Abstract
The binding of the Epstein-Barr virus glycoprotein gp350 by complement receptor type 2 (CR2) is critical for viral attachment to B lymphocytes. We set out to test hypotheses regarding the molecular nature of this interaction by developing an enzyme-linked immunosorbent assay (ELISA) for the efficient analysis of the gp350-CR2 interaction by utilizing wild-type and mutant forms of recombinant gp350 and also of the CR2 N-terminal domains SCR1 and SCR2 (designated CR2 SCR1-2). To delineate the CR2-binding site on gp350, we generated 17 gp350 single-site substitutions targeting an area of gp350 that has been broadly implicated in the binding of both CR2 and the major inhibitory anti-gp350 monoclonal antibody (MAb) 72A1. These site-directed mutations identified a novel negatively charged CR2-binding surface described by residues Glu-21, Asp-22, Glu-155, Asp-208, Glu-210, and Asp-296. We also identified gp350 amino acid residues involved in non-charge-dependent interactions with CR2, including Tyr-151, Ile-160, and Trp-162. These data were supported by experiments in which phycoerythrin-conjugated wild-type and mutant forms of gp350 were incubated with CR2-expressing K562 cells and binding was assessed by flow cytometry. The ELISA was further utilized to identify several positively charged residues (Arg-13, Arg-28, Arg-36, Lys-41, Lys-57, Lys-67, Arg-83, and Arg-89) within SCR1-2 of CR2 that are involved in the binding interaction with gp350. These experiments allowed a comparison of those CR2 residues that are important for binding gp350 to those that define the epitope for an effective inhibitory anti-CR2 MAb, 171 (Asn-11, Arg-13, Ser-32, Thr-34, Arg-36, and Tyr-64). The mutagenesis data were used to calculate a model of the CR2-gp350 complex using the soft-docking program HADDOCK.
Epstein-Barr virus (EBV; human herpesvirus type 4) is a member of the herpesvirus family and is found in a large proportion (>90%) of the world's population. While childhood EBV infection often is clinically silent, delayed infection until adolescence or later can result in the development of infectious mononucleosis. In immunodeficient individuals, EBV also has been linked to malignancies or disease states including African Burkitt's lymphoma, nasopharyngeal carcinoma, EBV-associated lymphoproliferative disease, non-Hodgkin's lymphoma, and oral hairy leukoplakia (6). EBV primarily infects two main cell types, B lymphocytes and epithelial cells, although under some circumstances the virus also can infect T cells, natural killer cells, smooth muscle cells, and astrocytes (27, 35). In B cells, the interaction between EBV and the cell surface is initially mediated by the predominant glycoprotein in the viral envelope, gp350/220, binding to complement receptor type 2 (CR2/CD21) (9, 10, 22, 31, 33, 41). Fusion with the cell membrane and the invasion of the host cell then is facilitated by the binding of a second viral glycoprotein, gp42, to HLA class II molecules and also by three additional viral glycoproteins, gB, gH, and gL (14, 15, 19, 28). Soluble forms of both CR2 and gp350 have been shown to inhibit the EBV invasion of peripheral blood mononuclear cells and B lymphocytes, respectively (32, 41). In addition, monoclonal antibodies (MAbs) generated against gp350 have been shown to effectively inhibit the infection of peripheral B lymphocytes with EBV (42), and some forms of this protein have been shown to act successfully as a vaccine against EBV-associated diseases in animal models and in human clinical trials (20, 25, 38). While recombinant EBV gp350 deletion strains have been shown to be capable of transforming human B lymphocytes, they do so with a significantly reduced efficiency (21), and there is little doubt that the interaction between CR2 and gp350 is the primary mechanism by which EBV is adsorbed to the cell surface.
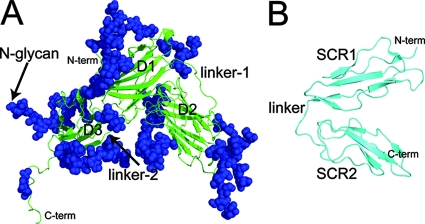
EBV gp350/220 is an extensively glycosylated polypeptide (907 residues) that is expressed as two alternatively spliced forms of approximately 350 and 220 kDa (4). The three-dimensional structure of a truncated form of gp350 comprising the amino-terminal 470 residues has been elucidated by X-ray crystallography at a 3.5-Å resolution (40). Three distinct domains were identified (residues 4 to 153, 165 to 305, and 317 to 426), each comprising an anti-parallel β-barrel structure and joined by two linker regions, each of 11 amino acid residues. These domains are packed tightly against each other, forming a distinctive L-shaped arrangement that is almost uniformly glycosylated (Fig. 1A). A limited multiple-site mutagenesis study, targeting a glycan-free area of gp350, suggested a putative CR2-binding site within a negatively charged region of this molecule that incorporates the two N-terminal domains and the linker region connecting them (residues 154 to 164) (40). Those mutations affecting CR2 binding also disrupted the ability of gp350 to be recognized by its major neutralizing MAb, 72A1. These data are consistent with two separate peptide mapping analyses that have identified a number of linear sequences of gp350 that are involved in EBV binding to CR2. These sequences are in close proximity to, or directly overlap with, the CR2-binding region identified from the mutagenesis and crystal studies (30, 43).
FIG. 1.
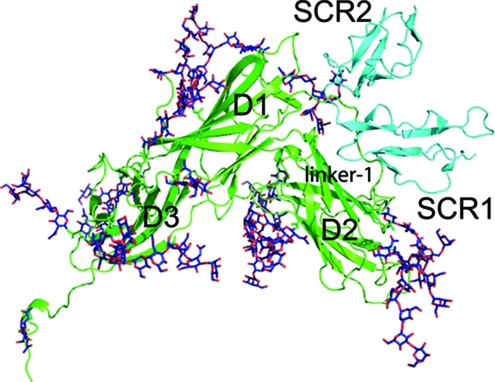
Schematic ribbon representations of (A) EBV gp350 and (B) CR2 SCR1-2, as determined by X-ray crystallography. (A) The secondary structure of the gp350 fragment corresponding to residues 4 to 443 of the native sequence of gp350 (B95-8 strain) is shown in green. Indicated are the three independent β-barrel domains comprising the major structural components: D1, corresponding to residues 4 to 153; D2 (residues 165 to 305); and D3 (residues 317 to 426). Also indicated are two structured linker regions, designated linker-1 (residues 154 to 164) and linker-2 (residues 306 to 316), connecting D1 and D2 as well as D2 and D3, respectively. A total of 14 glycan moieties identified in the crystal structure also are indicated in blue. (PDB accession code 2H6O) (40). (B) The secondary structure of the two amino-terminal domains (SCR1-2) of CR2 as identified in the cocrystal structure of the CR2-C3d complex are shown in cyan. Each SCR domain comprises five short β-strands and four cysteine residues forming disulfide bonds at either end of the domain. Indicated are SCR1 (residues 1 to 62), SCR2 (residues 71 to 129), and the linker region connecting both domains (residues 63 to 70) (PDB accession code 1GHQ) (39). N-term, N terminus; C-term, C terminus.
CR2 is a member of the regulators of the complement activation family of proteins (24). Members of this family are characterized by the presence of short repeating domains comprising approximately 60 amino acids, known as short consensus repeats (SCR) or complement control protein modules (37). Each of these compact units contains conserved residues including four cysteines and a nearly invariant tryptophan. The molecular structure of CR2 comprises a 15- or 16-SCR extracellular domain and short transmembrane and cytoplasmic regions. In addition to gp350, physiologically relevant ligands of CR2 (reviewed in reference 16) include proteolytic fragments of complement component C3, including C3d, C3dg, and iC3b (23, 45), alpha interferon (1, 7), and the low-affinity immunoglobulin E receptor CD23 (2, 3). The structures of CR2 N-terminal domains SCR1 and SCR2 (designated CR2 SCR1-2), in both free and C3d-complexed forms, have been determined by X-ray crystallography (34, 39). In both cases CR2 SCR1-2 adopts a compact conformation in which the two SCR domains are packed against each other, forming a tight V shape (Fig. 1B). Whether the tightly packed conformation of these domains as seen in the crystal structures reflects their conformation under native conditions is debatable; an analytical ultracentrifugation and X-ray scattering study carried out in conjunction with constrained modeling indicated that CR2 SCR1-2 is more extended in solution than it is in the crystals (11). Recently, we identified residues in CR2 that are involved in the ligation of gp350 by employing flow cytometry to measure the binding of recombinant wild-type gp350 to K562 erythroleukemia cells transfected with wild-type, or single-site mutant, full-length forms of CR2 (46). Our data suggested that gp350 binding is mediated primarily by positively charged residues within SCR1 of CR2.
