Abstract
Toll-like receptors (TLRs) are important sensors of microbial pathogens and mediators of innate immune responses. Although the signal transduction of TLRs is well elucidated, their basal regulation is largely unexplored. Here we show that the tumor suppressor p53 positively regulates the transcription of TLR3, a receptor for viral double-stranded RNA and poly(I-C), by binding to the p53 site in the TLR3 promoter. TLR3 expression was lower in HCT116 p53−/− cells than in HCT116 p53+/+ cells. Activation of p53 by 5-fluorouracil increased the TLR3 mRNA in epithelial cell lines with wild-type p53 but not in cell lines harboring mutant p53. Knockdown of p53 by small interfering RNA decreased the TLR3 expression. TLR3 mRNA was also lower in liver and intestine of p53−/− mice than in p53+/+ mice. Furthermore, the poly(I-C)-induced phosphorylation of IκB-α, nuclear translocation of NF-κB, and phosphorylation of interferon regulatory transcription factor 3, were drastically reduced in HCT116 p53−/− cells, indicating a dysregulation of the two signaling pathways governed by TLR3. Consequently, induction of interleukin-8 and beta interferon after poly(I-C) stimulation was impaired in HCT116 p53−/− cells. These results suggest that p53 influences TLR3 expression and function and highlight a role of p53 in innate immune response in epithelial cells.
The tumor suppressor p53 protein has been proven to regulate a network of biological processes such as cell cycle, differentiation, aging, and death by its role as a transcriptional regulator (32, 44). p53 regulates transcription by binding to DNA in a sequence-specific manner through a highly conserved DNA-binding domain. The importance of DNA-protein interactions in p53 function is emphasized by the fact that the majority of p53 mutations found in human tumors are clustered in the DNA-binding domain (reviewed in reference 20). The mutated p53 allele encodes defective protein that can no longer bind to DNA to activate transcription.
Inactivation of p53 protein can be induced by some viruses implicated in the development of cancer (6, 31) as one of the viral mechanisms to inhibit apoptosis and prolong the survival of the virus. However, viruses with no tumorigenic potential and double-stranded RNA (dsRNA) have also been shown to downregulate p53 (14, 25), suggesting the importance of p53 in host response to viruses. Further evidence of p53's role in antiviral defense came from the observation that p53 can be induced by interferon (IFN), an antiviral cytokine, to evoke apoptosis in virus-infected cells (38). These studies highlight the function of p53 not only in cancer but also in immunity.
An important arm in innate immunity is the recognition of viral and bacterial products mediated by pattern recognition receptors such as the Toll-like receptor (TLR) family, which consists of more than 10 members that respond to a variety of pathogen-associated molecular patterns (PAMPs) (1). A subfamily of TLR, TLRs 3, 7, 8, and 9, recognizes viral nucleic acids and induces type I IFN. TLR3 recognizes dsRNA and its synthetic analog poly(I-C), which has been extensively used to mimic dsRNA (2). TLR7 and TLR8 recognize single-stranded RNA (8, 16). TLR9 responds to viral DNA containing the CpG motif (reviewed in reference 1). These TLRs together constitute a powerful system to detect the genetic material of viruses. While numerous studies have already elucidated the signal transduction of these virus-sensing TLRs and how they regulate the antiviral response (reviewed in references 23 and 35), fewer studies have focused on their basal regulation.
Because p53 is a well-known transcription factor that is also involved in viral response, we explored the possibility of p53 being a regulator of TLRs. In a screening of various TLR ligands, we observed that poly(I-C), a ligand for TLR3, induced a cytokine response dependently on p53. Here we present evidence that p53 activates TLR3 transcription by binding to the p53 consensus site in the TLR3 promoter. TLR3 expression was decreased in colonic epithelial HCT116 p53−/− cells as well as in the liver and intestine of p53−/− mice. The downregulated expression of TLR3 in HCT116 p53−/− cells led to a dysfunction in both NF-κB and IFN regulatory transcription factor 3 (IRF-3) signaling pathways, which are governed by TLR3 (22), in response to poly(I-C) treatment and, consequently, a reduced induction of downstream cytokines upon stimulation with poly(I-C). These findings present a novel, direct role of p53 in regulating TLR3 and may have a significant implication for viral recognition mediated by TLR3.
MATERIALS AND METHODS
Reagents and antibodies.
Poly(I-C) and bacterial DNA (B-DNA) were purchased from InvivoGen (San Diego, CA). Peptidoglycan (PGN) from Staphylococcus aureus was obtained from Fluka (Buchs, Switzerland). Lipopolysaccharide (LPS) from Escherichia coli O111:B4 was purchased from Sigma (St. Louis, MO). R848 was from Alexis Biochemicals (San Diego, CA). 5-Fluorouracil (5-FU) was purchased from Wako (Osaka, Japan). IFN-β was supplied by TORAY (Tokyo, Japan). Antibodies for p65 (sc-8008), IRF-3 (sc-9082), normal mouse immunoglobulin G (IgG [sc-2025]), and γ-tubulin (sc-7396) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies for IκB-α (#9242), phosphorylated IκB-α (14D4) and phosphorylated IRF-3 (4D4G) were purchased from Cell Signaling Technology (Danvers, MA). Anti-TLR3 antibody (IMG-315A) was purchased from Imgenex (San Diego, CA). Anti-Hsc-70 antibody (SPA-815) was obtained from Stressgen Bioreagents (Canada). The horseradish peroxidase-conjugated secondary antibodies used in this study were purchased from Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA).
Cell culture, treatment, and transfection.
Human colorectal cancer cell line HCT116 (p53+/+ [wild type]) and HCT116 p53−/− cells were kindly provided by B. Vogelstein from Johns Hopkins University. These cells were maintained in Dulbecco's modified Eagle's medium-Ham's F-12 (DMEM/F12) medium supplemented with 10% fetal bovine serum (FBS). A549 lung adenocarcinoma cells and Caco2 colon adenocarcinoma cells were maintained in DMEM containing 10% FBS and antibiotics. Human hepatoma cells (HepG2) were maintained in minimum essential medium supplemented with 10% FBS and antibiotics. Calu-3 cells were cultured in DMEM/F12 with 15% FBS and antibiotics. These cell lines were obtained from the American Type Culture Collection and were cultured at 37°C in a humidified atmosphere of 5% CO2. Treatment of cells with 4 or 8 μM 5-FU was carried out for 24 h. For the HepG2, Caco2, and Calu-3 cell lines, treatment with 5-FU was for 8 h at a concentration of 400 or 800 μM. Stimulation of cells with 10 μg/ml TLR agonists in serum-free medium was performed for 6 h. Poly(I-C) treatment was carried out at the indicated time and concentration. At the time of treatment, the culture medium was replaced with fresh, serum-free DMEM/F12 containing poly(I-C). Transient transfections of plasmids were performed using Hilymax (Dojindo Laboratories, Kumamoto, Japan) following the manufacturer's recommendation. Specifically, 4 μl of Hilymax diluted in Opti-MEM (Gibco) was mixed with total DNA in a ratio of 1:4 (DNA-Hilymax) and applied to subconfluent cells in DMEM/F12. Small interfering RNA (siRNA) for p53 (si-p53) and TLR3 (si-TLR3) was transfected into HCT116 p53+/+ cells using Trans-IT TKO (Mirus, Madison, WI) according to the manufacturer's instructions. Fifty or 100 nM of p53 siRNA and 100 nM of TLR3 siRNA duplex were transfected into 70% confluent cells to knock down p53 and TLR3, respectively. GL2-luciferase (luc) siRNA duplex was used as a control. The cells were harvested 48 h after transfection.
RT-PCR analysis.
