Abstract
Poly(A)-binding protein (PABP) stimulates translation initiation by binding simultaneously to the mRNA poly(A) tail and eukaryotic translation initiation factor 4G (eIF4G). PABP activity is regulated by PABP-interacting (Paip) proteins. Paip1 binds PABP and stimulates translation by an unknown mechanism. Here, we describe the interaction between Paip1 and eIF3, which is direct, RNA independent, and mediated via the eIF3g (p44) subunit. Stimulation of translation by Paip1 in vivo was decreased upon deletion of the N-terminal sequence containing the eIF3-binding domain and upon silencing of PABP or several eIF3 subunits. We also show the formation of ternary complexes composed of Paip1-PABP-eIF4G and Paip1-eIF3-eIF4G. Taken together, these data demonstrate that the eIF3-Paip1 interaction promotes translation. We propose that eIF3-Paip1 stabilizes the interaction between PABP and eIF4G, which brings about the circularization of the mRNA.
Translational control is an important mechanism by which cells govern gene expression, providing a rapid response to growth and proliferation stimuli, stress, and nutrient availability (17, 18, 22). Initiation, the rate-limiting step of translation, is an important target of translational control. Initiation entails the recruitment of ribosomes to the mRNA, their traversing the 5′ untranslated region, and recognition of the initiation codon. Ribosome recruitment is mediated by eukaryotic translation initiation factors (eIFs). All nuclear-transcribed eukaryotic mRNAs possess a 5′ cap structure (m7GpppN, where N is any nucleotide and m is a methyl group), and most possess a 3′ poly(A) tail. The 5′ cap and the 3′ poly(A) tail synergistically enhance translation (25). The cap is bound by the eIF4F complex, which consists of eIF4E, eIF4A, and eIF4G. eIF4E binds directly to the mRNA 5′ cap; eIF4A is an RNA helicase; and eIF4G is a modular scaffolding protein that binds eIF4E, eIF4A, eIF3, and the poly(A)-binding protein (PABP). Mammalian eIF3, the largest initiation factor, is composed of 13 different subunits that are named eIF3a through eIF3m and range in mass from 170 to 25 kDa (8). While the individual functions of the different subunits are not yet well established, eIF3 plays an essential role in translation initiation by interacting with the 40S ribosomal subunit and by promoting the formation of the 43S preinitiation complex (21). Thus, eIF3 serves as a bridge between the mRNA-eIF4F complex and the ribosome (18).
The poly(A) tail is bound by several molecules of PABP, an essential protein that mediates the stimulatory effect of the poly(A) tail on translation initiation (25, 41). PABP is a multidomain protein containing four phylogenetically conserved RNA recognition motifs (RRMs) (1, 42). The C-terminal one-third of the protein (PABC) contains a docking site for several proteins (12, 29, 32, 40). We identified three partners of PABP, termed PABP-interacting proteins (Paips): Paip1 (10), Paip2A (29), and Paip2B (6). Paips bind to PABP via two distinct PABP-binding motifs (PAMs) (40) (Fig. 1A). PAM1 is an acidic region of approximately 25 amino acids that binds to RRM2 in the N terminus of PABP, while PAM2 is a well-defined and conserved region of approximately 15 amino acids that binds to the PABC of PABP (27, 32, 40). The PAM2 motif was subsequently identified in many proteins exhibiting diverse functions (2), suggesting that PAM2 may play a role in protein-protein interactions in a wide range of cellular processes.
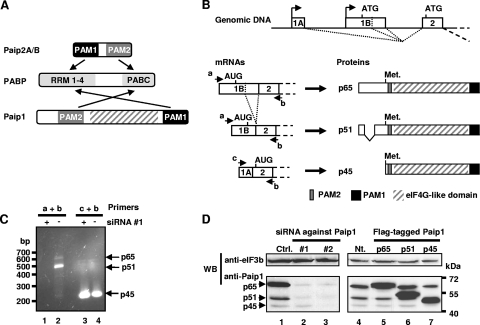
FIG. 1.
(A) Structural organization and interactions of PABP with Paip proteins. (B) Structural organization of the human Paip1 gene, mRNAs, and proteins. The p51 isoform is generated by alternative splicing of exon 1B and lacks amino acids 10 to 88. The p45 isoform is generated from a downstream AUG present in exon 2. Primers a, b, and c used for RT-PCR are indicated. (C) HeLa cells were treated with siRNA against Paip1 exon 1B. Total RNA was extracted, and RT-PCR experiments were performed with primers a and b for mRNA encoding p65 and p51 isoforms of Paip1 (lanes 1 and 2) and with primers c and b for mRNA encoding p45 isoform (lanes 3 and 4). Size marker and PCR products for the p65 (670 bp), the p51 (430 bp), and the p45 (220 bp) isoforms are indicated. (D) HeLa cells were treated with siRNA against exon 1B (lane 2, siRNA#1) or exon 5 (lane 3, siRNA#2) of the Paip1 gene or with control siRNA (Ctrl.) (left panel). HeLa cells were transfected with pCDNA3 Flag-tagged constructs expressing the p65, p51, or p45 isoform of Paip1 or left nontransfected (Nt.) (right panel). Extracts were subjected to SDS-PAGE and Western blotting (WB) using the indicated antibodies. Positions of molecular weight markers are indicated on the right.
Paip1 was discovered as a PABP-binding protein that stimulates translation of luciferase reporter mRNA in COS-7 cells (10). Paip1 possesses 39% similarity to the middle domain of eIF4G (10), spanning amino acids 616 to 1087 (9). In eIF4G, this region contains one of the two eIF4A binding sites and the eIF3 binding site (24, 37). Consistent with this homology, eIF4A coimmunoprecipitates with Paip1 (10). The mechanism by which Paip1 stimulates translation has not been previously elucidated. Here, we present evidence for an interaction between Paip1 and eIF3 and demonstrate that this interaction is important for the translational stimulatory activity of Paip1.
MATERIALS AND METHODS
Reagents.
Anti-Paip1 antibody was generated by immunizing rabbits with recombinant GST-Paip1 p65. Goat anti-human eIF3 antiserum was a kind gift from J. W. B. Hershey (University of California, Davis, CA) (3). Anti-eIF3g antibody was received from T. K. Tang (Academia Sinica, Taipei, Taiwan) (23). Anti-eIF3b and anti-eIF3e antibodies were from Santa Cruz Biotechnology. Anti-PABP, anti-α-tubulin, and anti-eIF4E antibodies were from Cell Signaling Technology. Anti-β-actin antibody was from Sigma-Aldrich. Anti-eIF3c, anti-eIF3f, and anti-eIF3k antibodies have been previously described (35). Anti-Paip2A antibody has been previously described (45). Recombinant full-length and truncated glutathione S-transferase (GST), GST-Paip1, GST-PABP, PABP, and eIF4GI were prepared as previously described (24, 37, 40). A similar protocol was applied for the production of GST-tagged eIF3b, eIF3e, eIF3f, eIF3g, and eIF3h. Purified rabbit reticulocyte eIF3 was a kind gift from W. C. Merrick (Case Western Reserve University, Cleveland, OH) and was purified as described previously (20), followed by gradient elution (100 to 450 mM KCl) from phosphocellulose.
Cell culture and transfection.
HeLa S3 cells were obtained from the American Tissue Culture Collection (CLL-2.2) and maintained in Dulbecco's minimum essential medium (Sigma) containing 10% fetal bovine serum and 5 U/ml penicillin-streptomycin solution (Gibco) in 5% CO2. DNA transfection was performed using Lipofectamine Plus reagent (Invitrogen) following the manufacturer's protocol.
siRNA transfection.