The goal of the current work was to accumulate sufficient data to underpin the reliable knowledge-based docking of an SCR1-2-gp350 complex by significantly increasing the available mutagenesis data. To achieve this, we developed an enzyme-linked immunosorbent assay (ELISA)-based analysis in which wild-type or mutant forms of SCR1-2 of CR2 were expressed as maltose-binding protein (MBP) fusion proteins using Escherichia coli. To minimize disruption to potential long-range electrostatic charge effects that have previously been associated with CR2-ligand interactions (29, 47), we decided to pursue an alanine substitution mutagenesis strategy, with only a single additional serine-to-proline mutation being generated. To delineate the corresponding CR2-binding site on gp350, we also employed an alanine-screening approach, targeting residues within the two N-terminal domains of gp350 and the connecting linker region using the ELISA described above. This ELISA-based characterization of the CR2-binding site on gp350 was verified by employing wild-type CR2-expressing K562 cells to measure the binding of our gp350 point mutants using flow cytometry. The resulting sets of ELISA- and cell-based mutagenesis data were utilized to generate a series of ambiguous interaction restraints (AIRs) that then were used to calculate a model of the CR2-gp350 complex (8). The validation of the model was provided by experiments in which our MBP-CR2 mutant library was used to characterize the structured epitope for an anti-CR2 MAb, 171, which strongly inhibits all known CR2-ligand interactions (1, 13, 46). Our data clearly show that the epitope for the 171 MAb on CR2 directly overlaps with a region of CR2 that interacts with the surface of the gp350 molecule.
MATERIALS AND METHODS
Production of the anti-CR2 MAbs 171 and 629.
Anti-CR2 MAbs 171 and 629 were obtained from the spent culture medium of hybridoma cells grown in RPMI 1640 supplemented with 2 mM l-glutamine, 100 IU penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum. Antibodies were purified by affinity chromatography using protein G Sepharose 4 fast flow resin (GE Healthcare Biosciences Corp.) according to the manufacturer's instructions. Purified MAbs 171 and 629 subsequently were exchanged into phosphate-buffered saline (PBS), pH 7.4 (containing 136.9 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4), and finally were concentrated to give a stock solution containing 1 mg/ml of antibody as determined by UV-visible spectrophotometry, and the product was stored at −20°C until needed.
Expression of wild-type and mutant MBP-CR2 SCR1-2 recombinant proteins in Escherichia coli.
DNA corresponding to residues 1 to 133 of wild-type CR2 (SCR1-2) was PCR amplified and then ligated into the prokaryotic expression vector pMAL-p2x (New England Biolabs), which encodes a maltose-binding protein (MBP) tag at the 5′ end of the inserted DNA, as previously described (46). Plasmid DNA subsequently was transformed into E. coli BL21 cells, and wild-type recombinant MBP-CR2 SCR1-2 was produced according to earlier protocols (46). To summarize, recombinant wild-type MBP-CR2 SCR1-2 protein was expressed from overnight cultures induced with isopropyl-β-d-thiogalactoside. Cultures then were harvested by centrifugation, and the resulting pellets were resuspended in a column buffer containing 20 mM Tris-HCl, pH 7.4, 0.2 M NaCl, 1 mM EDTA, and lysed by sonication. The lysate was clarified by centrifugation and then purified by successive affinity (amylose resin; New England Biolabs) and size-exclusion (Hiload S200 26/60; GE Biosciences) chromatography stages.
Recombinant L10A, N11A, R13A, S15P, Y16A, R28A, S32A, T34A, R36A, K41A, K50A, K57A, Y64A, K67A, Y68A, R83A, T86A, R89A, and M117A forms of CR2 SCR1-2 DNA were produced from wild-type MBP-CR2 SCR1-2 DNA by utilizing a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Plasmid DNA containing the mutant CR2 SCR1-2 insert then was transformed into E. coli BL21, and recombinant mutant CR2 SCR1-2 proteins were expressed and purified as described above.
Expression of recombinant gp350 proteins.
EBV genomic DNA was extracted from previously obtained cell supernatants of the marmoset B95-8 leukocyte cell line (ATCC) using a QIAamp UltraSens virus kit (Qiagen) as described previously (46). DNA corresponding to residues 1 to 470 of EBV gp350/220 and a fragment of the E. coli biotin carboxyl carrier protein (BCCP) corresponding to residues 70 to 156 were separately PCR amplified. PCR fragments then were ligated into the pSecTag2/Hygro B eukaryotic expression vector (Invitrogen), which encodes a myc epitope and a hexahistidine tag at the 3′ end of the inserted DNA. Plasmid DNA subsequently was transfected into human embryonic kidney 293f freestyle cells (Invitrogen) for the soluble expression of recombinant EBV gp350 into the medium. Resulting recombinant gp350 was concentrated and concurrently exchanged into a sodium phosphate column buffer for purification purposes. Recombinant gp350 protein was purified by utilizing successive immobilized metal affinity and size-exclusion chromatography steps. After being purified, gp350 either was aliquoted and then frozen at −70°C until required for ELISA or was biotinylated (gp350-biotin) using biotin ligase (Avidity). The resulting gp350-biotin was conjugated to phycoerythrin (PE)-NeutrAvidin (Molecular Probes), generating fluorochrome-tagged gp350 monomers for dual-color flow cytometric binding analysis.
Mutant forms of gp350 were generated to target a number of residues located within domain 1 (D1) and domain 2 (D2) of gp350 and also the 11-residue linker region (linker-1) connecting the two domains. Residues Glu-21, Asp-22, Asp-53, Glu-119, Tyr-151, Glu-155, Tyr-159, Ile-160, Trp-162, Asp-163, Glu-201, Asp-208, Glu-210, Glu-214, Asp-215, Glu-236, and Glu-296 were selected for alanine-screening studies to delineate the CR2-binding site on the gp350 molecule and also to characterize the nature of the interaction between the two molecules. Single-site mutant forms of recombinant EBV gp350 were produced from wild-type B95-8 gp350 DNA by utilizing a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Recombinant mutant gp350 proteins were expressed and purified as described above for the wild type.
CR2-gp350, CR2-171 MAb, and CR2-629 MAb ELISAs.
Plates were coated overnight at 4°C with 5 μg/ml of gp350, MAb 171, or MAb 629 in 20 mM sodium bicarbonate buffer, pH 8.8. After being coated, the plates were blocked using 0.1% bovine serum albumin (BSA) in a PBS solution, pH 7.4, for 1 h at room temperature. The plates then were washed and incubated with either wild-type or mutant MBP-CR2 SCR1-2 at concentrations ranging from 0.03125 to 2.0 μg/ml in PBS for 1 h at room temperature. After further washes, wild-type or mutant MBP-CR2 binding was detected using commercially available horseradish peroxidase-conjugated anti-MBP MAb (New England Biolabs) according to the manufacturer's instructions.
Flow cytometry.
Flow cytometric experiments were carried out using K562 erythroleukemia cells transfected with full-length wild-type CR2 as described previously (17, 46). Binding analyses were carried out using wild-type or mutant forms of gp350-biotin. For each condition, 5 × 105 human CR2-transfected K562 cells first were incubated with anti-CR2 HB5 MAb at 1 μg/ml on ice for 1 h. The HB5-coated cells subsequently were incubated with fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse polyclonal antibody (BD Biosciences) at 1 μg/ml on ice for 30 min. The primary epitope for HB5 has been identified within the N-terminal SCR3-4 domains of CR2 and accordingly does not interfere with ligand binding. During this incubation, 100 μl of gp350-biotin monomers in PBS-0.1% BSA-0.01% sodium azide were prepared for each condition by adding 0.5 μg of recombinant wild-type or mutant gp350-biotin and 0.4 μg PE-conjugated NeutrAvidin (Molecular Probes) and incubating them at room temperature for 30 min. Following the washing of the FITC-stained K562 cells, 100 μl of monomeric PE-conjugated gp350-biotin was added to each sample of cells and incubated for 30 min on ice. After being washed, the cells were fixed and analyzed by multicolor flow cytometry in the University of Colorado Cancer Center Flow Cytometry Core Facility (Denver). Cells were analyzed as previously described, with gating on either whole-cell populations and/or the intermediate 25% of CR2-expressing cells (FITC positive). Wild-type or mutant gp350-biotin binding was determined by PE mean channel fluorescence. A minimum of three separate experiments was carried out for each mutation.
