Abstract
Chromatin status is characterized in part by covalent posttranslational modifications of histones that regulate chromatin dynamics and direct gene expression. BORIS (brother of the regulator of imprinted sites) is an insulator DNA-binding protein that is thought to play a role in chromatin organization and gene expression. BORIS is a cancer-germ line gene; these are genes normally present in male germ cells (testis) that are also expressed in cancer cell lines as well as primary tumors. This work identifies SET1A, an H3K4 methyltransferase, and BAT3, a cochaperone recruiter, as binding partners for BORIS, and these proteins bind to the upstream promoter regions of two well-characterized procarcinogenic genes, Myc and BRCA1. RNA interference (RNAi) knockdown of BAT3, as well as SET1A, decreased Myc and BRCA1 gene expression but did not affect the binding properties of BORIS, but RNAi knockdown of BORIS prevented the assembly of BAT3 and SET1A at the Myc and BRCA1 promoters. Finally, chromatin analysis suggested that BORIS and BAT3 exert their effects on gene expression by recruiting proteins such as SET1A that are linked to changes in H3K4 dimethylation. Thus, we propose that BORIS acts as a platform upon which BAT3 and SET1A assemble and exert effects upon chromatin structure and gene expression.
Insulator DNA sequences partition the genome into functional chromosomal domains to regulate gene expression (2, 10). The compartmentalization of chromatin into distinct regulatory domains appears to alter interactions between target genes and nearby cis-acting enhancer elements (28). CTCF was the first human insulator DNA-binding protein identified. It is a ubiquitously expressed, highly conserved, zinc finger protein that has multiple roles in gene regulation (3), including regulation of the imprinted maternal H19 allele (22, 28). CTCF appears to bind to unmethylated H19 DNA, limiting access to the H19 enhancer and silencing H19 gene expression (14).
Less is known about the CTCF paralog CTCFL, or BORIS (brother of the regulator of imprinted sites). BORIS is also an insulator DNA-binding protein, but it is expressed predominantly in testis (26). It has also been detected in more than 100 cancer cell lines representing all major forms of human tumors (20). BORIS shares a homologous 11 zinc finger domain with CTCF (39); however, CTCF and BORIS differ significantly in their amino and carboxy termini, suggesting that these regions may interact with different binding partners that regulate gene expression by different mechanisms. Indeed, in contrast to CTCF, BORIS appears to be a methylation-independent DNA-binding protein (28b) that activates, rather than inhibits, gene expression (15). These results suggest that CTCF and BORIS use a similar DNA-binding sequence to elicit very different changes in gene expression: one is a suppressor and the other is an inducer. Thus, one critical question is how the differing N-terminal domains might recruit different protein complexes that either repress or activate gene expression.
Mammalian chromatin contains and transmits genetic information encoded within the DNA as well as epigenetic information carried by histone proteins in the form of reversible covalent modifications (21). These histone modifications occur at the unstructured histone “tails” that stick out between the gyres of nucleosomal DNA that encircle the histone core (27) and alter histones' interactions with DNA and nuclear proteins (18). These modifications may regulate access to the DNA and thus influence nuclear processes, including but not limited to transcription (23). Histones H3 and H4 have long tails protruding from the nucleosome that can be covalently modified at several places (35). Modifications of the tail include methylation, acetylation, phosphorylation, ubiquitination, sumoylation, citrullination, and ADP-ribosylation (18). Combinations of modifications are thought to constitute a so-called “histone code” (27). Histone modifications act in diverse biological processes such as gene regulation, DNA repair, and chromosome condensation (mitosis) (25, 38).
It has been proposed that insulator DNA-binding proteins may recruit specific cofactors and/or protein complexes to their binding sites and that the precise composition of these complexes may be critical to the subsequent mechanisms stabilizing long-range interactions (40). Therefore, the identification of the specific factors in these complexes is a critical first step in identifying the mechanism of action (21). Thus, we hypothesize that the specific protein complex assembled through mediation by BORIS might dictate gene expression and that changes in histone modifications may be, at least in part, one of the underlying mechanisms in this process.
In this work we identified BAT3, a protein recruitment factor, and SET1A, an H3K4 methyltransferase, as binding partners for BORIS and found that both are present at the promoter regions of Myc and BRCA1 as well as the H19 differentially methylated region (DMR). RNA interference (RNAi) knockdown of either BAT3 or SET1A decreased expression of Myc, BRCA1, and H19 and altered the dimethyl-H3K4/dimethyl-H3K9 ratio within the Myc and BRCA1 promoters, predominantly via a decrease in H3K4 dimethylation. Loss of either BAT3 or SET1A did not affect the binding properties of BORIS, but RNAi knockdown of BORIS prevented the assembly of BAT3, SET1A, and ASH2 at the Myc and BRCA1 promoters. Thus, we propose that BORIS acts as a scaffold upon which BAT3 and SET1A assemble and through which BAT3 and SET1A exert their effects upon chromatin structure and gene expression.
MATERIALS AND METHODS
Cell lines and cell culture.
HCT116 cells (human colon carcinoma) were cultured in McCoy's 5A, and COS-7 cells were grown in RPMI medium containing 10% heat-inactivated (56°C, 30 min) fetal bovine serum supplemented with penicillin (100 units/ml) and streptomycin (100 μg/ml).
Yeast two-hybrid analysis.
The two-hybrid screen was done in collaboration with Myriad Genetics and the NCI using the Center for Cancer Research core facilities. This automated process uses ProNet technology for the large-scale identification of protein-protein interactions based on nuclear yeast two-hybrid methodology (see methods in the supplemental material).
Preparation of nuclear cell extracts.
Nuclear extracts were prepared using a modification of a previously described method (33); no reducing agents were added to the extraction buffers. Following preparation and protein analysis, samples were stored at −80°C and thawed on ice immediately before use. Protein concentrations were determined using the Bradford assay (Bio-Rad Labs, Hercules, CA) and a Beckman DU-640 spectrophotometer, and immunoblotting was done as previously described (33).
IP, transient-transfection IP, His tag IP, and immunoblot analysis.
Immunoprecipitation (IP) with BAT3 and BORIS and Western analyses were done as previously described (28b). The control for these IP experiments is normalized to rabbit immunoglobulin G. Transient-transfection IPs were done by cotransfection of pCMV-GFP-BORIS and/or pCMV-BAT3 followed by IP with an anti-green fluorescent protein (anti-GFP) antibody (Santa Cruz, Inc.) after 36 h and Western analysis with an anti-BAT3 (see Fig. S3 in the supplemental material) antibody. His-tagged BAT3 was expressed in bacteria and isolated as described by Qiagen, Inc., and mixed with whole-cell extracts from HCT116 cells, and IP was performed using an anti-His antibody (Santa Cruz Biotechnology, Santa Cruz, CA) as described above. Samples were mixed with Laemmli lysis buffer, boiled for 5 min, and loaded into denaturing sodium dodecyl sulfate (SDS)-polyacrylamide gels for electrophoresis as described previously (11).
