Abstract
Schizosaccharomyces pombe Pfh1p is an essential member of the Pif family of 5′-3′ DNA helicases. The two Saccharomyces cerevisiae homologs, Pif1p and Rrm3p, function in nuclear DNA replication, telomere length regulation, and mitochondrial genome integrity. We demonstrate here the existence of multiple Pfh1p isoforms that localized to either nuclei or mitochondria. The catalytic activity of Pfh1p was essential in both cellular compartments. The absence of nuclear Pfh1p resulted in G2 arrest and accumulation of DNA damage foci, a finding suggestive of an essential role in DNA replication. Exogenous DNA damage resulted in localization of Pfh1p to DNA damage foci, suggesting that nuclear Pfh1p also functions in DNA repair. The absence of mitochondrial Pfh1p caused rapid depletion of mitochondrial DNA. Despite localization to nuclei and mitochondria in S. pombe, neither of the S. cerevisiae homologs, nor human PIF1, suppressed the lethality of pfh1Δ cells. However, the essential nuclear function of Pfh1p could be supplied by Rrm3p. Expression of Rrm3p suppressed the accumulation of DNA damage foci but not the hydroxyurea sensitivity of cells depleted of nuclear Pfh1p. Together, these data demonstrate that Pfh1p has essential roles in the replication of both nuclear and mitochondrial DNA.
The founding member of the Pif family of 5′-3′DNA helicases is Saccharomyces cerevisiae Pif1p (ScPif1p) . The genomes of most multicellular animals contain a single Pif1-like gene, but several single-celled eukaryotes, including S. cerevisiae, encode two Pif1-like proteins. Although ScPif1p and ScRrm3p are ∼40% identical in the helicase domain and both unwind double-stranded DNA with 5′-3′ polarity (22, 28), these paralogs have largely nonoverlapping functions (6).
ScPif1p was first identified because of its role in the repair and recombination of mitochondrial DNA (mtDNA) (15, 16). In the nucleus, ScPif1p is a catalytic inhibitor of telomerase that acts by removing telomerase from DNA ends (5, 44, 60). As a result, cells lacking ScPif1p have long telomeres. ScPif1p has additional, less well characterized roles in the replication (8, 23) and recombination (56) of chromosomal DNA. In vitro, ScPif1p preferentially unwinds RNA/DNA hybrids (7) and branched substrates (27). ScRRM3 was first identified as a suppressor of recombination in the ribosomal DNA (rDNA) (25). ScRrm3p moves with the replication fork (3) and facilitates its progression past stable, non-histone protein-DNA complexes (21, 51). ScRrm3p-sensitive sites, which are found throughout the genome (3), include tRNA genes, telomeres, centromeres, inactive replication origins, silent mating type loci, and multiple sites within rDNA (21-23). In the absence of ScRrm3p, replication forks stall and sometimes break at ScRrm3p-sensitive sites, leading to intra-S-phase checkpoint activation (21, 51) and dependency on replication fork restart activities for viability (40, 43, 50, 52). In the rDNA, which is particularly dependent on ScRrm3p to prevent replication fork stalling, ScPif1p antagonizes ScRrm3p action by helping to maintain the replication fork barrier (23). ScRrm3p and ScPif1p also impact mtDNA abundance in opposite fashion, since deletion of ScRRM3 partially reverses the mtDNA decrease observed in cells lacking ScPif1p (38, 49).
An important question emerging from these studies is whether the single Pif family member proteins in other eukaryotes carry out ScPif1p-like or ScRrm3p-like functions. Like ScPif1p, human PIF1 (hPIF1p) unwinds both DNA/DNA and DNA/RNA duplexes, and its overexpression in tissue culture cells may result in telomere shortening (59). In addition, both hPIF1p and mouse Pif1p (mPif1p) coimmunoprecipitate with telomerase (31, 45). However, even after four mouse generations, deletion of mPif1 (45) has no detectable effects on telomere length, cell cycle progression, or the DNA damage response.
To determine the functions of a Pif-like protein in an organism with a single member of this helicase family, we used the genetically tractable Schizosaccharomyces pombe. Fission yeast Pfh1p (for Pif1 homolog) has about equal similarity to ScPif1p and ScRrm3p (61) and, like the other Pif helicase family members, unwinds double-stranded DNA with 5′-3′ polarity (48, 61). However, unlike ScPIF1, ScRRM3, and mPif1, pfh1+ is essential. Spore progeny carrying a pfh1Δ allele arrest in G2 phase (61), mirroring the phenotype observed in cold-sensitive pfh1 strains at restrictive temperatures (48). Even at permissive temperatures, cold-sensitive pfh1 strains display severely reduced viability in the presence of hydroxyurea (HU) or methyl methanesulfonate, suggesting a role for Pfh1p in DNA repair (48). S. pombe strains that can tolerate the complete absence of telomeric or mtDNA still require pfh1+ for viability, suggesting that pfh1− lethality cannot be attributed solely to defects at either one of these loci (61).
We show here that there are multiple isoforms of Pfh1p that exhibit differential localization within the cell. Using separation-of-function alleles, we demonstrate that the helicase activity of Pfh1p was essential in both nuclei and mitochondria. In the absence of nuclear Pfh1p, cells arrested in G2 phase and contained DNA damage foci. In the absence of mitochondrial Pfh1p, mtDNA was rapidly depleted. Although ScPif1p, ScRrm3p, and hPIF1p localized to both mitochondria and nuclei when expressed in S. pombe, none of these homologs could supply all essential nuclear and mitochondrial functions of Pfh1p. However, expression of ScRrm3p, but not ScPif1p or hPIF1p, was able to supply the essential nuclear function(s) of Pfh1p. Although ScRrm3p did not suppress the HU sensitivity of Pfh1p-depleted cells, it did prevent the accumulation of DNA damage foci in these cells. We conclude that Pfh1p, like ScRrm3p, is important for the replication of chromosomal DNA, and its critical replication functions can be supplied by ScRrm3p. Like ScPif1p, Pfh1p has a key role in the maintenance of mtDNA and is needed for DNA repair, but neither of these functions can be replaced by expressing ScRrm3p or ScPif1p.