Generation by HADDOCK of a CR2-gp350 model.
Mutagenesis data from these and previous studies were used to derive a series of models of the CR2-gp350 complex utilizing version 1.3 of the soft-docking program HADDOCK (8). This employs a knowledge-based approach by which the program utilizes experimentally derived data, in conjunction with the available structures, to drive the docking of two macromolecules. Residues that have been experimentally implicated in a binding interaction and that are therefore likely to form part of the interface between two molecules are designated active. In this case, active residues for each molecule were defined as those residues that our mutagenesis data have identified as playing a significant role in the binding interaction between CR2 and EBV (46) and that possessed a main chain or side chain solvent accessibility of 50% or greater (as determined by the program NACCESS (S. Hubbard and J. Thornton). Selected active residues for CR2 were Arg-13, Ser-15, Arg-28, Lys-41, Lys-67, and Arg-83. For gp350, the selected active residues were Glu-21, Tyr-151, Glu-155, Ile-160, Trp-162, Asp-208, Glu-210, and Asp-296. Also used in the HADDOCK calculations are passive residues, defined as those residues in close proximity to active residues with high main chain or side chain solvent accessibility. This information is introduced in the form of AIRs consisting of calculated distance restraints between any atom within the active residues of one protein and all atoms within the active and passive residues on the respective binding partner (8). Standard analyses were performed by HADDOCK on 100 water-refined structures from an initial 2,000 structures calculated and include an analysis of the energy contributions from buried surface area and electrostatic interactions.
RESULTS
Single-site mutations were generated within the domains of EBV gp350 and human CR2, which mediate the interaction between these proteins, and were subjected to a functional assay. The aim was to accumulate sufficient data to accurately delineate the binding surfaces and to drive a reliable soft-docking exercise resulting in a robust model of the complex.
MBP-CR2 SCR1-2-gp350 mutant ELISA.
To characterize the CR2 SCR1-2-binding site on gp350, we generated single-site substitutions targeting a glycan-free area of this molecule that was broadly implicated in CR2-ligand binding by previous crystal-driven multiple-site mutagenesis studies and, separately, by peptide mapping studies (30, 43, 46). Our alanine substitutions targeted residues within the two N-terminal β-barrel domains (D1, residues 4 to 153; and D2, residues 165 to 305) and the 11-residue linker region that connects them (linker-1).
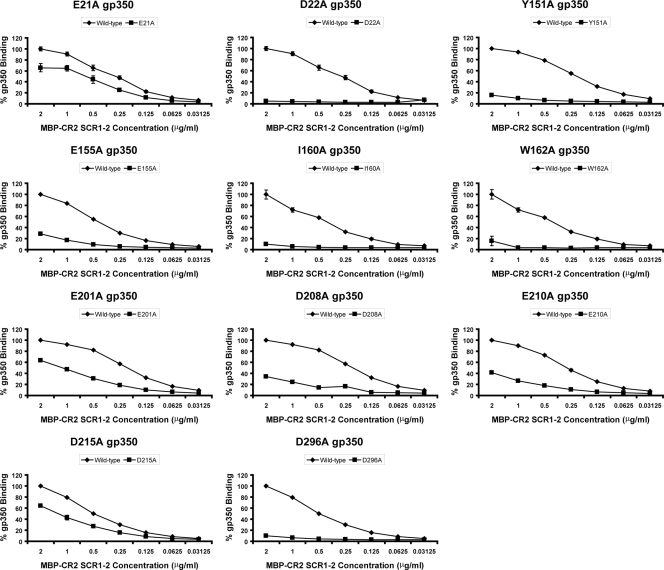
Within D1 of gp350, E21A (in the crystal structure of gp350 identified as Asp-21), D22A, and Y151A showed significant (greater than 20%) decreases in CR2 SCR1-2 binding relative to that of wild-type gp350, while D53A and E119A exhibited approximately wild-type-like levels of binding (Fig. 2, Table 1).
FIG. 2.
MBP-CR2 SCR1-2-gp350 ELISA data targeting residues within gp350. The abilities of MBP-CR2 SCR1-2 to bind plate-bound mutant forms of gp350 (E21A, D22A, Y151A, E155A, I160A, W162A, E201A, D208A, E210A D215A, and D296A) are shown. The averages and standard errors of the means of the normalized values relative to those of wild-type gp350 binding are given. Data are not shown for a number of gp350 mutants that exhibited wild-type-like (<20% reduction) binding: D53A, E119A, Y159A, D163A, E214A, and E236A (Table 1).
TABLE 1.
Summary of wild-type CR2 binding to mutant forms of gp350a
| gp350 mutation | ELISA
|
Cell-binding assay
|
||
|---|---|---|---|---|
| % gp350 binding by MBP-CR2 SCR1-2 (SEM) | Weighting | % gp350-biotin binding, MFI (SEM) | Weighting | |
| E21A | 65.6 (7.6) | ++ | 57.6 (3.2) | ++ |
| D22A | 5.2 (0.1) | − | 44.6 (3.4) | ++ |
| D53A | 89.3 (1.1) | +++ | 90.8 (2.6) | ++++ |
| E119A | 101.5 (0.1) | ++++ | 80.4 (5.3) | +++ |
| Y151A | 16.1 (1.5) | − | 57.5 (0.2) | ++ |
| E155A | 28.5 (1.4) | + | 67.6 (3.8) | ++ |
| Y159A | 82.8 (3.4) | +++ | 94.8 (2.6) | ++++ |
| I160A | 10.4 (8.3) | − | 50.1 (5.5) | ++ |
| W162A | 15.7 (8.9) | − | 13.3 (2.7) | − |
| D163A | 86.7 (1.4) | +++ | 83.7 (0.7) | +++ |
| E201A | 63.5 (2.5) | ++ | 103.9 (3.4) | ++++ |
| D208A | 34.5 (2.5) | + | 68.2 (12.3) | ++ |
| E210A | 41.6 (1.2) | ++ | 63.6 (4.2) | ++ |
| E214A | 98.7 (1.4) | ++++ | 76.2 (0.8) | +++ |
| D215A | 64.4 (1.4) | +++ | 86.5 (14.4) | +++ |
| E236A | 101.2 (1.1) | ++++ | 90.6 (4.6) | ++++ |
| D296A | 10.3 (0.4) | − | 61.4 (2.0) | ++ |
Given are the percentages of the CR2 binding of single-site mutant forms of gp350 relative to those of wild-type gp350 from both ELISA and cell-binding analyses. ++++, >90%; +++, 89.9 to 70%; ++, 69.9 to 40%; +, 39.9 to 20%; −, 19.9 to 0%. MFI, mean fluorescent intensity. Data are reported for concentrations of 2 μg/ml of MBP-CR2 SCR1-2 and 5 μg/ml of gp350 for the ELISA study and 5 × 105 full-length CR2-transfected K562 cells and 5 μg/ml of gp350-biotin for the cell-binding study. Mutants indicated in boldface demonstrate a greater-than 20% decrease in binding affinities relative to those of wild-type CR2 in both ELISA and the cell-binding assays. Residues (wild type) that have been underlined have been defined as active for the purposes of the soft-docking study.
Within linker-1 of gp350 we generated single-site mutations targeting those residues tentatively implicated as contributing to the gp350-CR2 interaction in a previous multiple-site mutagenesis study (40). The current single-substitution mutagenesis data identified Ile-160 and Trp-162 (which form part of the previously described multiple-site mutations) as well as Glu155 (which does not) as key players in the ligation of MBP-CR2 SCR1-2 to gp350, since all were unable to bind significant levels of CR2. On the other hand, Y159A and D163A mutant forms of gp350 retained the ability to bind wild-type-like levels of CR2 (Table 1).
Within D2, a double-site opposite-charge substitution targeting residues Asp-208 and Glu-210 (D208R/E210R) was previously shown to inhibit the binding of gp350 to CR2 and also to disrupt the epitope for MAb 72A1 (40). These data were used to direct alanine-screening experiments targeting residues Glu-201, Asp-208, Glu-210, Glu-214, Asp-215, Glu-236, and Asp-296. Plate-bound forms of E201A, D208A, E210A, D215A, and D296A exhibited a decreased ability to bind MBP-CR2 SCR1-2 (Fig. 2). The remaining D2-based mutants generated in the current study, E214A and E236A, which are spatially close to each other, exhibited no significant decreases in their capacity to bind MBP-CR2 SCR1-2 (Table 1).