Following separation by electrophoresis, samples were immunoblotted with anti-BAT3 or anti-BORIS (see Fig. S1 in the supplemental material) antibodies. Blots were incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA), analyzed using an ECL protocol (Amersham Biosciences, Piscataway, NJ), and visualized in a Fuji Las-3000 intelligent darkbox (FujiFilm Medical Systems, Stamford, CT). Figures are representative of the outcome in at least two independent experiments.
Cytomegalovirus (CMV) and GFP expression plasmids.
BAT3 was subcloned from a holding vector (Origene, Rockville, MD) into the pCIneo expression vector (Clontech, Mountain View, CA) to construct pCMV-His-BAT3. The BORIS and CTCF vectors were kind gifts from David Schrump (NCI). pCMV-His-Δ2P-BAT3 was created from pCMV-His-BAT3 using two primers (see methods in the supplemental material) and reverse transcription-PCR (RT-PCR) to remove amino acids (aa) 387 to 675. BORIS was subcloned into a pGFP expression vector to construct pGFP-BORIS.
ChIP.
Cells were cultured for 24 h and prepared using a chromatin IP (ChIP) assay kit from Upstate Biotechnology, Inc. (Lake Placid, NY) according to the manufacturer's recommendations as previously described (28b). Samples with DNA sheared to between 200 and 1,000 bp were used for subsequent ChIP reactions. ChIP was performed using CTCF (C-20) (Santa Cruz, CA), BORIS, BAT3, SET1A (Bethyl Laboratories), or ASH2 (Bethyl Laboratories) antibodies. ChIP-enriched DNA, along with input DNA, was purified by phenol-chloroform extraction and ethanol precipitation. Purified DNA samples were analyzed by real-time quantitative PCR (QPCR) using Sybr green PCR master mix (Applied Biosystems) with the ABI Prism 7500 detection system (Applied Biosystems) and primers to the promoter of Myc or BRCA1 or the H19 DMR (see the supplemental material). Data were collected and analyzed by comparative cycle threshold methods (28b).
EMSA.
Electrophoretic mobility shift assays (EMSAs) were performed as described previously described using a 32P-radiolabeled oligonucleotide corresponding to the consensus CTCF DNA-binding site (19). Nuclear extracts (10 μg) were incubated with poly(dI-dC) for 10 min on ice, followed by the addition of radiolabeled oligonucleotide (100,000 cpm of radiolabeled probe per reaction) and incubation at 25°C for 20 min. Samples were electrophoresed on a 6% nondenaturing polyacrylamide gel, dried, exposed to a phosphorimager screen, and analyzed using a Typhoon 860 phosphorimager (GE Healthcare, Piscataway, NJ) with ImageQuant software.
RNA extraction and real-time PCR.
Total cellular RNA was isolated, and reverse transcription and real-time PCR were carried out as previously described (5). RT-PCRs were done three times for each sample. Standard curves for tested genes and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were done for each RT-PCR, and the relative amounts of transcripts of the tested genes were normalized by GAPDH. The average ratios of relative amounts of transcripts in three replicate runs were calculated.
ChIP assay for histone H3K4 and H3K9 dimethylation.
ChIP was performed using either a histone H3 dimethyl K4 or a histone H3 dimethyl K9 antibody (Abcam) (24). Each experiment was repeated at least twice. Pulled-down DNA was analyzed using quantitative RT-PCR normalized by input DNA with primers that overlap the H19 DMR (see primer section in the supplemental material).
RESULTS
BAT3 is a novel BORIS-interacting protein.
We have previously shown that BORIS, but not CTCF, DNA binding is methylation independent (28b), strongly suggesting different cellular functions for these two insulator DNA-binding proteins. While BORIS and CTCF exhibit significant homology in their zinc finger DNA-binding domains, their N-terminal amino acid sequences are quite different (26). Thus, distinct protein binding partners and/or a multiprotein complex may associate with the different N-terminal regions of these two proteins, accounting for their different cellular functions.
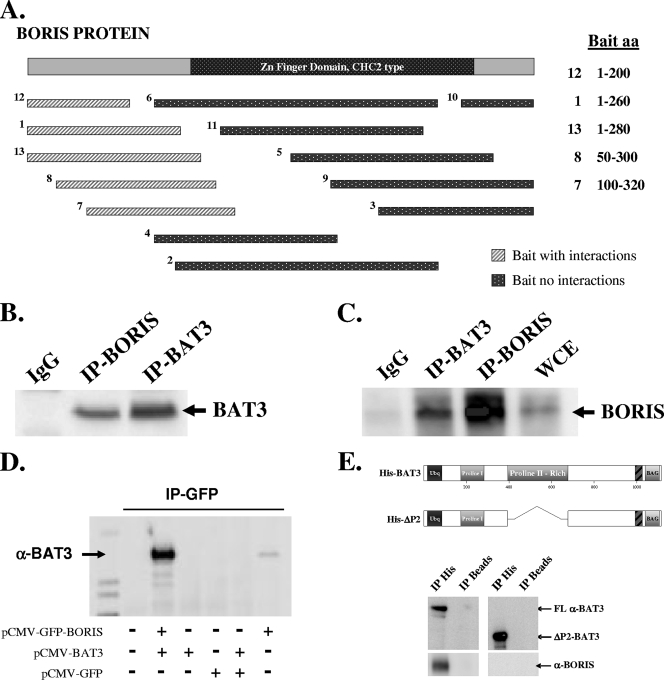
To identify binding partners of BORIS, a yeast two-hybrid screen was done using multiple BORIS N-terminal or zinc finger domain baits. Only segments in the unique N-terminal region of BORIS were identified as having binding partners; baits 1, 7, 8, 12, and 13 had at least one protein interaction each (Fig. 1A), whereas no binding partners were identified for baits 3 to 6 or 9 to 11. From a screening of independent transformants of a mouse embryonic day 11.5 whole-embryo cDNA library (Myriad, Inc.), 16 potential BORIS-interacting proteins were identified (Table 1), all of which targeted the unique N-terminal protein domain of BORIS (Fig. 1A).
FIG. 1.