MATERIALS AND METHODS
Growth conditions, strains, and complementation analysis.
Strain genotypes are listed in Table 1. Unless otherwise noted these strains are also ade6-M210/M216 leu1-32 his3-D1 ura4-D18. In addition, we used strain PR3474 (leu1-32 his3-D1 ura4-D18 rad22-RFP-kanMX6) (11). Cells were cultured in supplemented yeast extract (YES; Difco) or Edinburgh minimal media (EMM; Sunrise Science) at 30°C unless otherwise noted. Treatment with 40 μΜ camptothecin was carried out for 3 h at 30°C. For serial culture dilution experiments (see Fig. 3, 4, and 5), cells were grown in liquid EMM in the presence or absence of 30 μM thiamine, counted by using a hemacytometer, and diluted to 5 × 105 cells/ml every 12 h. For telomere length analysis, serial culture dilution experiments were performed as described above using a pfh1::ura4+-nmt1-pfh1-GFP strain (see Fig. 3D and E), a strain that has an almost identical growth curve response to thiamine as the pfh1::ura4+-nmt1-pfh1-mt strain characterized in Fig. 3A to C.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype |
|---|---|
| ySP4 | h−pfh1+ |
| ySP205 | h+/h−leu1-32/leu1-32::pJK148-pfh1+pfh1+/pfh1::ura4+ |
| ySP207 | h+/h−leu1-32/leu1-32::pJK148-pfh1-GFP pfh1+/pfh1::ura4+ |
| ySP211 | h+/h−leu1-32/leu1-32::pJK148-pfh1-m1-GFP pfh1+/pfh1::ura4+ |
| ySP215 | h+/h−leu1-32/leu1-32::pJK148-pfh1-m21-GFP pfh1+/pfh1::ura4+ |
| ySP289 | h+leu1-32::pJK148 pfh1::kanMX6 + pVS117b |
| ySP291 | h−leu1-32::pJK148 pfh1::kanMX6 + pVS117 |
| ySP293 | h+leu1-32::pJK148-pfh1+pfh1::kanMX6 + pVS117 |
| ySP295 | h−leu1-32::pJK148-pfh1+pfh1::kanMX6 + pVS117 |
| ySP301 | h+leu1-32::pJK148-pfh1-m1 pfh1::kanMX6 + pVS117 |
| ySP303 | h−leu1-32::pJK148-pfh1-m1 pfh1::kanMX6 + pVS117 |
| ySP308 | h+leu1-32::pJK148-pfh1-m21 pfh1::kanMX6 + pVS117 |
| ySP310 | h−leu1-32::pJK148-pfh1-m21 pfh1::kanMX6 + pVS117 |
| ySP318 | h−leu1-32::pJK148 pfh1::ura4+nmt-pfh1-m21 |
| ySP322 | h−leu1-32::pJK148-pfh1+ pfh1::ura4+nmt-pfh1-m21 |
| ySP326 | h−leu1-32::pJK148-pfh1-m1 pfh1::ura4+nmt-pfh1-m21 |
| ySP367 | h+leu1-32::pJK148-pfh1-K337A pfh1::kanMX6 + pVS117 |
| ySP370 | h−leu1-32::pJK148-pfh1-K337A pfh1::kanMX6 + pVS117 |
| ySP377 | h+leu1-32::pJK148-pfh1-m21,170,265,320-NES-GFP (mt*) pfh1::kanMX6 + pVS117 |
| ySP379 | h−leu1-32::pJK148-pfh1- m21,170,265,320-NES-GFP (mt*) pfh1::kanMX6 + pVS117 |
| ySP383 | h−pfh1::ura4+-nmt1-pfh1+-GFP |
| ySP391 | h+/h−leu1-32::pJK148-pfh1-GFP gar2+/gar2-mcherry-kanMX6 |
| ySP396 | h+/h−leu1-32::pJK148-pfh1-GFP rad22+/rad22-RFP-kanMX6 |
| ySP403 | h+leu1-32::pJK148-nmt1-ScPIF1-GFP |
| ySP405 | h+leu1-32::pJK148-nmt1-ScRRM3-GFP |
| ySP407 | h+leu1-32::pJK148-nmt1-hPIF1-GFP |
| ySP409 | h−leu1-32::pJK148 pfh1::ura4+nmt-pfh1-m21 rad22-RFP-kanMX6 |
| ySP413 | h−leu1-32::pJK148-pfh1+ pfh1::ura4+nmt-pfh1-m21 rad22-RFP-kanMX6 |
| ySP417 | h−leu1-32::pJK148-pfh1-m1 pfh1::ura4+nmt-pfh1-m21 rad22-RFP-kanMX6 |
| ySP425 | h+leu1-32::pJK148-pfh1+pfh1::loxP-pfh1-kanMX6-loxP |
| ySP433 | h−leu1-32::pJK148-nmt1-hPIF1 pfh1::loxP-pfh1-kanMX6-loxP |
| ySP437 | h−leu1-32::pJK148-nmt1-ScPIF1 pfh1::loxP-pfh1-kanMX6-loxP |
| ySP441 | h−leu1-32::pJK148-nmt1-ScRRM3 pfh1::loxP-pfh1-kanMX6-loxP |
| ySP469 | h−leu1-32::pJK148-nmt1-hPIF1 pfh1::loxP-pfh1-kanMX6-loxP his3-D1::pfh1-nuc |
| ySP472 | h−leu1-32::pJK148-nmt1-ScPIF1 pfh1::loxP-pfh1-kanMX6-loxP his3-D1::pfh1-nuc |
| ySP475 | h−leu1-32::pJK148-nmt1-ScRRM3 pfh1::loxP-pfh1-kanMX6-loxP his3-D1::pfh1-nuc |
| ySP478 | h−leu1-32::pJK148-nmt1-hPIF1 pfh1::loxP-pfh1-kanMX6-loxP his3-D1::pfh1-mt* |
| ySP481 | h−leu1-32::pJK148-nmt1-ScPIF1 pfh1::loxP-pfh1-kanMX6-loxP his3-D1::pfh1-mt* |
| ySP484 | h−leu1-32::pJK148-nmt1-ScRRM3 pfh1::loxP-pfh1-kanMX6-loxP his3-D1::pfh1-mt* |
| ySP496 | h+leu1-32::pJK148-pfh1-mt* pfh1::kanMX6 |
| SA31 | h+pfh1::loxP his3-D1::pfh1-mt* rad22-RFP-kanMX6 |
| SA36 | h+leu1-32::pJK148-ScRRM3 pfh1::loxP his3-D1::pfh1mt* rad22RFP-kanMX6 |
| SA54 | h+leu1-32::pJK148-pfh1-mt* pfh1::kanMX6 + pJR41XHa |
| SA55 | h+leu1-32::pJK148-pfh1-mt* pfh1::kanMX6 + pJR41XH-ScRRM3 |
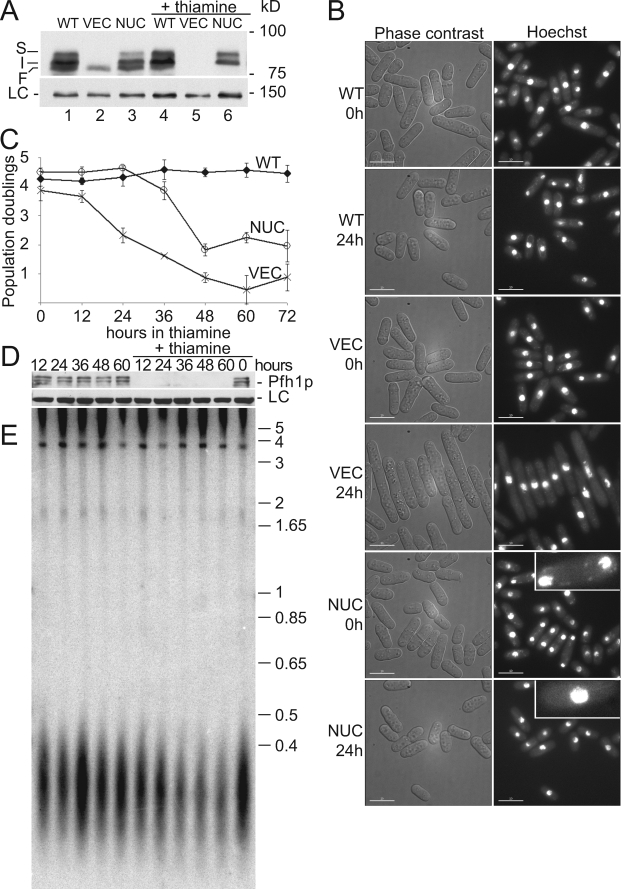
FIG. 3.
Nuclear Pfh1p is necessary and sufficient to prevent G2-phase arrest. (A) Anti-Pfh1p Western blot of nmt1-pfh1-mt strains that also express the indicated pfh1 alleles from the pfh1+ promoter. WT, pfh1+; VEC, empty vector; NUC, pfh1-nuc. Samples in lanes 4 to 6 are from cells grown in the presence of thiamine (24 h). LC, loading control blot probed with anti-Rdp1 polyclonal serum. (B) Phase-contrast and fluorescence microscopy of Hoechst 33342-stained cells at 0 and 24 h after the addition of thiamine. Insets in NUC photographs are enlarged ×3 to detect mitochondrial nucleoids. (C) Number of population doublings per 12-h period before (0 h) or after thiamine addition. Cells expressing endogenous Pfh1p (WT, ⧫), cells depleted for both mitochondrial and nuclear Pfh1p (VEC, ×), and cells expressing nuclear Pfh1p but depleted of mitochondrial Pfh1p (NUC, ○). (D) Anti-Pfh1p Western blot of a pfh1::ura4+-nmt1-pfh1-GFP strain grown with (+thiamine) or without thiamine for the indicated amounts of time. (E) Southern blot analysis of genomic DNA taken from the same cells examined in panel D for Pfh1p levels was digested with ApaI and probed with S. pombe telomeric DNA from pSPT16 (47). Molecular mass markers are indicated in kilobase pairs.
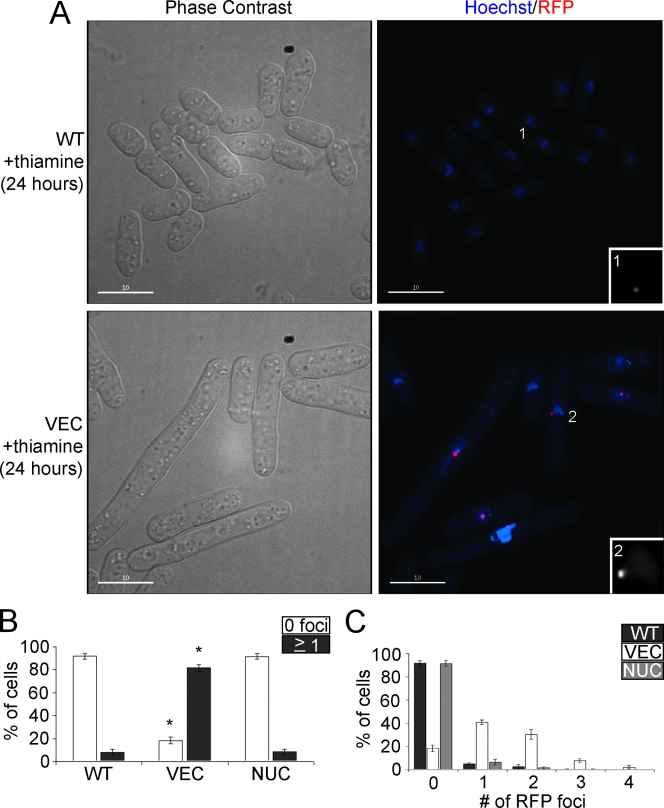
FIG. 4.
Depletion of nuclear Pfh1p results in the formation of DNA damage foci. (A) Cells were visualized by phase-contrast and fluorescence microscopy 24 h after the addition of thiamine to cells expressing endogenous levels of Pfh1p (WT) and cells depleted of both mitochondrial and nuclear Pfh1p (VEC). Both strains also expressed Rad22p-RFP to visualize DNA damage foci. Insets are ×3 enlargements of nuclei with Rad22p-RFP foci. (B) Percentage of WT, VEC, and NUC cells without (white) or with (dark gray) Rad22p-RFP foci after 24 h of thiamine addition. Asterisk denotes statistically significant difference (t test) from WT strain with P < 0.0001. (C) Percentage of WT (dark gray), VEC (white), and NUC (light gray) cells with zero to four DNA damage foci per cell.