All small interfering RNA (siRNA) duplexes were designed as 21-mers and were purchased from Dharmacon. The sequences of the siRNAs are as follows: siPaip1#1, 5′-CAUGUCGGACGGUUUCGAUdTdT-3′ (where dT is deoxyribosylthymine); siPaip1#2, 5′-GCUGCAAAAGGGGAUGAAGdTdT-3′; sieIF3a, 5′-GUCAACAGGUGAACAUAAAdTdT-3′; sieIF3b, 5′-GAGAGAAGGCGCACCAUGAdTdT-3′; sieIF3e, 5′-GGAUGCUCUUUGACUACCUdTdT-3′; sieIF3g, 5′-CCAUCCGUGUCACCAACUUdTdT-3′; sieIF4GI, 5′-AACGUUACGACCGUGAGUUdTdT-3′. siPABP and siPaip2A were previously described (45). siRNA 4E-T inverted (15) was used as a control. siRNA transfections were performed in six-well plates, 1 day before plasmid transfection, as described previously (45).
Plasmid construction.
Constructs encoding GST-HMK-Paip1 (where HMK is heart muscle kinase) p65 and fragments thereof were described previously (40). pcDNA3 encoding Flag-tagged Paip1 p65 isoform was also previously described (29). The p51 and p45 isoforms of Paip1 were amplified by reverse transcription-PCR (RT-PCR) using total RNA from HeLa cells. BamHI and XhoI sites were added to the 5′ and 3′ ends, respectively. Plasmids encoding GST-HMK-Paip1 fragments were digested with BamHI and XhoI and subcloned using the same restriction sites into pcDNA3-Flag-Paip1 as well as p51 and p45 isoforms of Paip1. Human Paip1 p65 isoform was subcloned using the same sites in pcDNA3 containing a hemagglutinin (HA) tag to create the pcDNA3-HA-Paip1.
Human eIF3b cDNA was amplified by PCR from pcDNA3-HA-eIF3b obtained from J. W. B. Hershey (University of California, Davis, CA). EcoRI and XhoI sites were added to the 5′ and 3′ ends, respectively. PCR fragments were inserted in pGEX-6P-1 (Pharmacia Biotech) to create GST-tagged eIF3b. Human eIF3e, eIF3f, eIF3g, and eIF3h cDNAs (35) were PCR amplified and subcloned using EcoRI sites into pGEX-4T-1. The resulting plasmids were used to produce recombinant GST-tagged eIF3e, eIF3f, eIF3g, and eIF3h proteins, respectively.
The Tet-off promoter fragment was amplified by PCR from the pUHD 10-3 plasmid (19). SpeI and HindIII sites were added to the 5′ and 3′ ends, respectively. The initial cytomegalovirus promoter from the pcDNA3-HA-Paip1 construct was replaced by the Tet-off promoter, using SpeI and HindIII sites, to create the inducible Tet-off HA-tagged Paip1 plasmid (pTet-HA-Paip1). Paip1 isoforms and fragments were subcloned into the pTet-HA-Paip1 using BamHI and XhoI restrictions sites as described for pcDNA3-Flag-Paip1.
The full-length eIF4GI cDNA clone DKFZp762O191Q3 (pSP4GI) (9) was a kind gift from Richard E. Lloyd (Baylor College of Medicine, Houston, TX). The HA-tagged full-length eIF4GI fragment consisting of residues 1 to 1599 was amplified from pSP4GI by PCR, digested with HindIII and XhoI, and inserted into the pcDNA3 vector [pcDNA3-HA-eIF4GI(1-1599)]. The cDNA encoding the N-terminal fragment of eIF4GI (eIF4GI N; amino acids 84 to 653) was amplified from pcDNA3-HA-eIF4GI(1-1599) and cloned in pGEX-6P-1 (Pharmacia Biotech) to generate pGEX-eIF4GI(84-653). The middle and C-terminal fragments of recombinant eIF4GI (eIF4GI M+C; amino acids 653 to 1600) have been described previously (amino acids 457 to 1404, in reference 24; amino acids 613 to 1560, in reference 37).
RNA extraction and RT-PCR.
Total RNA was extracted from HeLa cells using Trizol reagent (Invitrogen) according to manufacturer's instructions. Reverse transcription (RT) was performed using Superscript II (Invitrogen), oligo(dT) and 1 microgram of total RNA, according to the manufacturer's instructions. One microliter of RT template was incubated with specific primers and PWO polymerase (Roche) according to the supplier's instruction.
Quantitative RT-PCR.
Total RNA was treated with a Turbo DNA-free kit (Ambion) prior to RT. RT reactions were diluted in 80 μl of water, and 1 μl was used for each reaction of quantitative PCR. PCRs were performed in duplicate, in a total reaction volume of 10 μl in 96-well reaction plates. PCRs were carried out in a Mastercycler Realplex2 (Eppendorf) using iQ Sybr Green Supermix (Bio-Rad) according to the manufacturer's instructions. The amplification conditions consisted of an initial denaturation step of 2 min at 94°C followed by 40 cycles of 15 s at 94°C, 15 s at 55°C, and 30 s at 68°C. Renilla mRNA was quantified and normalized against β-actin mRNA. The ΔΔCT method (where CT is threshold cycle) was used to determine the relative variation of Renilla mRNA between experimental samples, using the repressed expression with tetracycline (Tet) as a reference. Results are presented as the luciferase mRNA ratio between the induced condition (no Tet) and the repressed condition (with Tet).
Primer sequences.
The primers used for cloning were as follows (restriction sites are underlined): for Paip1, 5′-TCAGGATCCGCTAAGCCCCAGGTGGTT-3′ (p45 sense), 5′-GGCTCGAGTTACTGTTTTCGCTTACG-3′ (Paip1 antisense), and as previously described (40); Tet-off promoter, 5′-ATTGACTAGTTGCATGCTCGAGTTTACCA-3′ (sense) and 5′-TGGAAGCTTATCGATGCGGCCGCGCTAGCA-3′ (antisense); eIF4GI(84-653), 5′-ATGCGGATCCCAAGTAATGATGATCCC-3′ (sense) and 5′-ATGCCTCGAGTTAATCCAGTGGCCGCAGTGGTGTTTT-3′ (antisense); eIF3b, 5′-CAGAATTCCAGGACGCGGAGAACGTGGC-3′ (sense) and 5′-TTCTCGAGTCACTCCTGATTCCCGAGGG-3′ (antisense). The primers used for RT-PCR were as follows: p65/p51 sense (primer a), 5′-GGAGAACTGGAAAGCCGAGGGTA-3′; p65/p51/p45 antisense (primer b), 5′-GTGTAACTGGAAGAATAACCTGAAGGG-3′; and p45 sense (primer c), 5′-ACGTCTCTCCTGAGAACTACCGAGT-3′. The primers used for the quantitative PCR were 5′-CAGTGGTGGGCCAGATGTAAACAA-3 (sense) and 5′-TAATACACCGCGCTACTGGCTCAA-3′ (antisense) for Renilla; β-actin sense and antisense primers were previously described (33).
GST pull-downs.
HeLa cells were grown to 80% confluence and collected in buffer A (20 mM HEPES-KOH, pH 7.4, 100 mM KCl, 0.1% NP-40, 0.5 mM EDTA, 10% glycerol, 25 mM β-glycerophosphate, 2 mM Na3VO4, 0.5 mM NaF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 5 μg/ml pepstatin A, 0.5 mM phenylmethylsulfonyl fluoride). Where indicated, extracts were subjected to RNase treatment, as previously described (44). Recombinant GST-Paip1 or GST alone (250 nM) was incubated with HeLa cell extract (5 mg of protein/ml) in the presence of glutathione-Sepharose beads for 3 h at 4°C. Beads were washed five times in buffer A, and bound proteins were eluted with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 0.1% bromophenol blue, 10% glycerol, 10 mM dithiothreitol, 5% β-mercaptoethanol). Eluates were subjected to SDS-PAGE and Western blotting. For mass spectrometry analysis, eluted proteins were separated by 4 to 15% SDS-PAGE. Bands which were present only in a GST-Paip1 pull-down sample were excised from the gel, stored in 1% acetic acid, and subjected to mass spectrometry analysis (Génome Québec Innovation Centre, Montréal, QC, Canada).