BAT3 physically interacts with BORIS in HCT116 cells. (A) Yeast two-hybrid design. Diagram of 13 bait sequences of BORIS used to identify binding partners. Light hatched rectangles represent baits that captured binding partners (Table 1), while dark dotted rectangles represent baits with no binding partners (see methods in the supplemental material). (B) BAT3 binds to BORIS in vitro. HCT116 cell lysates were subjected to IP with an anti-BORIS antibody (see Fig. S1 and S2 in the supplemental material), followed by Western analysis with an anti-BAT3 antibody. IgG, immunoglobulin G. (C) HCT116 cell lysates were subjected to IP with an anti-BAT3 antibody, followed by Western analysis with an anti-BORIS antibody. (D) COS-7 cells were transfected with pCMV-GFP, pCMV-GFP-BORIS, and/or pCMV-BAT3 and subjected to IP with an anti-GFP antibody. Immunoprecipitates were resolved by SDS-polyacrylamide gel electrophoresis and blotted with an anti-BAT3 antibody. (E) BORIS binds to the BAT3 proline-rich region. HCT116 cell lysates were incubated with purified wild-type His-tagged BAT3 or His-tagged ΔP2-BAT3, which contains a deletion in the proline-rich region (aa 387 to 675 deleted). Lysates were subjected to IP with an anti-His antibody (Santa Cruz, Inc.), and the His tag IP products were resolved and immunoblotted with either an anti-BAT3 (top panel) or anti-BORIS (lower panel) antibody.
TABLE 1.
BORIS-interacting proteins
| Bait | aa | Interacting protein(s) |
|---|---|---|
| 1 | 1-260 | BAT3,a CHD8,b ELF2,c POGZ,d HCFC2,a,c HCFC1,a,c MGA, TLK2,c NFAT5,c ZNF518,c ATF7,c CSTA, FSLZ1c,d |
| 12 | 1-200 | BAT3,a CHD8,b ELF2,c POGZ,d HCFC2,a,c HCFC1,a,c MGA, SRCAP,b MKL2,c CSTA, FSLZ1,b,c CSTA, FHL2 |
| 13 | 1-280 | BAT3,a CHD8,b ELF2,c POGZ,d HCFC2,a,c FSLZ1, TLK2,c SRCAPb |
| 8 | 50-300 | CHD8,b ELF2,c POGZd |
| 7 | 100-320 | CDH8b |
Testis-specific proteins.
Helicase-associated proteins.
Transcription factor proteins.
Chromatin-associated proteins.
These potential binding partners were subsequently categorized into four function groups: (i) testis-specific proteins, (ii) chromatin-associated proteins, (iii) helicase-associated proteins, and (iv) transcription factor proteins (Table 1). BAT3(Bcl-2-associated athanogene-6, Scythe) was chosen for further investigation based on several criteria. First, BAT3 is also a testis-specific protein (41), improving the likelihood of an interaction with BORIS. Second, BAT3 bound to multiple two-hybrid baits (Table 1, baits 1, 12, and 13), suggesting that it would be a legitimate binding partner. Third, both BAT3 (7, 30) and BORIS (26) have previously been identified as nuclear proteins, further suggesting a potential protein interaction. Finally, BAT3 has previously been shown to be a cochaperone or hinge protein, and it seemed reasonable to determine if it played a similar role via an interaction with BORIS (32).
To establish a physical interaction, HCT116 whole-cell lysates were subjected to IP with an anti-BORIS antibody (see Fig. S1 and S2 in the supplemental material), followed by Western analysis with an anti-BAT3 antibody (see Fig. S3 in the supplemental material). HCT116 cells were used because they express both BORIS (see Fig. S1 in the supplemental material) and BAT3 (see Fig. S3, lane 1, in the supplemental material). These experiments suggested a protein-protein interaction (Fig. 1B), as did a reciprocal BAT3 IP followed by Western analysis with an anti-BORIS antibody (Fig. 1C). The physical interaction between BAT3 and BORIS was also demonstrated using a transient-cotransfection system in COS-7 cells. Transient cotransfection of both BORIS (pCMV-GFP-BORIS) and BAT3 (pCMV-BAT3) in COS-7 cells followed by GFP IP and anti-BAT3 Western blotting also showed a physical interaction (Fig. 1D).
Finally, the yeast two-hybrid screen used to identify BORIS binding partners suggested that the internal region (aa 188 to 688) of BAT3 (data not shown) binds to the N terminus (aa 1 to 200) of BORIS (Fig. 1A). Examination of the BAT3 protein sequence (see Fig. S4 in the supplemental material) identified two proline-rich regions that might be a protein-protein interaction domain. Therefore, a bacterial expression vector that deleted both proline-rich BAT3 domains was constructed. HCT116 cell lysates were incubated with purified wild-type His-tagged BAT3 or His-tagged ΔP2-BAT3, which contained a deletion in the second proline-rich region (aa 387 to 675 deleted [see Fig. S4 in the supplemental material]), followed by the addition of an anti-His antibody (Santa Cruz, Inc.). The subsequent samples after IP were separated, and Western analysis was done using either an anti-BAT3 (Fig. 1E, upper panel) or an anti-BORIS (lower panel) antibody. These experiments demonstrated that the deletion of the internal proline-rich BAT3 domain is essential for binding to BORIS. Deletion of the N-terminal proline-rich domain had no effect on the interaction with BORIS (data not shown). The results of these experiments suggest that BAT3 physically interacts with BORIS via its central proline-rich domain.
BAT3 and BORIS bind to the promoter regions of BRCA1 and Myc and the H19 DMR.
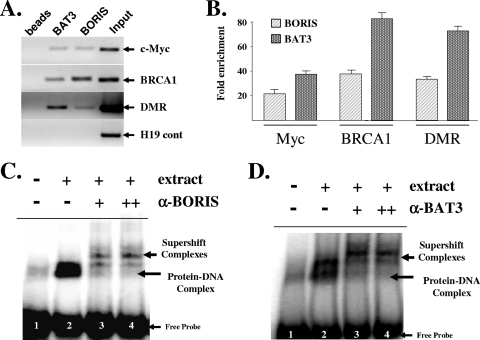
The promoter regions of Myc (12) and BRCA1 (4) (see Fig. S5 in the supplemental material) as well as the H19 DMR (36) have been previously shown to contain CTCF DNA-binding target sequences. To determine if BAT3 and BORIS bind to these regions, HCT116 cells were harvested for ChIP with either an anti-BAT3 or an anti-BORIS antibody. Purified DNA samples were analyzed by either PCR (Fig. 2A) or real-time QPCR (Fig. 2B) with primers that overlap the CTCF DNA-binding sites of the Myc or BRCA1 promoters or the H19 DMR (see primer section in the supplemental material). Each target region showed enrichment in the presence of BAT3 and BORIS. H19 exon 4/5 (17) was used as a negative control (Fig. 2A, bottom panel). The results of these experiments suggest that both BAT3 and BORIS are physically present at the promoter regions of Myc and BRCA1 as well as the H19 DMR.