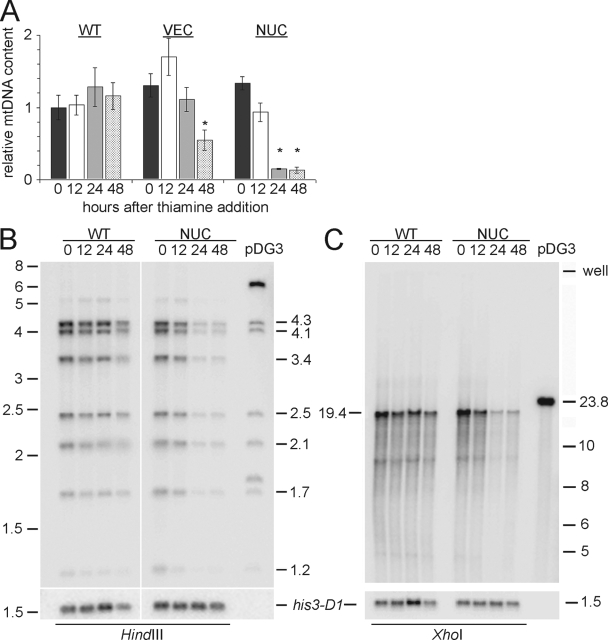
FIG. 5.
Depletion of mitochondrial Pfh1p results in mtDNA depletion. (A) Real-time PCR analysis of mtDNA content in WT, VEC, and NUC strains at 0 h (dark gray), 12 h (white), 24 h (light gray), and 48 h (spotted) after thiamine addition. Asterisk denotes statistically significant difference (t test) from corresponding 0 h value with P < 0.003. (B and C) Southern blots of genomic DNA digested with HindIII or XhoI probed with S. pombe mtDNA (in pBR322; pDG3) or the EcoNI-PstI fragment from pAF1 to detect sequence 3′ to the his3-D1 marker present in these strains. The rightmost lane in each blot shows the restriction pattern of pDG3. Molecular mass markers are given in kilobase pairs.
The gar2+ locus was modified by 3′ tagging by in-frame integration of a mCherry-kanMX6 cassette. Alleles of pfh1 were integrated at the leu1-32 locus using the pJK148 vector. Construction of the pfh1-mt* allele was carried out by successive site-directed mutagenesis (Stratagene) of methionine codons M265 and M320 to alanine codons. M170 was changed to a leucine rather than alanine codon to preserve the local secondary structure as predicted by jpred (www.compbio.dundee.ac.uk/∼www-jpred/). Separately, the methionine codon changes or the addition of a carboxy-terminal nuclear export sequence (NES) were insufficient to abolish the nuclear function of Pfh1p (data not shown). Therefore, these changes were combined to generate the pfh1-mt* allele. As expected, by cytological criteria, the NES-green fluorescent protein (GFP)-tagged Pfh1p encoded by the pfh1-mt* allele was detected only in mitochondria (data not shown). For complementation analysis, pfh1Δ strains with empty vector (VEC), pfh1+ (WT), pfh1-nuc (nuc), pfh1-mt (mt), pfh1-mt* (mt*), or pfh1-cd (cd) alleles of pfh1 integrated at leu1-32 and carrying the his3+ marked pBG2-pfh1+ plasmid were streaked twice onto YES plates (30 or 18°C) and grown to log phase in liquid YES. Colonies were replica plated onto EMM lacking histidine, and the percentage of colonies that lost the his3+ plasmid was determined. For plasmid loss experiments with haploid strains, three independent clones from each of two parental strains of opposite mating types were used. The results were comparable in both mating types (h−; data not shown for brevity). Like GFP-tagged WT Pfh1p, GFP-tagged Pfh1p-K338A localized correctly to nuclei and mitochondria (data not shown).
For heterologous complementation experiments, strains carrying the loxP-pfh1-kanMX6-loxP cassette at the endogenous pfh1 locus were grown to log phase in supplemented liquid EMM and transformed with either pREP82-cre (58) or pREP82-cre-Y324F, a point mutation that abolishes the catalytic activity of Cre recombinase, by using a rapid transformation protocol (36). Transformation plates with pfh1-mt* strains were first incubated at 18°C for 6 days and then at 30°C for 3 days. For drug sensitivity assays (see Fig. 7A), pfh1+ and pfh1-mt* strains carrying pJR41XH or pJR41XH-ScRRM3 plasmids were plated on EMM with glutamate (http://www-rcf.usc.edu/∼forsburg/media.html) containing the indicated concentrations of HU or bleomycin (BM) and incubated at 32°C for 4 days.
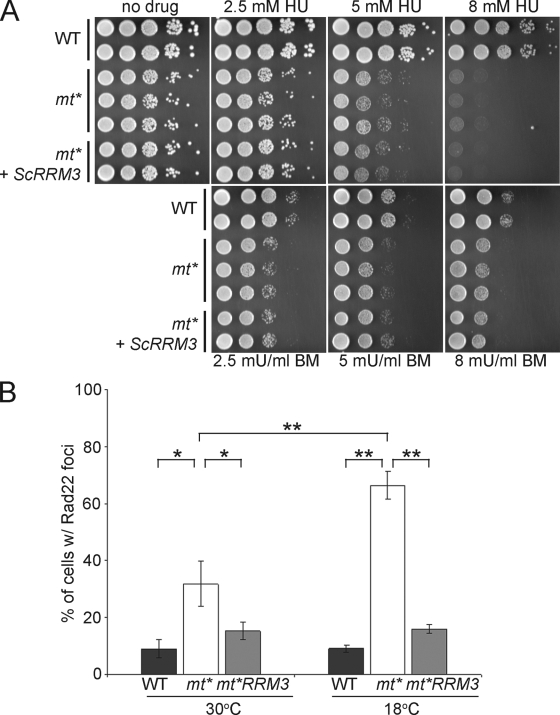
FIG. 7.
ScRrm3p suppresses spontaneous DNA damage foci but not DNA damage sensitivity. (A) Log-phase WT, pfh1-mt* (mt*) and pfh1-mt* expressing ScRrm3p (mt*+ScRRM3) cells were spotted at 10-fold serial dilutions on plates with the indicated concentrations of HU or BM. (B) Log-phase WT, pfh1-mt* (mt*), and pfh1-mt* expressing ScRrm3p (mt*+RRM3) cells were visualized by fluorescence microscopy, and the percentage of cells with Rad22p-RFP foci was calculated. Brackets with asterisks denote statistically significant differences (Student t test) between strains: P < 0.02 (*) and P < 0.002 (**).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis, immunoblotting, and Southern analysis.
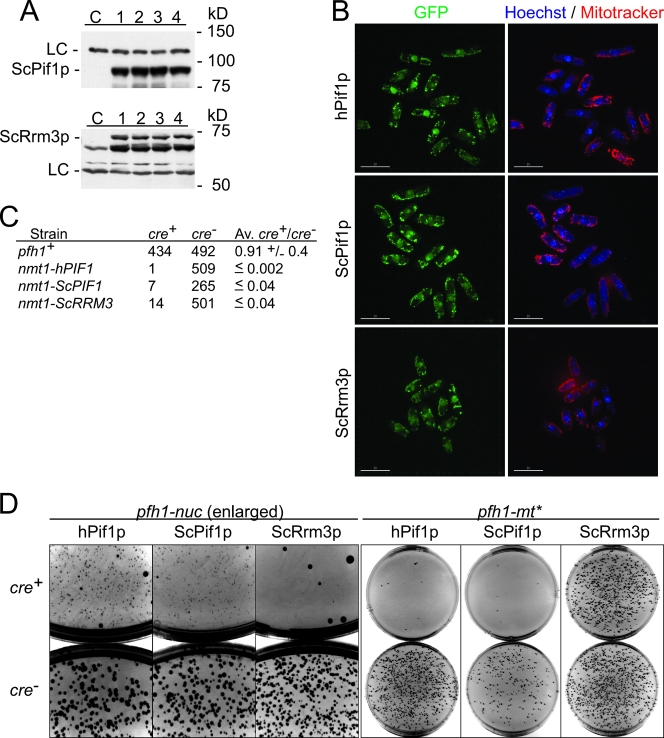
Whole-cell extracts were prepared by glass-bead lysis in modified HB buffer (25 mM HEPES [pH 7.2], 60 mM β-glycerophosphate, 15 mM MgC12, 15 mM EGTA, 1 mM dithiothreitol, 0.1 mM sodium vanadate, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride) containing protease inhibitor cocktail (Roche). Blots were probed with rabbit anti-Pfh1p polyclonal serum raised against the helicase domain of Pfh1p (61) or rabbit anti-Rdp1 polyclonal serum (Abcam) and horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G polyclonal serum (Bio-Rad). For heterologous complementation analyses, expression of ScPif1p and ScRrm3p was confirmed by Western analysis (see Fig. 6A) using anti-Pif1p and anti-Rrm3p polyclonal sera (23). Expression of untagged hPIF1p could not be confirmed by Western analysis since it comigrated with a nonspecific background band (data not shown). However, hPIF1p-GFP was readily detected by fluorescence microscopy (see Fig. 6B).
FIG. 6.
ScRrm3p supplies the essential nuclear function(s) of Pfh1p. (A) Anti-ScPif1p and anti-ScRrm3p Western blots of independent clones (lanes 1 to 4) expressing ScPif1p, ScRrm3p, or no exogenous protein (lane C). The loading control (LC) is a nonspecific background band. (B) Fluorescence microscopy of cells expressing hPIF1p-GFP, ScPif1p-GFP, or ScRrm3p-GFP. GFP, green; Hoechst 33342-stained DNA (blue), and MitoTracker CM-H2Xros-stained mitochondria (red). (C) Cross complementation assay in pfh1::loxP-pfh1-kanMX6-loxP strain with Pfh1p (pfh1+)-, hPIF1p (nmt1-hPIF1)-, ScPif1p (nmt1-ScPIF1)-, and ScRrm3p (nmt1-ScRRM3)-expressing strains after transformation with plasmid expressing either cre (cre+) or cre-Y324F (cre−) recombinase. The numbers of colonies and averages derived from at least three independent trials are indicated. (D) Contrast-inverted pictures of transformation plates with cre+ plates on the top and cre− plates on the bottom. The heterologous expression constructs (hPIF1p, ScPif1p, or ScRrm3p) and pfh1 alleles are indicated.