To perform GST pull-downs using purified proteins, recombinant GST-Paip1 or GST was incubated with recombinant PABP, recombinant eIF4GI, or purified eIF3 (all at 100 nM), as indicated in Fig. 5, in buffer B (20 mM HEPES-KOH, pH 7.4, 15 mM NaCl, 140 mM KCl, 0.5% NP-40, 2.5 mM MgCl2), supplemented with 0.1 mg/ml bovine serum albumin, in the presence of glutathione-Sepharose beads for 2 h at 4°C. For experiments using the recombinant GST-tagged eIF3 subunit, 40 ng of p51 Paip1 was incubated in 200 μl of buffer B with 0.1 mg/ml bovine serum albumin with 1 μg of GST-eIF3b, -eIF3e, -eIF3f, -eIF3g, or -eIF3h or GST alone. Beads were washed five times in buffer B, and bound proteins were eluted with SDS-PAGE loading buffer. Eluates were subjected to SDS-PAGE and Western blotting.
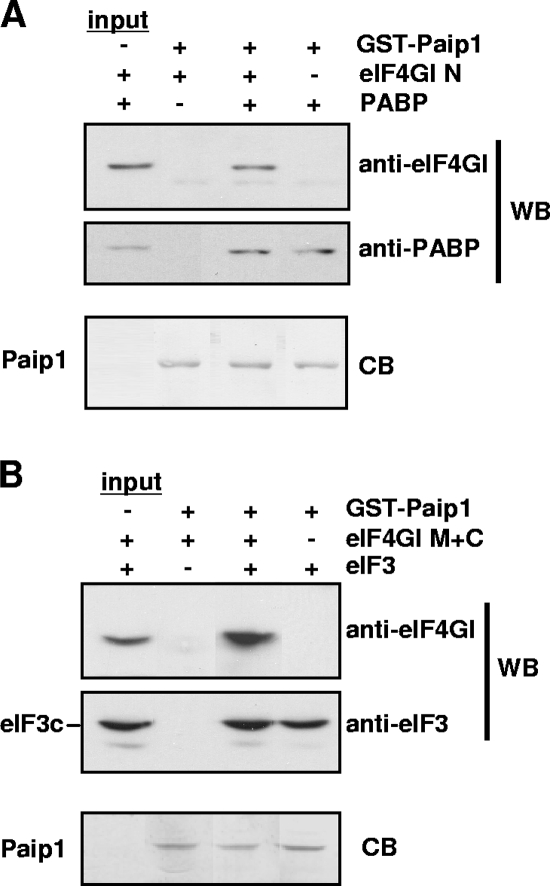
FIG. 5.
Simultaneous binding of Paip1, eIF3, PABP, and eIF4GI. GST pull-downs were conducted with GST-Paip1 under the following conditions: in the presence of eIF4GI N and PABP (A) and of eIF4G M+C and eIF3 (B). Eluates were subjected to SDS-PAGE and Western blotting (WB) with the indicated antibodies. Blots were then stained with Coomassie blue stain (CB).
Immunoprecipitation.
HeLa cell extracts (1 mg of protein/ml) were used for immunoprecipitation, as previously described (34). The antibodies indicated in Fig. 2C and 4C were added to the extract together with protein G-Sepharose beads and incubated for 90 min at 4°C. Bound proteins were eluted with SDS-PAGE loading buffer and subjected to SDS-PAGE and Western blotting.
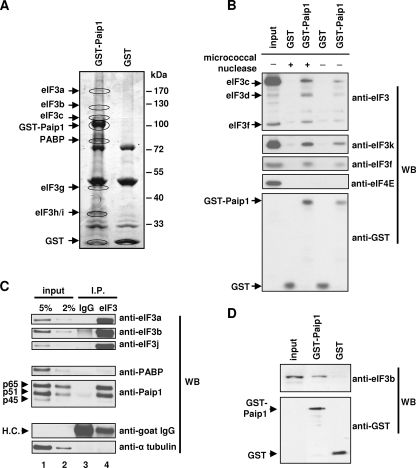
FIG. 2.
RNA-independent interaction between Paip1 and eIF3. (A) GST pull-down experiments were conducted with GST-Paip1 or GST on HeLa cell extracts. Eluates were subjected to SDS-PAGE. Bands unique to GST-Paip1 (circled) were excised and subjected to mass spectrometry analysis. Identified proteins are indicated next to the analyzed bands. Protein sequence coverage is as follows: eIF3a, 15.4%; eIF3b, 14.4%; eIF3c, 20.2%; PABP, 56.0%; eIF3g, -h, and -i, ∼10%. (B) GST pull-downs were conducted with GST-Paip1 or GST in untreated or micrococcal nuclease-treated HeLa cell extracts. Eluates were subjected to SDS-PAGE and Western blotting (WB) using the indicated antibodies. (C) Immunoprecipitation using anti-eIF3 antibody (eIF3) or preimmune goat serum (IgG) was conducted using a HeLa cell extract. Extract (Input) and eluates (IP) were subjected to SDS-PAGE and Western blotting using the indicated antibodies. The Paip1 isoforms and IgG heavy chain (HC) are indicated. (D) GST pull-downs were conducted with GST or GST-Paip1 together with purified eIF3. Eluates were subjected to SDS-PAGE and Western blotting using the indicated antibodies.
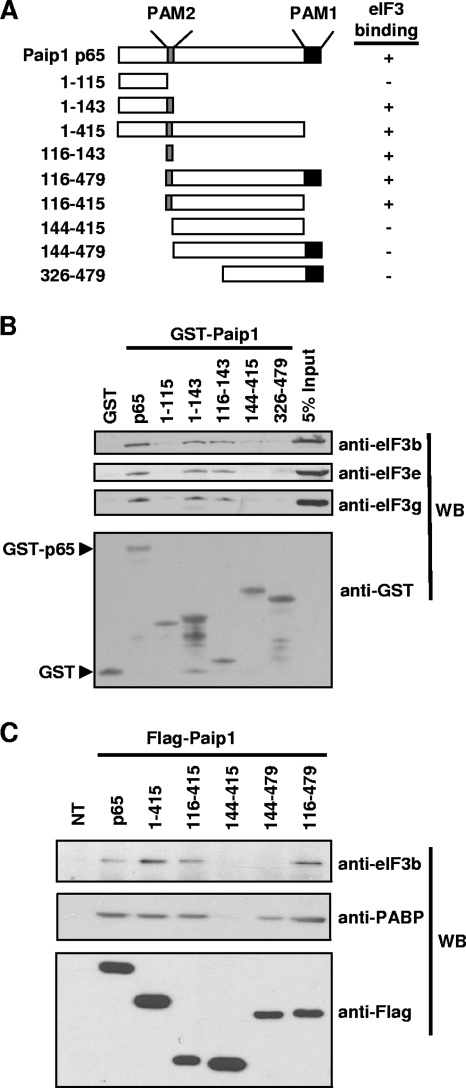
FIG. 4.
Identification of the eIF3-binding domain of Paip1. (A) Schematic representation of Paip1 fragments used in GST pull-downs and Flag immunoprecipitation. Relative binding was evaluated visually. (B) GST pull-downs were conducted with the indicated GST-Paip1 fragments in HeLa cell extracts. Eluates were subjected to SDS-PAGE and Western blotting (WB) with the indicated antibodies. The GST and GST-tagged p65 proteins are indicated. (C) HeLa cells were transfected with plasmids expressing the indicated Flag-tagged Paip1 fragments or left untransfected (NT). Immunoprecipitations were performed 48 h posttransfection. Eluates were subjected to SDS-PAGE and Western blotting with the indicated antibodies.