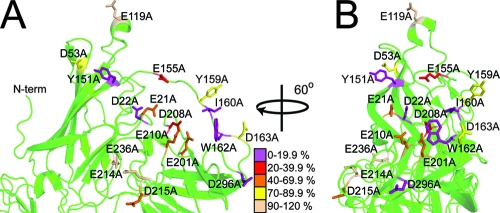
Mapping the results of the gp350 mutant ELISA onto the crystal structure of gp350 reveals a single binding surface for CR2 SCR1-2 on the rigid β-barrel domains comprising D1 and D2 (Fig. 3). This region is dominated by negatively charged residues. However, additional contributions to the CR2-binding interaction are provided by residues within linker-1. These include some significant non-charge-dependent interactions arising from Ile-160 and Trp-162.
FIG. 3.
Effect of gp350 mutagenesis on wild-type CR2 binding (ELISA). (A) Alanine substitutions mapped onto a ribbon representation of the region comprising D1 and D2 of gp350. The scheme used to color residues represents the percent binding of wild-type MBP-CR2 SCR1-2 to mutant forms of gp350 (at a concentration of 5 μg/ml of mutant gp350 and 2 μg/ml of wild-type MBP-CR2 SCR1-2). The image in panel B was constructed similarly to that for panel A, except the molecule has been rotated about the y axis by 60°. N-term, N terminus.
EBV gp350-biotin-K562 erythroleukemia cell-binding assay.
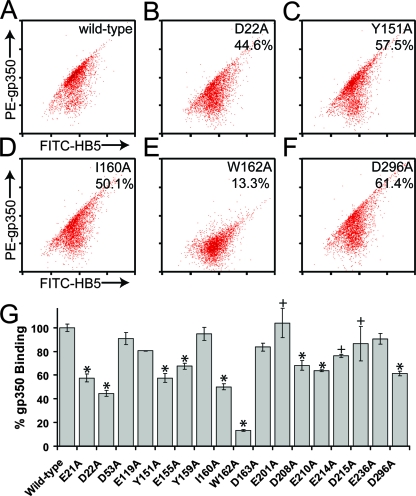
K562 erythroleukemia cells expressing wild-type CR2 initially were incubated with the noninhibitory anti-CR2 MAb HB5 and subsequently were FITC-labeled by incubation with FITC-conjugated goat anti-mouse immunoglobulin G. They then were assessed for their capacity to bind PE-conjugated wild-type or mutant forms of gp350-biotin using flow cytometry. For the most part, the data obtained were in excellent agreement with the MBP-CR2 SCR1-2-gp350 ELISA results described above (Fig. 4, Table 1). For example, D21A, E22A, Y151A, E155A, I160A, W162A, D208A, E210A, and D296A mutant forms of gp350-biotin all exhibited decreases (>20%) in their capacity to bind to full-length CR2 SCR1-15, although the decrease in binding was less marked than that observed by ELISA. Again, consistently with the ELISA analysis, D53A, E119A, Y159A, D163A, and E236A mutant forms of gp350-biotin demonstrated wild-type-like levels of binding. Three of the 17 mutants tested, E201A, E214A, and D215A, were exceptions in that they displayed differing levels of binding to those observed in the ELISA analysis: E201A and D215A both retained wild-type-like levels of binding (103.4 and 86.5%, respectively), while E214A (76.2%) exhibited only a slight variation in binding from the wild-type-like binding curve seen in the ELISA.
FIG. 4.
K562 cell-binding flow cytometry analysis of the wild-type CR2-mutant gp350-biotin interaction. (A to G) The capacity of wild-type CR2 SCR1-15-expressing cell populations, which have been labeled with FITC, to bind wild-type or mutant forms of PE-conjugated gp350-biotin. (A to F) Representative whole-cell populations of K562 erythroleukemia cells expressing wild-type CR2 to bind wild-type (A), D22A (B), Y151A (C), I160A (D), W162A (E), and D296A (F) forms of gp350. (G) Bar chart of the normalized values of the intermediate CR2-expressing population (25%) for binding to all 17 gp350-biotin mutants generated in this study. Averages and standard errors of the normalized values for the mean fluorescence intensity of the intermediate CR2-expressing population (25%) are shown. An asterisk indicates that plate-bound forms of mutant gp350 also exhibited a decreased ability to bind MBP-CR2 SCR1-2 in the ELISA study. A plus indicates that data are inconsistent with those observed in the MBP-CR2 SCR1-2 ELISA study (Table 1).
MBP-CR2 SCR1-2 mutant-gp350 ELISA.
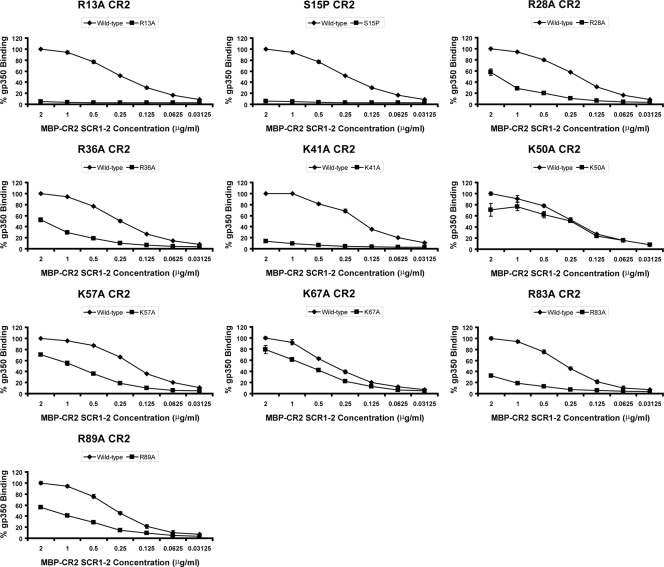
Mutations within the two N-terminal SCR domains of CR2 were selected on the basis of previous experimental work that implicated certain residues as being important in EBV or gp350 binding (26, 46). The strategy also was influenced by the fact that SCR1 of CR2 is characterized by a large number of positively charged residues, and the interaction between CR2 and gp350 has been demonstrated to be charge dependent in nature (12, 36). In the current work, mutations directed at residues Arg-13 (R13A), Arg-28 (R28A), Arg-36 (R36A), Lys-41 (K41A), and Lys-57 (K57A) resulted in the significantly decreased capacity of recombinant MBP-CR2 SCR1-2 to bind gp350 (Fig. 5, Table 2). In addition, a single mutation targeting Ser-15 (S15P) within the first intercysteine region of SCR1 (chosen on the basis of a comparison to the mouse orthologue of CR2) also resulted in a major reduction in gp350 binding. Additional mutations targeting Leu-10 (L10A), Asn-11 (N11A), Tyr-16 (Y16A), Ser-32 (S32A), Thr-34 (T34A), and Lys-50 (K50A) all failed to inhibit gp350 binding (Table 2). These data strongly reinforce previous cell-binding studies that suggested a SCR1-dominant contribution to gp350 attachment and also expand the amount of surface area of this domain that has been mutated and analyzed. Only one substitution, Y16A, appeared to exhibit levels of gp350 binding that were different from that previously observed in the cell-binding study (46) (66.7% in the cell-binding study and 95.1% in the ELISA) (Table 2).
FIG. 5.
MBP-CR2 SCR1-2-gp350 ELISA data targeting residues within CR2 SCR1-2. The ability of mutant forms of MBP-CR2 SCR1-2 to bind plate-bound wild-type gp350 is shown. Data are shown for R13A, S15P, R28A, R36A, K41A, K50A, K57A, K67A, R83A, and R89A. The averages and standard errors of the means of the normalized values relative to those for wild-type gp350 binding are given. Data are not shown for a number of MBP-CR2 SCR1-2 mutants that exhibited wild-type-like levels of binding (<20% reduction): L10A, N11A, Y16A, S32A, T34A, Y64A, Y68A, T86A, and M117A (Table 2).
TABLE 2.