FIG. 2.
(A and B) BAT3 and BORIS bind to the promoter regions of BRCA1 and Myc and to the H19 DMR. HCT116 cells were fixed with 1% formaldehyde to cross-link protein-DNA interactions and sonicated, and fixed cells were subjected to IP with either an anti-BAT3 or an anti-BORIS antibody. DNA was eluted and purified before analysis by either PCR (A) or QPCR (B) with primers to either the promoters of BRCA1 or Myc or the H19 DMR. All ChIP experiments were done in triplicate, and error bars represent one standard deviation about the arithmetic mean. Fold enrichment for all ChIP experiments was normalized to the immunoglobulin G control IP. (C and D) BAT3 and BORIS bind to a CTCF-binding element. HCT116 cells were harvested, and nuclear cell extracts were used for EMSA with a 32P-labeled oligonucleotide containing a consensus CTCF-binding sequence. Lane 1 is probe alone and lanes 2 to 4 were incubated with cellular extract. Supershift assays were done by preincubating extracts with increasing concentrations (+ or ++) of either anti-BORIS (C) or anti-BAT3 (D) antibodies (lanes 3 and 4). Sections of fluorograms from native gels using a Typhoon phosphorimager are shown. Arrows indicate the supershifted complexes as well as the protein-CTCF-DNA complex and unbound (free) oligonucleotide probe.
While these experiments demonstrate that BORIS and BAT3 physically bind to DNA, they do not a priori identify the specific DNA-binding sequences. This was determined by EMSA and supershift experiments using HCT116 nuclear lysates mixed with a 32P-labeled oligonucleotide containing a CTCF DNA-binding consensus sequence (see primer section in the supplemental material). Supershift experiments with either an anti-BORIS (Fig. 2C, lanes 3 and 4) or an anti-BAT3 (Fig. 2D, lanes 3 and 4) antibody demonstrated that both BAT3 and BORIS bind to a CTCF DNA-binding sequence. The BAT3 antibody does not bind to the oligonucleotide alone (see Fig. S6, lane 1, in the supplemental material), and both anti-NF-κB and anti-Fos antibodies failed to supershift the CTCF oligonucleotide-protein complex band (lanes 3 and 4, respectively). Finally, bacterially expressed His-tagged, column-purified BAT3 protein did not bind to the CTCF DNA-binding site, suggesting that BAT3 likely binds to BORIS and not directly to the DNA (data not shown). The results of these experiments provide initial results suggesting that BORIS may function as a platform or scaffold protein to assemble a multiprotein transcriptional complex.
siRNA knockdown of BAT3 decreases Myc, BRCA1, and H19 gene expression.
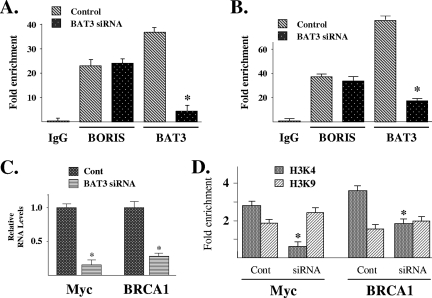
Since BAT3 interacts indirectly with DNA through contact with BORIS, we wanted to investigate BAT3's role in altering the expression of BORIS/CTCF-regulated genes. First, we determined if BAT3 was required for BORIS DNA binding. Toward this end, HCT116 cells were transfected with a small interfering RNA (siRNA) (Dharmacon, GENOME SMARTpool) to BAT3 (see lower panel of Fig. S7A in the supplemental material), followed by ChIP analysis with an anti-BORIS or an anti-BAT3 antibody. Purified DNA samples were analyzed by QPCR with primers (see primer section in the supplemental material) to the promoter regions of either Myc (Fig. 3A) or BRCA1 (Fig. 3B). As expected following BAT3 knockdown, a significant decrease in BAT3 DNA binding was observed compared to cells transfected with a control siRNA. A similar result was observed for the H19 DMR (see Fig. S7A and B in the supplemental material). However, the level of BORIS DNA binding remained unchanged, strongly suggesting that although BAT3 and BORIS physically interact at the DMR, BAT3 is not required for BORIS DNA binding.
FIG. 3.
siRNA knockdown of BAT3 eliminates BAT3 binding to the promoter regions of BRCA1 and Myc, decreases gene expression, and alters H3K4 dimethylation. (A and B) BAT3 is not required for BORIS DNA binding. HCT116 cells were treated with either nonsense (control) or BAT3 siRNA, and ChIP analysis was done followed by QPCR to with primers to the Myc (A) or BRCA1 (B) promoters (see Fig. S5 in the supplemental material). (C) siRNA knockout of BAT3 decreases BRCA1 and MycR RNA levels. HCT116 cells were treated with nonsense (Cont) or BAT3 siRNA (in duplicate); RNA was isolated; and BRCA1 and Myc expression was determined by RT-PCR that was done in triplicate. Error bars represent one standard deviation about the arithmetic mean, and statistical significance was established by Student's t test. *, P < 0.05. (D) siRNA knockout of BAT3 decreases histone H3K4 dimethylation. HCT116 cells were treated with nonsense (Cont) or BAT3 siRNA and harvested for ChIP using either an H3K4 or H3K9 antibody followed by QPCR with primers to either the Myc (left panel) or BRCA1 (right panel) promoter. All ChIP experiments were done in triplicate, and error bars represent one standard deviation about the arithmetic mean. Statistical significance was established by Student's t test. *, P < 0.05 by t test.
Since BAT3 did not alter BORIS DNA binding, we next determined whether BAT3 might have any effects on gene expression. RNA was isolated from HCT116 cells transfected with either control or BAT3 siRNA, followed by RT-PCR with primers for Myc and BRCA1. The genes regulated by the H19 DMR, IGF2 and H19, are complex due to a combination of tissue-specific mechanism that includes multiple enhancers, silencers, and boundary elements. Since imprinting is clearly due to more than the DMR, we limited our experiments to BAT3 and BORIS binding to the DMR and did not examine changes in IGF2 or H19 gene expression (10, 31). The results of these experiments demonstrated that siRNA knockdown of BAT3 decreased the expression of Myc and BRCA1 (Fig. 3C).
siRNA knockdown of BAT3 decreases H3K4 dimethylation.