Total RNA was isolated from cells and mouse tissues (liver, spleen, and intestine) with TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Conventional (or semiquantitative) reverse transcriptase PCR (RT-PCR) was carried out with an RT-PCR kit (TaKaRa, Japan) by the recommended protocol. The oligonucleotide primers used for PCR are shown in Table 1.
TABLE 1.
Primers used for semiquantitative RT-PCR
| Gene | Primer sequence
|
|
|---|---|---|
| Sense | Antisense | |
| TLR3 | 5′-CCTGCAGCTGACTAGGAACTCCTTTG-3′ | 5′-TGCTGCAAATCGAGAATTTCTAG-3′ |
| IFN-β | 5′-CACGACAGCTCTTTCCATGA-3′ | 5′-AGCCAGTGCTCGATGAATCT-3′ |
| IL-8 | 5′-AACATGACTTCCAAGCTGGCC-3′ | 5′-TTATGAATTCTCAGCCTTCTTC-3′ |
| p21 | 5′-ATGTCAGAACCGGCTGGGGATGTCCGTC-3′ | 5′-TTAGGGCTTCCTCTTGGAGAAGATCAGCC-3′ |
| p53 | 5′-CCTCAACAAGATGTTTTGCCAACTG-3′ | 5′-GAGTCTTCCAGTGTGATGATGGTGAC-3′ |
| GAPDH | 5′-CGGGAAGCTTGTGATCAATGG-3′ | 5′-GGCAGTGATGGCATGGACTG-3′ |
Real-time quantitative RT-PCR analyses for TLR3, interleukin-8 (IL-8), IFN-β, hypoxanthine phosphoribosyltransferase (HPRT), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were carried out with Sybr green master mix (Applied Biosystems, CA) according to the manufacturer's instructions. PCR amplifications were performed as described previously (36). The threshold cycle values for each gene amplification were normalized by subtracting the threshold cycle value calculated for GAPDH (internal control for cells) or HPRT (internal control for mice tissues). The normalized gene expression values were expressed as the relative quantity of gene-specific mRNA (TLR3, IL-8, and IFN-β). The oligonucleotide primers used in the real-time quantitative PCR amplifications are shown in Table 2.
TABLE 2.
Primers used for real-time quantitative RT-PCR
| Gene | Primer sequence
|
|
|---|---|---|
| Sense | Antisense | |
| Human TLR3 | 5′-ACCCGATGATCTACCCACAAAC-3′ | 5′-GTTGGCGGCTGGTAATCTTC-3′ |
| IFN-β | 5′-CACGACAGCTCTTTCCATGA-3′ | 5′-AGCCAGTGCTCGATGAATCT-3′ |
| IL-8 | 5′-CTGGCCGTGGCTCTCTTG-3′ | 5′-CCTTGGCAAAACTGCACCTT-3′ |
| GAPDH | 5′-CGGGAAGCTTGTGATCAATGG-3′ | 5′-GGCAGTGATGGCATGGACTG-3′ |
| Mouse TLR3 | 5′-TTGTCTTCTGCACGAACCTG-3′ | 5′-CGCAACGCAAGGATTTTATT-3′ |
| HPRT | 5′-GTTGGATACAGGCCAGACTTTGTTG-3′ | 5′-GATTCAACTTGCGCTCATCTTAGGC-3′ |
Plasmids and luciferase assay.
Cloning of the TLR3 genomic 5′ region and construction of luciferase reporter gene plasmids were performed as described previously (40). Briefly, the 2 kb upstream of TLR3 gene and part of its 5′ untranscribed region were amplified with TaKaRa LA Taq polymerase and the primers listed in Table 3 using the genomic DNA prepared from HCT116 p53+/+ cells as a template. The PCR product was cloned into the pCR2.1 vector with the TA cloning kit (Invitrogen, Carlsbad, CA). TLR3 promoter-luc reporter plasmid was constructed by subcloning TLR3 promoter fragment (from −2006 to +48 [−2006/+48]) into the KpnI-BglII sites of pGL3-basic vector (pGL3b [Promega, Madison, WI]). Deletion mutants of the TLR3 promoter were constructed using primers (Table 3) and the −2006/+48 construct as a template and inserting the amplified fragments into pGL3b. All of the point-mutated constructs were prepared using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The sequences of primers used for mutagenesis are shown in Table 4. All mutant plasmids were made using the −2006/+48 construct as a template. The IFN-β-luc construct (−300 bp) was cloned in pCR2.1 and subcloned into pGL3b vector in the same manner described above using genomic DNA isolated from HCT116 cells. The primers used for cloning the IFN-β promoter are listed in Table 3. All of the plasmids used in this study were sequenced at the genomics facility of Macrogen (Seoul, South Korea). The reporter construct IL-8-luc contains the luciferase gene under the regulation of the IL-8 promoter, as described previously (34). The p53 expression plasmid was cloned in the pCDM8 expression vector as described in reference 39.
TABLE 3.
Primers used for cloning of TLR3 genomic 5′ region and promoter constructs
| Primer | Sequence |
|---|---|
| 5′TLR3 prom (bp −2006 KpnI) | 5′-GGGGTACCATGACTCAATGCAAACTTGAATTTGCC-3′ |
| 5′TLR3 prom (bp −987 KpnI) | 5′-GGGGTACCTATAAAGCGGTCTAGCTGAAGCTGGAG-3′ |
| 5′TLR3 prom (bp −504 KpnI) | 5′-GGGGTACCTCTCAGCTTTGCCATGTTTGGC-3′ |
| 3′TLR3 prom (BglII) | 5′-GCGCAGATCTCATTTCATCAGGGAAGTGTGTGGC-3′ |
| 5′IFN-β prom (KpnI) | 5′-GGGGTACCTTGAATTCTCAGGTCGTTTGC-3′ |
| 3′IFN-β prom (BglII) | 5′-GCGGAGATCTAAAGGTTGCAGTTAGAATGTCC-3′ |
TABLE 4.
Primers used for the generation of mutant TLR3 promoter constructs
| Primer | Sequence |
|---|---|
| TLR3 prom MT1 (−1929)_sense | 5′-GCTGAGACTATAGGGATCCACCAACATGCCCAGC-3′ |
| TLR3 prom MT1 (−1929)_antisense | 5′-GCTGGGCATGTTGGTGGATCCCTATAGTCTCAGC-3′ |
| TLR3 prom MT2 (−1313)_sense | 5′-CCCACTGTTCAAAGCTAGTCGGATCCCCAAGCCTAGAGTCAGCG-3′ |
| TLR3 prom MT2 (−1313)_antisense | 5′-CGCTGACTCTAGGCTTGGGGATCCGACTAGCTTTGACCAGTGGG-3′ |
For luciferase assays, HCT116 cells seeded onto 12-well plates were transfected with 0.2 μg of the TLR3-luc, IFN-β-luc, or IL-8-luc promoter construct, together with Renilla luciferase plasmid (phRG-TK; Promega Corp., Madison, WI), p53 expression plasmid, or si-p53 duplex. pcDNA3.1 vector and si-GL2 duplex were used as controls. Luciferase activity was determined using a dual-luciferase reporter assay system (Promega) as described previously (37). The siRNA oligonucleotide sequences are listed in Table 5.
TABLE 5.
siRNA oligonucleotide sequences
| Oligonucleotide | Sequence |
|---|---|
| hTLR3 siRNA_sense | 5′-GGUAUAGCCAGCUAACUAGTT-3′ |
| hTLR3 siRNA_antisense | 5′-CUAGUUAUCUGGCUAUACCTT-3′ |
| hp53 siRNA_sense | 5′-GACUCCAGUGGUAAUCUACTT-3′ |
| hp53 siRNA_antisense | 5′-GUAGAUUACCACUGGAGUCTT-3′ |
| GL2 siRNA_sense | 5′-CGUACGCGGAAUACUUCGATT-3′ |
| GL2 siRNA_antisense | 5′-UCGAAGUAUUCCGCGUACGTT-3′ |
ChIP assay.