Far-Western analysis.
The procedure for far-Western analysis was previously described in detail (28). Forty micrograms of purified rabbit reticulocyte eIF3 was resolved by 8% SDS-PAGE. 32P-labeled HMK-p45 Paip1 was used as a probe at 250,000 cpm/ml of hybridization solution.
Translation assays.
HeLa cells were seeded in six-well tissue culture dishes 1 day prior to transfection. Cells were cotransfected with 250 ng of pTet-HA-Paip1 or control vector, 250 ng of pUHD-15-1 which expresses the Tet-controlled transactivator (tTA) (19), and 50 ng of pRL-CMV (Promega) per well as a Renilla luciferase reporter construct. pBI-L vector (Stratagene) expressing firefly luciferase was used as a control vector. Four hours after transfections, the medium was replaced and supplemented with Tet at a final concentration of 0 or 300 ng/ml. Tet at 300 ng/ml caused a complete inhibition of the Tet-off promoter (19). Consequently, this was used as the repressed condition, and medium without Tet was used as the induced condition. Cells were harvested 48 h after transfection and lysed using 400 μl/well of 1× passive lysis buffer (Promega). Four microliters of extract was used to quantify Renilla luciferase activity with a Dual-Luciferase Reporter Assay system (Promega). Protein concentration was determined using 4 μl of HeLa extract and Bio-Rad protein assay reagent according to the manufacturer's instruction. Renilla luciferase activity was corrected based on protein concentration. The relative induction for each construct was determined by calculating the ratio of Renilla luciferase activity between the induced condition (no Tet) and the repressed condition (300 ng/ml Tet). Extracts were subjected to SDS-PAGE and Western blotting.
RESULTS
Paip1 exists in three different isoforms.
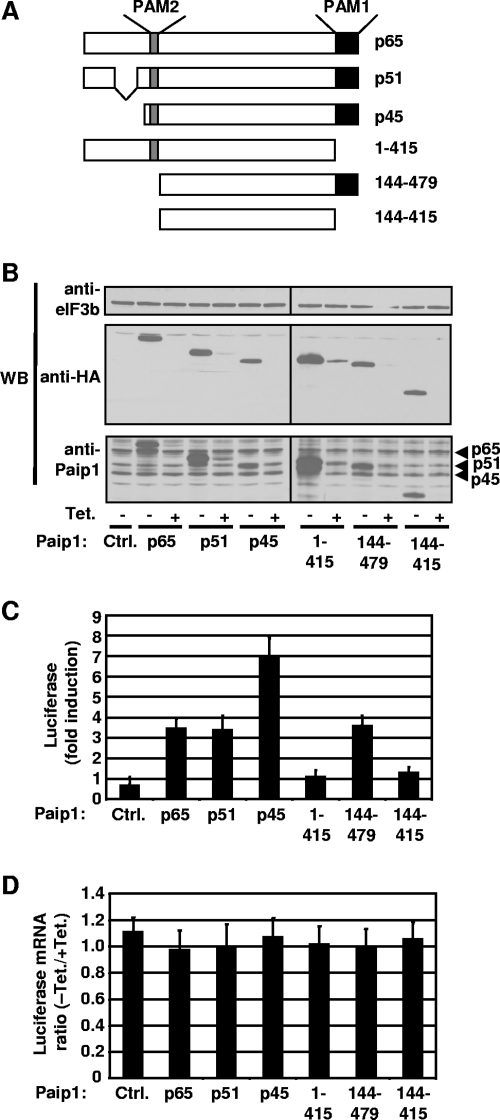
Human Paip1 was originally described as a ∼70-kDa protein (10). Subsequently, two closely related cDNAs were entered into the GenBank database. The related cDNAs have most probably arisen from alternative splicing and promoter usage (see description below and Fig. 1B). We developed an anti-Paip1 antibody that recognizes three different Paip1 isoforms at 65, 51 and 45 kDa after resolution of a HeLa cell extract by SDS-PAGE (Fig. 1D, lane 1). The full-length isoform (p65) corresponds to the originally described Paip1 (accession number NM_006451) (10), while the p51 isoform lacks amino acids 10 to 88 due to alternative splicing within exon 1B (accession number NM_182789) (Fig. 1B). The p45 isoform consists of amino acids 113 to 479 of the p65 isoform (Fig. 1B). The mRNA encoding the p45 isoform is produced from an upstream promoter and uses an alternate first exon (Fig. 1B, exon 1A), resulting in a different 5′ untranslated region and consequently the utilization of a downstream in-frame AUG (accession number NM_183323). The existence of the three different Paip1 mRNAs described in the GenBank database was confirmed by RT-PCR (Fig. 1C, lanes 2 and 4) and sequence analysis. To further correlate the three cDNA database entries with the three isoforms of Paip1 proteins, HeLa cells were treated with two different siRNAs against Paip1 mRNAs. The first siRNA targets exon 1B of the mRNAs encoding the p65 and p51 isoforms while the second targets exon 5, which is common to all three isoforms. The second siRNA, which targets all three mRNAs, caused a dramatic reduction in the amount of all three protein isoforms (Fig. 1D, lane 3). In contrast, the first siRNA, which targets p65 and p51 mRNAs, dramatically reduced the expression of only the p65 and p51 isoforms (Fig. 1D, lane 2). In addition, mRNAs expressing p65 and p51 isoforms of Paip1 are no longer detectable by RT-PCR in HeLa cells treated with the first siRNA (Fig. 1C, lane 1), whereas the mRNA expressing the Paip1 p45 isoform was amplified (Fig. 1C, lane 3).
Transfection of a DNA vector expressing an N-terminal Flag-tagged Paip1 isoform (p65, p51, or p45) into HeLa cells leads to the expected expression of a slightly higher-molecular-weight protein (due to the presence of the Flag-tagged peptide) than the corresponding Paip1 isoform (Fig. 1D, lanes 5 to 7). No increase in expression of the lower-molecular-weight Paip1 p45 isoform was observed when Flag-tagged p65 or p51 Paip1 vector was transfected (Fig. 1D, lanes 5 and 6). Therefore, the downstream in-frame AUG initiating p45 Paip1 translation is not used in p65 and p51 Paip1-encoding mRNAs. Taken together, these data demonstrate that the three isoforms of Paip1 are translated from three distinct mRNAs.
Paip1 interacts with eIF3.
To understand how Paip1 stimulates translation, we wished to identify Paip1-interacting proteins. A GST pull-down assay was performed using a HeLa cell extract, and binding partners of the GST-Paip1 p65 isoform (referred to as GST-Paip1) were identified by mass spectrometry. The p65 isoform was chosen because it was the only isoform known at the time of the experiment to bind PABP and to stimulate translation (10). GST-Paip1 specifically coeluted with several eIF3 subunits, together with PABP (Fig. 2A). The association of eIF3 with Paip1 was confirmed by Western blotting using a goat anti-human eIF3 antiserum (Fig. 2B, upper panel) and antibodies against the eIF3f and eIF3k subunits (Fig. 2B). Importantly, the Paip1-eIF3 interaction also occurred in an RNase-treated cell extract, suggesting that the binding of eIF3 to Paip1 is RNA independent (Fig. 2B). eIF4E failed to coelute with GST-Paip1, further indicating that the pull-down was not due to tethering through RNA (Fig. 2B). To confirm the eIF3-Paip1 interaction, eIF3 was immunoprecipitated from a HeLa cell extract using a goat anti-human eIF3 antiserum. All three isoforms of Paip1 (p65, p51, and p45) were coimmunoprecipitated with eIF3 (Fig. 2C, lane 4). PABP was found to be associated with eIF3 at very low stoichiometry (much less than 2%) (Fig. 2C, lanes 2 and 4) compared to Paip1 (more than 5%). The coimmunoprecipitation specificity was confirmed by Western blotting using anti-α-tubulin and anti-goat immunoglobulin G (IgG) antibodies (Fig. 2C). These results demonstrate that eIF3 binds to all Paip1 isoforms, independently of PABP. Furthermore, GST-Paip1 interacted with purified rabbit reticulocyte eIF3 in a GST pull-down assay as determined by Western blotting with eIF3b antibody (Fig. 2D). Taken together, these data show that the interaction between eIF3 and Paip1 is specific and RNA independent.