Phenol-chloroform extraction of genomic DNA was carried out as previously described (4). HindIII (NEB)-digested DNA was resolved on 1% agarose gels in Tris-borate-EDTA. XhoI (NEB)-digested DNA was resolved on 0.6% agarose gels in Tris-acetate-EDTA as previously described (14). Blots were hybridized with radiolabeled pDG3 (12), which contains the entire S. pombe mtDNA in pBR322, or with radiolabeled EcoNI-PstI fragment from pAF1 (39), which contains the 3′ end of the his3+ gene.
Microscopy.
For phase contrast and fluorescence microscopy using a DeltaVision microscope workstation (Applied Precision), cells were grown in supplemented EMM to log phase and stained with 1 μg of Hoechst 33342 dye (Invitrogen)/ml and 0.1 μg of MitoTracker Red CM-H2XRos (Invitrogen)/ml for 30 min before imaging. A total of 24 sections at 0.15-μm intervals were collected and deconvolved, and 16 central sections (2.4 μm) were flattened into a single image using the maximum-intensity algorithm (softWoRx; Applied Precision) to optimize sensitivity. For imaging of Rad22p-RFP foci, cells were grown to log phase at 30°C or shifted to 18°C for 18 h (Fig. 7B). Fourteen sections at 0.15-μm intervals were collected, deconvolved, and flattened (2.1 μm) into a single image using the average intensity algorithm to permit visualization of DNA damage foci while detecting general nuclear fluorescence.
Quantitative PCR.
Genomic DNA extraction was carried out as previously published (20). Quantitation of relative mtDNA content was performed on an iCycler iQ real-time PCR detection system (Bio-Rad). mtDNA levels were assayed by amplification of cox1 sequence and normalized to nuclear DNA, represented by amplification of rga7 sequence (primer sequences are available on request).
RESULTS
Pfh1p exists in multiple isoforms.
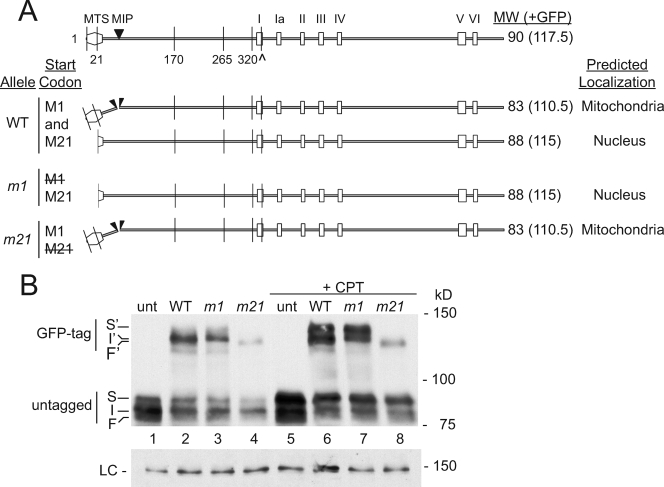
ScPIF1 gives rise to two isoforms of distinct functions by virtue of alternative translational initiation at the first or second in-frame AUG codon (44, 60). A mitochondrial targeting signal (MTS) is present between the first and second AUG. As a result, translation from the first AUG results in a ScPif1p isoform that is targeted to mitochondria, while translation from the second produces the nuclear isoform. Since sequence analysis of the pfh1+ gene predicted an MTS in the amino terminus of the Pfh1p polypeptide (61) (Fig. 1A), we mutated the first and second AUG codons to see whether there are multiple isoforms of Pfh1p.
FIG. 1.
Pfh1p is present in three distinct isoforms. (A) Schematic of Pfh1p ORF and predicted isoforms drawn to scale. MTS, predicted mitochondrial targeting signal; MIP, predicted mitochondrial import processing site. Methionine codons upstream of helicase domain are denoted by amino acid positions 1, 21, 170, 265, and 320. Helicase motifs are indicated by roman numerals; the invariant lysine K338 in helicase motif I (denoted by ^) is mutated to A338 in the catalytically dead (cd) allele. The predicted translation products from methionine codons M1 and M21 in WT and mutant strains are indicated. (B) Whole-cell anti-Pfh1p Western blot in untagged and GFP-tagged pfh1 strains. unt, pfh1+; WT, pfh1+-GFP; m1, pfh1-m1-GFP; m21, pfh1-m21-GFP. Here and in subsequent figures, S, I, and F denote top, middle, and bottom Pfh1p bands. S′, I′, and F′ denote top, middle, and bottom Pfh1p-GFP bands. The faint band below (F) is seen in a subset of gels and is likely a degradation product. +CPT, camptothecin. LC, loading control blot probed with anti-Rdp1 polyclonal serum. Molecular mass markers here and in subsequent figures are given in kilodaltons.
Translation of full-length Pfh1p from the first AUG codon (M1) will generate an ∼90-kDa precursor with an MTS, whose cleavage upon import into mitochondria will yield an ∼83-kDa mitochondrial isoform (Fig. 1A). If the second in-frame AUG codon (M21) acts as an alternative translation initiation site, the resulting protein of ∼88 kDa will lack a functional MTS and be excluded from mitochondria. We used site-directed mutagenesis to change either the first (M1) or the second (M21) methionine codon in the Pfh1p open reading frame (ORF) to alanine codons to generate, respectively, the pfh1-m1 and pfh1-m21 alleles (Fig. 1A). The wild-type (WT), m1, and m21 alleles were expressed as carboxy-terminal fusions to GFP, adding 27.5 kDa to their mass. Thus, the predicted molecular masses of the fusion proteins were ∼110.5 kDa for the WT and m21 strains (translated from M1) and ∼115 kDa for the WT and m1 strains (translated from M21) (Fig. 1A). These fusions were integrated at the leu1-32 locus but expressed from the pfh1+ promoter in a strain that also expressed untagged Pfh1p.
In the strain containing only untagged pfh1+ (Fig. 1B, lane 1, unt), immunoblotting revealed a major band of intermediate mobility (I), as well as incompletely resolved slower (S)- and faster (F)-migrating forms, with smearing between the bands (Fig. 1B, lane 1, unt). The intensity of band S increased dramatically after induction of DNA damage (camptothecin treated, lane 5, +CPT) (57). GFP-tagged WT Pfh1p (lane 2, WT) was also detected as a broad band. The fusion protein expressed from the m1 allele (lane 3, m1) produced a more discrete, band with the mobility of the upper half (I′) of the broad WT Pfh1p-GFP band (lane 2), while the fusion protein in the m21 strain migrated faster (F′) (lane 4, m21). These data suggest that the broad band generated by WT Pfh1p-GFP (lane 2) is a composite of at least two species with different mobilities (F′ and I′). The small differences in the mobility of the isoforms were more evident after DNA damage (+CPT; see lanes 7 and 8). Because the slow-migrating S′ form was detected in the m1 strain (lane 7, m1) but not in the m21 strain (lane 8, m21) and the intensity of S and S′ forms increased after DNA damage (lanes 5 to 8), S and S′ are likely nonmitochondrial forms of Pfh1p that are posttranslationally modified in response to DNA damage. This hypothesis is supported by additional experiments as described below.
Pfh1p localizes to nuclei and mitochondria.
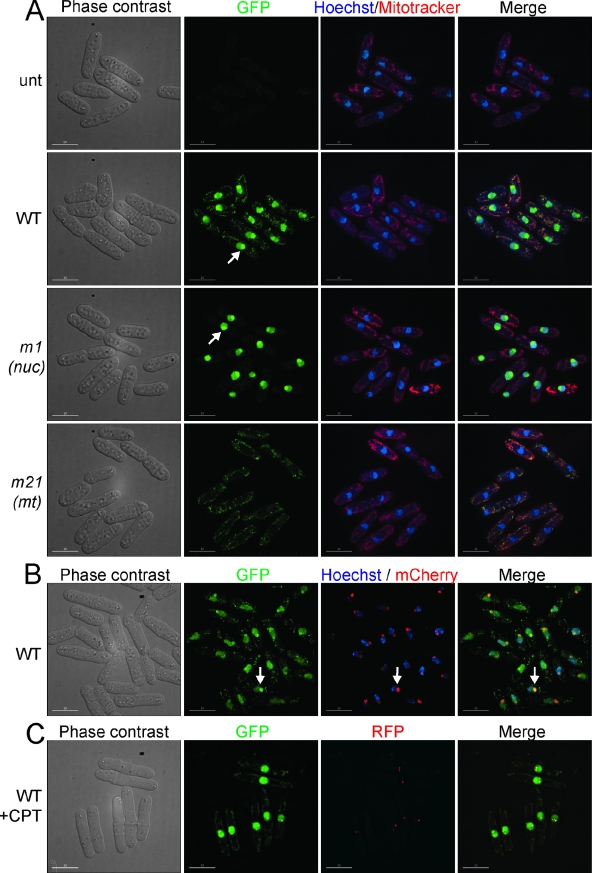
We determined the subcellular localization of GFP-tagged Pfh1p generated from the WT, pfh1-m1, and pfh1-m21 alleles in living cells (Fig. 2A). As in the Western analysis (Fig. 1B), all strains also expressed untagged WT Pfh1p. In the strain expressing GFP-tagged WT Pfh1p, fluorescence was detected in both nuclei and mitochondria (Fig. 2A, WT). In the m1 strain, fluorescence was detected exclusively in nuclei, whereas in the m21 strain, fluorescence was detected only in mitochondria (Fig. 2A, compare m1 to m21). These results indicate that alternative start codon usage generates two Pfh1p isoforms that localize to different subcellular compartments. To reflect their respective localization, the pfh1-m1 allele is hereafter called pfh1-nuc (nuclear) and the pfh1-m21 allele is called pfh1-mt (mitochondrial).