Summary of wild-type or mutant MBP-CR2 SCR1-2-gp350 binding ELISA dataa
| CR2 mutation | ELISA
|
Cell-binding assayb
|
||
|---|---|---|---|---|
| % gp350 binding by MBP-CR2 SCR1-2 (SEM) | Weighting | % gp350-biotin binding, MFI (SEM)b | Weighting | |
| L10A | 98.2 (5.1) | ++++ | NA | NA |
| N11A | 93.6 (0.6) | ++++ | 112.6 (11.6) | ++++ |
| R13A | 4.9 (0.1) | − | 7.0 (0.4) | − |
| S15P | 5.8 (0.5) | − | 5.4 (0.4) | − |
| Y16A | 95.1 (0.1) | ++++ | 66.7 (4.0) | ++ |
| R28A | 58.0 (5.6) | ++ | 21.9 (1.8) | + |
| S32A | 94.8 (1.0) | ++++ | 108 (12.9) | ++++ |
| T34A | 106.6 (5.4) | ++++ | NA | NA |
| R36A | 52.2 (3.1) | ++ | 29 (4.8) | + |
| K41A | 13.6 (0.7) | − | 20.8 (1.8) | + |
| K50A | 70.8 (11.4)c | +++c | 59.2 (1.9)c | ++c |
| K57A | 70.7 (2.3) | +++ | 36.5 (3.7) | + |
| Y64A | 93.6 (1.0) | ++++ | 84.5 (3.4) | +++ |
| K67A | 79.5 (7.4) | +++ | 42.1 (3.9) | +++ |
| Y68A | 87.5 (2.7) | ++++ | 111.4 (7.4) | ++++ |
| R83A | 32.1 (0.8) | + | 32.9 (4.9) | + |
| T86A | 91.9 (0.5) | ++++ | NA | NA |
| R89A | 56.1 (0.7) | ++ | NA | NA |
| M117A | 96.5 (4.1) | ++++ | NA | NA |
Given are percentages of gp350 binding to mutant CR2 relative to that of wild-type CR2. ++++, >90%; +++, 89.9 to 70%; ++, 69.9 to 40%;+, 39.9 to 20%; −, 19.9 to 0%.
Binding data and weighting were obtained from a previously reported cell-binding analysis of the CR2-gp350 binding interaction (46). NA, not applicable. Mutants indicated in boldface demonstrate a greater than 20% decrease in binding affinities relative to that of wild-type gp350 in both ELISA and the cell-binding assay (where available).
While the K50A mutant demonstrates a reduction in binding at the protein concentrations described in this table, at other values recorded for both the ELISA and cell-binding analyses, K50A binding approximates that of wild-type CR2 (Fig. 5) (1, 46). Residues (wild type) that have been underlined have been defined as active to drive the soft-docking study.
Three residues were targeted for mutagenesis within the eight-residue (63-EYFNKYSS-70) linker region connecting SCR1 and SCR2; Tyr-64, Lys-67, and Tyr-68 each were replaced with an alanine residue (Y64A, K67A, and Y68A). Only the K67A substitution demonstrated any significant effect on gp350 binding, with an approximately 20% decrease (Fig. 5).
Within SCR2, a total of four residues were selected for mutagenesis screening: Arg-83 (R83A), Thr-86 (T86A), Arg-89 (R89A), and Met-117 (M117A). Arg-83 previously has been suggested to play an important role in gp350 binding and also to be essential in the interaction between CR2 and its major physiologic ligand, C3d (17, 39, 46). Of the four alanine substitutions generated in SCR2, only the R83A and R89A mutants exhibited decreased binding affinities for gp350 (Fig. 5).
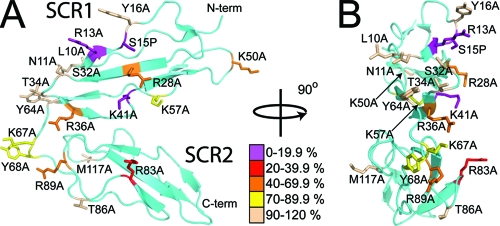
When mapped onto the crystal structure of CR2 SCR1-2, the data obtained using the MBP-CR2 SCR1-2 mutants delineates a binding interface that consists of a contiguous positively charged surface spread over SCR1 and SCR2, with two hot spots centered around Arg-83 and Arg-89 on SCR2 and Arg-13, Ser-15, Arg-28, Arg-36, and Lys-41 on SCR1 (Fig. 6) that are essential for the attachment of gp350.
FIG. 6.
Effect of CR2 mutagenesis on wild-type gp350 binding (ELISA). (A) Alanine and proline substitutions mapped onto a ribbon representation of CR2 SCR1-2. The scheme used to color residues represents the percent binding of wild-type gp350 to mutant forms of MBP-CR2 SCR1-2 (at a concentration of 5 μg/ml gp350 and 2 μg/ml of mutant MBP-CR2 SCR1-2). The image in panel B was constructed similarly to that for panel A, except the molecule has been rotated about the y axis by 90°. N-term, N terminus; C-term, C terminus.
MBP-CR2 SCR1-2-171 MAb and MBP-CR2 SCR1-2-629 MAb ELISA.
The excellent correlation observed between our cell-binding studies and the ELISA analyses provides proof of principle that the MBP-CR2 SCR1-2 ELISA is an effective means of characterizing CR2-ligand interactions. We therefore utilized our 19 MBP-CR2 SCR1-2 mutants to describe the structured epitope for the anti-CR2 SCR1-2 MAb 171, which inhibits the interaction with gp350 as well as all other known CR2 ligands (1, 13, 46). ELISA revealed that single-site mutants targeting a patch of residues located within the first intercysteine region of SCR1 of CR2, comprising Asn-11, Arg-13, Ser-32, Thr-34, and Arg-36 (N11A, R13A, S32A, T34A, and R36A), and also a single residue within the linker region connecting SCR1 and SCR2, Tyr-64 (Y64A), disrupted the epitope for MAb 171 (Table 3). This epitope overlaps extensively with the binding surface on CR2 described above for gp350 and also with the previously described binding sites for C3dg and C3d (17, 46). By contrast, none of these MBP-CR2 SCR1-2 mutants were found to have any deleterious effects on the binding of a noninhibitory anti-CR2 SCR1-2 MAb, 629 (13). This suggests that the epitope for MAb 629 is located on a surface discrete from the ligand-binding sites described above for gp350 and also for the previously characterized C3 fragments C3dg and C3d (Table 3). A single alanine substitution targeting Tyr-16 (Y16A) demonstrated increased binding (∼150%) to MAb 629 relative to that of wild-type MBP-CR2 SCR1-2.
TABLE 3.
Summary of wild-type or mutant MBP-CR2 SCR1-2 binding to the anti-CR2 SCR1-2 MAbs 171 and 629a
| CR2 mutation | MAb 171
|
MAb 629
|
||
|---|---|---|---|---|
| % Binding (SEM) | Weighting | % Binding (SEM) | Weighting | |
| L10A | 94.7 (3.2) | ++++ | 113.2 (6.4) | ++++ |
| N11A | 24.1 (1.4) | + | 87.5 (2.1) | +++ |
| R13A | 15.6 (2.1) | − | 108.1 (0.8) | ++++ |
| S15P | 108.5 (1.6) | ++++ | 91.7 (7.3) | ++++ |
| Y16A | 81.4 (3.7) | +++ | 154.4 (4.0) | ++++ |
| R28A | 103.8 (1.7) | ++++ | 103.1 (1.8) | ++++ |
| S32A | 77.5 (8.9) | +++ | 117.8 (3.4) | ++++ |
| T34A | 12.4 (1.4) | − | 84.9 (5.8) | +++ |
| R36A | 19.8 (1.7) | − | 113.9 (1.5) | ++++ |
| K41A | 97.3 (1.7) | ++++ | 97.5 (0.8) | ++++ |
| K50A | 94.4 (3.5) | ++++ | 87.5 (3.2) | +++ |
| K57A | 91.9 (0.8) | ++++ | 85.5 (0.7) | +++ |
| Y64A | 5.8 (0.1) | − | 82.0 (1.5) | +++ |
| K67A | 87.7 (3.2) | +++ | 107.2 (4.5) | ++++ |
| Y68A | 92.1 (4.8) | ++++ | 102.3 (4.1) | ++++ |
| R83A | 96.9 (2.4) | ++++ | 94.4 (3.9) | ++++ |
| T86A | 89.6 (5.2) | +++ | 85.6 (3.3) | +++ |
| R89A | 93.9 (7.8) | ++++ | 87.7 (2.1) | +++ |
| M117A | 97.2 (4.1) | ++++ | 113.8 (2.9) | ++++ |
HADDOCK-derived CR2-gp350 model.
