitreous water across the holes in the carbon supporting film were transferred into the microscope in a 626 70° cryoholder maintained at −176°C. Grids were imaged at a magnification of ×15,000 by using a JEOL 2200FS microscope operated at 200 keV and equipped with an Omega energy filter. Cryo-ET tilt series data were imaged with a 20-eV slit to remove inelastically scattered electrons, and data frames were recorded on a slow-scan 4K charge-coupled device (CCD) camera (UltraScan 895; GATAN, Inc.) using ∼0.8 electrons/Å2/CCD frame.
Single-axis tilt series were collected at 2° steps over a ±68° tilt interval. Full 4K CCD frames were collected, and typically, 80 to 89 data frames were collected from each tilt series. Data were recorded under low-dose conditions by using SerialEM (27) to conduct automatic tilting, tracking, focusing, and image acquisition. The tilt increment was inversely proportional to cosine (α)1/3, where α is the tilt angle.
Image processing.
Tilt series were processed and the tomograms were reconstructed using IMOD (28). Images in the tilt series were aligned using gold beads as fiducial markers, and the final tomograms were calculated by a weighted back projection algorithm. Full tomograms were of 4,096 by 4,096 by 600 voxels. Individual RVFV MP-12 particles were boxed from the full tomograms by using the TRIMVOL program contained in IMOD.
3D single-particle averaging.
Subtomograms of RVFV MP-12 particles were normalized to an average density of zero and similar standard deviations. Particles were centered using the AUTOCOR option within the PROC3D program in the EMAN software package (25). Forty-six subtomograms were aligned relative to one another by using the TOMO-HUNTER program (40) and averaged together to minimize artifacts arising from incomplete data in the tomograms due to the limited range of tilt angles in EM (the “missing-wedge” problem). Tomograms were initially low-pass-filtered to 60-Å resolution to reduce high-frequency noise artifacts.
Visualization of the 3D cryo-ET reconstructions was done using Chimera (34). The orientation of the virus particle was determined using self-rotation functions calculated with the program GLRF (48). Structure factors were calculated to 60 Å by using the SFALL program from the CCP4 software package (9) and assuming space group P1 with the following unit cell dimensions: a = b = c = 1,514.52 Å, and α = β = γ = 90°. The calculated structure factors were then used in self-rotation functions searching for five-, three-, and twofold symmetry axes (i.e., with search angle κ fixed at 72, 120, and 180 degrees, respectively). All rotation functions were calculated using a 600-Å radius of integration. Based on the results of the self-rotation function, the tomograms were rotated so that the icosahedral symmetry axes corresponded to the EMAN convention, and the rotated tomograms were then icosahedrally averaged using the EMAN program PROC3D. RobEM (http://cryoEM.ucsd.edu/programs.shtm) was used to generate radial density projections.
RESULTS
Concentration and purification of RVFV MP-12.
Several studies that describe bunyavirus morphology recognize the instability of these viruses (31, 37, 50). Initial preparations of RVFV MP-12 contained large amounts of distorted and broken particles, possibly reflecting the fragility of the particles. To overcome this difficulty yet avoid fixatives such as glutaraldehyde which may distort the virus architecture, the isosmotic reagent iodixanol (OptiPrep) was utilized as a gradient medium. This methodology resulted in reproducibly similar preparations of concentrated unfixed intact particles.
Ultrastructure of RVFV MP-12.
Purified RVFV MP-12 was initially visualized by negative staining in a TEM (Philips EM 201) by using 2% UA (Fig. 1). As is typical for bunyaviruses (26), negative staining showed pleomorphic virus particles (Fig. 1A), although some particles were roughly spherical (Fig. 1B). The dimensions of 84 virus particles were measured, and these particles were observed to have an average diameter ± standard deviation of 95 ± 9 nm (Fig. 1C). The virus surface was distinct, sharply defined, and covered predominantly with round to hexagonal capsomeres (Fig. 1B). The individual capsomeres were generally uniform in appearance and apparently organized on the virus surface in a tightly packed regular lattice. The sizes and morphologies of the RVFV MP-12 particles and their capsomeres were similar to those in earlier descriptions of negatively stained wild-type RVFV (13, 23, 26).