Since BORIS is an insulator DNA-binding protein and the results above suggest that BAT3 binds to BORIS as well as altering gene expression, it seemed reasonable to determine if changes in chromatin might accompany the changes in Myc and BRCA1 expression. Dimethylation of histone H3 at residue K4 (H3K4) or at residue K9 (H3K9) in upstream promoters has been shown to define distinct chromatin regions that are permissive or nonpermissive for gene expression, respectively (13, 34). To determine if BAT3 alters chromatin arrangement, HCT116 cells were transfected with BAT3 siRNA and examined for histone H3 dimethylation by ChIP analysis with either an anti-H3K4 or an anti-H3K9 antibody. In HCT116 cells transfected with a control siRNA, the promoter regions of Myc and BRCA1 were slightly enriched in dimethyl-H3K4 compared to dimethyl-H3K9 (Fig. 3D). In contrast, cells transfected with BAT3 siRNA showed a marked and statistically significant decrease in dimethyl-H3K4 without a statistically significant change in dimethyl-H3K9. Similar results were observed for the H19 DMR (see Fig. S8 in the supplemental material). These results suggest that BAT3 may selectively influence H3K4 dimethylation via its binding to BORIS and that this may be one mechanism accounting for the changes in Myc and BRCA1 gene expression. However, since BAT3 is not a histone methyltransferase, it seems unlikely that BAT3 directly alters H3K4 dimethylation.
SET1A and ASH2 bind to the Myc and BRCA1 promoters and the H19 DMR.
BAT3 is part of a family of proteins thought to modulate, either positively or negatively, the functions of other proteins, suggesting that these proteins are cochaperones (9). In addition, BAT3 has been shown to form a complex with p300 that enhances the efficiency of p300 recruitment to and acetylation of p53 (32). In this work it was strongly suggested that BAT 3 is a hinge or recruitment factor to assemble a more complex and functional protein complex. These results suggested that BAT3 might indirectly alter H3K4 dimethylation and gene expression via its role as an assembly factor. Thus, we used software from the Ingenuity Pathway Analysis (Ingenuity Systems, Inc.) to identify additional potential BAT3/BORIS-interacting proteins (see Fig. S9 in the supplemental material). This analysis identified SET1A as one potential BORIS complex protein via a secondary interaction with host cell factor 1, which was identified as a direct BORIS-interacting protein in our two-hybrid screen; however, other candidate genes were considered (Table 1). In addition, it has previously been shown that SET1 physically interacts with host cell factor 1 (28a). Furthermore, SET1A interacts with ASH2 to form a SET1A/ASH2 histone H3K4 methyltransferase complex (42). Therefore, it seemed reasonable to determine if SET1A/ASH2 interacts with BORIS and if this methyltransferase complex alters the dimethylation status of the Myc and BRCA1 promoters.
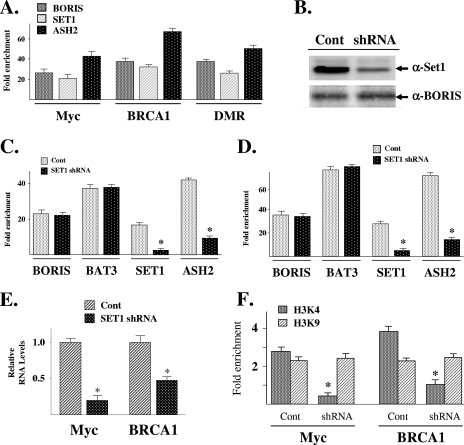
HCT116 cells were harvested for ChIP with either an anti-SET1A or an anti-ASH2 antibody, followed by QPCR with primers that overlap the promoter regions of BRCA1 and Myc. SET1A and ASH2 were also shown to bind to the H19 DMR (see Fig. S10 in the supplemental material). These results showed enrichment of the targeted regions in the presence of either SET1A or ASH2 (Fig. 4A), suggesting that both proteins are physically present at these DNA regions. Finally, reciprocal IP with either an anti-BORIS or an anti-SET1A antibody followed by Western analysis with the corresponding antibody demonstrated a physical interaction between SET1A and BORIS (see Fig. S10 in the supplemental material).
FIG. 4.
SET1A and ASH2 bind to the Myc and BRCA1 promoters and the H19 DMR, and shRNA knockdown of SET1A decreases both H3K4 dimethylation and gene expression. (A) SET1A and ASH2 bind to the promoter regions of Myc and BRCA1 as well as the H19 DMR. HCT116 cells were harvested and subjected to IP with either an anti-BORIS, an anti-SET1A (Santa Cruz, Inc.), or an anti-ASH2 (Santa Cruz, Inc.) antibody. DNA was eluted and purified before analysis with primers to BRCA1, Myc, or the H19 DMR. All ChIP experiments were done in triplicate, and error bars represent one standard deviation about the arithmetic mean. (B) HCT116 cells were transfected with either vector only (Cont) or a SET1A shRNA and then harvested, separated by SDS-polyacrylamide gel electrophoresis, transferred onto nitrocellulose, and immunoblotted using an anti-SET1A antibody (Bethyl Laboratories). (C and D) shRNA knockdown of SET1A decreases SET1A and ASH2, but not BORIS or BAT3, binding to the Myc (C) or BRCA1 (D) promoter. HCT116 cells were treated with nonsense (Cont) or SET1A shRNA, and ChIP was done with an anti-BORIS, anti-BAT3, anti-SET1A, or anti-ASH2 antibody followed by QPCR (as described above). (E) shRNA knockdown of SET1A decreases BRCA1, Myc, and H19 gene expression. HCT116 cells were treated with nonsense (Cont) or SET1A shRNA (in duplicate), RNA was isolated, and BRCA1, Myc, and H19 RNA levels were determined by RT-PCR that was done in triplicate. Error bars represent one standard deviation about the arithmetic mean. *, P < 0.05 by t test. (F) shRNA knockdown of SET1A decreases promoter histone H3K4 dimethylation. HCT116 cells were treated with nonsense (Cont) or SET1A shRNA, harvested for ChIP with either an H3K4 or H3K9 antibody, and analyzed by QPCR with primers to either the Myc or BRCA1 promoter. All ChIP experiments were done in triplicate, and error bars represent one standard deviation about the arithmetic mean. Statistical significance was established by Student's t test. *, P < 0.05.
shRNA knockdown of SET1A decreases both gene expression and H3K4 dimethylation.
HCT116 cells were transfected with a short hairpin RNA (shRNA) expression vector targeting SET1A (OriGene Technologies) (Fig. 4B), followed by ChIP with antibodies to BORIS, BAT3, SET1A, or ASH2. QPCR with primers that overlap the CTCF DNA-binding site of Myc showed that SET1A knockdown reduces the binding of both SET1A and ASH2 with no change in the binding characteristics of either BORIS or BAT3 (Fig. 4C). Similar results were observed for BRCA1 (Fig. 4D) and the H19 DMR (see Fig. S11A in the supplemental material). The results of these experiments suggest that SET1A is not required for either BORIS DNA binding or the protein-protein interaction between BAT3 and BORIS but is required for bringing ASH2 to this protein complex.