FIG. 2.
Pfh1p localizes to the nucleus and mitochondria. (A) Phase-contrast and fluorescence microscopy of cells expressing WT or mutant Pfh1p fused to GFP (green). DNA is visualized with Hoechst 33342 (blue), and mitochondria are visualized with MitoTracker CM-H2Xros (red). unt, pfh1+; WT, pfh1+-GFP; m1 (also referred to as pfh1-nuc), pfh1-m1-GFP; m21 (also referred to as pfh1-mt), pfh1-m21-GFP. Here and in subsequent figures, white bars in the lower left corners denote 10 μm. (B) Phase-contrast and fluorescence microscopy of Pfh1p-GFP (green) and Gar2p-mCherry (red) for visualization of nucleolus and DNA (Hoechst 33342) (blue). Arrows indicate nucleolar Pfh1p-GFP signal. (C) Phase-contrast and fluorescence microscopy of Pfh1p-GFP (green) and Rad22p-RFP (red). The apparent reduction in the nucleolar signal here compared to panel B is likely due to a different z-stack projection algorithm (see Materials and Methods). +CPT, camptothecin.
In many cells carrying the WT and nuc alleles (Fig. 2A, WT and m1), fluorescence was concentrated in a section of the nucleus that was poorly stained by Hoechst, a known characteristic of the S. pombe nucleolus (Fig. 2A, WT, m1, arrows) (54). To determine whether nuclear Pfh1p-GFP was concentrated in nucleoli, we generated a fusion between the nucleolar protein Gar2p and mCherry. Since the two fusion proteins, Gar2p-mCherry and WT Pfh1p-GFP, colocalized (Fig. 2B), we conclude that nuclear Pfh1p is found throughout the nucleus but is concentrated in the nucleolus.
ScPif1p, but not ScRrm3p, localizes to Rad52p DNA damage foci (56). To determine whether Pfh1p also localizes to DNA damage foci, we used a strain that expressed both GFP-tagged WT Pfh1p and RFP-tagged Rad22p. Like its S. cerevisiae homolog, Rad52p, the S. pombe Rad22p plays a critical role in double-stranded break repair (13) and forms DNA damage foci in response to DNA damage (34). After camptothecin treatment, WT Pfh1p-GFP colocalized with Rad22p-RFP in distinct nuclear foci (Fig. 2C). This result suggests that nuclear Pfh1p functions in DNA repair.
Pfh1p is essential in nuclei and mitochondria.
Since pfh1Δ spore progeny (61) and strains with cold-sensitive pfh1 alleles grown at restrictive temperatures (48) exhibit a G2 cell cycle arrest, we expected the nuclear function(s) of Pfh1p to be essential. However, it is not clear whether the mitochondrial isoform is also essential. To identify the essential function(s) of Pfh1p, we used a plasmid loss assay (19). We tested for the ability of a pfh1 allele integrated at leu1-32 to support growth of pfh1Δ cells after loss of a covering pfh1+ his3+plasmid. Integration of the empty vector control (VEC) resulted in exclusively His+ colonies (Table 2, line 1), indicating that these cells do not retain viability if they lose the covering plasmid. This result confirms the essential nature of pfh1+ (61). As expected, integration of pfh1+ (WT) generated exclusively His− colonies (Table 2, line 2). Also, as expected, the catalytically dead (cd) pfh1-K338A allele (Fig. 1A) did not complement the pfh1Δ strain (Table 2, line 3). These results are consistent with previously published data and validate the plasmid loss assay (61).
TABLE 2.
Pfh1p is essential in the nucleus and the mitochondria
| Line | Strain (leu1-32::) | Temp (°C) | No. of colonies
|
% His− coloniesa | Complementationb | |
|---|---|---|---|---|---|---|
| His+ | Total | |||||
| 1 | VEC | 30 | 216 | 216 | ≤0.5 | N |
| 2 | WT | 30 | 0 | 511 | ≥99.8 | Y |
| 3 | cd | 30 | 156 | 161 | ≤3.7 | N |
| 4 | nuc | 30 | 117 | 118 | ≤1.7 | N |
| 5 | mt | 30 | 0 | 458 | ≥99.8 | Y |
| 6 | mt* | 30 | 413 | 688 | 40 ± 3 | I |
| 7 | mt* | 18 | 402 | 407 | ≤2.0 | N |
| 8 | WT | 18 | 0 | 377 | ≥99.7 | Y |
| 9 | mt*×VEC | 18 | 651 | 672 | ≤3.3 | N |
| 10 | mt*×mt* | 18 | 734 | 747 | ≤1.9 | N |
| 11 | mt*×WT | 18 | 0 | 558 | ≥99.8 | Y |
| 12 | mt*×nuc | 18 | 0 | 670 | ≥99.9 | Y |
| 13 | mt*×cd | 18 | 531 | 531 | ≤0.2 | N |
| 14 | nuc×VEC | 30 | 182 | 182 | ≤0.6 | N |
| 15 | nuc×WT | 30 | 0 | 510 | ≥99.8 | Y |
| 16 | nuc×cd | 30 | 152 | 159 | ≤5.0 | N |
Percentages of colonies that lost the his3+ marked pBG2-pfh1+ plasmid were rounded to the nearest 1/10 of a percent.
Y, yes; N, no; I, intermediate.
The pfh1-nuc allele (nuc) did not complement the pfh1Δ allele, indicating that the mitochondrial isoform of Pfh1p is essential (Table 2, line 4). In contrast, the pfh1-mt allele (mt) did complement pfh1Δ (Table 2, line 5). At face value, this result argues that the nuclear isoform of Pfh1p is not essential, a surprising result given that pfh1− cells arrest in G2 phase (48, 61). To resolve this contradiction, we considered that the pfh1-mt allele might give rise to some nuclear Pfh1p, even though nuclear GFP was not detected in cells expressing this allele (Fig. 2A, pfh1-mt). To test this possibility, we further modified the pfh1-mt allele with the goal of reducing its potential to generate nuclear localized Pfh1p. We mutated three additional methionine codons upstream of the helicase domain to alanine (M265 and M320) or leucine (M170) codons. In addition, we inserted the NES from the human immunodeficiency virus Rev protein, which functions in S. pombe (26), between the pfh1+ and GFP ORFs. This multiply altered allele is called pfh1-mt*.
When pfh1-mt* cells were grown at 30°C, they produced an intermediate phenotype in the plasmid loss assay (40% His− colonies; Table 2, line 6). However, when this strain was grown at 18°C, the pfh1-mt* allele did not complement pfh1Δ (402/407 His+ cells; Table 2, line 7). In contrast, pfh1+ cells (WT) readily lost the covering plasmid at 18°C (0 of 377 colonies were His+; Table 2, line 8). These data suggest that viability is reduced when nuclear Pfh1p activity is very low, and this reduced viability is exacerbated at low temperatures. To provide further support for this hypothesis, we tested the functions of the pfh1 mutant alleles at 18°C in diploid strains that coexpressed two different alleles (Table 2, lines 9 to 16). As predicted, combination of the pfh1-mt* allele with the empty vector (mt* × VEC) or a second pfh1-mt* allele (mt* × mt*) did not complement the pfh1Δ strain at 18°C (Table 2, lines 9 and 10). Also, as expected, complementation was seen in the mt* × WT diploid. Importantly, combination of the pfh1-mt* allele with the pfh1-nuc allele (mt* × nuc) fully complemented the pfh1Δ strain at 18°C, arguing that the pfh1-mt* strain was indeed deficient in nuclear Pfh1p function (Table 2, line 12). In addition, because the pfh1-mt* allele did not complement the catalytically inactive pfh1-K338A allele (mt* × cd) (Table 2, line 13), the ATPase/helicase activity of Pfh1p was essential in the nucleus. Finally, combination of the pfh1-nuc allele with the pfh1+ allele (nuc × WT) complemented the pfh1Δ allele (Table 2, line 15), but the pfh1-nuc allele in combination with the pfh1-K338A allele (nuc × cd) did not (Table 2, line 16). Therefore, the ATPase/helicase activity of Pfh1p was also essential in mitochondria. We conclude that the catalytic activity of Pfh1p is required for viability in both nuclei and mitochondria.
Nuclear Pfh1p is sufficient to permit cell cycle progression.
To extend the analysis of the phenotypes of cells lacking different Pfh1p isoforms, we developed a regulated Pfh1p expression system, based on the thiamine-repressible nmt1 promoter (33). To minimize the basal level of Pfh1p, we exploited the observation that the original pfh1-mt allele generated a small amount of nuclear Pfh1p (Table 2), as well as normal levels of mitochondrial Pfh1p (Fig. 1B and 2A). The endogenous pfh1+ locus was modified by integration of the pfh1-mt allele under the control of the weakest nmt1 promoter (35). In addition, empty vector, pfh1+, or pfh1-nuc was integrated at the leu1-32 locus. Hereafter, these strains are referred to by the pfh1 allele integrated at the leu1-32 locus (VEC, WT, or NUC).