FIG. 1.
Morphology of RVFV MP-12 particles revealed by electron microscopy. (A) Negative-stain TEM micrograph showing pleomorphic RVFV MP-12 particles. (B) Negative-stain TEM micrograph showing spherical RVFV MP-12 particles with a distinct surface structure composed of morphological units with a central cavity. (C) Histogram representing the size distribution of negatively stained RVFV MP-12 particles, with an average diameter of 95 ± 9 nm (n = 84). The scale bar represents 100 nm.
Architecture of RVFV MP-12 determined by cryo-ET.
Cryo-ET was used to visualize the 3D structure of RVFV MP-12 particles. This approach allowed 3D reconstruction of individual virions without averaging over different particles, which was required to avoid potential heterogeneity of the particles in size, shape, and structure. Six low-dose tilt series of frozen-hydrated RVFV MP-12 were collected by using a JEOL 2200FS microscope equipped with an in-column energy filter (Omega) to eliminate inelastically scattered electrons which would degrade image quality. Each tilt series was separately processed and reconstructed using the IMOD software package (20) as described in Materials and Methods (Fig. 2A). A preliminary examination of individual particles within tomograms suggested that the virus was more structurally homogeneous than previously recognized. Forty-six individual RVFV MP-12 particles were extracted from tomograms as separate volumes of density and used for subsequent analysis and averaging.
FIG. 2.
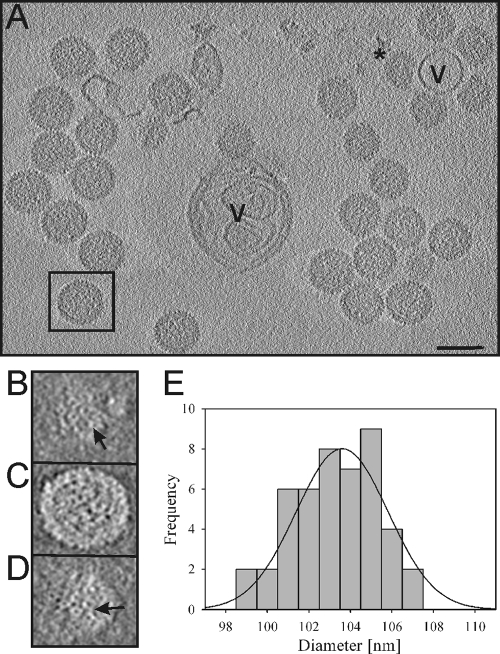
Cryo-ET analysis and preliminary assessment of tomograms. (A) Slice from a tomogram of vitrified-hydrated RVFV MP-12 particles corresponding to the tilt stage position normal to the electron beam. Vesicles present in this tomogram are labeled “V.” An example of a distorted particle can be seen in close contact to the vesicle in the upper right corner (labeled with an asterisk). (B to D) Top (B), center (C), and bottom (D) slices from a tomogram of an isolated virus particle. This particle is identified by the black box in panel A. In the top slice, individual capsomeres are visible and indicated by the arrow. In a central slice, distinct densities corresponding to the three RNP complexes were not identified. In a bottom slice, bridging densities among capsomeres are indicated by the arrow. (E) Histogram representing the size distribution of frozen-hydrated RVFV MP-12 particles, with an average diameter of 103 ± 2 nm (n = 46). The scale bar represents 100 nm.
Each RVFV MP-12 tomogram contained several particles. Although all reconstructed RVFV MP-12 particles were of the same diameter in 3D, they had a range of sizes in slices through representative tomograms (Fig. 2A) since each slice intersected particles embedded in ice at different heights. Interestingly, vesicles were observed in the tomograms (Fig. 2A). For example, the upper right corner of Fig. 2A shows a vesicle surrounded by five virus particles. When the virus particles were examined in 3D, it was clear that they did not make contact with the vesicle, although they were in close proximity to it. In contrast, the larger vesicle visible in the center of Fig. 2A appears to make direct contact with the adjacent RVFV MP-12 particles. Most likely, the few observed vesicles comigrated with the RVFV MP-12 particles during the iodixanol gradient purification. As one could expect, RVFV MP-12 particles that made contact with other virus particles, vesicles, or objects were often distorted in shape close to the contact region.