Paip1 binds directly to the g subunit of eIF3 (p44).
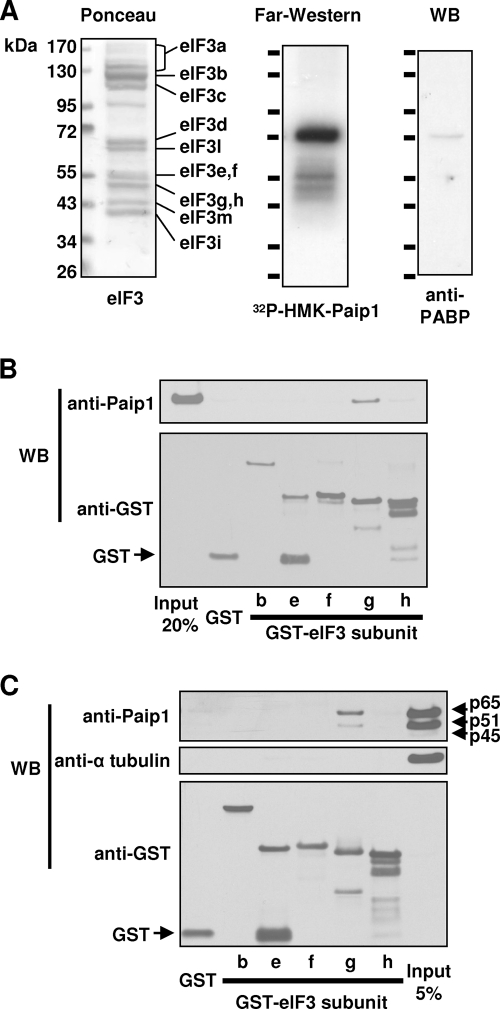
To determine whether the Paip1-eIF3 interaction is direct and also to identify the eIF3 subunit that interacts with Paip1, purified rabbit reticulocyte eIF3 was resolved by SDS-PAGE and transferred onto a nitrocellulose membrane. Ponceau S staining revealed the presence of 11 major bands corresponding to the known eIF3 subunits (Fig. 3A, left panel) (note that eIF3a is partially degraded as observed in many other reports) (39). The membrane was used for far-Western analysis with 32P-labeled HMK-p45 Paip1 as a probe. One strong band at 72 kDa and two weaker bands around 55 kDa were detected by autoradiography (Fig. 3A, middle panel). Ponceau S staining did not reveal any major protein migrating around 72 kDa, indicating that this Paip1-interacting protein has a very high affinity for the 32P-labeled probe (Fig. 3A). Considering the size of this protein and the strength of this interaction, it is highly likely that PABP is the minor contaminating protein, which is present at very low stoichiometry. The Paip1-PABP interaction is extremely strong and has an apparent Kd of 1.9 nM (10, 40). Therefore, a duplicate membrane was used for Western blotting with anti-PABP antibody, which demonstrated the presence of residual PABP in the eIF3 preparation (Fig. 3A, right panel). We estimate that there is >0.1% of PABP in the eIF3 preparation. Additional Western blotting experiments also demonstrated a similar level of contamination of the purified eIF3 with Paip1 and eIF4GI (data not shown). To identify the two weaker bands, duplicate membranes were used for Western blotting with antibodies raised against various eIF3 subunits. eIF3e, eIF3f, eIF3g, and eIF3h are the only eIF3 subunits at the corresponding size of the autoradiographic signal (7, 30, 39). Recombinant GST-tagged eIF3e, eIF3f, eIF3g, and eIF3f were produced in E. coli, purified, and employed for GST pull-down experiments against recombinant p51 Paip1. GST-eIF3b and GST were used as negative controls. Paip1 interacted only with the GST-eIF3g (Fig. 3B). A GST pull-down was performed using a HeLa cell extract and the same GST-tagged proteins. Only GST-eIF3g pulled down Paip1 from the cell extract (Fig. 3C). α-Tubulin was not associated with Paip1, demonstrating the specificity of the interaction (Fig. 3C). Taken together, these results demonstrate that Paip1 interacts directly with the g subunit (p44) of eIF3. This interaction occurs in vitro and in vivo.
FIG. 3.
Identification of Paip1-interacting subunit of eIF3. (A) Purified eIF3 was subjected to SDS-PAGE and transferred onto a nitrocellulose membrane. Membrane was subjected to Ponceau S staining and far-Western blotting using 32P-labeled HMK-Paip1 p45 as a probe. A duplicate membrane was subjected to Western blotting (WB) with PABP antibody. eIF3 subunits and fragments of eIF3a are indicated. (B) GST pull-downs were conducted on recombinant p51 Paip1 with GST-eIF3b, GST-eIF3e, GST-eIF3f, GST-eIF3g, GST-eIF3h, or GST. Eluates were subjected to SDS-PAGE and Western blotting (WB) using Paip1 or GST antibodies. (C) GST pull-downs were conducted with GST-eIF3b, GST-eIF3e, GST-eIF3f, GST-eIF3g, GST-eIF3h, or GST on a HeLa cell extract. Eluates were subjected to SDS-PAGE and Western blotting (WB) using the indicated antibodies.
The eIF3-binding domain in Paip1 spans amino acids 116 to 143.
To map the eIF3-binding domain in Paip1, GST-Paip1 pull-downs were performed using deletion mutants of Paip1 (Fig. 4A). eIF3 interacted only with fragments containing amino acids 116 to 143 of Paip1 (Fig. 4B). The interaction was observed with antibodies raised against several eIF3 subunits: eIF3b, eIF3e, and eIF3g (Fig. 4B). Similarly, when Flag-tagged Paip1 fragments were expressed in HeLa cells, only those containing amino acids 116 to 143 copurified with eIF3 (Fig. 4C).
Simultaneous binding of Paip1 with eIF4G, PABP, and eIF3.
To gain insight into the functional significance of the eIF3-Paip1 interaction, we examined whether eIF3, Paip1, PABP, and eIF4G form complexes. GST pull-downs were conducted using GST-Paip1 as bait in the presence of recombinant PABP, eIF4GI proteins, and purified eIF3. Because it is difficult to produce intact full-length recombinant eIF4GI (24), eIF4GI fragments were employed for these experiments: the eIF4GI N fragment, which possesses PABP but not eIF3-binding activity (24), and the eIF4GI M+C fragment, which possesses eIF3 but not PABP-binding activity (37). eIF4GI N was pulled down by GST-Paip1 only when PABP was present (Fig. 5A), indicating that PABP interacts simultaneously with eIF4GI and Paip1. Similarly, eIF4GI M+C was pulled down by GST-Paip1 only in the presence of eIF3, indicating that eIF3 interacts simultaneously with Paip1 and eIF4GI (Fig. 5B). These results thus demonstrate that ternary complexes composed of Paip1-PABP-eIF4G and Paip1-eIF3-eIF4G can form in vitro.
Stimulation of translation by Paip1 isoforms and fragments.
