Abstract
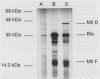
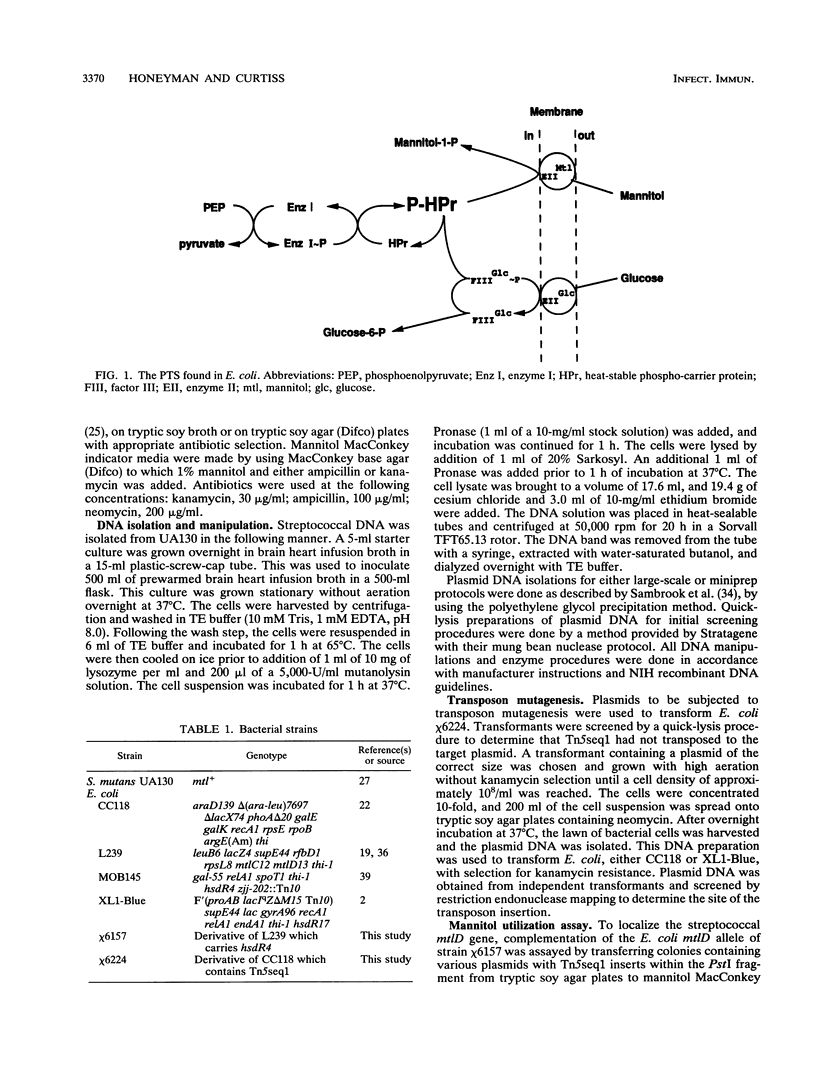
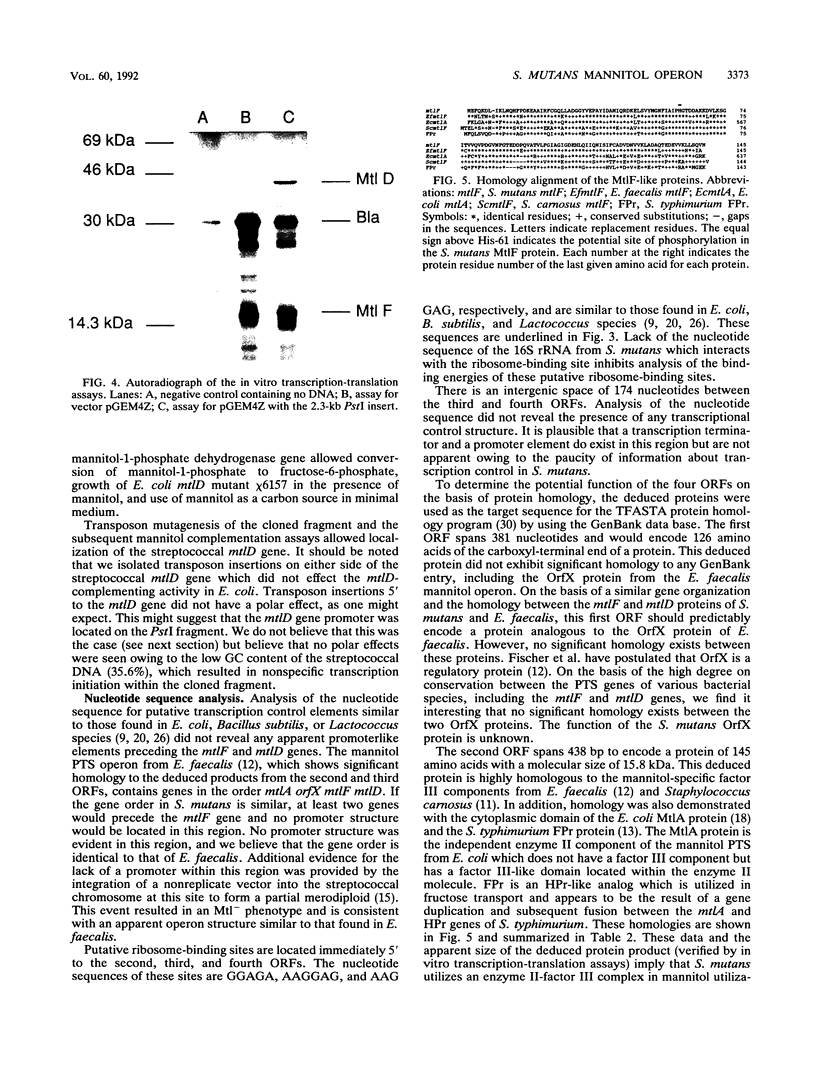
Streptococcus mutans, the causative agent of dental caries, utilizes carbohydrates by means of the phosphoenolpyruvate-dependent phosphotransferase system (PTS). The PTS facilitates vectorial translocation of metabolizable carbohydrates to form the corresponding sugar-phosphates, which are subsequently converted to glycolytic intermediates. The PTS consists of both sugar-specific and sugar-independent components. Complementation of an Escherichia coli mtlD mutation with a streptococcal recombinant DNA library allowed isolation of the mannitol-1-phosphate dehydrogenase gene (mtlD) and the adjacent sugar-specific mannitol factor III gene (mtlF) from S. mutans. Subsequent transposon mutagenesis of the complementing DNA fragment with Tn5seq1 defined the region that encodes the mtlD-complementing activity, the streptococcal mtlD gene. Nucleotide sequence analysis of this region revealed two complete open reading frames (ORFs) from within the streptococcal mannitol PTS operon. One ORF encodes the mtlD gene product, a 43.0-kDa protein which exhibits similarity to the E. coli and Enterococcus faecalis mannitol-1-phosphate dehydrogenases. The second ORF encodes a 15.8-kDa protein which exhibits similarity to mannitol factor III proteins from several bacterial species. In vitro transcription-translation assays were used to produce proteins of the sizes predicted by the streptococcal ORFs. These data indicate that the S. mutans mannitol PTS utilizes an enzyme II-factor III complex similar to the mannitol system found in other gram-positive organisms, as opposed to that of E. coli, which utilizes an independent enzyme II system.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown A. T., Patterson C. E. Ethanol production and alcohol dehydrogenase activity in Streptococcus mutans. Arch Oral Biol. 1973 Jan;18(1):127–131. doi: 10.1016/0003-9969(73)90027-7. [DOI] [PubMed] [Google Scholar]
- CURTIS S. R., 3rd CHROMOSOMAL ABERRATIONS ASSOCIATED WITH MUTATIONS TO BACTERIOPHAGE RESISTANCE IN ESCHERICHIA COLI. J Bacteriol. 1965 Jan;89:28–40. doi: 10.1128/jb.89.1.28-40.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. M., Ye Q. Z., Zhu Z. M., Wanner B. L., Walsh C. T. Molecular biology of carbon-phosphorus bond cleavage. Cloning and sequencing of the phn (psiD) genes involved in alkylphosphonate uptake and C-P lyase activity in Escherichia coli B. J Biol Chem. 1990 Mar 15;265(8):4461–4471. [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Davis T., Yamada M., Elgort M., Saier M. H., Jr Nucleotide sequence of the mannitol (mtl) operon in Escherichia coli. Mol Microbiol. 