Frozen-hydrated RVFV MP-12 particles were spherical (Fig. 2A), in contrast to the frequently observed pleomorphic appearance of negatively stained particles. The 46 analyzed particles had similar sizes and appearances, with a mean particle diameter of 103 ± 2 nm. Regular lattices of closely packed surface spikes were visible on the virus surface and on the periphery of a continuous layer, an arrangement which was interpreted to represent viral glycoproteins within the lipid membrane bilayer (Fig. 2A).
Top and bottom slices through an RVFV MP-12 tomogram resolved individual capsomeres (Fig. 2B and D). As previously noted for negatively stained particles (Fig. 1B), individual capsomeres were cylindrical to hexagonal (Fig. 2B and D). Bridging densities connecting individual capsomeres were visible on the virus surface (Fig. 2D). In central slices through individual particles, distinct densities corresponding to the three RNP complexes within the centers of the particles were not resolved in our tomograms (Fig. 2C).
Averaging of individual RVFV MP-12 particle subtomograms.
Because of the observed similarity between individual virus particles in the tomograms, it was possible to average data from multiple particles. The averaging of several individual particle images extracted from tomograms can improve the signal-to-noise ratio and thus improve the resolution of the resulting map and the visibility of common features in these particles. Since a specimen in an EM cannot be tilted to ±90° to completely sample 3D space, some data were missing in the collected tilt series for RVFV MP-12. This problem, commonly termed the missing wedge in electron tomography, leads to anisotropic resolution in different directions, with the most severe impact on the vertical direction (17). Missing-wedge limitations can be overcome by averaging similar parts of different tomograms because, in general, each particle in the tomogram is randomly oriented in space and, thus, the particles are complementary to one another. Unfortunately, the missing wedge may dominate the particle alignment, giving false correlation peaks and leading to blurred, or washed-out, features in the averages (40). To overcome misalignment problems due to the missing wedge, weighted cross-correlation in Fourier space was used to orient particles relative to one another (40); this approach minimized alignment artifacts from noise present in the missing wedges of individual transforms.
A total of 46 RVFV MP-12 particles were extracted from three tomograms and inspected for distortions. Nineteen intact particles were selected for initial alignment and averaging, and a particle showing the most detailed features was used as the initial reference for aligning RVFV MP-12 particle subtomograms. Aligned particles were averaged to generate an improved reference, which was used for aligning the remaining 27 RVFV MP-12 particle images that showed some slight distortions. All 46 aligned particle images were averaged together to generate the final RVFV MP-12 3D map (Fig. 3A).
FIG. 3.
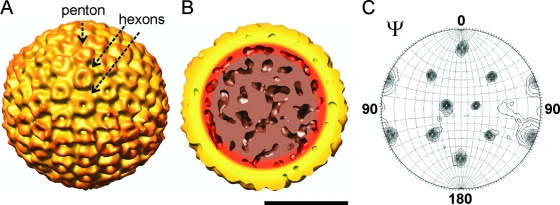
Structure of RVFV MP-12. Images were color coded according to radial distance. All particles are oriented along the threefold axis. (A) Shaded isosurface representation of RVFV MP-12 reconstructed from tilt series data at a 7.5-nm resolution. The structure was generated by averaging data for 46 individual particles extracted from three tomograms. The missing-wedge effect was greatly reduced because individual particle tomograms adopted different random orientations. Pentons and hexons are indicated by the arrows. (B) Cut-away view of the averaged RVFV MP-12 structure. No distinct densities were observed in the core of the particle that could be interpreted as RNP complexes. (C) Stereographic projection of the threefold (κ = 120°) self-rotation function calculated for the map reconstructed from tomographic data. The number and relative positions of peaks corresponded to those expected for an icosahedral particle. The peaks were sharp, implying strong symmetry relationships in the structure. The scale bar represents 50 nm.