The mutagenesis data for gp350 and CR2 obtained in the current study were used to generate a series of AIRs that subsequently were employed by the soft-docking program HADDOCK (8) to derive a model of the CR2 SCR1-2-gp350 complex. Solvent-accessible (50% or greater) residues on CR2 and gp350, the mutation of which disrupts complex formation according to both our ELISA and cell-binding assays, were designated active residues for the docking calculations (Tables 1 and 2). Starting from the randomly oriented structures of CR2 and gp350, active residues were used by HADDOCK to direct the docking process toward the most plausible complex of CR2 and gp350, which also satisfied the data previously recorded (Fig. 7). A cluster analysis of the resulting 100 water-refined structures, employing a 1.5-Å cutoff limit, revealed that 99 of these models fell into a single cluster. For the 100 water-refined structures analyzed, the backbone root mean square deviation (RMSD) at the intermolecular interface for all structures was 1.5 Å (1.9 Å for all backbone atoms in all structures). The average buried surface area for the 100 water-refined structures was 2,510 ± 192 Å2. The structures calculated are in good agreement with experimental evidence, as determined by an average of 0.04 AIR violations for >0.5 Å per structure. Our models of the gp350-CR2-SCR1-2 complex also are highly structurally uniform and are in broad agreement with results generated using an incomplete set of AIRs (18). The backbone interface RMSD values obtained for our model of the CR2-gp350 complex are consistent with those of other HADDOCK-derived models of protein complexes that compare favorably with the corresponding atomic resolution structures (44). Reported in the fourth and fifth rounds of the critical assessment of the prediction of interactions, these models exhibited backbone interface RMSD values ranging from 0.3 to 2.0 Å (44). Moreover, the CR2 SCR1-2-binding site on gp350 is highly plausible, being located within a negatively charged region on the surface of this molecule that is devoid of the glycan moieties that cover much of the surface.
FIG. 7.
Ribbon representative of the HADDOCK-derived model of the CR2-gp350 complex generated utilizing the mutagenesis data described in Tables 1 and 2 and the crystal structures of CR2 SCR1-2 and gp350.
DISCUSSION
The interaction between the two N-terminal extracellular SCR domains of CR2 and the major EBV envelope protein gp350 is an integral part of the adsorption of virions onto the surface of B lymphocytes. Knowledge of the precise nature of this interaction is required to understand the mechanism whereby the engagement of the N terminus of CR2 by EBV progresses to invasion. In the current work, data derived from single-site mutant forms of both CR2 and gp350 were used to drive a working model of the CR2-gp350 complex that is consistent with the mutagenesis data. This model also is consistent with previous experimental data pertaining to EBV-CR2 interactions and with the results of other experiments reported here in which we have characterized the structured epitope of a major inhibitory anti-CR2 SCR1-2 MAb.
An effective ELISA was developed that has allowed us, for the first time, to map out the binding interface for the CR2 SCR1-2-EBV gp350 interaction on both molecules. Our ELISA data are supported by cell-binding studies in which PE-conjugated gp350-biotin mutants were incubated with K562 cells transfected with human CR2 SCR1-15. Indeed, the cell-binding assays delineate an identical CR2-binding surface on gp350, with only two mutants, E201A and D215A, behaving differently from what would be expected from the ELISA. Both of these point mutants exhibited wild-type levels of binding in the cell-binding assay and approximately 60 to 70% activity by ELISA. Interestingly, Asp-215 is remote from the major CR2 interaction center within gp350, and it is likely that Glu-201 and Asp-215 play only a peripheral or negligible role in the docking of gp350. A likely explanation for any observed differences in binding utilizing the two methodologies employed in this study may be attributable to a lower sensitivity of the cell-binding approach or to the differing solution conditions used for the experiments.
Of the 17 single-site gp350 mutants generated in the current study, targeting D1, D2, and linker-1 of gp350, 10 consistently exhibited compromised ability to bind CR2: E21A, D22A, Y151A, E155A, I160A, W162A, E201A, D208A, D210A, and D296A. The remaining seven (D53A, E119A, Y159A, D163A, E214A, D215A, and E236A) demonstrated wild-type-like or only slightly reduced levels of gp350 binding. When mapped onto the gp350 crystal structure, the mutations that affect binding correspond to a predominantly electronegative binding surface within D1 and D2 (Fig. 3); this appears to be complementary to the positively charged binding region identified in SCR1-2 of CR2.
Strikingly, both ELISA and cell-binding data identify a significant additional contribution from residues within the gp350 D1-D2 linker. A total of five point mutations were generated within the linker-1 region, three of which appeared to critically affect CR2 binding (E155A, I160A, and W162A) and two of which did not (Y159A and D163A). In support of these data, the side chains of Glu-155, Ile-160, and Trp-162 are oriented toward the putative binding site delineated by Glu-21, Asp-22, Tyr-151, Asp-208, Glu-210, and Asp-296 in the gp350 crystal structure, while the side chains of Tyr-159 and Asp-163 are oriented away from this region (Fig. 3). Taking these findings together, it is unlikely that the reduction in binding observed for those deleterious mutations targeting the D1-D2 linker are the result of a significant reorientation. From the crystal structure of gp350, Glu-155, Ile-160, and Trp-162 all possess side chain solvent accessibilities of greater than 50% (according to NACCESS) and do not appear to significantly contribute to the overall fold of the gp350 molecule.
With regard to delineating the gp350-binding site on CR2, 19 single-site CR2 SCR1-2 mutants were engineered as soluble recombinant MBP fusion proteins, and binding was measured using the ELISA described above. The data obtained were considered in conjunction with a previously published study in which wild-type or mutant full-length CR2-transfected K562 cells were incubated with wild-type gp350 and binding interactions assessed by flow cytometry (46). The ELISA-derived data delineate a contiguous positively charged gp350-binding surface upon CR2 that is consistent with the cell-binding assay (46). Arg-83 and Arg-89 on SCR2 both play important roles in the ligation of gp350. Of the other mutations within SCR2, Thr-86 is in close proximity to the putative gp350-binding site outlined in Fig. 6, but T86A binds gp350 with wild-type-like affinity. Notably, the side chain of this amino acid residue extends downwards and away from the plane formed by residues 83 and 89. Met-117, on the other hand, is located on the opposite face of the domain and, unsurprisingly, the M117A mutant binds gp350 with the same affinity as wild-type CR2. Point mutations that target CR2-SCR1 indicate that residues Arg-13, Ser-15, Arg-28, Arg-36, and Lys-41 play an important role in the interaction between CR2 and gp350, consistent with previous work (46). The R13A, S15P, and K41A mutations, in particular, have a deleterious effect on gp350 binding. The S15P substitution is likely to induce a localized structural perturbation of the viral binding surface on CR2, while the R13A and K41A mutations (as well as the R36A and R28A substitutions) are likely to interfere with the well-established charge dependence of the CR2-gp350 interaction (12, 36). In previous work, R13A, S15P, R28A, R36A, K41A, K57A, K67A, and R83A MBP-CR2 SCR1-2 demonstrated reduced or zero capacity to pull down concentrated intact virus; this supports the case for the recombinant gp350-binding interface identified on CR2 SCR1-2, reflecting that of intact gp350/220 expressed on the surface of enveloped EBV virions (46).
It has previously been suggested that the available crystal structures of CR2 SCR1-2 do not reflect the physiological orientation of the two amino-terminal domains with respect to one another. Both crystallized forms of the protein were expressed using Pichia pastoris and then deglycosylated prior to crystallization. Prota et al. suggested that the presence of the nonnative N-glycan moiety attached to Asn-107 would sterically force the two SCR domains apart in the native protein (34). We have demonstrated in the current study, however, that the data obtained using E. coli-derived recombinant MBP-CR2 SCR1-2, with no posttranslational processing, are similar to those obtained from a K562 mammalian cell expression system that produced glycosylated protein (5). These data suggest that glycosylation within the two N-terminal domains of human CR2 does not significantly contribute to its structure-function relationships. With regard to the CR2 SCR1-2 architecture under solution conditions, an analytical ultracentrifugation and solution-scattering study, carried out in conjunction with molecular modeling by Perkins and coworkers, suggested that the domain packing of CR2 SCR1-2 is more extended than that observed in the available crystal elucidations, but that the molecule still adopts an overall constrained V-shaped conformation (11).