To examine the effect of Paip1-eIF3 interaction on Paip1 translational stimulatory activity, in vivo translation assays were performed. DNA vectors expressing different Paip1 isoforms and fragments (Fig. 6A), under the control of the Tet-off promoter (19), were transfected into HeLa cells along with constructs expressing the Renilla luciferase and the tTA. Each construct yielded comparable amounts of protein (Fig. 6B). The Paip1 isoforms were expressed at levels about two- to threefold higher than the endogenous protein (Fig. 6B). The relative induction of luciferase activity mediated by Paip1 was determined by calculating the ratio of Renilla luciferase activity between induced (without Tet) and repressed (300 ng/ml Tet) expression of HA-tagged Paip1. The p65 form of Paip1 enhanced translation by approximately 3.5-fold, as previously reported (10), and the p51 isoform enhanced translation to the same extent (Fig. 6C). Strikingly, the p45 isoform enhanced translation approximately sevenfold (Fig. 6C). Overexpression of Paip1 isoforms had no effect on Renilla mRNA levels (Fig. 6D). Thus, the p45 isoform of Paip1 is a more potent translational enhancer than the p65 or p51 isoform. The possible reasons for these differences among the different Paip1 isoforms are addressed in the Discussion. To determine which region of Paip1 is required for translational enhancement, constructs expressing different Paip1 fragments were transfected into HeLa cells. Paip1 consisting of residues 1 to 415, lacking PAM1, failed to stimulate luciferase activity (Fig. 6C), as previously reported (10), showing that binding of Paip1 to PABP via this domain is required for Paip1 activity. Paip1 has a higher affinity for the RRM1 to RRM4 of PABP (Kd of 0.56 nM) (40) than the PABC domain (Kd of 4.7 nM) (40), indicating that the PAM1 domain binds more strongly to PABP than the PAM2 domain (10). Paip1 consisting of residues 144 to 479, lacking the eIF3-binding domain, exhibited reduced stimulatory activity (∼50% compared to p45). These data confirm the importance of PAM1, and thus PABP binding, to Paip1 translational activity (10) and also demonstrate that the eIF3-Paip1 interaction is required for maximal Paip1 activity in translational enhancement.
FIG. 6.
Paip1-dependent translation stimulation in vivo. (A) Schematic representation of HA-tagged Paip1 fragments expressed in HeLa cells. (B) Cells were transfected with the indicated pTet-HA-Paip1 plasmids together with constructs expressing the Renilla luciferase and the tTA. Cells were placed in medium containing 0 or 300 ng/ml of Tet. Extracts were subjected to SDS-PAGE and Western blotting (WB) with the indicated antibodies. (C) Renilla luciferase activity was quantified in HeLa cell extracts from panel B and normalized on the total protein level. Relative induction of the luciferase reporter was determined by calculating the ratio of Renilla luciferase activity between induced (without Tet) and repressed (300 ng/ml Tet) expression of the indicated HA-tagged Paip1. Error bars denote the standard error of the mean for three independent experiments. (D) Renilla luciferase mRNA was measured by quantitative RT-PCR from total RNA extracted from duplicate HeLa cells from panel B. Total RNA was DNase-treated and reverse transcribed. Using RT products, Renilla mRNA was quantified and normalized against β-actin mRNA. Luciferase mRNA ratio is calculated by the ΔΔCT method using the repressed expression (+Tet) for each Paip1 construct as a reference. Error bars denote the standard error of the mean for three independent experiments.
Paip1 requires eIF3 and PABP to exert its full effect.
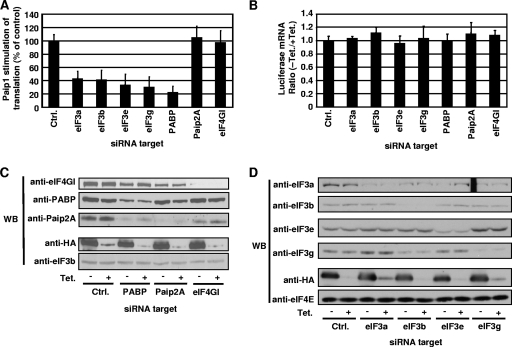
To confirm the importance of eIF3 and PABP for Paip1 activity, the expression of PABP, as well as that of several eIF3 subunits, was silenced by siRNA, and the effect on Paip1 activity was examined. Because p45 is the most translationally active isoform of Paip1, it was employed in these experiments. Renilla luciferase mRNA levels were not affected by siRNA treatments (Fig. 7B). Similarly, we failed to observe any reduction in HA-Paip1 p45 expression or induction (Fig. 7C and D), which could have been responsible for the attenuation in the stimulation of Renilla luciferase activity. Paip1 translational enhancement was reduced by PABP silencing (Fig. 7A) whereas control vector expression was not affected and was similar to untreated cells (data not shown and Fig. 6C). Since PABP knockdown induces the proteasome-mediated degradation of Paip2A (45), we also silenced Paip2A (Fig. 7C), which had no effect on Paip1 activity (Fig. 7A). Knockdown of eIF4GI had no effect on Paip1 function (Fig. 7A and C), probably due to the redundant function of eIF4GII, which is also expressed in HeLa cells (43). The lack of effect in knockdowns of Paip2A and eIF4GI indicates that the PABP knockdown-mediated effect on Paip1 function is specific. These data demonstrate that PABP is required for optimal Paip1 activity.
FIG. 7.
Effect of eIF3 and PABP on Paip1-dependent translation stimulation in vivo. (A) HeLa cells were transfected with the indicated siRNAs, and siRNA 4E-T inverted (15) was used as a control (Ctrl). At 24 h after siRNA transfection, cells were transfected as described in the legend of Fig. 6B using the pTet-HA-Paip1 p45 construct. Cells were placed in medium containing 0 or 300 ng/ml of Tet. Data are expressed as the percentage of the relative increase in induction in cells transfected with control siRNA (set at 100%). Error bars denote the standard error of the mean of three independent experiments. (B) Renilla luciferase mRNA was measured by quantitative RT-PCR from total RNA extracted from duplicate HeLa cells from panel A. Total RNA was DNase treated and reverse transcribed. By using RT reactions, Renilla mRNA was quantified and normalized against β-actin mRNA. The luciferase mRNA ratio is calculated by the ΔΔCT method using the repressed expression (+ Tet) for each siRNA as a reference. Error bars denote the standard error of the mean for three independent experiments. (C and D) Extracts from panel A were subjected to SDS-PAGE and Western blotting (WB) with the indicated antibodies.
Paip1-dependent translation stimulation was decreased to similar levels upon silencing of eIF3a, eIF3b, eIF3e, and eIF3g (Fig. 7A). Renilla luciferase mRNA levels and HA-Paip1 p45 expression were not affected by the knockdown of eIF3 subunits (Fig. 7B and D). Silencing of eIF3b, eIF3e, and eIF3g caused reduced levels of eIF3a (Fig. 7D) because silencing of these subunits may alter the folding, function, or stability of other eIF3 subunits including eIF3a (16, 35). Thus, destabilization of eIF3 can potentially lead to the proteasome-mediated degradation of eIF3a (4). eIF3 function requires core subunits, without which eIF3 cannot be properly folded or function in translation (35, 38). In mammalian cells and in Saccharomyces cerevisiae, eIF3a is a core component (21, 35). In conclusion, Paip1 requires eIF3 core subunits to exert its full activity.