The averaged RVFV MP-12 map had a nominal resolution of 7.5 nm, as estimated by the Fourier shell correlation function using a 0.5 threshold. Significantly, symmetry was not imposed during the reconstruction of the tomograms and particle image alignment and averaging. However, the averaged map showed an icosahedral arrangement of the glycoprotein spikes on the surface of RVFV MP-12 (Fig. 3A). To verify that the averaged particle structure had an icosahedral geometry, the required five-, three- and twofold axes of symmetry were unambiguously identified in self-rotation functions calculated from the averaged particle by using the program GLRF (48). The self-rotation functions showed all peaks expected for an icosahedral particle. Stereographic projections of the rotation function of the particle showed sharp peaks, an indication of strong symmetry elements (Fig. 3C). A cut-away view of the averaged RVFV MP-12 particle structure did not show distinct densities that could be interpreted as individual RNP complexes (Fig. 3B). Due to the missing arrangement of RNP complexes as symmetrical structural features, the sum of densities in the averaged map will not reflect the true RNP arrangement and interpretations at this resolution would be speculative.
Glycoprotein structure and organization.
Although averaging provided definitive proof that the RVFV MP-12 particle was icosahedral, the map still suffered from an uneven distribution of orientations from the limited number of particle subtomograms that were averaged. To improve the quality of the reconstruction, the icosahedral symmetry indicated by the visual inspection of the averaged map and quantitatively characterized by self-rotation functions was imposed (Fig. 4A). The resulting icosahedrally averaged map had an isotropic resolution of 6.1 nm, without missing-wedge density degradation effects. The particle diameter was 100 nm, and the volume was ∼2.3 × 105 nm3. Cross-sectional views through the capsomeres showed them to be hollow cylinders of ∼12 nm in height with a central cavity of 4 nm in diameter (Fig. 4B). Hexons were ∼14 nm in diameter, and their centers were ∼16 nm apart. Pentons in the map were smaller, with a diameter of ∼12 nm. The gap between two capsomeres measured ∼2.2 nm. The central section of the icosahedrally averaged map showed the RNP core to be ∼66 nm in diameter and exhibited a gap between the lipid bilayer and the RNP core of ∼4.5 nm (Fig. 4C).
FIG. 4.
RVFV MP-12 glycoprotein spike organization on an icosahedral lattice. Images were color coded according to radial distance. All particles are oriented along the threefold axis. (A) Shaded isosurface representation of RVFV MP-12 after the imposition of icosahedral symmetry onto the averaged tomogram shown in Fig. 3A. The icosahedral asymmetric unit is indicated by the black triangle. (B) Interior of the virus particle after the imposition of icosahedral symmetry. The central disordered RNP region was removed, thus exposing the inner surface of the lipid bilayer. (C) Central section through the map after the imposition of icosahedral symmetry as described in the legend to panel A. The dotted circles enclose the gap (white density) between the glycoprotein shell and the RNP core (gray density). Protein densities are represented in black. (D) Radial projection of the map after the imposition of icosahedral symmetry. Bridging densities among capsomeres were visible. Protein density is represented in black. The scale bar corresponds to 50 nm.
The 12 pentameric and 110 hexameric capsomeres were arranged on the surface of the RVFV MP-12 virus as an icosahedral lattice with a T=12 triangulation number (4). To our knowledge, a T=12 triangulation number has been described previously only for the recently published tomographic reconstruction of UUKV, another member of the Bunyaviridae family (31). A radial projection through the surface of the icosahedrally averaged map of RVFV MP-12 showed the capsomere arrangement (Fig. 4D).
High-resolution structures of bunyavirus glycoproteins are not available, and the glycoprotein sequences are not sufficiently homologous to known structures to enable reliable homology modeling of RVFV MP-12 GC and GN proteins. Thus, pseudoatomic modeling to determine possible locations for the glycoproteins within the virus envelope was not possible. An initial estimation of the hexon volume was ∼1,420 nm3, suggesting that the hexameric capsomere could accommodate a protein mass of ∼600 kDa (assuming ∼40% solvent) or 12 glycoproteins (with GN of 47 kDa and GC of 51 kDa for the extracellular domains). A similar analysis suggested that the pentameric capsomeres could contain 10 glycoproteins. This analysis correlated with the virus containing 720 glycoproteins of each type, GN and GC. The limited resolution of the RVFV MP-12 structure and the similar molecular masses of the GN and GC glycoproteins have made it difficult to unambiguously locate the glycoproteins within the capsomere. For example, the 12 glycoproteins in the hexon may be GC or GN monomers or homodimers or GN-GC heterodimers.