For the chromatin immunoprecipitation (ChIP) assay, HCT116 p53+/+ cells were cross-linked using formaldehyde (1% final concentration) added directly to the cell culture media at 37°C for 15 min, and the reaction was stopped by adding glycine (0.125 M final concentration). Cells were rinsed with cold phosphate-buffered saline and resuspended in cell lysis buffer, consisting of 5 mM PIPES [piperazine-N,N′-bis(ethanesulfonic acid), pH 8.0], 85 mM KCl, 0.5% NP-40, and 1% protease inhibitor (PI) cocktail (Sigma). This mixture was incubated on ice for 10 min and then Dounce homogenized. The nuclei were resuspended in nucleus lysis buffer (1% sodium dodecyl sulfate [SDS], 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], and 1% PI cocktail) and incubated on ice for 10 min. The samples were sonicated on ice with an Ultrasonic Disruptor (TOMY, Japan) to an average length of ∼1,000 bp and microcentrifuged. The chromatin solution was precleared using Staphylococcus aureus protein A-positive cells (Pansorbin, #507862; Calbiochem). Precleared chromatin was incubated with 4 μg of anti-p53 mouse monoclonal antibody or mouse IgG at 4°C for 24 h and microcentrifuged. We performed the washing, cross-link reversal, and DNA extraction following the procedure described in http://www.genomecenter.ucdavis.edu/expression_analysis/. Samples were analyzed by PCR using LA Taq polymerase (TaKaRa) according to the recommended protocol. The primers used (Table 6) recognize a fragment of the human TLR3 promoter, p21 promoter, or GAPDH promoter (45).
TABLE 6.
Primers used for the ChIP assay
| Primer | Sequence |
|---|---|
| TLR3 prom_p53 binding site_sense | 5′-GCACTCATGACTCAATGCAAAC-3′ |
| TLR3 prom_p53 binding site_antisense | 5′-CACTTTAGGAGATTGCTTGAGC-3′ |
| p21 prom_p53 binding site_sense | 5′-CCAGCCCTTTGGATGGTTT-3′ |
| p21 prom_p53 binding site_antisense | 5′-GCCTCCTTTCTGTGCCTGA-3′ |
| GAPDH prom_sense | 5′-AAAAGCGGGGAGAAAGTAGG-3′ |
| GAPDH prom_antisense | 5′-CTAGCCTCCCGGGTTTCTCT-3′ |
Western blotting and immunoprecipitation.
To analyze the TLR3 expression level, HCT116 p53+/+ and p53−/− cells were lysed with radioimmunoprecipitation assay buffer (50 mM Tris-HCl, 150 mM NaCl, 1 mg/ml sodium deoxycholate, 1% NP-40) containing 1% PI cocktail. Cell lysates were incubated for 12 h at 4°C with 2 μg anti-TLR3 antibody or mouse IgG immobilized in protein G Sepharose beads (Amersham Bioscience, Sweden). After washing, immunoprecipitates were eluted in 2× SDS sample buffer, boiled, loaded on 6% SDS-polyacrylamide gel electrophoresis gel, and blotted onto polyvinylidene difluoride membrane (Millipore Corp., MA). Blots were probed with anti-TLR3 antibody or anti-Hsc70 antibody (from the input fraction) for internal control. For Western blotting analysis of p65, IκB-α, and IRF-3, HCT116 p53+/+ and HCT116 p53−/− cells were stimulated with poly(I-C). p65 was detected in nuclear extracts obtained as described previously (24). IκB-α, phosphorylated IκB-α, IRF-3, and phosphorylated IRF-3 were assessed in cytosolic fractions extracted by lysing cells with radioimmunoprecipitation assay buffer containing 1% PI cocktail and rotated overnight at 4°C. Equal amounts of samples were fractionated on a 7.5% SDS-polyacrylamide gel electrophoresis gel and transferred to polyvinylidene difluoride membrane. After blocking, the membrane was probed with the appropriate antibodies, and blots were visualized using SuperSignal (Pierce, Rockford, IL).
Animals.
p53 knockout (p53−/−) mice characterized previously (41) were kindly provided by N. Araki from Kumamoto University, Kumamoto, Japan. The p53+/+, p53+/−, and p53−/− mice used in this study were housed in a vivarium in accordance with the guidelines of the animal facility center of Kumamoto University. The animals were fed with chow ad libitum. All experiments were performed according to the protocols approved by the Animal Welfare Committee of Kumamoto University (#A19-115).
Statistical analysis.
For statistical analysis, the data were analyzed by Student's t test or one-way analysis of variance (ANOVA) with either Tukey-Kramer's or Dunnett's multiple comparison test (JMP software; SAS Institute), as indicated in each figure legend. A P value of <0.05 is considered statistically significant.
RESULTS
p53 positively regulates TLR3 expression in human epithelial cells.
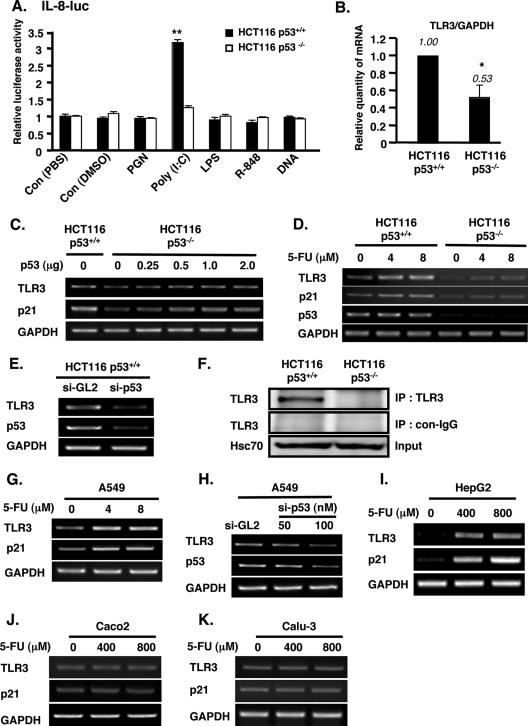
p53 has been implicated in the innate response to pathogens (30, 31), and while TLRs are well known for mediating the recognition and response to microbial products, the relationship between p53 and TLRs is largely unexplored. To assess the involvement of p53 in TLRs' response to PAMPs, we stimulated HCT116 p53+/+ and p53−/− cells with various TLR ligands, such as PGN, poly(I-C), LPS, B-DNA, and R848, and we assayed the promoter activity of IL-8, a general downstream cytokine of TLRs, in cells treated with these TLR ligands. We observed that only poly(I-C), a TLR3 ligand, significantly activated the promoter of IL-8 in p53+/+ cells (Fig. 1A). Interestingly, this activation was dependent on p53 because poly(I-C) had no effect on IL-8 activity in HCT116 p53−/− cells (Fig. 1A). Based on these results and the previous report that TLR3 is highly expressed in human colon epithelia (11), we focused on TLR3 and its molecular link with p53.