1988 May;2(3):405–412. doi: 10.1111/j.1365-2958.1988.tb00045.x. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drucker D. B., Melville T. H. Fermentation end-products of cariogenic and non-cariogenic streptococci. Arch Oral Biol. 1968 May;13(5):565–570. doi: 10.1016/0003-9969(68)90117-9. [DOI] [PubMed] [Google Scholar]
- Fischer R., Eisermann R., Reiche B., Hengstenberg W. Cloning, sequencing and overexpression of the mannitol-specific enzyme-III-encoding gene of Staphylococcus carnosus. Gene. 1989 Oct 30;82(2):249–257. doi: 10.1016/0378-1119(89)90050-4. [DOI] [PubMed] [Google Scholar]
- Fischer R., von Strandmann R. P., Hengstenberg W. Mannitol-specific phosphoenolpyruvate-dependent phosphotransferase system of Enterococcus faecalis: molecular cloning and nucleotide sequences of the enzyme IIIMtl gene and the mannitol-1-phosphate dehydrogenase gene, expression in Escherichia coli, and comparison of the gene products with similar enzymes. J Bacteriol. 1991 Jun;173(12):3709–3715. doi: 10.1128/jb.173.12.3709-3715.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerse R. H., Izzo F., Postma P. W. The PEP: fructose phosphotransferase system in Salmonella typhimurium: FPr combines enzyme IIIFru and pseudo-HPr activities. Mol Gen Genet. 1989 Apr;216(2-3):517–525. doi: 10.1007/BF00334399. [DOI] [PubMed] [Google Scholar]
- Hamada S., Slade H. D. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980 Jun;44(2):331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Wu L. F., Tomich J., Saier M. H., Jr, Niehaus W. G. Corrected sequence of the mannitol (mtl) operon in Escherichia coli. Mol Microbiol. 1990 Nov;4(11):2003–2006. doi: 10.1111/j.1365-2958.1990.tb02050.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee C. A., Saier M. H., Jr Mannitol-specific enzyme II of the bacterial phosphotransferase system. III. The nucleotide sequence of the permease gene. J Biol Chem. 1983 Sep 10;258(17):10761–10767. [PubMed] [Google Scholar]
- Lengeler J. Mutations affecting transport of the hexitols D-mannitol, D-glucitol, and galactitol in Escherichia coli K-12: isolation and mapping. J Bacteriol. 1975 Oct;124(1):26–38. doi: 10.1128/jb.124.1.26-38.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesche W. J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986 Dec;50(4):353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryanski J. H., Wittenberger C. L. Mannitol transport in Streptococcus mutans. J Bacteriol. 1975 Dec;124(3):1475–1481. doi: 10.1128/jb.124.3.1475-1481.