DISCUSSION
The present study revealed the spatial architecture of RVFV vaccine strain MP-12 by cryo-ET. Viral glycoproteins were found to be unambiguously organized on an icosahedral lattice with a triangulation number of T=12. This unusual organization has been described previously only for the UUKV, another member of the genus Phlebovirus within the family Bunyaviridae (31). UUKV particles are more heterogeneous in size than RVFV MP-12 particles; the UUKV pH 7 structure has a diameter of 125 nm, and the pH 6 structure has a diameter of ∼115 nm, which are both larger than the diameter of the RVFV MP-12 structure. Moreover, UUKV particles were reported to display various degrees of pleomorphy, in contrast to the more homogenous RVFV MP-12 particles that allowed for averaging over all particles to improve the signal-to-noise ratio of the averaged RVFV MP-12 structure. The heterogeneity and pleomorphy of UUKV particles relative to RVFV MP-12 particles may be attributed to the glutaraldehyde cross-linking procedure or the pelleting step used when purifying UUKV. Our results, along with those in other studies presenting cryo-EM images of frozen-hydrated bunyaviruses (e.g., La Crosse virus [47], Hantaan virus [genus Hantaviridae] [2], and UUKV [31]), suggest that bunyaviruses are spherical, contrary to the earlier view that they are pleomorphic. It is likely that all phleboviruses will be found to assemble into T=12 icosahedrons, and perhaps all bunyaviruses may adopt icosahedral symmetry.
Our map of RVFV MP-12 revealed a virus envelope consisting of a ∼4-nm-thick membrane into which were inserted ∼12-nm surface glycoproteins. The glycoproteins assembled into clear pentameric and hexameric capsomeres. However, it was not possible to discern the exact glycoprotein organization of the capsomeres. Findings in previous studies of several members of the Phlebovirus genus, such as Punta Toro virus (PTV), UUKV, and RVFV, have suggested that the two glycoproteins GN and GC form heterodimers (6, 7, 15, 33). However, homodimers of GC of PTV and of GN and GC of UUKV have also been described previously (6, 38). The volume of the hexameric and pentameric capsomeres indicates that they may contain 12 and 10 glycoproteins, respectively, although the exact capsomere composition could not be resolved.
Two distinct pH-dependent conformations of the glycoproteins were revealed in the 3D structure of UUKV. At neutral pH, the glycoproteins appeared to adopt an extended conformation, whereas at pH 6, a more compact conformation was observed (31). This conformational difference was suggested to reflect pH-dependent conformational changes within the GC glycoprotein, a putative class II fusion protein (14), although the transition to a compact flat glycoprotein at low pH differs from conformational changes observed previously for other pH-dependent fusion proteins (19). Interestingly, the structure of unfixed RVFV MP-12 particles isolated and purified at pH 7.2 resembled the structure of UUKV particles isolated at pH 6. Future structural studies at different pHs are necessary to determine if RVFV MP-12 undergoes pH-dependent conformational changes similar to those reported for UUKV.
In all bunyaviruses, the M segment encodes the GN and GC glycoproteins (39). The M segment of mosquito- and sand fly-vectored phleboviruses encodes an additional NSm protein, whereas the tick-borne phleboviruses (e.g., UUKV) are lacking the NSm protein. The RVFV M segment encodes a 14-kDa NSm protein and a 78-kDa protein, and the 78-kDa protein consists of the GN protein and the product of a region upstream of that encoding GN. Although the 78-kDa protein was reported previously to be a structural protein, as various small amounts in virus preparations have been noted (36, 45), we did not detect any band corresponding to the 78-kDa protein in the Western blot analysis. The direct identification and determination of the glycoprotein arrangement on the virus surface, as well as the possible presence of the NSm and 78-kDa proteins, will likely require higher-resolution reconstructions and antibody labeling.