Depending on the medium, the VEC strain will express mitochondrial Pfh1p (no thiamine) or no Pfh1p at all (+ thiamine). The NUC strain will produce both nuclear and mitochondrial Pfh1p (no thiamine) or only nuclear Pfh1p (+ thiamine). WT cells will express nuclear and mitochondrial Pfh1p regardless of thiamine supplementation. These predictions were supported by Western analysis (Fig. 3A): the VEC strain expressed only the mitochondrial isoform (F) (no thiamine, lane 2) or no Pfh1p (+ thiamine, lane 5), the NUC strain expressed all isoforms (S, I, and F) in the absence of thiamine (lane 3) but did not express the mitochondrial isoform (F) in thiamine supplemented medium (lane 6), and the WT strain expressed all three Pfh1p isoforms (S, I, and F) in both growth conditions (lanes 1 and 4). This result provides further evidence that the fastest-migrating species (F) is the mitochondrial isoform, while I and S are nuclear isoforms.
Each of the strains was also examined microscopically at 0 and 24 h after addition of thiamine (Fig. 3B). As expected, the morphology of the WT strain did not change in the presence of thiamine (Fig. 3B, WT). Although VEC cells were normal at 0 h, at 24 h, they were elongated with a single nucleus (Fig. 3B, VEC), a morphology similar to that of pfh1Δ spore progeny (61) or cold-sensitive pfh1 cells at restrictive temperature (48). This result supports the interpretation that cells expressing the pfh1-mt allele produce enough nuclear localized Pfh1p to prevent cell cycle arrest since these cells did not elongate in the absence of thiamine (Fig. 3B, VEC, 0 h). Moreover, since the NUC strain, which expressed only nuclear Pfh1p in the presence of thiamine (Fig. 3A), did not display an elongated cell phenotype, nuclear Pfh1p was sufficient to prevent the G2 arrest observed in VEC cells. Finally, although NUC cells did not elongate after ≥24 h in thiamine (Fig. 3B, data not shown), the intensity of mitochondrial nucleoids was greatly diminished after growth for 24 h in thiamine (insets, Fig. 3B). This observation was further supported by real-time PCR and Southern blotting results as described below (Fig. 5).
We also determined the growth rates of these strains (Fig. 3C). In the absence of thiamine, WT, VEC, and NUC strains had similar growth rates with ∼4 population doublings in the 12-h period preceding thiamine addition. The growth rate of the WT strain did not change even after 72 h in thiamine. Consistent with the G2 arrest observed at 24 h (Fig. 3C), the growth rate of the VEC strain declined precipitously after only 12 h in thiamine and, at 72 h, its division rate was ∼1 doubling in 12 h. The growth rate of the NUC strain did not decline until 36 h in thiamine and then dropped dramatically until it plateaued at ∼1.8 population doublings per 12-h period. Thus, in cells expressing only nuclear Pfh1p (NUC), the decline in growth rate was preceded by the loss of mitochondrial nucleoids (Fig. 3B). Although the growth rate of cells that lack Pfh1p altogether (VEC) declined earlier than cells expressing only nuclear Pfh1p (NUC), the terminal phenotype for both strains was very slow growth.
Telomere length does not change in Pfh1p-depleted cells.
Because S. cerevisiae Pif1p removes telomerase from DNA ends (5), pif1Δ cells have long telomeres (44). Although ScRrm3p affects fork progression through telomeric DNA, its depletion results in only a very modest telomere lengthening and, unlike pif1Δ, does not increase the rate of telomere addition to double-strand breaks (22). We used Southern blotting to determine whether telomere length was affected in cells depleted of Pfh1p. Although by Western blotting nuclear Pfh1p was undetectable after 12 h of growth in thiamine medium (Fig. 3D), telomere length was not markedly affected, even after 60 h without Pfh1p (Fig. 3E).
Nuclear Pfh1p is required to prevent and/or to repair DNA damage.
Cold-sensitive pfh1 strains are sensitive to HU and methyl methanesulfonate, suggesting that Pfh1p plays a role in DNA repair (48). To test whether the G2 arrest in cells depleted of nuclear Pfh1p is associated with persistent DNA damage, we combined our regulated Pfh1p expression system with a rad22-RFP fusion (11). We monitored Rad22p foci in living WT, VEC (Fig. 4A), and NUC cells (data not shown) 24 h after the addition of thiamine. Consistent with published reports (34), Rad22p foci were rare in WT cells since fewer than 10% of WT cells had foci, and these foci were faint (inset, Fig. 4A). Rad22p foci were similarly rare in NUC cells, which expressed nuclear Pfh1p. In contrast, 80% of VEC cells had one to four intense Rad22p foci (Fig. 4B and C) (see inset, Fig. 4A). Based on their elongated cellular morphology, and large number of Rad22 foci, we conclude that cells depleted of nuclear Pfh1p suffer persistent DNA damage, and this damage triggers a G2-phase arrest.
Mitochondrial Pfh1p is required for the maintenance of mtDNA.
We used real-time PCR to determine the amount of mtDNA in Pfh1p-depleted cells. Loss of mitochondrial Pfh1p in the NUC strain resulted in a rapid decrease in mtDNA since NUC cells contained less than 20% of WT levels within 24 h of thiamine addition (Fig. 5A, NUC). mtDNA content also decreased in the VEC strain, which lacked all Pfh1p, although the rate of decline was slower than in the NUC strain (Fig. 5A, VEC). This slower decline is probably due to the earlier onset of slow growth after thiamine addition in VEC cells (Fig. 3B).
In S. cerevisiae, the mtDNA in respiratory-deficient cells is often rearranged (10). To examine the structure of the remaining mtDNA in cells depleted of mitochondrial Pfh1p, we isolated DNA from WT and NUC cells at different times after thiamine addition. The DNA was analyzed by Southern blotting after digestion with HindIII (cleaves 10 times in S. pombe mtDNA; Fig. 5B) or with XhoI (cleaves once; Fig. 5C). For the remaining mtDNA in cells depleted of mitochondrial Pfh1p the DNA cleavage pattern was indistinguishable from that of WT mtDNA. In addition, no mtDNA was detected in the wells in the XhoI blot, as would be expected for unresolved mtDNA recombination intermediates (14). Thus, in the absence of mitochondrial Pfh1p, mtDNA is lost but not rearranged.
ScRrm3p provides the essential nuclear function(s) of Pfh1p.
When expressed in S. pombe, GFP-tagged hPIF1p, ScPif1p, and ScRrm3p (Fig. 6A) each localized to both nuclei and mitochondria (Fig. 6B). To determine whether the human or the S. cerevisiae Pif-like proteins could provide the essential functions of S. pombe pfh1+, we expressed untagged hPIF1p, ScPif1p, and ScRrm3p from the intermediate-strength nmt1 promoter (35) and integrated the constructs at the leu1-32 locus. In these cells, S. pombe Pfh1p was expressed from its endogenous locus, where it was flanked by loxP sites. Recombination between the two loxP sites can be induced by expression of the Cre recombinase, which will excise the DNA between the sites to generate a pfh1Δ cell. A ura4+ plasmid carrying Cre (58) was introduced by transformation into cells expressing hPIF1p, ScPif1p or ScRrm3p. If any of these helicases were able to complement the pfh1Δ allele, Ura+ colonies will be produced after introduction of the cre+ plasmid. To control for transformation efficiency, each strain was independently transformed with the ura4+ plasmid carrying a catalytically inactive Cre (called cre−).
Strains expressing Pfh1p, hPIF1p, ScPif1p, or ScRrm3p were transformed with either the cre+ or cre− plasmids and plated onto medium lacking uracil. In the positive control strain (pfh1+), comparable numbers of Ura+ transformants were recovered after transformation with either plasmid (Fig. 6C). However, strains expressing hPIF1p, ScPif1p, or ScRrm3p produced large numbers of transformants only when transformed with the cre− plasmid (Fig. 6C). Thus, despite their correct subcellular localization (Fig. 6B), neither hPIF1p, ScPif1p, nor ScRrm3p could supply all essential Pfh1p functions in S. pombe.
Next we tested whether hPIF1p, ScPif1p, or ScRrm3p could perform either the nuclear or the mitochondrial functions of Pfh1p. To this end, pfh1+ (WT), pfh1-nuc, and pfh1-mt* alleles were integrated at his3-D1 to supply either both Pfh1p functions (WT) or only its nuclear (nuc) or mitochondrial function(s) (mt*). All strains produced comparable numbers of Ura+ colonies after transformation with the cre+ and cre− plasmids if they carried the pfh1+ gene (data not shown). Strains with the pfh1-nuc allele did not produce cre+ transformants if they expressed ScRrm3p (Fig. 6D) but produced many small colonies when they expressed hPIF1p or ScPif1p. However, these colonies did not grow upon restreaking (data not shown). Higher-level expression of hPIF1p or ScPif1p from an extrachromosomal plasmid produced slightly larger colonies after transformation, but again these colonies were not viable after restreaking (data not shown). Finally, hPIF1p or ScPif1p did not produce cre+ transformants in the pfh1-mt* strain, indicating that they could not provide the essential nuclear function(s) of Pfh1p. However, expression of ScRrm3p in pfh1-mt* cells produced many cre+ transformants. Thus, when expressed in S. pombe, ScRrm3p can provide the essential nuclear function(s) of Pfh1p (Fig. 6D).
ScRrm3p suppresses spontaneous DNA damage in cells lacking nuclear Pfh1p but not sensitivity to DNA-damaging agents.
To determine the functional overlap of ScRrm3p and nuclear Pfh1p, we tested whether ScRrm3p suppressed the phenotypes of pfh1-mt* cells. Like previously characterized cold-sensitive pfh1 mutants (48), the pfh1-mt* strain, which is deficient in nuclear Phf1p, was sensitive to HU and BM (Fig. 7A). Although ScRrm3p expression suppressed the cold sensitivity of pfh1-mt* cells (Fig. 6D), it did not suppress their HU and BM sensitivity (Fig. 7A). Therefore, ScRrm3p is unable to supply the DNA repair function of nuclear Pfh1p.