FIG. 1.
p53 positively regulates the expression of TLR3 in epithelial cells. (A) HCT116 p53+/+ and p53−/− cells were transfected with IL-8 promoter plasmid and treated with 10 μg/ml each of the indicated TLR agonists for 6 h. IL-8 promoter activity was measured 48 h after transfection of IL-8 promoter. The data shown are means ± standard errors from triplicate platings and represent three independent experiments. **, P < 0.001 versus control (Con; phosphate-buffered saline [HCT116 p53+/+]) assessed by ANOVA with Dunnett's procedure. (B) TLR3 mRNA in HCT116 p53+/+ and p53−/− cells was determined by real-time quantitative PCR. The TLR3 mRNA level was normalized to GAPDH (internal control). The data shown are means ± standard deviations from triplicate determinations from three independent experiments. *, P < 0.05 assessed by Student's t test. (C) TLR3 mRNA was examined in HCT116 p53+/+ and in HCT116 p53−/− cells untransfected or transfected with increasing amounts of p53 expression plasmid. (D) TLR3 mRNA was determined in HCT116 p53+/+ and p53−/− cells treated for 24 h with 4 or 8 μM 5-FU. For panels C and D, p21 served as a positive control. GAPDH was used as an internal control for RT-PCR analyses. (E) HCT116 p53+/+ cells were transfected with p53 siRNA (si-p53) or control siRNA duplex (si-GL2), and total RNA from these cells was isolated for analysis of TLR3 and p53 mRNA by RT-PCR. (F) The TLR3 protein level was determined in lysates of HCT116 p53+/+ and p53−/− cells by immunoprecipitation (IP) and Western blotting using anti-TLR3 antibody. Hsc70 was used as internal control. con-IgG, control IgG. (G) A549 cells were treated with 4 or 8 μM 5-FU for 24 h. (H) si-GL2 or the indicated amount of p53 siRNA duplex was transfected in A549 cells. Twenty-four hours later, RNA was extracted. (I to K) HepG2 (I), Caco2 (J), and Calu-3 (K) cells were treated with 400 or 800 μM 5-FU for 8 h. For panels G to K, total RNA was extracted and analyzed by RT-PCR for the expression of TLR3, p21 (positive control), or p53. GAPDH served as the internal control.
We first investigated the effect of p53 on TLR3 mRNA expression by performing quantitative real time RT-PCR using HCT116 p53+/+ and p53−/− cells. The normalized mRNA level of TLR3 was significantly decreased to half in p53−/− cells compared with that in p53+/+ cells (Fig. 1B). Transient transfection of the p53 expression plasmid in HCT116 p53−/− cells increased the level of TLR3 mRNA, as determined by conventional RT-PCR (Fig. 1C). Activation of p53 in HCT116 p53+/+ cells by treatment with 5-FU notably increased the expression of TLR3, but it had minimal effect on the TLR3 level in p53−/− cells (Fig. 1D). p21, a known target of p53, was used as a positive control in these experiments. Conversely, the knockdown of p53 by siRNA in HCT116 p53+/+ cells downregulated the expression of TLR3 (Fig. 1E). Consistent with the mRNA data, the endogenous protein expression of TLR3 was also higher in p53+/+ cells than in p53−/− cells, as determined by immunoprecipitation followed by Western blotting analysis (Fig. 1F).
Furthermore, we investigated the expression of TLR3 in other epithelial cell lines that contain the wild-type p53 gene. In A549 cells, a lung epithelial cell line, the mRNA expression level of TLR3 was also increased upon activation of p53 by 5-FU treatment, similar to that of p21 (Fig. 1G). As confirmatory evidence, the knockdown of endogenous p53 in A549 cells by p53 siRNA decreased the expression of TLR3 (Fig. 1H). In the hepatoma cell line HepG2, we found that although the baseline levels of TLR3 and p21 were barely detectable, the activation of p53 by 5-FU induced the mRNA expression of TLR3 and p21 (Fig. 1I). Treatment of HepG2 cells with 5-FU was carried out at a higher concentration (400 or 800 μM) and for a shorter period (8 h), unlike the treatment conditions (4 or 8 μM and 24 h) for HCT116 p53+/+ or A549 cells, because the basal level of p53 in HepG2 cells is lower than that in the other two cell lines (unpublished data). In contrast to the results of 5-FU treatment obtained in these cell lines carrying wild-type p53, we observed that in cell lines harboring mutant p53, such as those of the colon carcinoma line Caco2 (9) and lung carcinoma line Calu-3 (10), the expression of TLR3 (and of p21) was unaffected by 5-FU treatment even at a high dose (Fig. 1J and K). Taken together, these data indicated that p53 upregulates TLR3 expression in epithelial cells.
p53 transactivates TLR3 by binding to the TLR3 promoter region.
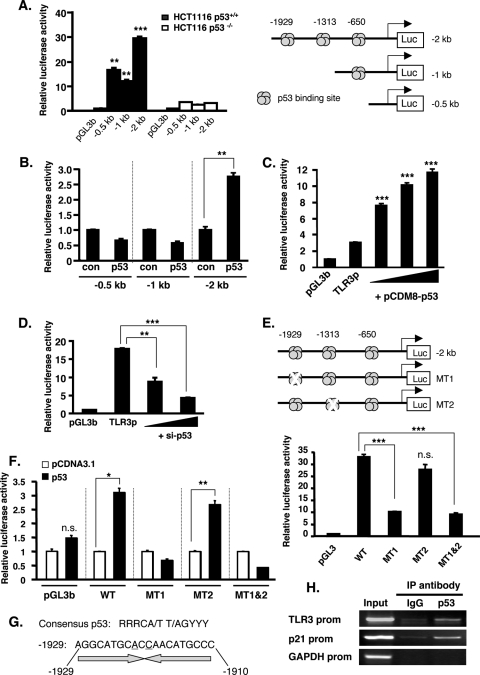
Because the effect of p53 on TLR3 expression was observed at the mRNA level, we focused on the TLR3 promoter activity to clarify the mechanism of how p53 activates TLR3 expression. In silico analysis of the TLR3 promoter using the Directory of p53 Consensus DNA Binding Site database (http://linkage.rockefeller.edu/p53/index.html) revealed a few putative binding sites for p53 contained within the 2 kb upstream from the transcription start site. To investigate the important region of TLR3 promoter that is regulated by p53, we used three lengths of TLR3 promoter constructs labeled −2 kb (−2006/+48), −1 kb (−987/+48), and −0.5 kb (−504/+48), containing 3, 1, or no p53 binding sites, respectively (Fig. 2A, right panel). These constructs were transfected in HCT116 p53+/+ and p53−/− cells. Reporter assays revealed that the TLR3 promoter has high activity in HCT116 p53+/+ cells but minimal activity in HCT116 p53−/− cells, suggesting that p53 is important in the activation of TLR3 promoter (Fig. 2A, left panel). Interestingly, the 0.5-kb promoter construct that does not contain p53 binding site was still responsive, with an activity of ∼15-fold in p53 wild-type cells, indicating that other factors might be necessary to activate the TLR3 promoter (40). The −2 kb promoter construct, which contains three binding sites for p53, showed the highest activity (∼30-fold) in HCT116 p53+/+ cells (Fig. 2A, left panel). To confirm the importance of the p53 binding sites and their responsiveness to p53, we reconstituted the HCT116 p53−/− cells with wild-type p53 by transient transfection of p53 plasmid together with the TLR3 promoter constructs. Consistent with the data above, albeit at a lower magnitude, the −2 kb promoter was significantly responsive to the transfected p53 compared with the −0.5 kb and −1 kb promoter constructs (Fig. 2B). Moreover, the activity of the −2 kb TLR3 promoter in HCT116 p53−/− cells was increased by transfection of p53 in a dose-dependent manner (Fig. 2C). Conversely, the knockdown of p53 by siRNA in HCT116 p53+/+ cells dose dependently suppressed the activity of the −2 kb TLR3 promoter (Fig. 2D). Collectively, these data indicated that p53 positively affects the TLR3 promoter activity.