Capsomeres have been observed on the surfaces of viruses from several members of the different genera within the Bunyaviridae family (26). However, viruses within each genus may have distinct surface capsomere structures (26). For example, negative-stain images of nairoviruses (e.g., Crimean-Congo hemorrhagic fever virus) have shown very small and closely packed surface units, whereas members of the Orthobunyavirus genus (e.g., La Crosse and Bunyamwera viruses) have small, knob-like morphological units with no apparent organizational pattern. Other phleboviruses (e.g., PTV and Karimabad virus) have distinct, sharply defined surface capsomeres similar to those of RVFV (43). Hantaviruses display a unique surface organization, with capsomeres arranged in a square, grid-like pattern (26). Future structural analyses of bunyaviruses from different genera will help elucidate differences in bunyavirus structure, morphology, and capsomere glycoprotein arrangement.
In contrast to other RNA viruses, bunyaviruses lack a structural matrix protein linking the viral envelope with the virus core (12). The results of mutational studies of tomato spotted wilt virus, UUKV, and Bunyamwera virus (32, 42, 44) have indicated that a direct interaction exists between the glycoprotein cytoplasmic tails and the RNP complexes. In the absence of structural matrix proteins, the bridging densities observed among capsomeres of RVFV MP-12 may help assemble and stabilize the virus particles. Similar bridging densities among surface spikes in the UUKV structure were observed previously (31).
The structure of the bunyavirus RVFV MP-12 demonstrates that members of the Phlebovirus genus are icosahedral. This observation provides an impetus for future high-resolution studies on bunyaviruses by using single-particle cryo-EM and icosahedral averaging. Higher-resolution structures will help elucidate the assembly patterns and plasticity of this important virus family and ultimately may help identify antiviral and vaccine targets.
Acknowledgments
We thank the Sealy Center for Structural Biology and Molecular Biophysics and the Cryo-Electron Microscopy Center for Macromolecular Systems Imaging (UTMB; Keck Foundation grant) for the use of cryo-EMs. Further, we thank John Morrill (UTMB) for providing us with RVFV MP-12, Robert Tesh (UTMB) for providing us with an anti-RVFV mouse polyclonal antibody, the staff of the EM laboratory at UTMB for support in EM analysis of negatively stained specimens, and Michael Schmid (Baylor College of Medicine, Houston, TX) for help with the alignment and averaging of subtomograms. We also thank C. J. Peters and Shinji Makino (UTMB) for helpful discussions.
This work was supported by a training fellowship from the W. M. Keck Foundation to the Gulf Coast Consortia through the Keck Center for Virus Imaging (A.N.F.) and by grants from the NIH/NIAID Western Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research (subaward from U54 AI057156 to M.B.S.) and the Welch Foundation (S.J.W.).
Footnotes
Published ahead of print on 20 August 2008.
REFERENCES
- 1.Baumeister, W. 2002. Electron tomography: towards visualizing the molecular organization of the cytoplasm. Curr. Opin. Struct. Biol. 12679-684. [DOI] [PubMed] [Google Scholar]
- 2.Boudko, S. P., R. J. Kuhn, and M. G. Rossmann. 2007. The coiled-coil domain structure of the Sin Nombre virus nucleocapsid protein. J. Mol. Biol. 3661538-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caplen, H., C. J. Peters, and D. H. Bishop. 1985. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J. Gen. Virol. 662271-2277. [DOI] [PubMed] [Google Scholar]
- 4.Caspar, D. L., and A. Klug. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 271-24. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2000. Update: outbreak of Rift Valley Fever—Saudi Arabia, August-November 2000. MMWR Morb. Mortal. Wkly. Rep. 49982-985. [PubMed] [Google Scholar]