Although SET1A does not alter the DNA-binding characteristics of either BORIS or BAT3, it is reasonable to suggest that it might alter gene expression and histone methylation. Thus, RNA was subsequently isolated from HCT116 cells transfected with either control or SET1A shRNA vector, followed by RT-PCR with primers to either Myc and BRCA1. The results of these experiments demonstrated that shRNA knockdown of SET1A decreased the expression of Myc and BRCA1 (Fig. 4E).
SET1A has previously been shown to be an H3K4 methyltransferase, so we investigated the effects of shRNA knockdown of SET1A on histone dimethylation. HCT116 cells were transfected with a SET1A shRNA expression vector and subsequently examined for histone dimethylation by ChIP analysis with either an anti-H3K4 or an anti-H3K9 antibody. QPCR with primers to the promoter regions of Myc and BRCA1 showed a statistically significant decrease in dimethyl-H3K4 (Fig. 4F) without a statistically significant change in dimethyl-H3K9. Similar results were observed for the H19 DMR (see Fig. S11A in the supplemental material). As a control, the upstream regulator regions of two additional genes were examined for any effects on gene expression from SET1 knockdown. shRNA knockdown of SET1A had no effect on p21 or p14 expression (see Fig. S12A in the supplemental material), suggesting that depletion of SET1A decreases the expression of specific genes while having no effect on others, as has been shown by others (8). ChIP with an anti-H3K4 antibody followed by QPCR with primers overlapping the upstream promoter regions p21 and p14, but not adjacent to any nearby CTCF binding site, also showed no change in histone dimethylation (see Fig. S12B in the supplemental material). These results are consistent with those published by others showing that knockdown of SET1A selectively alters promoter histone methylation (16). These results suggest that SET1A may influence H3K4 dimethylation via its binding to BORIS and that this interaction may, at least in part, influence both gene expression and chromatin status.
shRNA knockdown of BORIS decreases SET1A and ASH2 binding, Myc and BRCA1 expression, and H3K4 dimethylation.
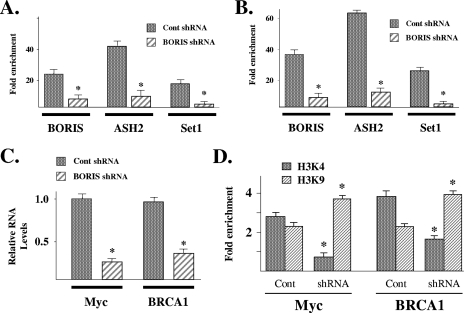
The results presented above suggest that BORIS may act as a scaffold upon which protein complexes form for the purpose of modulating gene expression and modifying chromatin. Thus, shRNA knockdown of BORIS should prevent the binding of SET1A and ASH2 to the Myc and BRCA1 promoters as well as alter Myc and BRCA1 expression. HCT116 cells were transfected with either control or BORIS shRNA (OriGene Technologies), and cells were harvested for ChIP with anti-BORIS, anti-ASH2, or anti-SET1A antibodies. Purified DNA samples were analyzed by QPCR with primers that overlap either the Myc (Fig. 5A) or BRCA1 (Fig. 5B) promoter regions.
FIG. 5.
shRNA knockdown of BORIS eliminates BORIS, SET1A, and ASH2 DNA binding, decreases Myc and BRCA1 expression, and reduces H3K4 dimethylation. (A and B) shRNA knockdown of BORIS decreases SET1A and ASH2 binding to the Myc and BRCA1 promoters. HCT116 cells were transfected with either nonsense (Cont) or BORIS shRNA, and ChIP was done with an anti-BORIS, anti-SET1A, or anti-ASH2 antibody. DNA was eluted and purified before analysis with primers to either the Myc (A) or BRCA1 (B) promoter. (C) shRNA knockdown of BORIS decreased Myc and BRCA1 expression. HCT116 cells were transfected with either nonsense (Cont) or BORIS shRNA (in duplicate), RNA was isolated, and Myc and BRCA1 expression was determined by RT-PCR in triplicate. Error bars represent one standard deviation about the arithmetic mean. (D) shRNA knockdown of BORIS decreases promoter histone H3K4 dimethylation. HCT116 cells were treated with nonsense (Cont) or BORIS shRNA, harvested for ChIP with either an H3K4 or H3K9 antibody, and analyzed by QPCR. All ChIP experiments were done in triplicate, and error bars represent one standard deviation about the arithmetic mean. Statistical significance was established by Student's t test. *, P < 0.05.
The results of these experiments suggest that BORIS is required for the recruitment of both SET1A and ASH2. In addition, shRNA knockdown of BORIS decreased the expression of both Myc and BRCA1 (Fig. 5C). In contrast, shRNA knockdown of BORIS had no effect on the expression of either p21 or p14 (see Fig. S12A in the supplemental material), suggesting that the induction of gene expression by BORIS is promoter specific. Finally, HCT116 cells were transfected with BORIS shRNA and examined for histone H3 dimethylation by ChIP analysis with either an anti-H3K4 or an anti-H3K9 antibody. QPCR with primers to the promoter regions of Myc and BRCA1 showed a statistically significant decrease in dimethyl-H3K4 (Fig. 5D). These results suggest that BORIS serves as a scaffold that establishes a protein assembly that influences H3K4 dimethylation via the recruitment of proteins such as SET1A. The proteins recruited by BORIS regulate gene expression by altering chromatin structure.
DISCUSSION
In this work, we propose that the insulator DNA-binding protein BORIS assembles a transcriptional regulatory complex that modulates gene expression via a mechanism that involves changes in promoter chromatin status. ChIP analysis using antibodies to BAT3, SET1A, and BORIS demonstrated that all three proteins bind to upstream promoter regions of Myc and BRCA1 as well as to the H19 DMR. All three of these DNA regions contain BORIS (CTCF DNA-binding site) target sequences (see Fig. S5 in the supplemental material) (17, 29). Furthermore, RNA knockdown experiments suggest that both SET1A and BAT3 bind to BORIS but do not alter the DNA-binding profile of BORIS. The results of these experiments suggest a potential mechanism whereby BORIS, anchored by its binding to DNA, establishes a platform upon which other proteins can assemble to promote transcription. Our understanding of the function of BORIS as a scaffold will be expanded by further investigations of the proteins suggested to interact with BORIS, including testis-specific proteins, chromatin-associated proteins, helicase-associated proteins, and transcription factors identified by the two-hybrid screen as BORIS binding partners (Table 1).