FIG. 2.
p53 binds to and transactivates the TLR3 promoter. (A) HCT116 p53+/+ and p53−/− cells were transiently transfected with the indicated TLR3 promoter constructs (0.2 μg). Luciferase activity was determined 48 h after transfection of plasmids and is expressed as activation (fold) over that of the pGL3b vector. Values are means ± standard errors from triplicate platings. The data shown are representative of three independent experiments. ** and ***, P < 0.001 and P < 0.0001, respectively, against the corresponding promoter length in HCT116 p53−/− cells, as determined by Student's t test. (Right panel) Schematic diagram of the TLR3 promoter constructs containing the p53 binding sites. (B) HCT116 p53−/− cells were transiently transfected with the indicated TLR3 promoter constructs (0.2 μg) and 0.025 μg p53 plasmid or pcDNA3.1 empty vector (con), and luciferase activity was assayed 48 h posttransfection. Values are means ± standard errors from triplicate platings. Data represent three independent experiments. **, P < 0.001, as determined by Student's t test. (C) HCT116 p53−/− cells were transiently transfected with pGL3b vector or −2 kb TLR3 promoter (TLR3p), cotransfected with p53 expression plasmid at increasing amounts (6.25, 12.5, and 25 ng), and assayed for luciferase activity. Values are means ± standard errors from triplicate platings. The data shown are representative of three independent experiments. ***, P < 0.0001 versus TLR3p, as assessed by ANOVA with Dunnett's procedure. (D) HCT116 p53+/+ cells were transiently transfected with the −2 kb TLR3 promoter and cotransfected with si-GL2 or si-p53 (50 or 100 nM) oligonucleotide, and luciferase activity was assayed 48 h posttransfection. Values are means ± standard errors from triplicate platings. The data represent three independent experiments. ** and ***, P < 0.001 and P < 0.0001, respectively, against TLR3p, as determined by ANOVA with Dunnett's test. (E) Wild-type or mutant (mutated p53-binding sites) TLR3 promoter (−2 kb) was transfected into HCT116 p53+/+ cells, and luciferase activity was assayed 48 h after transfection. Values are means ± standard errors from triplicate platings. Data represent three independent experiments. ***, P < 0.0001 against the wild-type (WT) TLR3 promoter, as determined by ANOVA with Tukey-Kramer's test. n.s., not significant. (Upper panel) Schematic diagram of the −2 kb TLR3 promoter with the indicated position of the mutated p53 binding site. (F) HCT116 p53−/− cells were transfected with pGL3b and wild-type (WT) or mutant TLR3 promoter and cotransfected with pCDNA3.1 empty vector or p53 plasmid, and luciferase activity was assessed 48 h after transfection. Values are means ± standard errors from triplicate platings. The data represent three independent experiments. * and **, P < 0.01 and P < 0.001, respectively, against pCDNA3.1 vector, as assessed by Student's t test. (G) Consensus site of p53 and the sequence of p53 binding site in the TLR3 promoter at −1929. Underlined bases denote sequence variation from the consensus p53 element. (H) Representative result of the ChIP analysis in HCT116 p53+/+ cells using p53 antibody or mouse IgG for immunoprecipitation (IP) and primers of the indicated promoter region for PCR.
Because the activity of the −1 kb TLR3 promoter construct containing one p53 binding site (at bp −650) was not increased by p53 transfection (Fig. 2B), we deduced that the important sites for p53 regulation of the TLR3 promoter are at bp −1929 and −1313. To investigate this possibility, we created point mutants at these sites (Fig. 2E, upper panel) and transfected the mutated constructs in HCT116 p53+/+ cells. As shown in Fig. 2E (lower panel), the mutation of bp −1929 (MT1) substantially reduced the promoter activity of TLR3. On the other hand, point mutation at bp −1313 (MT2) barely affected the activity of TLR3 promoter. When both sites (bp −1929 and −1313 [MT1&2]) were mutated, the promoter activity was decreased to the level similar to that of MT1, indicating that bp −1929 is an important site for TLR3 promoter response to p53. To confirm the importance of bp −1929 for the responsiveness to p53, we transfected HCT116 p53−/− cells with wild-type or mutant TLR3 promoter together with an empty vector or p53-expressing plasmid. Wild-type TLR3 and MT2 promoters were responsive to exogenous p53, but the MT1 and MT1&2 promoters were not activated by p53 (Fig. 2F), confirming that bp −1929 (MT1) is necessary for the TLR3 promoter responsiveness to p53.
The tetrameric p53 protein binds to a palindromic consensus DNA sequence arranged as inverted repeats (18). This putative p53 binding sequence is present within the TLR3 promoter at bp −1929 to −1910, as shown in Fig. 2G. To test whether p53 binds to this region in the TLR3 promoter, we performed a ChIP experiment in HCT116 p53+/+ cells using primers that recognize the p53 binding site at bp −1929. Immunoprecipitation with p53 antibody, but not with control mouse IgG antibody and subsequent PCRs revealed the association of p53 with the promoter region of TLR3 (Fig. 2H). Similarly, as a positive control, p53 also binds to the p21 promoter. On the other hand, p53 binding was not detected in the GAPDH promoter, which served as a negative control. Taken together, these data suggested that p53 activates TLR3 transcription through the binding of p53 to the TLR3 promoter.
The TLR3 signaling pathway is impaired in HCT116 p53−/− cells.
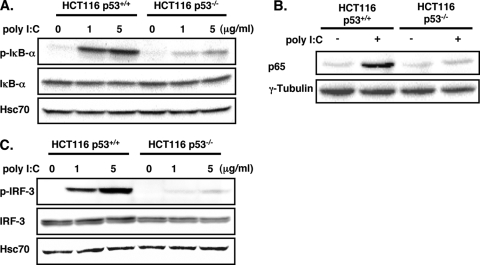
After elucidating the mechanism of how p53 regulates TLR3 expression, we next asked whether the effect of p53 is relevant in terms of the activity of TLR3, which utilizes two signaling pathways, NF-κB and IRF-3, to induce antiviral cytokines IL-8 and IFN-β, respectively, in response to viral dsRNA or its ligand, poly(I-C) (recently reviewed in reference 43). Activation of NF-κB is marked by the phosphorylation and subsequent degradation of its inhibitor, IκB-α. This is followed by the nuclear translocation of NF-κB (15). As we expected, stimulation of HCT116 p53+/+ cells with poly(I-C) induced the phosphorylation of IκB-α and the nuclear translocation of NF-κB subunit p65 (Fig. 3A and B). However, the poly(I-C)-induced IκB-α phosphorylation and p65 nuclear translocation were drastically reduced in HCT116 p53−/− cells (Fig. 3A and B). Similarly, the phosphorylation of IRF-3 caused by poly(I-C) treatment was also severely diminished in HCT116 p53−/− cells compared with that in p53+/+ cells (Fig. 3C). These data indicated that in the absence of p53, the TLR3 signaling pathways become unresponsive to poly(I-C) stimulation.
FIG. 3.
Activation of TLR3 signaling pathway by poly(I-C) is impaired in HCT116 p53−/− cells. (A to C) Cytoplasmic lysates (A and C) or nuclear extracts (B) from HCT116 p53+/+ and p53−/− cells unstimulated or stimulated for 3 h with poly(I-C) at the indicated concentration (A and C) or 5 μg/ml (B) were analyzed by Western blotting for phosphorylated IκB-α (p-IκB-α) and basal IκB-α (A), p65 (B), phosphorylated IRF-3 (p-IRF-3), and basal IRF-3 (C) expression. Hsc70 was used as loading control for panels A and C. γ-Tubulin was used as the loading control for panel B.
The poly(I-C)-induced expression of cytokines downstream of TLR3 is decreased in HCT116 p53−/− cells.