- 6.Chen, S. Y., and R. W. Compans. 1991. Oligomerization, transport, and Golgi retention of Punta Toro virus glycoproteins. J. Virol. 655902-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. Y., Y. Matsuoka, and R. W. Compans. 1992. Assembly of G1 and G2 glycoprotein oligomers in Punta Toro virus-infected cells. Virus Res. 22215-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiu, W. 1986. Electron microscopy of frozen, hydrated biological specimens. Annu. Rev. Biophys. Biophys. Chem. 15237-257. [DOI] [PubMed] [Google Scholar]
- 9.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50760-763. [DOI] [PubMed] [Google Scholar]
- 10.Dewey, R. A., L. C. Semorile, O. Grau, and J. V. Crisci. 1997. Cladistic analysis of Tospovirus using molecular characters. Mol. Phylogenet. Evol. 811-32. [DOI] [PubMed] [Google Scholar]
- 11.Dubochet, J., M. Adrian, J. J. Chang, J. C. Homo, J. Lepault, A. W. McDowall, and P. Schultz. 1988. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21129-228. [DOI] [PubMed] [Google Scholar]
- 12.Elliott, R. M. 1990. Molecular biology of the Bunyaviridae. J. Gen. Virol. 71501-522. [DOI] [PubMed] [Google Scholar]
- 13.Ellis, D. S., P. V. Shirodaria, E. Fleming, and D. I. Simpson. 1988. Morphology and development of Rift Valley fever virus in Vero cell cultures. J. Med. Virol. 24161-174. [DOI] [PubMed] [Google Scholar]
- 14.Garry, C. E., and R. F. Garry. 2004. Proteomics computational analyses suggest that the carboxyl terminal glycoproteins of Bunyaviruses are class II viral fusion protein (beta-penetrenes). Theor. Biol. Med. Model. 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerrard, S. R., and S. T. Nichol. 2007. Synthesis, proteolytic processing and complex formation of N-terminally nested precursor proteins of the Rift Valley fever virus glycoproteins. Virology 357124-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunewald, K., O. Medalia, A. Gross, A. C. Steven, and W. Baumeister. 2003. Prospects of electron cryotomography to visualize macromolecular complexes inside cellular compartments: implications of crowding. Biophys. Chem. 100577-591. [DOI] [PubMed] [Google Scholar]
- 17.Hoppe, W., and R. Hegerl. 1980. Three-dimensional structure determination by electron microscopy, p. 127-186. In P. W. Hawkes (ed.), Computer processing of electron microscope images. Springer-Verlag, Heidelberg, Germany.
- 18.Isaacson, M. 2001. Viral hemorrhagic fever hazards for travelers in Africa. Clin. Infect. Dis. 331707-1712. [DOI] [PubMed] [Google Scholar]
- 19.Kielian, M., and F. A. Rey. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 467-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 11671-76. [DOI] [PubMed] [Google Scholar]
- 21.Kuismanen, E., B. Bang, M. Hurme, and R. F. Pettersson. 1984. Uukuniemi virus maturation: immunofluorescence microscopy with monoclonal glycoprotein-specific antibodies. J. Virol. 51137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuismanen, E., K. Hedman, J. Saraste, and R. F. Pettersson. 1982. Uukuniemi virus maturation: accumulation of virus particles and viral antigens in the Golgi complex. Mol. Cell. Biol. 21444-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levitt, J., W. D. Naude, and A. Polson. 1963. Purification and electron microscopy of pantropic Rift Valley fever virus. Virology 20530-533. [DOI] [PubMed] [Google Scholar]
- 24.Lucic, V., F. Forster, and W. Baumeister. 2005. Structural studies by electron tomography: from cells to molecules. Annu. Rev. Biochem. 74833-865. [DOI] [PubMed] [Google Scholar]
- 25.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 12882-97. [DOI] [PubMed] [Google Scholar]
- 26.Martin, M. L., H. Lindsey-Regnery, D. R. Sasso, J. B. McCormick, and E. Palmer. 1985. Distinction between Bunyaviridae genera by surface structure and comparison with Hantaan virus using negative stain electron microscopy. Arch. Virol. 8617-28. [DOI] [PubMed] [Google Scholar]
- 27.Mastronarde, D. N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 15236-51. [DOI] [PubMed] [Google Scholar]
- 28.Mastronarde, D. N. 1997. Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120343-352. [DOI] [PubMed] [Google Scholar]
- 29.McIntosh, R., D. Nicastro, and D. Mastronarde. 2005. New views of cells in 3D: an introduction to electron tomography. Trends Cell Biol. 1543-51. [DOI] [PubMed] [Google Scholar]