DISCUSSION
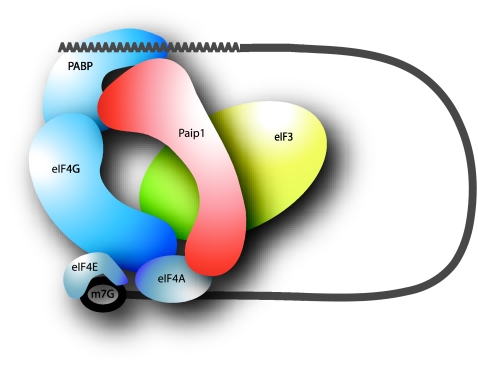
Here, we provide evidence that Paip1 stimulates translation by interacting with eIF3. The interaction is direct and RNA independent and mediated by eIF3g (p44). Deletion of the eIF3-binding domain in Paip1, as well as siRNA-mediated silencing of eIF3 subunits, resulted in diminished Paip1-mediated translational enhancement of a luciferase reporter in HeLa cells. We therefore propose that the interaction of Paip1 with eIF3 stabilizes the circular mRNA conformation, which is formed by the eIF4G-PABP interaction (Fig. 8). Consistent with this model, simultaneous interactions between Paip1-eIF3-eIF4GI and Paip1-PABP-eIF4GI were documented (Fig. 5). The identification of eIF3 as a Paip1 binding partner adds to our earlier finding that Paip1 interacts with eIF4A (10) in that, like eIF4G, Paip1 can interact with both eIF3 and eIF4A. Paip1 possesses homology to the middle domain of eIF4GI (amino acids 616 to 1087) (10), which contains binding sites for eIF4A and eIF3 (24, 31, 37). Thus, Paip1 is expected to interact with eIF3 and eIF4A. The binding domain for eIF3 in Paip1 is contained within the eIF4G-homologous region (10) (minimal proposed sequence, amino acids 712 to 742 in eIF4GI), although the eIF3-binding domain in eIF4G is much larger (amino acids 711 to 1128) (37); we note that a smaller domain exhibiting weaker affinity, spanning amino acids 1014 to 1117, has also been reported (31). This suggests that Paip1 does not employ an eIF3-binding domain identical to that of eIF4G. Indeed, the GST pull-down experiments using purified proteins (Fig. 5B) showed that Paip1 and eIF4G can bind to eIF3 simultaneously rather than competing for binding. Thus, it is possible that Paip1-eIF3 binding might affect or be affected by the eIF3-eIF4G interaction.
FIG. 8.
Model for the mechanism of Paip1-mediated translation stimulation. Paip1 stabilizes the interaction between eIF4G and PABP by binding to eIF3, which in turn binds to eIF4G and the 40S ribosome (not shown in the figure). m7G, cap structure.
The eIF3-binding domain in Paip1 overlaps with a region where one of the PABP-binding domains, PAM2, resides (40); therefore, PABP and eIF3 can potentially compete for Paip1 binding via the PAM2 domain. However, PABP could remain associated with Paip1 through its PAM1 domain because the Paip1 PAM1-PABP RRM interaction is much stronger (Kd of 0.56 nM) (10, 40) than the PAM2-PABC interaction (Kd of 4.7 nM) (40). This should favor the formation of the ternary complex between PABP, Paip1, and eIF3 and therefore stabilize the circular mRNA conformation together with eIF4G.
Paip1 interacts directly with eIF3g (p44) as determined by GST pull-down (Fig. 3B). eIF3g/Tif35 is one of the core subunits in Saccharomyces cerevisiae (21, 38) and associates strongly with the central part of mammalian eIF3 (11); however, it is not a core subunit of the reconstituted mammalian eIF3 (35). eIF3g is not required for the stable interaction of the 40S ribosome-initiation factor complex with the mRNA at the AUG codon (35), suggesting its involvement in translational regulation. Human eIF3g interacts strongly with eIF3a and more weakly with eIF3b and eIF3c (7). A recent mass spectrometry report also supports these interactions by showing that human eIF3g is part of a stable eIF3 subcomplex composed of eIF3a, eIF3b, eIF3g, eIF3i, and eIF3c, which also exists in yeast eIF3 (46). eIF3g is phosphorylated on Thr41 and Ser42 (5, 11, 36), and this phosphorylation is increased by about threefold upon serum stimulation (11). As recently suggested, phosphorylation of eIF3 subunits could stabilize eIF3 complex (46) and consequently favor peripheral interactions. This raises the possibility that phosphorylation of eIF3g might stimulate the interaction between Paip1 and eIF3. The physiological conditions under which the Paip1-eIF3 interaction is modulated remain to be studied.
The far-Western analysis demonstrated a visually weak interaction between eIF3 subunits and Paip1 compared to the PABP-Paip1 interaction (Fig. 3A). However, the proper refolding of proteins on the membrane is a crucial element for protein-protein interaction, but it is not always efficient (13, 14). The lack of proper refolding might be an explanation of why we failed to observe a stronger signal in the far-Western experiment. The best folding for eIF3g might be achieved only when it is incorporated into eIF3. It is also possible that the eIF3-Paip1 interaction occurs through several subunits.
In the in vivo luciferase reporter translation assays, the p45 isoform was more active than p65 or p51 (approximately twofold) (Fig. 6C). Because of the N-terminal location of the eIF3 binding site on the p45 isoform (Fig. 1B), it is possible that the PAM2 domain is more accessible, and, thus, its binding to eIF3 is increased. Indeed, many PAM2-containing proteins possess the PAM motif near the N or C terminus (2). Paip1 isoforms are differentially expressed (Fig. 1D), which renders difficult any direct comparison of affinity based on coimmunoprecipitation (Fig. 2C) or GST pull-down experiments (Fig. 3C). Isothermal titration microcalorimetry measurements will have to be performed to determine the Kd and the stoichiometry of the interaction between eIF3g and Paip1 isoforms (45).
Silencing of various eIF3 subunits by siRNA blocked the Paip1-dependent translation enhancement, demonstrating that eIF3 is required for Paip1 activity (Fig. 7A). Paip1-eIF3 is therefore likely to stabilize the interaction of eIF4G with PABP (Kd of 20 nM) (26) and, consequently, the circular mRNA configuration. Thus, Paip1 might act as a proxy through which eIF3 mediates PABP stimulation of translation.
Acknowledgments
We thank L. W. Ler for helpful comments on the manuscript, W. C. Merrick for purified eIF3, J. W. B. Hershey for the goat anti-human eIF3 antiserum, and T. K. Tang for anti-eIF3g antibody.
This work was funded by a National Institutes of Health grant to N.S. (RO1 GM066157) and to A.-B.S. (RO1 GM46454) and a Canadian Institutes of Health Research grant to K.G. (MOP-14219). Y.M. was supported by a McGill Cancer Centre Postdoctoral Fellowship. A.Y. is the recipient of Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad.
Footnotes
Published ahead of print on 25 August 2008.