BAT3, also referred to as BAG-6 (Bcl-2-associated athanogene-6) or the human homolog of Scythe is part of the BAG family of proteins initially identified by their similarity to the founding member of this family, BAG-1, which was discovered through a screen for Bcl-2-binding proteins (37). Several members of the BAG family interact with heat shock protein 70 (Hsc/Hsp70) and modulate, either positively or negatively, the function of chaperone proteins, suggesting that BAG family proteins are cochaperones (9). Interestingly, BAT3 is a nuclear protein that is expressed predominantly in testicular germ cells, similar to the case for BORIS (30, 41).
The results presented here show that RNAi knockdown of BAT3 alters Myc and BRCA1 gene expression as well as the H3K4/H3K9 dimethylation ratio. While statistically significant changes in H3K4 dimethylation were observed by altering intracellular BAT3 levels, relatively little change in H3K9 dimethylation was observed. In this regard, BAT3 may determine the specific proteins binding to BORIS, and the subsequent protein complex may preferentially modulate histone H3 dimethylation at K4. While the role of BAT3 in the regulation of H3K4 dimethylation has still to be fully determined, it has been shown that BAT3 is essential for p300-mediated acetylation of p53 (32). In this work, BAT3 formed a complex with p300 and enhanced the efficiency of p300 recruitment to and acetylation of p53. Therefore, it was suggested that BAT3 is a cochaperone or hinge protein that may stabilize a functional p300 catalytic acetylation complex. Since BAT3 is thought to be a cochaperone, its intracellular function may be to establish and/or recruit specific protein complexes that play a more direct role in altering either protein posttranslation modification or possibly gene expression via histone modification.
To begin to address this idea, we identified several potential BORIS-interacting protein using software from the Ingenuity Pathway Analysis (Ingenuity Systems, Inc.), our two hybrid data, and a previous publication (28a). It has previously been shown that SET1A interacts with ASH2 to form a SET1A/ASH2 histone H3K4 methyltransferase complex (42). While other potential proteins were considered, the histone H3K4 methyltransferase activity of SET1 matched our knockdown results with both BORIS and BAT3. Thus, it seemed logical to determine if SET1A mediated the changes in H3K4 dimethylation observed after BAT3 knockdown. ChIP demonstrated that SET1A and its binding partner, ASH2, bind together to BORIS at the promoters of Myc and BRCA1. shRNA knockdown of SET1A decreased its own binding as well as that of ASH2, which has previously been shown as a binding partner (43), without altering the promoter occupancy of either BAT3 or BORIS. In addition, SET1A knockdown decreased H3K4 dimethylation at the Myc and BRCA1 promoters as well as gene expression. This was as expected, since lysine methyltransferases usually modify a single lysine on a single histone and this posttranslational modification has been previously shown to be involved in activation or repression of transcription (1). Lastly, knockdown of BAT3 had no effect on SET1A promoter occupancy (data not shown), suggesting that BAT3 likely has an indirect stabilizing effect on SET1A activity. These results suggest that SET1 is part of a chromatin protein complex that includes BORIS and BAT3; however, it seems likely that SET1 will have considerable genome-wide effects on the methylation of histones independent of this interaction. Finally, we propose that SET1A and BAT3, are recruited to BORIS as part of a transcriptional complex that increases both H3K4 lysine dimethylation and gene expression.
Supplementary Material
Acknowledgments
This work was supported by grant CA65145 (to A.P.F.) from the NCI and also (in part) by the Intramural Research Program of the NIH, the NCI, and the CCR.
We thank Melissa Stauffer of Scientific Editing Solutions for editorial assistance.
Footnotes
Published ahead of print on 2 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bannister, A. J., and T. Kouzarides. 2005. Reversing histone methylation. Nature 4361103-1106. [DOI] [PubMed] [Google Scholar]
- 2.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405482-485. [DOI] [PubMed] [Google Scholar]
- 3.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98387-396. [DOI] [PubMed] [Google Scholar]
- 4.Butcher, D. T., D. N. Mancini-DiNardo, T. K. Archer, and D. I. Rodenhiser. 2004. DNA binding sites for putative methylation boundaries in the unmethylated region of the BRCA1 promoter. Int. J. Cancer 111669-678. [DOI] [PubMed] [Google Scholar]
- 5.Cui, H., I. L. Horon, R. Ohlsson, S. R. Hamilton, and A. P. Feinberg. 1998. Loss of imprinting in normal tissue of colorectal cancer patients with microsatellite instability. Nat. Med. 41276-1280. [DOI] [PubMed] [Google Scholar]
- 6.Cui, H., P. Onyango, S. Brandenburg, Y. Wu, C. L. Hsieh, and A. P. Feinberg. 2002. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 626442-6446. [PubMed] [Google Scholar]
- 7.Desmots, F., H. R. Russell, Y. Lee, K. Boyd, and P. J. McKinnon. 2005. The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Mol. Cell. Biol. 2510329-10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diehl, F., L. Rossig, A. M. Zeiher, S. Dimmeler, and C. Urbich. 2007. The histone methyltransferase MLL is an upstream regulator of endothelial-cell sprout formation. Blood 1091472-1478. [DOI] [PubMed] [Google Scholar]
- 9.Doong, H., A. Vrailas, and E. C. Kohn. 2002. What's in the ‘BAG’?—a functional domain analysis of the BAG-family proteins. Cancer Lett. 18825-32. [DOI] [PubMed] [Google Scholar]
- 10.Fedoriw, A. M., P. Stein, P. Svoboda, R. M. Schultz, and M. S. Bartolomei. 2004. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science 303238-240. [DOI] [PubMed] [Google Scholar]
- 11.Gius, D., X. M. Cao, F. J. Rauscher III, D. R. Cohen, T. Curran, and V. P. Sukhatme. 1990. Transcriptional activation and repression by Fos are independent functions: the C terminus represses immediate-early gene expression via CArG elements. Mol. Cell. Biol. 104243-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gombert, W. M., S. D. Farris, E. D. Rubio, K. M. Morey-Rosler, W. H. Schubach, and A. Krumm. 2003. The c-myc insulator element and matrix attachment regions define the c-myc chromosomal domain. Mol. Cell. Biol. 239338-9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grewal, S. I., and D. Moazed. 2003. Heterochromatin and epigenetic control of gene expression. Science 301798-802. [DOI] [PubMed] [Google Scholar]