Further evidence that the gp350-binding site located within SCR1 of CR2 identified by our mutagenesis studies is indeed a primary site of EBV attachment is provided by our mutagenesis screening of the MAb 171 and MAb 629 epitopes on CR2 SCR1-2. MAb 171 is an effective inhibitor of all CR2-ligand interactions (1, 13, 46). Our data demonstrate that the structured epitope for MAb 171 is located within a patch of residues within the first intercysteine region of SCR1 and also the initial part of the eight-residue linker region connecting SCR1 and SCR2 (Asn-11, Arg-13, Ser-32, Thr-34, Arg-36, and Tyr-64) (Table 3). Residues important in the binding of 171 MAb overlap directly with residues that we have identified as being present at the CR2-gp350 interface (Table 2) (46) and that also have been shown to play a significant role in C3d/C3dg ligation (Arg-13 and Arg-36) by both site-directed mutagenesis (17) and, more recently, heteronuclear nuclear magnetic resonance spectroscopy (J. M. Kovacs, J. P. Hannan, E. Z. Eissenmesser, and V. M. Holers, unpublished data). Our data delineating the MAb 171 structured epitope differ significantly from those obtained by overlapping peptide-based approaches, which have demarcated the linear epitope of MAb 171 to residues 86-TPYRH-90 located within SCR2 of CR2 (13). However, it is likely that the 171 epitope delineated by our mutagenesis mapping is a better reflection of the experimentally defined inhibitory properties that have been described for MAb 171, since all competition studies have utilized folded proteins expressed either on cell surfaces or as soluble recombinant molecules (1, 13, 46). Notably, of the 86-TPYRH-90 sequence described by the use of mimotope sequences displayed on pins, Tyr-88 and His-90 are buried beneath the protein surface in both of the available crystal structures of CR2 (34, 39). Interestingly, when our mutant MBP-CR2 SCR1-2 library was employed to screen a control noninhibitory anti-CR2 antibody, MAb 629, none of our mutants targeting the CR2 ligand-binding sites were found to disrupt binding to any significant extent (Table 3). It therefore is likely that the structured epitope for 629 is located on a surface discrete from the characterized gp350 and C3d/C3dg binding sites (Table 3).
A goal of assembling this extensive set of mutagenesis data was to drive a plausible model of the CR2-gp350 complex using a molecular docking algorithm. Our best-fit model of the complex generated by HADDOCK locates the CR2 SCR1-2 binding site within a glycan-free area of gp350. These data are consistent with previous studies demonstrating that the numerous N-linked glycans coating gp350 do not play a significant role in the binding interaction between CR2 and gp350 (40). The nature of the binding pocket for CR2 SCR1-2 observed within gp350 is consistent with a conformation of the CR2 SCR1-2 molecule in which the domain orientation approximates a V-shaped topography. Whether the two SCR domains make contact with each other, as reflected in the two crystallographic studies (34, 39), or adopt a more open but still V-shaped conformation, as observed in solution studies, does not have a significant bearing on the model (11). The complex predicted by HADDOCK aligns SCR1 of CR2 with the electronegative region on gp350 delineated primarily by Glu-201, Asp-208, Glu-210, and Asp-296 and SCR2 oriented toward Glu-21, Asp-22, and Glu-155. Another notable feature of the interaction is the presence of aromatic and nonpolar residues dominating the center of the complex. In all, our CR2-gp350 model appears to describe a classic protein-protein interaction, whereupon opposite charges on each molecule direct complex initiation and formation and surround a hydrophobic/nonpolar core that is buried within the complex.
In summary, we have carried out a site-directed mutagenesis study to provide the basis of a structural understanding of the initial contact that occurs between EBV and B lymphocytes. We have extensively characterized the gp350-binding site on CR2 and compared this binding site to the structured epitope of an effective inhibitory anti-CR2 MAb, 171. Furthermore, we have provided the first detailed delineation of the CR2-binding site on gp350, describing a predominantly negatively charged binding surface within the two N-terminal domains of gp350, and identified additional side chain contributions from the linker region between them. Furthermore, we have identified nonpolar and hydrophobic residues on gp350 that are necessary for efficient binding. Finally, our mutagenesis data have been used as the basis for generating a testable constraint-derived model of the CR2-gp350 complex. In toto, these data provide a structural basis for developing inhibitors of this important virus-receptor interaction.
Acknowledgments
This work was supported by National Institutes of Health grant R0-1 CA053615 to V.M.H. and was assisted by the University of Colorado Denver Cancer Center Flow Cytometry Core.
Footnotes
Published ahead of print on 10 September 2008.
REFERENCES
- 1.Asokan, R., J. Hua, K. A. Young, H. J. Gould, J. P. Hannan, D. M. Kraus, G. Szakonyi, G. J. Grundy, X. S. Chen, M. K. Crow, and V. M. Holers. 2006. Characterization of human complement receptor type 2 (CR2/CD21) as a receptor for IFN-alpha: a potential role in systemic lupus erythematosus. J. Immunol. 177383-394. [DOI] [PubMed] [Google Scholar]
- 2.Aubry, J. P., S. Pochon, J. F. Gauchat, A. Nueda-Marin, V. M. Holers, P. Graber, C. Siegfried, and J. Y. Bonnefoy. 1994. CD23 interacts with a new functional extracytoplasmic domain involving N-linked oligosaccharides on CD21. J. Immunol. 1525806-5813. [PubMed] [Google Scholar]
- 3.Aubry, J. P., S. Pochon, P. Graber, K. U. Jansen, and J. Y. Bonnefoy. 1992. CD21 is a ligand for CD23 and regulates IgE production. Nature 358505-507. [DOI] [PubMed] [Google Scholar]
- 4.Beisel, C., J. Tanner, T. Matsuo, D. Thorley-Lawson, F. Kezdy, and E. Kieff. 1985. Two major outer envelope glycoproteins of Epstein-Barr virus are encoded by the same gene. J. Virol. 54665-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boackle, S. A., V. M. Holers, X. Chen, G. Szakonyi, D. R. Karp, E. K. Wakeland, and L. Morel. 2001. Cr2, a candidate gene in the murine Sle1c lupus susceptibility locus, encodes a dysfunctional protein. Immunity 15775-785. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, J. I. 2005. Clinical aspects of Epstein-Barr virus infection, p. 35-54. In E. S. Robertson (ed.), Epstein-barr virus, Horizon Scientific Press, Wymondham, United Kingdom.
- 7.Delcayre, A. X., F. Salas, S. Mathur, K. Kovats, M. Lotz, and W. Lernhardt. 1991. Epstein Barr virus/complement C3d receptor is an interferon alpha receptor. EMBO J. 10919-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominguez, C., R. Boelens, and A. M. Bonvin. 2003. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 1251731-1737. [DOI] [PubMed] [Google Scholar]
- 9.Fingeroth, J. D., J. J. Weis, T. F. Tedder, J. L. Strominger, P. A. Biro, and D. T. Fearon. 1984. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc. Natl. Acad. Sci. USA 814510-4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frade, R., M. Barel, B. Ehlin-Henriksson, and G. Klein. 1985. gp140, the C3d receptor of human B lymphocytes, is also the Epstein-Barr virus receptor. Proc. Natl. Acad. Sci. USA 821490-1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert, H. E., J. T. Eaton, J. P. Hannan, V. M. Holers, and S. J. Perkins. 2005. Solution structure of the complex between CR2 SCR 1-2 and C3d of human complement: an X-ray scattering and sedimentation modelling study. J. Mol. Biol. 346859-873. [DOI] [PubMed] [Google Scholar]
- 12.Guthridge, J. M., J. K. Rakstang, K. A. Young, J. Hinshelwood, M. Aslam, A. Robertson, M. G. Gipson, M. R. Sarrias, W. T. Moore, M. Meagher, D. Karp, J. D. Lambris, S. J. Perkins, and V. M. Holers. 2001. Structural studies in solution of the recombinant N-terminal pair of short consensus/complement repeat domains of complement receptor type 2 (CR2/CD21) and interactions with its ligand C3dg. Biochemistry 405931-5941. [DOI] [PubMed] [Google Scholar]
- 13.Guthridge, J. M., K. Young, M. G. Gipson, M. R. Sarrias, G. Szakonyi, X. S. Chen, A. Malaspina, E. Donoghue, J. A. James, J. D. Lambris, S. A. Moir, S. J. Perkins, and V. M. Holers. 2001. Epitope mapping using the X-ray crystallographic structure of complement receptor type 2 (CR2)/CD21: identification of a highly inhibitory monoclonal antibody that directly recognizes the CR2-C3d interface. J. Immunol. 1675758-5766. [DOI] [PubMed] [Google Scholar]
- 14.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290106-114. [DOI] [PubMed] [Google Scholar]
- 15.Haddad, R. S., and L. M. Hutt-Fletcher. 1989. Depletion of glycoprotein gp85 from virosomes made with Epstein-Barr virus proteins abolishes their ability to fuse with virus receptor-bearing cells. J. Virol. 634998-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hannan, J. P., G. Szakonyi, R. Asokan, X. Chen, and V. M. Holers. 2005. Complement receptor CR2/CD21 and CR2-C3d complexes, p. 143-160. In D. Morikis and J. D. Lambris (ed.), Structural biology of the complement system, CRC Press, Boca Raton, FL.