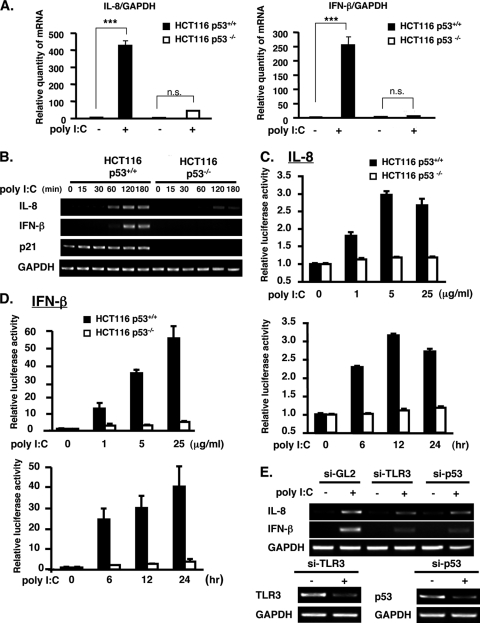
Having determined the requirement of p53 in TLR3 signaling, we next confirmed the effect of p53 after poly(I-C) stimulation on the production of cytokines IL-8 and IFN-β. Quantification of IL-8 and IFN-β mRNA revealed a dramatic difference between poly(I-C)-treated HCT116 p53+/+ and p53−/− cells. A robust induction of IL-8 and IFN-β was observed in poly(I-C)-treated HCT116 p53+/+ cells, whereas only a slight induction of cytokines occurred in p53−/− cells after stimulation with poly(I-C) for 3 h (Fig. 4A). Response to poly(I-C) was observed after 60 min for IL-8 transcription and after 120 min for IFN-β transcription in p53+/+ cells as determined by conventional RT-PCR (Fig. 4B). In contrast, neither IL-8 nor IFN-β mRNA was detected in poly(I-C)-treated HCT116 p53−/− cells within this period (Fig. 4B). Moreover, consistent with our preliminary investigation of IL-8 (Fig. 1A), the promoter activities of IL-8 (Fig. 4C) and IFN-β (Fig. 4D) were increased by poly(I-C) stimulation in HCT116 p53+/+ cells but not in p53−/− cells in a dose-dependent (upper panels) and time-dependent (lower panels) manner.
FIG. 4.
The response of cytokines IL-8 and IFN-β, which are downstream of TLR3, to poly(I-C) stimulation requires p53. (A) Real-time quantitative PCR analysis of IL-8 and IFN-β mRNA was performed on HCT116 p53+/+ and p53−/− cells untreated or treated with 5 μg/ml poly(I-C) for 3 h. mRNA expression was normalized to GAPDH. Values are means ± standard deviations of triplicate measurements. ***, P < 0.0001, as analyzed by ANOVA with Tukey-Kramer's test. (B) The mRNA level of IL-8 and IFN-β was determined in HCT116 p53+/+ and p53−/− cells stimulated with 5 μg/ml poly(I-C) for the indicated time. p21 and GAPDH served as the positive control and internal control, respectively. (C and D) Promoter activity of IL-8 (C) and IFN-β (D) was examined in HCT116 p53+/+ and p53−/− cells stimulated with poly(I-C) at the indicated concentration for 6 h (upper panels) or with 5 μg/ml poly(I-C) for the indicated time (lower panels). Values are means ± standard errors from triplicate platings. The data shown are representative of two to three independent experiments. (E) The mRNA expression of IL-8 and IFN-β was analyzed in HCT116 p53+/+ cells unstimulated or stimulated with poly(I-C) and untransfected or transfected with siRNA duplex for TLR3 (si-TLR3) or p53 (si-p53) or with si-GL2 (for control). Expression of TLR3 and p53 was knocked down by the transfection of their respective siRNAs in p53+/+ cells (lower panels).
To confirm that the cytokine activation after poly(I-C) treatment was through TLR3, we knocked down the expression of TLR3 in HCT116 p53+/+ cells using si-TLR3. IL-8 and IFN-β induction after poly(I-C) stimulation was attenuated by transfection of si-TLR3 (Fig. 4E), indicating that the effect of poly(I-C) on the induction of IL-8 and IFN-β was through TLR3. As expected, the suppression of p53 by p53 siRNA also inhibited the cytokine production in poly(I-C)-treated HCT116 cells (Fig. 4E). The efficiency of each siRNA was confirmed by RT-PCR (Fig. 4E, lower panels). Altogether, these results suggest that p53, through its regulatory function on TLR3 expression, is necessary for the activation by poly(I-C) of cytokines downstream of TLR3.
p53 affects TLR3 expression in mice liver and intestine.
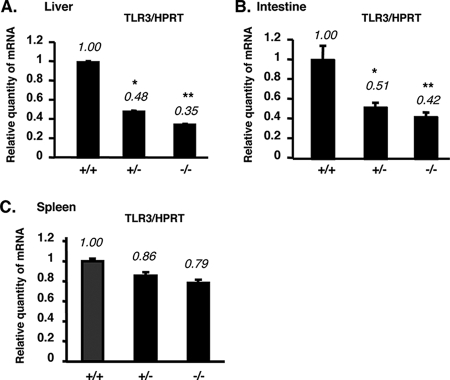
We partially extended our in vitro observations to the in vivo system. We compared the TLR3 mRNA expression in some tissues of p53 wild type (+/+), p53 heterozygous (+/−), and p53 knockout (−/−) mice by real-time quantitative RT-PCR. Similar to the results in vitro, the TLR3 mRNA expression was decreased in liver tissue and intestine of p53−/− mice in comparison with p53+/+ mice (Fig. 5A and B). However, the TLR3 expression levels in mouse spleen were similar between p53+/+ and p53−/− mice (Fig. 5C). In addition, we found that TLR3 mRNA expression in macrophages of p53−/− mice was similar to that in p53+/+ mice (data not shown). These data suggest that p53 also upregulates TLR3 in vivo, but this regulation is specific to certain tissues.
FIG. 5.
The TLR3 mRNA level is downregulated in tissues of p53−/− mice. (A to C) Total RNA isolated from liver (A), intestine (B), and spleen (C) of p53+/+, p53+/−, and p53−/− mice was analyzed for the expression of TLR3 by real-time quantitative RT-PCR. TLR3 mRNA levels were normalized to HPRT (internal control). The results represent means ± standard deviations (n = 3). * and **, P < 0.01 and P < 0.001, respectively, against p53 wild-type mice (+/+), as assessed by ANOVA with Dunnett's test.
DISCUSSION
Several studies have highlighted the role of the transcription factor p53 in antiviral responses (7, 25, 30); however, the molecular mechanism remains to be clarified. Because the initiation of antiviral response is mediated, in part, by TLRs, which are important receptors that recognize microbial pathogens, we focused on the expression of TLRs and their possible regulation by p53. A principal finding in this study is that p53 positively affects the expression of TLR3 in epithelial cells (HCT116, HepG2, and A549). The present study is the first report on the transcriptional control of a TLR family member by p53 in epithelial cells and in some mouse tissues (liver and intestine). We have presented the following lines of evidence that TLR3 is a bona fide target of p53 (32) in epithelial cells: (i) a p53 response element (RE) is present in the TLR3 promoter; (ii) TLR3 mRNA and protein levels are upregulated in HCT116 p53+/+ cells but not in p53−/− cells; (iii) the TLR3 promoter containing the p53 RE could activate the luciferase reporter, but this activity was compromised when the RE was absent or mutated; and (iv) endogenous p53 binds to the RE in the TLR3 promoter region, as determined by the ChIP assay.