- 14.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405486-489. [DOI] [PubMed] [Google Scholar]
- 15.Hong, J. A., Y. Kang, Z. Abdullaev, P. T. Flanagan, S. D. Pack, M. R. Fischette, M. T. Adnani, D. I. Loukinov, S. Vatolin, J. I. Risinger, M. Custer, G. A. Chen, M. Zhao, D. M. Nguyen, J. C. Barrett, V. V. Lobanenkov, and D. S. Schrump. 2005. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 657763-7774. [DOI] [PubMed] [Google Scholar]
- 16.Huang, J., J. R. Kent, B. Placek, K. A. Whelan, C. M. Hollow, P. Y. Zeng, N. W. Fraser, and S. L. Berger. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 805740-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishihara, K., M. Oshimura, and M. Nakao. 2006. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol. Cell 23733-742. [DOI] [PubMed] [Google Scholar]
- 18.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 19.Karimpour, S., J. Lou, L. L. Lin, L. M. Rene, L. Lagunas, X. Ma, S. Karra, C. M. Bradbury, S. Markovina, P. C. Goswami, D. R. Spitz, K. Hirota, D. V. Kalvakolanu, J. Yodoi, and D. Gius. 2002. Thioredoxin reductase regulates AP-1 activity as well as thioredoxin nuclear localization via active cysteines in response to ionizing radiation. Oncogene 216317-6327. [DOI] [PubMed] [Google Scholar]
- 20.Klenova, E. M., H. C. Morse III, R. Ohlsson, and V. V. Lobanenkov. 2002. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin. Cancer Biol. 12399-414. [DOI] [PubMed] [Google Scholar]
- 21.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 22.Kurukuti, S., V. K. Tiwari, G. Tavoosidana, E. Pugacheva, A. Murrell, Z. Zhao, V. Lobanenkov, W. Reik, and R. Ohlsson. 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA 10310684-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 24.Li, J., Q. Lin, H. G. Yoon, Z. Q. Huang, B. D. Strahl, C. D. Allis, and J. Wong. 2002. Involvement of histone methylation and phosphorylation in regulation of transcription by thyroid hormone receptor. Mol. Cell. Biol. 225688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, Y., G. D. Kao, B. A. Garcia, J. Shabanowitz, D. F. Hunt, J. Qin, C. Phelan, and M. A. Lazar. 2006. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 202566-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loukinov, D. I., E. Pugacheva, S. Vatolin, S. D. Pack, H. Moon, I. Chernukhin, P. Mannan, E. Larsson, C. Kanduri, A. A. Vostrov, H. Cui, E. L. Niemitz, J. E. Rasko, F. M. Docquier, M. Kistler, J. J. Breen, Z. Zhuang, W. W. Quitschke, R. Renkawitz, E. M. Klenova, A. P. Feinberg, R. Ohlsson, H. C. Morse III, and V. V. Lobanenkov. 2002. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc. Natl. Acad. Sci. USA 996806-6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellor, J. 2006. It takes a PHD to read the histone code. Cell 12622-24. [DOI] [PubMed] [Google Scholar]
- 28.Murrell, A., S. Heeson, and W. Reik. 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36889-893. [DOI] [PubMed] [Google Scholar]
- 28a.Narayanan, A., W. T. Ruyechan, and T. M. Kristie. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. USA 10410835-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28b.Nguyen, P., H. Cui, K. S. Bisht, L. Sun, K. Patel, R. S. Lee, H. Kugoh, M. Oshimura, A. P. Feinberg, and D. Gius. 2008. CTCFL/BORIS is a methylation-independent DNA-binding protein that preferentially binds to the paternal H19 differentially methylated region. Cancer Res. 685546-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohlsson, R., R. Renkawitz, and V. Lobanenkov. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17520-527. [DOI] [PubMed] [Google Scholar]
- 30.Ozaki, T., E. Hanaoka, M. Naka, A. Nakagawara, and S. Sakiyama. 1999. Cloning and characterization of rat BAT3 cDNA. DNA Cell Biol. 18503-512. [DOI] [PubMed] [Google Scholar]
- 31.Pant, V., S. Kurukuti, E. Pugacheva, S. Shamsuddin, P. Mariano, R. Renkawitz, E. Klenova, V. Lobanenkov, and R. Ohlsson. 2004. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell. Biol. 243497-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki, T., E. C. Gan, A. Wakeham, S. Kornbluth, T. W. Mak, and H. Okada. 2007. HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev. 21848-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smart, D. K., K. L. Ortiz, D. Mattson, C. M. Bradbury, K. S. Bisht, L. K. Sieck, M. W. Brechbiel, and D. Gius. 2004. Thioredoxin reductase as a potential molecular target for anticancer agents that induce oxidative stress. Cancer Res. 646716-6724. [DOI] [PubMed] [Google Scholar]
- 34.Spotswood, H. T., and B. M. Turner. 2002. An increasingly complex code. J. Clin. Investig. 110577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 35a.Sun, L., L. Huang, P. Nguyen, K. S. Bisht, G. Bar-Sela, A. S. Ho, C. M. Bradbury, W. Yu, H. Cui, S. Lee, J. B. Trepel, A. P. Feinberg, and D. Gius. 2008. DNA methyltransferase 1 and 3B activate BAG-1 expression via recruitment of CTCFL/BORIS and modulation of promoter histone methylation. Cancer Res. 682726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takai, D., F. A. Gonzales, Y. C. Tsai, M. J. Thayer, and P. A. Jones. 2001. Large scale mapping of methylcytosines in CTCF-binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum. Mol. Genet. 102619-2626. [DOI] [PubMed] [Google Scholar]
- 37.Takayama, S., K. Kochel, S. Irie, J. Inazawa, T. Abe, T. Sato, T. Druck, K. Huebner, and J. C. Reed. 1996. Cloning of cDNAs encoding the human BAG1 protein and localization of the human BAG1 gene to chromosome 9p12. Genomics 35494-498. [DOI] [PubMed] [Google Scholar]
- 38.van Attikum, H., and S. M. Gasser. 2005. The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 6757-765. [DOI] [PubMed] [Google Scholar]
- 39.Vatolin, S., Z. Abdullaev, S. D. Pack, P. T. Flanagan, M. Custer, D. I. Loukinov, E. Pugacheva, J. A. Hong, H. Morse III, D. S. Schrump, J. I. Risinger, J. C. Barrett, and V. V. Lobanenkov. 2005. Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res. 657751-7762. [DOI] [PubMed] [Google Scholar]
- 40.Wallace, J. A., and G. Felsenfeld. 2007. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17400-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, R., and C. C. Liew. 1994. The human BAT3 ortholog in rodents is predominantly and developmentally expressed in testis. Mol. Cell Biochem. 13649-57. [DOI] [PubMed] [Google Scholar]
- 42.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokoyama, A., Z. Wang, J. Wysocka, M. Sanyal, D. J. Aufiero, I. Kitabayashi, W. Herr, and M. L. Cleary. 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 245639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.