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf W. W., Wanner B. L. Involvement of the Escherichia coli phn (psiD) gene cluster in assimilation of phosphorus in the form of phosphonates, phosphite, Pi esters, and Pi. J Bacteriol. 1991 Jan;173(2):587–600. doi: 10.1128/jb.173.2.587-600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Murchison H. H., Barrett J. F., Cardineau G. A., Curtiss R., 3rd Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect Immun. 1986 Nov;54(2):273–282. doi: 10.1128/iai.54.2.273-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag D. K., Huang H. V., Berg D. E. Bidirectional chain-termination nucleotide sequencing: transposon Tn5seq1 as a mobile source of primer sites. Gene. 1988 Apr 15;64(1):135–145. doi: 10.1016/0378-1119(88)90487-8. [DOI] [PubMed] [Google Scholar]
- Pas H. H., Robillard G. T. S-phosphocysteine and phosphohistidine are intermediates in the phosphoenolpyruvate-dependent mannitol transport catalyzed by Escherichia coli EIIMtl. Biochemistry. 1988 Aug 9;27(16):5835–5839. doi: 10.1021/bi00416a002. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma P. W., Lengeler J. W. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol Rev. 1985 Sep;49(3):232–269. doi: 10.1128/mr.49.3.232-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiche B., Frank R., Deutscher J., Meyer N., Hengstenberg W. Staphylococcal phosphoenolpyruvate-dependent phosphotransferase system: purification and characterization of the mannitol-specific enzyme IIImtl of Staphylococcus aureus and Staphylococcus carnosus and homology with the enzyme IImtl of Escherichia coli. Biochemistry. 1988 Aug 23;27(17):6512–6516. doi: 10.1021/bi00417a047. [DOI] [PubMed] [Google Scholar]
- Saier M. H., Jr, Yamada M., Erni B., Suda K., Lengeler J., Ebner R., Argos P., Rak B., Schnetz K., Lee C. A. Sugar permeases of the bacterial phosphoenolpyruvate-dependent phosphotransferase system: sequence comparisons. FASEB J. 1988 Mar 1;2(3):199–208. doi: 10.1096/fasebj.2.3.2832233. [DOI] [PubMed] [Google Scholar]
- Schachtele C. F., Mayo J. A. Phosphoenolpyruvate-dependent glucose transport in oral streptococci. J Dent Res. 1973 Nov-Dec;52(6):1209–1215. doi: 10.1177/00220345730520060801. [DOI] [PubMed] [Google Scholar]
- Solomon E., Lin E. C. Mutations affecting the dissimilation of mannitol by Escherichia coli K-12. J Bacteriol. 1972 Aug;111(2):566–574. doi: 10.1128/jb.111.2.566-574.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood D. O., Atkinson W. H., Sikorski R. S., Winkler H. H. Expression of the Rickettsia prowazekii citrate synthase gene in Escherichia coli. J Bacteriol. 1983 Jul;155(1):412–416. doi: 10.1128/jb.155.1.412-416.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]