Importantly, the regulation of TLR3 expression by p53 impacts the TLR3 signaling pathways and the subsequent induction of cytokines downstream of TLR3 in response to poly(I-C) (Fig. 3 and 4). Response to poly(I-C) and dsRNA could also be mediated by the cytoplasmic viral sensors, the RNA helicases retinoic inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA-5) (13, 28). It was recently reported by Hirata et al. that the recognition of poly(I-C) in intestinal epithelial cells is mediated by RIG-I but not by TLR3 (17). The different cell lines used may account for the conflicting findings: whereas, we used the HCT116 cell line, which expresses wild-type p53, Hirata et al. utilized cell lines with mutated p53 (17). We observed that HCT116 p53+/+ and p53−/− cells showed the same expression levels of RIG-I and MDA-5, which indicated that the regulation of their expression might not be dependent on p53 (data not shown). In our study, the specific knockdown of TLR3 by siRNA in HCT116 p53+/+ cells drastically reduced the IFN-β and IL-8 mRNA induction by poly(I-C) (Fig. 4E), which demonstrated the essential role of TLR3 in the cell's response to poly(I-C). Several studies have confirmed the importance of TLR3, RIG-I, or MDA-5 in poly(I-C) recognition (19, 21, 26, 27), and the question of whether RIG-I or TLR3 predominantly recognizes poly(I-C) in cells remains unresolved. Based on our results that p53 affects the expression of TLR3 but not that of MDA-5 or RIG-I, it is tempting to speculate that the activation of these pattern recognition receptors by poly(I-C) may, in part, depend on the p53 status of cells or tissues.
Although we did not observe a difference in the responses of the IL-8 promoter activity to other TLR ligands (PGN, LPS, CpG, and R-848) between p53+/+ and p53−/− cells (Fig. 1A), we could not rule out the possibility that p53 affects the steady-state level of other TLRs and the induction of cytokines other than IL-8, especially in light of studies proving that p53 is necessary in mounting a response against a variety of viruses, some of which may not be recognized by TLR3 (25, 30). Interestingly, our preliminary investigations showed that the mRNA expression of TLR7 and TLR8 was downregulated in HCT116 p53−/− cells (data not shown), which hints at the possibility that p53 may also regulate the basal transcription of TLR7/8. Further investigations may clarify this issue.
It is noteworthy that hepatitis C virus (HCV) can induce anomalies in p53 function (3) and that TLR3 expression is downregulated in chronic HCV infection through an unknown mechanism (4, 33). Although HCV is a single-stranded virus, which is susceptible to detection by TLR7/8, its genome also encodes regions of extensive secondary dsRNA structure that could be engaged by dsRNA-sensing receptor such as TLR3 (12). Based on these observations, it may be likely that certain viruses, which can induce downregulation of p53, may also in part circumvent TLR3 antiviral functions (and cause persistent infection) due to TLR3 being a molecular target of p53.
It has been reported that primary induction of IFNs by virus infection transcriptionally activates p53 to trigger apoptosis in infected cells (29, 38). While these previous studies clearly indicate that host protection provided by p53 is dependent on its role as inducer of apoptosis (5, 42), our present results suggest a novel mechanism of p53's function in antiviral signaling which is through its transactivation of TLR3 expression (and possibly that of other TLRs as well). Considering together the previous and current findings, it is likely that a positive feedback loop may exist between p53, TLR3, and IFN-β for antiviral host defense.
In conclusion, our results here first demonstrate a molecular link between p53 and a virus-sensing molecule.
Acknowledgments
We are grateful to B. Vogelstein (Johns Hopkins University) for generously providing the HCT116 p53+/+ and HCT116 p53−/− cells and to N. Araki (Kumamoto University) for the p53 knockout mice.
This work was supported by grants from the Ministry of Education, Science, Sports and Culture (MEXT) of Japan and from the Global COE Program (Cell Fate Regulation Research and Education Unit), MEXT, Japan.
Footnotes
Published ahead of print on 8 September 2008.
REFERENCES
- 1.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124783-801. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413732-738. [DOI] [PubMed] [Google Scholar]
- 3.Anzola, M., and J. J. Burgos. 2003. Hepatocellular carcinoma: molecular interactions between hepatitis C virus and p53 in hepatocarcinogenesis. Expert Rev. Mol. Med. 51-16. [DOI] [PubMed] [Google Scholar]
- 4.Atencia, R., F. J. Bustamante, A. Valdivieso, A. Arrieta, M. Rinon, A. Prada, and N. Maruri. 2007. Differential expression of viral PAMP receptors mRNA in peripheral blood of patients with chronic hepatitis C infection. BMC Infect. Dis. 7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castedo, M., J. L. Perfettini, M. Piacentini, and G. Kroemer. 2005. p53—a pro-apoptotic signal transducer involved in AIDS. Biochem. Biophys. Res. Commun. 331701-706. [DOI] [PubMed] [Google Scholar]
- 6.Collot-Teixeira, S., J. Bass, F. Denis, and S. Ranger-Rogez. 2004. Human tumor suppressor p53 and DNA viruses. Rev. Med. Virol. 14301-319. [DOI] [PubMed] [Google Scholar]
- 7.Dharel, N., N. Kato, R. Muroyama, H. Taniguchi, M. Otsuka, Y. Wang, A. Jazag, R. X. Shao, J. H. Chang, M. K. Adler, T. Kawabe, and M. Omata. 2008. Potential contribution of tumor suppressor p53 in the host defense against hepatitis C virus. Hepatology 471136-1149. [DOI] [PubMed] [Google Scholar]
- 8.Diebold, S. S., T. Kaisho, H. Hemmi, S. Akira, and C. Reis e Sousa. 2004. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 3031529-1531. [DOI] [PubMed] [Google Scholar]
- 9.Djelloul, S., M. E. Forgue-Lafitte, B. Hermelin, M. Mareel, E. Bruyneel, A. Baldi, A. Giordano, E. Chastre, and C. Gespach. 1997. Enterocyte differentiation is compatible with SV40 large T expression and loss of p53 function in human colonic Caco-2 cells. Status of the pRb1 and pRb2 tumor suppressor gene products. FEBS Lett. 406234-242. [DOI] [PubMed] [Google Scholar]
- 10.Forbes, S., J. Clements, E. Dawson, S. Bamford, T. Webb, A. Dogan, A. Flanagan, J. Teague, R. Wooster, P. A. Futreal, and M. R. Stratton. 2006. Cosmic 2005. Br. J. Cancer 94318-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furrie, E., S. Macfarlane, G. Thomson, and G. T. Macfarlane. 2005. Toll-like receptors-2, -3 and -4 expression patterns on human colon and their regulation by mucosal-associated bacteria. Immunology 115565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436939-945. [DOI] [PubMed] [Google Scholar]
- 13.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 1038459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groskreutz, D. J., M. M. Monick, T. O. Yarovinsky, L. S. Powers, D. E. Quelle, S. M. Varga, D. C. Look, and G. W. Hunninghake. 2007. Respiratory syncytial virus decreases p53 protein to prolong survival of airway epithelial cells. J. Immunol. 1792741-2747. [DOI] [PubMed] [Google Scholar]