Cells lacking nuclear Pfh1p often contain spontaneous DNA damage foci (Fig. 4). To determine whether ScRrm3p could suppress the formation of these foci, we generated a pfh1-mt* rad22-RFP strain that also expressed ScRrm3p. As seen before, Rad22p-RFP foci were rare in WT pfh1+ cells (Fig. 7B) and frequent in pfh1-mt* cells, especially at 18°C. At both 18 and 30°C, ScRrm3p expression reduced the number of DNA damage foci in pfh1-mt* cells to near WT levels (Fig. 7B). The frequent presence of DNA damage foci in Pfh1p-depleted cells suggests that some aspect of chromosomal DNA replication is Pfh1p dependent and that it is this Pfh1p activity, not its repair function, that can be carried out by ScRrm3p.
DISCUSSION
We demonstrate that there are two differently sized isoforms of S. pombe Pfh1p that are likely generated by alternative start codon usage (Fig. 1). The smaller isoform, produced by translation from the first AUG codon, localized to mitochondria (Fig. 1 and 2), while the larger isoform, generated from the second AUG codon, localized to nuclei (Fig. 1 and 2). A similar mechanism is responsible for generating the nuclear and mitochondrial isoforms of ScPif1p (44, 60). Moreover, the ATPase/helicase activity of Pfh1p is essential in both nuclei and mitochondria (Table 2). The pfh1-mt allele, which should produce only the smaller mitochondrial isoform, was limited to mitochondria by cytological criteria (Fig. 2A), and yet it produced enough nuclear protein to supply its essential function(s) in this compartment. Although it is unclear how pfh1-mt gives rise to nuclear localized Pfh1p, this behavior is reminiscent of the residual nuclear function of the pif1-m2 “mitochondrial only” S. cerevisiae allele (37, 44).
A slower-migrating species (S) of nuclear Pfh1p increased in abundance in response to DNA damage (Fig. 1). Similarly, slow-mobility forms of ScPif1p are seen after DNA damage in S. cerevisiae (L. Vega and V. A. Zakian, unpublished data). We propose that these isoforms (S and S′) are generated by posttranslational modification of nuclear Pfh1p and function in DNA repair. A role for Pfh1p in DNA repair is also suggested by the colocalization of GFP-tagged WT Pfh1p to DNA damage foci in response to exogenous DNA damage (Fig. 2). In S. cerevisiae, Pif1p, but not Rrm3p, localizes to DNA damage foci after gamma irradiation (56). Therefore, Pfh1p and ScPif1p may perform similar functions in DNA repair. The repair function of Pfh1p is probably not essential because expression of ScRrm3p in cells lacking nuclear Pfh1p function restored viability (Fig. 6D) but did not suppress HU sensitivity (Fig. 7A).
The absence of nuclear Pfh1p resulted in a G2-phase arrest and induction of spontaneous DNA damage foci in the absence of exogenous DNA damage (Fig. 3 and 4). This result suggests that replication of chromosomal DNA in the absence of Pfh1p creates lesions that are detected as damaged DNA. Thus, nuclear Pfh1p likely functions not only in DNA repair but also in replication of chromosomal DNA. Spontaneous DNA damage foci are also observed in S. cerevisiae rrm3Δ cells but not in pif1Δ cells (1). Pfh1p depletion had little or no effect on telomere length, suggesting that Pfh1p, unlike ScPif1p, does not inhibit telomerase (Fig. 3E). However, the essential role of Pfh1p in chromosome replication might mask a nonessential telomere function, given that many mutations that affect telomere length show phenotypic lag (30).
The best clue to the nature of the essential nuclear function(s) of Pfh1p is that it can be supplied by ScRrm3p (Fig. 6D). Moreover, ScRrm3p expression suppressed the formation of spontaneous DNA damage foci in Pfh1p-depleted cells (Fig. 7B). Taken together, these data suggest that Pfh1p plays an essential ScRrm3p-like role in the replication of chromosomal DNA. In S. cerevisiae, Rrm3p promotes fork progression past over 1,000 stable protein-DNA complexes. Although ScRrm3p-sensitive sites are scattered throughout the genome, more than half of these sites are in the rDNA (21). The concentration of nuclear Pfh1p in nucleoli (Fig. 2) suggests that Pfh1p, like ScRrm3p, may be particularly important for rDNA replication. Alternatively (or in addition), nuclear Pfh1p may play a role in Okazaki fragment processing, since S. pombe pfh1+, as well as S. cerevisiae PIF1 and RRM3, interact genetically with dna2+/DNA2 (8, 9, 42), which encodes an essential 5′-3′ DNA helicase/endonuclease involved in 5′ flap cleavage (24).
Pfh1p was also essential in mitochondria (Table 2), where its depletion resulted in rapid loss of mtDNA and very slow growth (Fig. 3 and 5). Of the eight helicases that localize to S. pombe mitochondria (32), six are likely RNA helicases (2). Therefore, Pfh1p may be one of the (or even the only) replicative helicase for S. pombe mtDNA. In S. cerevisiae, pif1Δ cells frequently generate respiratory deficient cells and lose mtDNA altogether at high temperatures (28, 55). Nonetheless, ScPif1p was unable to substitute for mitochondrial Pfh1p, even though it localized properly to S. pombe mitochondria (Fig. 6). However, the mitochondrial genomes of these two yeasts are very different both in size and in AT content (17, 29), which may explain why ScPif1p cannot provide the mitochondrial Pfh1p functions. The hPIF1p helicase localizes to human (18) and S. pombe mitochondria (Fig. 6) and was partially able to supply the mitochondrial functions of Pfh1p (Fig. 6). Although mPif1p cannot be essential for replication of mtDNA since mpif1−/− mice are viable (45), mammalian Pif1-like proteins may contribute to replication of mtDNA, perhaps in concert with Twinkle, a 5′-3′ DNA helicase that is thought to be the replicative helicase for mammalian mtDNA (46, 53).
We conclude that the S. pombe Pfh1p DNA helicase is not functionally identical to either ScRrm3p or ScPif1p but rather shares functions with both. Like ScPif1p, Pfh1p is required for maintenance of mtDNA, while both ScRrm3p and Pfh1p have important roles in chromosomal replication. Although Rrm3p function is not essential in S. cerevisiae, the essential nuclear function(s) of Pfh1p in S. pombe can be supplied by ScRrm3p (Fig. 6). An appealing model is that replication of one or more loci in S. pombe is absolutely dependent on a ScRrm3p-like role in bypassing protein complexes. For example, ScRrm3p helps replication forks move through the ∼125-bp S. cerevisiae centromeres (21). If Pfh1p has a similar function, cells might not be able to compensate for its loss since the heterochromatic S. pombe centromeres are ≥35 kb in size (41).
Acknowledgments
We thank M. Rose and S. Clark for the mCherry-kanMX6 construct, C. Webb for assistance in constructing the gar2-mcherry strain, and N. Sedighi for initiating the heterologous complementation experiments. We thank J. C. Ribas for the pJR vector series; P. Russell for the rad22-RFP strain; T. Carr for the pREP82-cre plasmid; M. Mateyak for the hPIF1 cDNA; and A. Azvolinsky, C. Tuzon, and C. Webb for their critical readings of the manuscript.
This study was supported in part by a predoctoral fellowship to S.F.P. from the Susan G. Komen Breast Cancer Foundation, an NIH Cancer Training grant 5T32CA009528-22 (S.D.A.), and NIH grant R37GM 026938 to V.A.Z.
Footnotes
Published ahead of print on 25 August 2008.