- 17.Hannan, J. P., K. A. Young, J. M. Guthridge, R. Asokan, G. Szakonyi, X. S. Chen, and V. M. Holers. 2005. Mutational analysis of the complement receptor type 2 (CR2/CD21)-C3d interaction reveals a putative charged SCR1 binding site for C3d. J. Mol. Biol. 346845-858. [DOI] [PubMed] [Google Scholar]
- 18.Hannan, J. P., K. A. Young, A. P. Herbert, X. S. Chen, P. N. Barlow, and V. M. Holers. 2007. Characterizing the complement receptor type 2 (CR2)- Epstein-Barr virus interaction: a site-directed mutagenesis and molecular docking approach. Mol. Immunol. 443929. [Google Scholar]
- 19.Hutt-Fletcher, L. M. 2007. Epstein-Barr virus entry. J. Virol. 817825-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackman, W. T., K. A. Mann, H. J. Hoffmann, and R. R. Spaete. 1999. Expression of Epstein-Barr virus gp350 as a single chain glycoprotein for an EBV subunit vaccine. Vaccine 17660-668. [DOI] [PubMed] [Google Scholar]
- 21.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H. J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 7410142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 10116286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalli, K. R., J. M. Ahearn, and D. T. Fearon. 1991. Interaction of iC3b with recombinant isotypic and chimeric forms of CR2. J. Immunol. 147590-594. [PubMed] [Google Scholar]
- 24.Kirkitadze, M. D., and P. N. Barlow. 2001. Structure and flexibility of the multiple domain proteins that regulate complement activation. Immunol. Rev. 180146-161. [DOI] [PubMed] [Google Scholar]
- 25.Mackett, M., C. Cox, S. D. Pepper, J. F. Lees, B. A. Naylor, N. Wedderburn, and J. R. Arrand. 1996. Immunisation of common marmosets with vaccinia virus expressing Epstein-Barr virus (EBV) gp340 and challenge with EBV. J. Med. Virol. 50263-271. [DOI] [PubMed] [Google Scholar]
- 26.Martin, D. R., A. Yuryev, K. R. Kalli, D. T. Fearon, and J. M. Ahearn. 1991. Determination of the structural basis for selective binding of Epstein-Barr virus to human complement receptor type 2. J. Exp. Med. 1741299-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menet, A., C. Speth, C. Larcher, W. M. Prodinger, M. G. Schwendinger, P. Chan, M. Jager, F. Schwarzmann, H. Recheis, M. Fontaine, and M. P. Dierich. 1999. Epstein-Barr virus infection of human astrocyte cell lines. J. Virol. 737722-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 746324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morikis, D., and J. D. Lambris. 2004. The electrostatic nature of C3d-complement receptor 2 association. J. Immunol. 1727537-7547. [DOI] [PubMed] [Google Scholar]
- 30.Nemerow, G. R., R. A. Houghten, M. D. Moore, and N. R. Cooper. 1989. Identification of an epitope in the major envelope protein of Epstein-Barr virus that mediates viral binding to the B lymphocyte EBV receptor (CR2). Cell 56369-377. [DOI] [PubMed] [Google Scholar]
- 31.Nemerow, G. R., C. Mold, V. K. Schwend, V. Tollefson, and N. R. Cooper. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 611416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemerow, G. R., J. J. Mullen III, P. W. Dickson, and N. R. Cooper. 1990. Soluble recombinant CR2 (CD21) inhibits Epstein-Barr virus infection. J. Virol. 641348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nemerow, G. R., R. Wolfert, M. E. McNaughton, and N. R. Cooper. 1985. Identification and characterization of the Epstein-Barr virus receptor on human B lymphocytes and its relationship to the C3d complement receptor (CR2). J. Virol. 55347-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prota, A. E., D. R. Sage, T. Stehle, and J. D. Fingeroth. 2002. The crystal structure of human CD21: implications for Epstein-Barr virus and C3d binding. Proc. Natl. Acad. Sci. USA 9910641-10646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 36.Sarrias, M. R., S. Franchini, G. Canziani, E. Argyropoulos, W. T. Moore, A. Sahu, and J. D. Lambris. 2001. Kinetic analysis of the interactions of complement receptor 2 (CR2, CD21) with its ligands C3d, iC3b, and the EBV glycoprotein gp350/220. J. Immunol. 1671490-1499. [DOI] [PubMed] [Google Scholar]
- 37.Soares, D. C., and P. N. Barlow. 2005. Complement control protein modules in the regulators of complement activation, p. 19-62. In D. Morikis and J. D. Lambris (ed.), Structural biology of the complement system, CRC Press, Boca Raton, FL.
- 38.Sokal, E. M., K. Hoppenbrouwers, C. Vandermeulen, M. Moutschen, P. Leonard, A. Moreels, M. Haumont, A. Bollen, F. Smets, and M. Denis. 2007. Recombinant gp350 vaccine for infectious mononucleosis: a phase 2, randomized, double-blind, placebo-controlled trial to evaluate the safety, immunogenicity, and efficacy of an Epstein-Barr virus vaccine in healthy young adults. J. Infect. Dis. 1961749-1753. [DOI] [PubMed] [Google Scholar]
- 39.Szakonyi, G., J. M. Guthridge, D. Li, K. Young, V. M. Holers, and X. S. Chen. 2001. Structure of complement receptor 2 in complex with its C3d ligand. Science 2921725-1728. [DOI] [PubMed] [Google Scholar]
- 40.Szakonyi, G., M. G. Klein, J. P. Hannan, K. A. Young, R. Z. Ma, R. Asokan, V. M. Holers, and X. S. Chen. 2006. Structure of the Epstein-Barr virus major envelope glycoprotein. Nat. Struct. Mol. Biol. 13996-1001. [DOI] [PubMed] [Google Scholar]
- 41.Tanner, J., Y. Whang, J. Sample, A. Sears, and E. Kieff. 1988. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J. Virol. 624452-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thorley-Lawson, D. A., and K. Geilinger. 1980. Monoclonal antibodies against the major glycoprotein (gp350/220) of Epstein-Barr virus neutralize infectivity. Proc. Natl. Acad. Sci. USA 775307-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Urquiza, M., R. Lopez, H. Patino, J. E. Rosas, and M. E. Patarroyo. 2005. Identification of three gp350/220 regions involved in Epstein-Barr virus invasion of host cells. J. Biol. Chem. 28035598-35605. [DOI] [PubMed] [Google Scholar]
- 44.van Dijk, A. D., S. J. de Vries, C. Dominguez, H. Chen, H. X. Zhou, and A. M. Bonvin. 2005. Data-driven docking: HADDOCK's adventures in CAPRI. Proteins 60232-238. [DOI] [PubMed] [Google Scholar]
- 45.Weis, J. J., T. F. Tedder, and D. T. Fearon. 1984. Identification of a 145,000 Mr membrane protein as the C3d receptor (CR2) of human B lymphocytes. Proc. Natl. Acad. Sci. USA 81881-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, K. A., X. S. Chen, V. M. Holers, and J. P. Hannan. 2007. Isolating the Epstein-Barr virus (EBV) GP350/220 binding site on complement receptor type 2 (CR2/CD21). J. Biol. Chem. 28236614-36625. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, L., B. Mallik, and D. Morikis. 2007. Immunophysical exploration of C3d-CR2(CCP1-2) interaction using molecular dynamics and electrostatics. J. Mol. Biol. 369567-583. [DOI] [PubMed] [Google Scholar]