- 15.Hayden, M. S., and S. Ghosh. 2008. Shared principles in NF-kappaB signaling. Cell 132344-362. [DOI] [PubMed] [Google Scholar]
- 16.Heil, F., H. Hemmi, H. Hochrein, F. Ampenberger, C. Kirschning, S. Akira, G. Lipford, H. Wagner, and S. Bauer. 2004. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 3031526-1529. [DOI] [PubMed] [Google Scholar]
- 17.Hirata, Y., A. H. Broquet, L. Menchen, and M. F. Kagnoff. 2007. Activation of innate immune defense mechanisms by signaling through RIG-I/IPS-1 in intestinal epithelial cells. J. Immunol. 1795425-5432. [DOI] [PubMed] [Google Scholar]
- 18.Hoh, J., S. Jin, T. Parrado, J. Edington, A. J. Levine, and J. Ott. 2002. The p53MH algorithm and its application in detecting p53-responsive genes. Proc. Natl. Acad. Sci. USA 998467-8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, Z., M. Zamanian-Daryoush, H. Nie, A. M. Silva, B. R. Williams, and X. Li. 2003. Poly(I-C)-induced Toll-like receptor 3 (TLR3)-mediated activation of NFkappa B and MAP kinase is through an interleukin-1 receptor-associated kinase (IRAK)-independent pathway employing the signaling components TLR3-TRAF6-TAK1-TAB2-PKR. J. Biol. Chem. 27816713-16719. [DOI] [PubMed] [Google Scholar]
- 20.Joerger, A. C., and A. R. Fersht. 2008. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 77557-582. [DOI] [PubMed] [Google Scholar]
- 21.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441101-105. [DOI] [PubMed] [Google Scholar]
- 22.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7131-137. [DOI] [PubMed] [Google Scholar]
- 23.Krishnan, J., K. Selvarajoo, M. Tsuchiya, G. Lee, and S. Choi. 2007. Toll-like receptor signal transduction. Exp. Mol. Med. 39421-438. [DOI] [PubMed] [Google Scholar]
- 24.Lu, Z., K. A. Kim, M. A. Suico, T. Shuto, J. D. Li, and H. Kai. 2004. MEF up-regulates human beta-defensin 2 expression in epithelial cells. FEBS Lett. 561117-121. [DOI] [PubMed] [Google Scholar]
- 25.Marques, J. T., D. Rebouillat, C. V. Ramana, J. Murakami, J. E. Hill, A. Gudkov, R. H. Silverman, G. R. Stark, and B. R. G. Williams. 2005. Down-regulation of p53 by double-stranded RNA modulates the antiviral response. J. Virol. 7911105-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsukura, S., F. Kokubu, M. Kurokawa, M. Kawaguchi, K. Ieki, H. Kuga, M. Odaka, S. Suzuki, S. Watanabe, T. Homma, H. Takeuchi, K. Nohtomi, and M. Adachi. 2007. Role of RIG-I, MDA-5, and PKR on the expression of inflammatory chemokines induced by synthetic dsRNA in airway epithelial cells. Int. Arch. Allergy Immunol. 143(Suppl. 1)80-83. [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto, M., and T. Seya. 2008. TLR3: interferon induction by double-stranded RNA including poly(I:C). Adv. Drug Deliv. Rev. 60805-812. [DOI] [PubMed] [Google Scholar]
- 28.Meylan, E., J. Tschopp, and M. Karin. 2006. Intracellular pattern recognition receptors in the host response. Nature 44239-44. [DOI] [PubMed] [Google Scholar]
- 29.Moiseeva, O., F. A. Mallette, U. K. Mukhopadhyay, A. Moores, and G. Ferbeyre. 2006. DNA damage signaling and p53-dependent senescence after prolonged beta-interferon stimulation. Mol. Biol. Cell 171583-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Munoz-Fontela, C., M. A. Garcia, I. Garcia-Cao, M. Collado, J. Arroyo, M. Esteban, M. Serrano, and C. Rivas. 2005. Resistance to viral infection of super p53 mice. Oncogene 243059-3062. [DOI] [PubMed] [Google Scholar]
- 31.Oster, B., E. Kofod-Olsen, B. Bundgaard, and P. Hollsberg. 2008. Restriction of human herpesvirus 6B replication by p53. J. Gen. Virol. 891106-1113. [DOI] [PubMed] [Google Scholar]
- 32.Riley, T., E. Sontag, P. Chen, and A. Levine. 2008. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9402-412. [DOI] [PubMed] [Google Scholar]
- 33.Sato, K., T. Ishikawa, A. Okumura, T. Yamauchi, S. Sato, M. Ayada, E. Matsumoto, N. Hotta, T. Oohashi, Y. Fukuzawa, and S. Kakumu. 2007. Expression of Toll-like receptors in chronic hepatitis C virus infection. J. Gastroenterol. Hepatol. 221627-1632. [DOI] [PubMed] [Google Scholar]
- 34.Seki, Y., M. A. Suico, A. Uto, A. Hisatsune, T. Shuto, Y. Isohama, and H. Kai. 2002. The ETS transcription factor MEF is a candidate tumor suppressor gene on the X chromosome. Cancer Res. 626579-6586. [PubMed] [Google Scholar]
- 35.Severa, M., and K. A. Fitzgerald. 2007. TLR-mediated activation of type I IFN during antiviral immune responses: fighting the battle to win the war. Curr. Top. Microbiol. Immunol. 316167-192. [DOI] [PubMed] [Google Scholar]
- 36.Shuto, T., T. Furuta, M. Oba, H. Xu, J. D. Li, J. Cheung, D. C. Gruenert, A. Uehara, M. A. Suico, T. Okiyoneda, and H. Kai. 2006. Promoter hypomethylation of Toll-like receptor-2 gene is associated with increased proinflammatory response toward bacterial peptidoglycan in cystic fibrosis bronchial epithelial cells. FASEB J. 20782-784. [DOI] [PubMed] [Google Scholar]
- 37.Suico, M. A., H. Yoshida, Y. Seki, T. Uchikawa, Z. Lu, T. Shuto, K. Matsuzaki, M. Nakao, J. D. Li, and H. Kai. 2004. Myeloid Elf-1-like factor, an ETS transcription factor, up-regulates lysozyme transcription in epithelial cells through interaction with promyelocytic leukemia protein. J. Biol. Chem. 27919091-19098. [DOI] [PubMed] [Google Scholar]
- 38.Takaoka, A., S. Hayakawa, H. Yanai, D. Stoiber, H. Negishi, H. Kikuchi, S. Sasaki, K. Imai, T. Shibue, K. Honda, and T. Taniguchi. 2003. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature 424516-523. [DOI] [PubMed] [Google Scholar]
- 39.Tamura, T., N. Aoyama, H. Saya, H. Haga, S. Futami, M. Miyamoto, T. Koh, T. Ariyasu, M. Tachi, M. Kasuga, et al. 1995. Induction of Fas-mediated apoptosis in p53-transfected human colon carcinoma cells. Oncogene 111939-1946. [PubMed] [Google Scholar]
- 40.Tanabe, M., M. Kurita-Taniguchi, K. Takeuchi, M. Takeda, M. Ayata, H. Ogura, M. Matsumoto, and T. Seya. 2003. Mechanism of up-regulation of human Toll-like receptor 3 secondary to infection of measles virus-attenuated strains. Biochem. Biophys. Res. Commun. 31139-48. [DOI] [PubMed] [Google Scholar]
- 41.Tsukada, T., Y. Tomooka, S. Takai, Y. Ueda, S. Nishikawa, T. Yagi, T. Tokunaga, N. Takeda, Y. Suda, S. Abe, I. Matsuo, Y. Ikawa, and S. Aizawa. 1993. Enhanced proliferative potential in culture of cells from p53-deficient mice. Oncogene 83313-3322. [PubMed] [Google Scholar]
- 42.Turpin, E., K. Luke, J. Jones, T. Tumpey, K. Konan, and S. Schultz-Cherry. 2005. Influenza virus infection increases p53 activity: role of p53 in cell death and viral replication. J. Virol. 798802-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vercammen, E., J. Staal, and R. Beyaert. 2008. Sensing of viral infection and activation of innate immunity by Toll-like receptor 3. Clin. Microbiol. Rev. 2113-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vousden, K. H., and D. P. Lane. 2007. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 8275-283. [DOI] [PubMed] [Google Scholar]
- 45.Yugawa, T., K. Handa, M. Narisawa-Saito, S.-I. Ohno, M. Fujita, and T. Kiyono. 2007. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol. Cell. Biol. 273732-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]