REFERENCES
- 1.Alvaro, D., M. Lisby, and R. Rothstein. 2007. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 3e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aslett, M., and V. Wood. 2006. Gene Ontology annotation status of the fission yeast genome: preliminary coverage approaches 100%. Yeast 23913-919. [DOI] [PubMed] [Google Scholar]
- 3.Azvolinsky, A., S. Dunaway, J. Torres, J. Bessler, and V. A. Zakian. 2006. The Saccharomyces cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 203104-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beach, D. H., and A. J. Klar. 1984. Rearrangements of the transposable mating-type cassettes of fission yeast. EMBO J. 3603-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boule, J., L. Vega, and V. Zakian. 2005. The Yeast Pif1p helicase removes telomerase from DNA. Nature 43857-61. [DOI] [PubMed] [Google Scholar]
- 6.Boule, J. B., and V. A. Zakian. 2006. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 344147-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boule, J. B., and V. A. Zakian. 2007. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 355809-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budd, M. E., C. C. Reis, S. Smith, K. Myung, and J. L. Campbell. 2006. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol. Cell. Biol. 262490-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budd, M. E., A. H. Tong, P. Polaczek, X. Peng, C. Boone, and J. L. Campbell. 2005. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 1e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contamine, V., and M. Picard. 2000. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 64281-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulon, S., E. Noguchi, C. Noguchi, L. L. Du, T. M. Nakamura, and P. Russell. 2006. Rad22Rad52-dependent repair of ribosomal DNA repeats cleaved by Slx1-Slx4 endonuclease. Mol. Biol. Cell 172081-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Giudice, L. 1981. Cloning of mitochondrial DNA from the petite negative yeast Schizosaccharomyces pombe in the bacterial plasmid pBR322. Mol. Gen. Genet. 184465-470. [DOI] [PubMed] [Google Scholar]
- 13.Doe, C. L., F. Osman, J. Dixon, and M. C. Whitby. 2004. DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res. 325570-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doe, C. L., F. Osman, J. Dixon, and M. C. Whitby. 2000. The Holliday junction resolvase SpCCE1 prevents mitochondrial DNA aggregation in Schizosaccharomyces pombe. Mol. Gen. Genet. 263889-897. [DOI] [PubMed] [Google Scholar]
- 15.Foury, F., and J. Kolodynski. 1983. pif mutation blocks recombination between mitochondrial rho+ and rho− genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 805345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foury, F., and A. Lahaye. 1987. Cloning and sequencing of the PIF gene involved in repair and recombination of yeast mitochondrial DNA. EMBO J. 61441-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foury, F., T. Roganti, N. Lecrenier, and B. Purnelle. 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440325-331. [DOI] [PubMed] [Google Scholar]
- 18.Futami, K., A. Shimamoto, and Y. Furuichi. 2007. Mitochondrial and nuclear localization of human Pif1 helicase. Biol. Pharm. Bull. 301685-1692. [DOI] [PubMed] [Google Scholar]
- 19.Hayles, J., and P. Nurse. 1992. Genetics of the fission yeast Schizosaccharomyces pombe. Annu. Rev. Genet. 26373-402. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57267-272. [DOI] [PubMed] [Google Scholar]
- 21.Ivessa, A. S., B. A. Lenzmeier, J. B. Bessler, L. K. Goudsouzian, S. L. Schnakenberg, and V. A. Zakian. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell 121525-1536. [DOI] [PubMed] [Google Scholar]
- 22.Ivessa, A. S., J.-Q. Zhou, V. P. Schulz, E. M. Monson, and V. A. Zakian. 2002. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and sub-telomeric DNA. Genes Dev. 161383-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivessa, A. S., J.-Q. Zhou, and V. A. Zakian. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100479-489. [DOI] [PubMed] [Google Scholar]
- 24.Kao, H. I., and R. A. Bambara. 2003. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit. Rev. Biochem. Mol. Biol. 38433-452. [DOI] [PubMed] [Google Scholar]
- 25.Keil, R. L., and A. D. McWilliams. 1993. A gene with specific and global effects on recombination of sequences from tandemly repeated genes in Saccharomyces cerevisiae. Genetics 135711-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kudo, N., S. Khochbin, K. Nishi, K. Kitano, M. Yanagida, M. Yoshida, and S. Horinouchi. 1997. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J. Biol. Chem. 27229742-29751. [DOI] [PubMed] [Google Scholar]
- 27.Lahaye, A., S. Leterme, and F. Foury. 1993. PIF1 DNA helicase from Saccharomyces cerevisiae: biochemical characterization of the enzyme. J. Biol. Chem. 26826155-26161. [PubMed] [Google Scholar]
- 28.Lahaye, A., H. Stahl, D. Thines-Sempoux, and F. Foury. 1991. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 10997-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang, B. F. 1993. The mitochondrial genome of Schizosaccharomyces pombe, p. 3118-3119. In S. J. O'Brien (ed.), Genetic maps, 6th ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Lustig, A. J., and T. D. Petes. 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 831398-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mateyak, M., and V. Zakian. 2006. Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle 232796-2804. [DOI] [PubMed] [Google Scholar]
- 32.Matsuyama, A., R. Arai, Y. Yashiroda, A. Shirai, A. Kamata, S. Sekido, Y. Kobayashi, A. Hashimoto, M. Hamamoto, Y. Hiraoka, S. Horinouchi, and M. Yoshida. 2006. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24841-847. [DOI] [PubMed] [Google Scholar]
- 33.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123127-130. [DOI] [PubMed] [Google Scholar]
- 34.Meister, P., M. Poidevin, S. Francesconi, I. Tratner, P. Zarzov, and G. Baldacci. 2003. Nuclear factories for signalling and repairing DNA double strand breaks in living fission yeast. Nucleic Acids Res. 315064-5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno, M. B., A. Duran, and J. C. Ribas. 2000. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast 16861-872. [DOI] [PubMed] [Google Scholar]
- 36.Morita, T., and K. Takegawa. 2004. A simple and efficient procedure for transformation of Schizosaccharomyces pombe. Yeast 21613-617. [DOI] [PubMed] [Google Scholar]
- 37.Myung, K., C. Chen, and R. D. Kolodner. 2001. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 4111073-1076. [DOI] [PubMed] [Google Scholar]
- 38.O'Rourke, T. W., N. A. Doudican, H. Zhang, J. S. Eaton, P. W. Doetsch, and G. S. Shadel. 2005. Differential involvement of the related DNA helicases Pif1p and Rrm3p in mtDNA point mutagenesis and stability. Gene 35486-92. [DOI] [PubMed] [Google Scholar]
- 39.Ohi, R., A. Feoktistova, and K. L. Gould. 1996. Construction of vectors and a genomic library for use with his3-deficient strains of Schizosaccharomyces pombe. Gene 174315-318. [DOI] [PubMed] [Google Scholar]
- 40.Ooi, S. L., D. D. Shoemaker, and J. D. Boeke. 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35277-286. [DOI] [PubMed] [Google Scholar]
- 41.Pidoux, A. L., and R. C. Allshire. 2005. The role of heterochromatin in centromere function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360569-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryu, G. H., H. Tanaka, D. H. Kim, J. H. Kim, S. H. Bae, Y. N. Kwon, J. S. Rhee, S. A. MacNeill, and Y. S. Seo. 2004. Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast. Nucleic Acids Res. 324205-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt, K. H., and R. D. Kolodner. 2004. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 243213-3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schulz, V. P., and V. A. Zakian. 1994. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76145-155. [DOI] [PubMed] [Google Scholar]
- 45.Snow, B., M. Mateyak, J. Paderova, A. Wakeham, C. Iorio, V. Zakian, J. Squire, and L. Harrington. 2007. Murine pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol. Cell. Biol. 271017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spelbrink, J. N., F. Y. Li, V. Tiranti, K. Nikali, Q. P. Yuan, M. Tariq, S. Wanrooij, N. Garrido, G. Comi, L. Morandi, L. Santoro, A. Toscano, G. M. Fabrizi, H. Somer, R. Croxen, D. Beeson, J. Poulton, A. Suomalainen, H. T. Jacobs, M. Zeviani, and C. Larsson. 2001. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 28223-231. [DOI] [PubMed] [Google Scholar]
- 47.Sugawara, N. 1989. DNA sequences at the telomeres of the fission yeast Schizosaccharomyces pombe. Ph.D. thesis. Harvard University, Cambridge, MA.
- 48.Tanaka, H., G. H. Ryu, Y. S. Seo, K. Tanaka, H. Okayama, S. A. MacNeill, and Y. Yuasa. 2002. The fission yeast pfh1+ gene encodes an essential 5′ to 3′ DNA helicase required for the completion of S-phase. Nucleic Acids Res. 304728-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor, S., H. Zhang, J. Eaton, M. Rodeheffer, M. Lebedeva, T. O'rourke, W. Siede, and G. Shadel. 2005. The conserved Mec1/Rad53 nuclear checkpoint pathway regulates mitochondrial DNA copy number in Saccharomyces cerevisiae. Mol. Biol. Cell 163010-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 2942364-2368. [DOI] [PubMed] [Google Scholar]
- 51.Torres, J. Z., J. B. Bessler, and V. A. Zakian. 2004. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the Saccharomyces cerevisiae DNA helicase Rrm3p. Genes Dev. 18498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torres, J. Z., S. L. Schnakenberg, and V. A. Zakian. 2004. The Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra S phase checkpoint and fork restart activities. Mol. Cell. Biol. 243198-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tyynismaa, H., H. Sembongi, M. Bokori-Brown, C. Granycome, N. Ashley, J. Poulton, A. Jalanko, J. N. Spelbrink, I. J. Holt, and A. Suomalainen. 2004. Twinkle helicase is essential for mtDNA maintenance and regulates mtDNA copy number. Hum. Mol. Genet. 133219-3227. [DOI] [PubMed] [Google Scholar]
- 54.Uzawa, S., and M. Yanagida. 1992. Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci. 101267-275. [DOI] [PubMed] [Google Scholar]
- 55.Van Dyck, E., F. Foury, B. Stillman, and S. J. Brill. 1992. A single-stranded DNA binding protein required for mitochondrial DNA replication in Saccharomyces cerevisiae is homologous to Escherichia coli SSB. EMBO J. 113421-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner, M., G. Price, and R. Rothstein. 2006. The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics 174555-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wan, S., H. Capasso, and N. C. Walworth. 1999. The topoisomerase I poison camptothecin generates a Chk1-dependent DNA damage checkpoint signal in fission yeast. Yeast 15821-828. [DOI] [PubMed] [Google Scholar]
- 58.Werler, P. J., E. Hartsuiker, and A. M. Carr. 2003. A simple Cre-loxP method for chromosomal N-terminal tagging of essential and nonessential Schizosaccharomyces pombe genes. Gene 304133-141. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, D. H., B. Zhou, Y. Huang, L. X. Xu, and J. Q. Zhou. 2006. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 341393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou, J.-Q., E. M. Monson, S.-C. Teng, V. P. Schulz, and V. A. Zakian. 2000. The Pif1p helicase, a catalytic inhibitor of telomerase lengthening of yeast telomeres. Science 289771-774. [DOI] [PubMed] [Google Scholar]
- 61.Zhou, J.-Q., H. Qi, V. Schulz, M. Mateyak, E. Monson, and V. Zakian. 2002. Schizosaccharomyces pombe pfh1+ encodes an essential 5′ to 3′ DNA helicase that is a member of the PIF1 subfamily of DNA helicases. Mol. Biol. Cell 132180-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]