- 30.Murphy, F. A., A. K. Harrison, and S. G. Whitfield. 1973. Bunyaviridae: morphologic and morphogenetic similarities of Bunyamwera serologic supergroup viruses and several other arthropod-borne viruses. Intervirology 1297-316. [DOI] [PubMed] [Google Scholar]
- 31.Overby, A. K., R. F. Pettersson, K. Grunewald, and J. T. Huiskonen. 2008. Insights into bunyavirus architecture from electron cryotomography of Uukuniemi virus. Proc. Natl. Acad. Sci. USA 1052375-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Overby, A. K., R. F. Pettersson, and E. P. Neve. 2007. The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J. Virol. 813198-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persson, R., and R. F. Pettersson. 1991. Formation and intracellular transport of a heterodimeric viral spike protein complex. J. Cell Biol. 112257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 251605-1612. [DOI] [PubMed] [Google Scholar]
- 35.Prins, M., and R. Goldbach. 1998. The emerging problem of tospovirus infection and nonconventional methods of control. Trends Microbiol. 631-35. [DOI] [PubMed] [Google Scholar]
- 36.Rice, R. M., B. J. Erlick, R. R. Rosato, G. A. Eddy, and S. B. Mohanty. 1980. Biochemical characterization of Rift Valley fever virus. Virology 105256-260. [DOI] [PubMed] [Google Scholar]
- 37.Robeson, G., L. H. el Said, W. Brandt, J. Dalrymple, and D. H. Bishop. 1979. Biochemical studies on the Phlebotomus fever group viruses (Bunyaviridae family). J. Virol. 30339-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronka, H., P. Hilden, C. H. Von Bonsdorff, and E. Kuismanen. 1995. Homodimeric association of the spike glycoproteins G1 and G2 of Uukuniemi virus. Virology 211241-250. [DOI] [PubMed] [Google Scholar]
- 39.Schmaljohn, C. S., and S. T. Nichol. 2007. Bunyaviridae, p. 1741-1790. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Wolters Kluwer, Philadelphia, PA. [Google Scholar]
- 40.Schmid, M. F., and C. R. Booth. 2008. Methods for aligning and for averaging 3D volumes with missing data. J. Struct. Biol. 161243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sherman, M. B., R. H. Guenther, F. Tama, T. L. Sit, C. L. Brooks, A. M. Mikhailov, E. V. Orlova, T. S. Baker, and S. A. Lommel. 2006. Removal of divalent cations induces structural transitions in red clover necrotic mosaic virus, revealing a potential mechanism for RNA release. J. Virol. 8010395-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi, X., A. Kohl, P. Li, and R. M. Elliott. 2007. Role of the cytoplasmic tail domains of Bunyamwera orthobunyavirus glycoproteins Gn and Gc in virus assembly and morphogenesis. J. Virol. 8110151-10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, J. F., and D. Y. Pifat. 1982. Morphogenesis of sandfly viruses (Bunyaviridae family). Virology 12161-81. [DOI] [PubMed] [Google Scholar]
- 44.Snippe, M., J. Willem Borst, R. Goldbach, and R. Kormelink. 2007. Tomato spotted wilt virus Gc and N proteins interact in vivo. Virology 357115-123. [DOI] [PubMed] [Google Scholar]
- 45.Struthers, J. K., R. Swanepoel, and S. P. Shepherd. 1984. Protein synthesis in Rift Valley fever virus-infected cells. Virology 134118-124. [DOI] [PubMed] [Google Scholar]
- 46.Subramaniam, S., A. Bartesaghi, J. Liu, A. E. Bennett, and R. Sougrat. 2007. Electron tomography of viruses. Curr. Opin. Struct. Biol. 17596-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Talmon, Y., B. V. Prasad, J. P. Clerx, G. J. Wang, W. Chiu, and M. J. Hewlett. 1987. Electron microscopy of vitrified-hydrated La Crosse virus. J. Virol. 612319-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong, L., and M. G. Rossmann. 1997. Rotation function calculations with GLRF program. Methods Enzymol. 276594-611. [PubMed] [Google Scholar]
- 49.Vialat, P., R. Muller, T. H. Vu, C. Prehaud, and M. Bouloy. 1997. Mapping of the mutations present in the genome of the Rift Valley fever virus attenuated MP12 strain and their putative role in attenuation. Virus Res. 5243-50. [DOI] [PubMed] [Google Scholar]
- 50.von Bonsdorff, C. H., and R. Pettersson. 1975. Surface structure of Uukuniemi virus. J. Virol. 161296-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]