REFERENCES
- 1.Adam, S. A., T. Nakagawa, M. S. Swanson, T. K. Woodruff, and G. Dreyfuss. 1986. mRNA polyadenylate-binding protein: gene isolation and sequencing and identification of a ribonucleoprotein consensus sequence. Mol. Cell. Biol. 62932-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht, M., and T. Lengauer. 2004. Survey on the PABC recognition motif PAM2. Biochem. Biophys. Res. Commun. 316129-138. [DOI] [PubMed] [Google Scholar]
- 3.Asano, K., T. G. Kinzy, W. C. Merrick, and J. W. Hershey. 1997. Conservation and diversity of eukaryotic translation initiation factor eIF3. J. Biol. Chem. 2721101-1109. [DOI] [PubMed] [Google Scholar]
- 4.Baugh, J. M., and E. V. Pilipenko. 2004. 20S proteasome differentially alters translation of different mRNAs via the cleavage of eIF4F and eIF3. Mol. Cell 16575-586. [DOI] [PubMed] [Google Scholar]
- 5.Beausoleil, S. A., M. Jedrychowski, D. Schwartz, J. E. Elias, J. Villen, J. Li, M. A. Cohn, L. C. Cantley, and S. P. Gygi. 2004. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. USA 10112130-12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlanga, J. J., A. Baass, and N. Sonenberg. 2006. Regulation of poly(A) binding protein function in translation: characterization of the Paip2 homolog, Paip2B. RNA 121556-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Block, K. L., H. P. Vornlocher, and J. W. Hershey. 1998. Characterization of cDNAs encoding the p44 and p35 subunits of human translation initiation factor eIF3. J. Biol. Chem. 27331901-31908. [DOI] [PubMed] [Google Scholar]
- 8.Browning, K. S., D. R. Gallie, J. W. Hershey, A. G. Hinnebusch, U. Maitra, W. C. Merrick, and C. Norbury. 2001. Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem. Sci. 26284. [DOI] [PubMed] [Google Scholar]
- 9.Byrd, M. P., M. Zamora, and R. E. Lloyd. 2002. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 224499-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig, A. W., A. Haghighat, A. T. Yu, and N. Sonenberg. 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392520-523. [DOI] [PubMed] [Google Scholar]
- 11.Damoc, E., C. S. Fraser, M. Zhou, H. Videler, G. L. Mayeur, J. W. Hershey, J. A. Doudna, C. V. Robinson, and J. A. Leary. 2007. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol. Cell Proteomics 61135-1146. [DOI] [PubMed] [Google Scholar]
- 12.Deo, R. C., N. Sonenberg, and S. K. Burley. 2001. X-ray structure of the human hyperplastic discs protein: an ortholog of the C-terminal domain of poly(A)-binding protein. Proc. Natl. Acad. Sci. USA 984414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Einarson, M. B. 2001. Protein interaction technologies, p. 18.48-18.53. In J. Sambrook and D. W. Russell (ed.), Molecular cloning: a laboratory manual, 3rd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 14.Einarson, M. B., and J. R. Orlinick. 2002. Identification of protein-protein interactions with Glutathione-S-Transferase Fusion Proteins, p. 37-52. In E. Golemis (ed.), Protein-protein interactions: a molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 15.Ferraiuolo, M. A., S. Basak, J. Dostie, E. L. Murray, D. R. Schoenberg, and N. Sonenberg. 2005. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 170913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser, C. S., J. Y. Lee, G. L. Mayeur, M. Bushell, J. A. Doudna, and J. W. Hershey. 2004. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J. Biol. Chem. 2798946-8956. [DOI] [PubMed] [Google Scholar]
- 17.Gingras, A. C., and B. Raught. 2007. Signaling to translation initiation, p. 369-400. In M. Mathews, N. Sonenberg, and J. W. B. Hershey (ed.), Translational control in biology and medicine, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68913-963. [DOI] [PubMed] [Google Scholar]
- 19.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 895547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grifo, J. A., S. M. Tahara, M. A. Morgan, A. J. Shatkin, and W. C. Merrick. 1983. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 2585804-5810. [PubMed] [Google Scholar]
- 21.Hinnebusch, A. G. 2006. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31553-562. [DOI] [PubMed] [Google Scholar]
- 22.Holcik, M., and N. Sonenberg. 2005. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell. Biol. 6318-327. [DOI] [PubMed] [Google Scholar]
- 23.Hou, C. L., C. Tang, S. R. Roffler, and T. K. Tang. 2000. Protein 4.1R binding to eIF3-p44 suggests an interaction between the cytoskeletal network and the translation apparatus. Blood 96747-753. [PubMed] [Google Scholar]
- 24.Imataka, H., and N. Sonenberg. 1997. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 176940-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kahvejian, A., G. Roy, and N. Sonenberg. 2001. The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 66293-300. [DOI] [PubMed] [Google Scholar]
- 26.Karim, M. M., Y. V. Svitkin, A. Kahvejian, G. De Crescenzo, M. Costa-Mattioli, and N. Sonenberg. 2006. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc. Natl. Acad. Sci. USA 1039494-9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khaleghpour, K., A. Kahvejian, G. De Crescenzo, G. Roy, Y. V. Svitkin, H. Imataka, M. O'Connor-McCourt, and N. Sonenberg. 2001. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell. Biol. 215200-5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khaleghpour, K., S. Pyronnet, A. C. Gingras, and N. Sonenberg. 1999. Translational homeostasis: eukaryotic translation initiation factor 4E control of 4E-binding protein 1 and p70 S6 kinase activities. Mol. Cell. Biol. 194302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khaleghpour, K., Y. V. Svitkin, A. W. Craig, C. T. DeMaria, R. C. Deo, S. K. Burley, and N. Sonenberg. 2001. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell 7205-216. [DOI] [PubMed] [Google Scholar]
- 30.Kolupaeva, V. G., A. Unbehaun, I. B. Lomakin, C. U. Hellen, and T. V. Pestova. 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11470-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korneeva, N. L., B. J. Lamphear, F. L. Hennigan, and R. E. Rhoads. 2000. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J. Biol. Chem. 27541369-41376. [DOI] [PubMed] [Google Scholar]
- 32.Kozlov, G., J. F. Trempe, K. Khaleghpour, A. Kahvejian, I. Ekiel, and K. Gehring. 2001. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. USA 984409-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Bacquer, O., E. Petroulakis, S. Paglialunga, F. Poulin, D. Richard, K. Cianflone, and N. Sonenberg. 2007. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E-BP1 and 4E-BP2. J. Clin. Investig. 117387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, K. A., S. S. Schalm, A. Romanelli, K. L. Keon, and J. Blenis. 2001. Ribosomal S6 kinase 2 inhibition by a potent C-terminal repressor domain is relieved by mitogen-activated protein-extracellular signal-regulated kinase kinase-regulated phosphorylation. J. Biol. Chem. 2767892-7898. [DOI] [PubMed] [Google Scholar]
- 35.Masutani, M., N. Sonenberg, S. Yokoyama, and H. Imataka. 2007. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 263373-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molina, H., D. M. Horn, N. Tang, S. Mathivanan, and A. Pandey. 2007. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 1042199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phan, L., X. Zhang, K. Asano, J. Anderson, H. P. Vornlocher, J. R. Greenberg, J. Qin, and A. G. Hinnebusch. 1998. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol. 184935-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pisarev, A. V., A. Unbehaun, C. U. Hellen, and T. V. Pestova. 2007. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 430147-177. [DOI] [PubMed] [Google Scholar]
- 40.Roy, G., G. De Crescenzo, K. Khaleghpour, A. Kahvejian, M. O'Connor-McCourt, and N. Sonenberg. 2002. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol. Cell. Biol. 223769-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sachs, A. 2000. Physical and functional interactions between the mRNA cap structure and the poly(A) tail, p. 447-466. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Sachs, A. B., M. W. Bond, and R. D. Kornberg. 1986. A single gene from yeast for both nuclear and cytoplasmic polyadenylate-binding proteins: domain structure and expression. Cell 45827-835. [DOI] [PubMed] [Google Scholar]
- 43.Svitkin, Y. V., A. Gradi, H. Imataka, S. Morino, and N. Sonenberg. 1999. Eukaryotic initiation factor 4GII (eIF4GII), but not eIF4GI, cleavage correlates with inhibition of host cell protein synthesis after human rhinovirus infection. J. Virol. 733467-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svitkin, Y. V., and N. Sonenberg. 2004. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol. Biol. 257155-170. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida, M., K. Yoshida, G. Kozlov, N. S. Lim, G. De Crescenzo, Z. Pang, J. J. Berlanga, A. Kahvejian, K. Gehring, S. S. Wing, and N. Sonenberg. 2006. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 251934-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, M., A. M. Sandercock, C. S. Fraser, G. Ridlova, E. Stephens, M. R. Schenauer, T. Yokoi-Fong, D. Barsky, J. A. Leary, J. W. Hershey, J. A. Doudna, and C. V. Robinson. 1 July 2008, posting date. Special feature: mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl. Acad. Sci. USA. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed]