Abstract
Avian reovirus, an important avian pathogen, expresses eight structural and four nonstructural proteins. The structural σA protein is a major component of the inner capsid, clamping together λA building blocks. σA has also been implicated in the resistance of avian reovirus to the antiviral action of interferon by strongly binding double-stranded RNA in the host cell cytoplasm and thus inhibiting activation of the double-stranded RNA-dependent protein kinase. We have solved the structure of bacterially expressed σA by molecular replacement and refined it using data to 2.3-Å resolution. Twelve σA molecules are present in the P1 unit cell, arranged as two short double helical hexamers. A positively charged patch is apparent on the surface of σA on the inside of this helix and mutation of either of two key arginine residues (Arg155 and Arg273) within this patch abolishes double-stranded RNA binding. The structural data, together with gel shift assay, electron microscopy, and sedimentation velocity centrifugation results, provide evidence for cooperative binding of σA to double-stranded RNA. The minimal length of double-stranded RNA required for σA binding was observed to be 14 to 18 bp.
Avian reovirus belongs to the Orthoreovirus genus of the Reoviridae family. It has been linked to various diseases in birds, which generate considerable economic losses in the poultry industry (17). The virus has a double concentric icosahedral capsid (2). The inner capsid is made up of the λA, λC, and σA proteins, which encapsulate a double-stranded RNA (dsRNA) genome consisting of 10 segments and up to 12 copies of the RNA-dependent RNA polymerase. The RNA polymerase consists of proteins λB and μA. Proteins μB, σB, and σC are components of the outer shell. The virus also encodes four nonstructural proteins (2), which are involved in cell fusion (p10), viral assembly (μNS and σNS), or as-yet-undetermined functions (p17). Up to now, only the crystal structure of the primary cell attachment protein σC has been reported (14).
The object of our present study, the protein σA, has two functions in avian reovirus biology. On one hand, it plays a structural role, forming part of the inner capsid. In this respect it is the structural equivalent of mammalian reovirus σ2. The structure of the mammalian reovirus inner capsid has been solved by X-ray crystallography (33). σ2 is present in 150 copies per core, in three different locations (σ2-i, σ2-ii, and σ2-iii), acting as a stabilizing clamp on the outside of the inner capsid. The protein interacts extensively with λ1 (the mammalian reovirus equivalent of λA) at two different binding sites (the binding sites on λ1 for σ2-ii and σ2-iii are similar). There are only sparse contacts between σ2 monomers and between σ2 and the other core protein λ2 (the equivalent of avian reovirus λC). Cryo-electron microscopy of avian reovirions confirmed that avian reovirus contains 150 σA “nodules” contacting mainly λA, with minor contacts formed with the λC turrets and other σA molecules (40). σA also contacts the μB proteins of the outer capsid (equivalent to μ1 in mammalian reovirus).
Apart from its structural role in the virus, σA has been shown to strongly bind dsRNA, and in this way it has been implicated in the virus's resistance to interferon by preventing the activation of the dsRNA-dependent protein kinase (PKR) (13, 25). This interaction would presumably take place in the host cell cytoplasm (outside the virus). Here, σA appears to play a role analogous to that of the mammalian reovirus outer capsid protein σ3, as this protein (the structural equivalent of avian reovirus σB) also exhibits dsRNA binding activity (39), albeit not as strongly as avian reovirus σA (25). Here we report the crystal structure of recombinant σA and show that it self-assembles in a helical fashion. Mutational studies support the hypothesis that dsRNA may bind to the inside of this helix.
MATERIALS AND METHODS
For structural studies, the avian reovirus S1133 protein σA was expressed as a maltose binding protein fusion, cleaved using the protease factor Xa, purified, and crystallized as described previously (15). Data collection, processing, and scaling were performed as described in the same paper. Molecular replacement was performed using a model based on the structure of mammalian reovirus σ2 (33). To construct the model used in molecular replacement, alignment of the σA and σ2 sequences was performed with CLUSTALW (4); this alignment and the σ2 structure were input into the program CHAINSAW (6), which retains conserved amino acids but prunes nonconserved residues to the C-gamma atom. Molecular replacement itself was performed with MOLREP (35), using the option that adapts the search model by recalculating the temperature factor of each atom as a function of its surface area. Model adjustment was done using COOT (11) and O (18), refinement was performed with REFMAC (27), and water molecules were built using ARP (23). Model validation was performed with PROCHECK (24), WHATCHECK (36), MOLPROBITY (7), and MOLEMAN (19).
Site-directed mutagenesis was performed using the QuikChange kit (Stratagene, Cultek, Madrid, Spain), following the manufacturer's instructions. σA and mutated σA were purified as described previously (15). Gel shift assays were performed using σA at 0.6 mg/ml in TE buffer (10 mM Tris-HCl, pH 8.5, 1 mM EDTA [disodium EDTA]) and a dsRNA ladder which contains seven dsRNAs of different lengths (500, 300, 150, 80, 50, 30, and 21 bp) at 0.5 mg/ml (New England Biolabs, Izasa S.A., Barcelona, Spain). Increasing amounts of σA (0, 0.6, 1.2, 3.0, and 6.0 μg) were incubated with 0.5 μg of dsRNA ladder in TE buffer-150 mM sodium chloride for 10 min on ice, and then samples were analyzed by nondenaturing polyacrylamide gel electrophoresis (see the legend to Fig. 3B for details). The dsRNA was stained with ethidium bromide and visualized under UV light. To determine the minimum dsRNA length that σA can bind, gel shift assays were performed using σA in phosphate-buffered saline (PBS) buffer (137 mM sodium chloride, 2.7 mM potassium chloride, 8 mM disodium hydrogen phosphate, 15 mM potassium dihydrogen phosphate, pH 7.4), supplemented with 2 mM magnesium chloride and three dsRNAs of different lengths (18, 14, and 10 bp). Experiments were performed in which increasing concentrations of σA (0, 16.0, and 38 μM) were added to fixed concentrations of dsRNA (1.6 μM of 18-bp dsRNA, 3.3 μM of 14-bp dsRNA, and 3.6 μM of 10-bp dsRNA) in a final volume of 12.2 μl leading to σA-to-dsRNA ratios indicated in the figures. Mixtures were incubated for 10 min at room temperature and analyzed on a 15% polyacrylamide gel in TBE buffer (89 mM Tris base, 89 mM boric acid, 2 mM EDTA, pH 8.0). The dsRNA was visualized as described above.
FIG. 3.
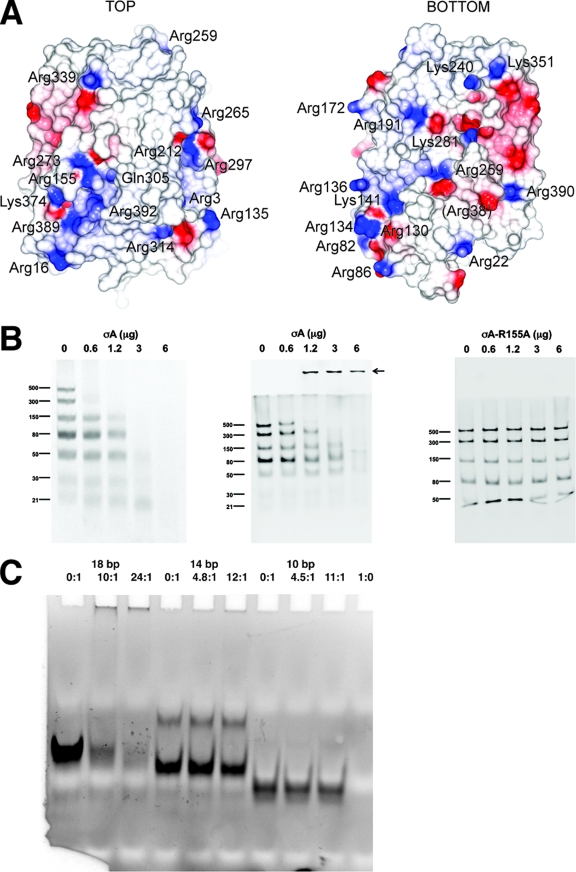
dsRNA binding of the σA protein. (A) Electrostatic surface of σA. On the left the orientation is as in Fig. 1A; on the right the view has been rotated 180° around a vertical axis centered on the apex. Positively charged residues on the molecular surface are labeled. The location of Gln305 is also shown, whereas Arg38 is shown in parentheses, as it is barely surface exposed. (B) dsRNA gel shift assays. The assays were performed using σA at 0.6 mg/ml in TE buffer (10 mM Tris-HCl, pH 8.5, 1 mM EDTA) and a dsRNA ladder which contains seven types of dsRNA of different lengths (500, 300, 150, 80, 50, 30, and 21 bp) at 0.5 mg/ml. Increasing amounts of σA (0, 0.6, 1.2, 3.0, and 6.0 μg) were incubated with 0.5 μg of dsRNA ladder in TE buffer-150 mM NaCl for 10 min on ice. Then, samples were analyzed by nondenaturing gel electrophoresis. (Left) A 10% polyacrylamide gel in TBE buffer was used. (Middle and right) Discontinuous gels as described by Laemmli (22) were used, with a 10% polyacrylamide separating gel and omission of sodium dodecyl sulfate from the sample, the gel, and the running buffer (13). Nonmutated σA protein was used in the left and middle panels. The right panel shows a gel shift assay performed with the Arg155-to-Ala mutant. Identical results were obtained with the Arg273-to-Ala mutant (data not shown). An arrow in the middle panel indicates the high-molecular-weight complexes observed. (C) Binding of σA to small dsRNA fragments as observed by gel shift assay. In the first three lanes an 18-bp dsRNA was used, in the next three lanes a 14-bp dsRNA was used, and in lanes 7 to 9 a 10-bp dsRNA was used. In lane 10 σA alone was loaded. σA/dsRNA ratios are indicated above the lanes.
Sedimentation velocity analysis was carried out with a Beckman Optima XL-A analytical ultracentrifuge equipped with absorbance optics, using an An-50Ti rotor and standard 12-mm optical path double-sector centerpieces of Epon-charcoal. σA protein was prepared as described before; an RNA oligonucleotide with the sequence 5′-ACA CUG UGA UCA GCA GGC GCC AGC UGA UCA CAG UGU-3′ was obtained from Nedken (Barcelona, Spain). dsRNA was prepared from this self-complementary oligonucleotide by heating and slow cooling. We used PBS buffer for all experiments, supplemented with 2 mM magnesium chloride. Samples of 0.4 ml with σA and dsRNA at different ratios were centrifuged at 40,000 rpm and 20°C. The concentration of σA in samples with excess protein was 4.9 μM, while dsRNA was added at 0.0, 1.0, 2.0, and 3.0 μM (duplex concentration), leading to σA-to-dsRNA ratios of 1:0.0, 1:0.21, 1:0.41, and 1:0.62, respectively. The concentration of dsRNA in samples with excess dsRNA was 1.77 μM (duplex concentration), while σA was added at 0.0, 1.71, 0.58, 0.18, and 0.058 μM, leading to σA-to-dsRNA ratios of 0:1.0, 1:1.0, 1:3.1, 1:9.3, and 1:31, respectively. For calculation of the concentrations, the calculated molar extinction coefficients of σA and the dsRNA based on their sequence were used (95,590 M−1 cm−1 at 280 nm for σA, 347,100 M−1 cm−1 at 260 nm for the dsRNA). Radial absorbance scans were taken at 280 nm (experiment 1) or 243 nm (experiment 2), and data were analyzed using the c(s) continuous distribution of Lamm equation solutions with the software SEDFIT (34).
Theoretical values of the sedimentation coefficient s20,w were estimated with the HYDROPRO program (12), using models composed of one, two, or three copies of chain G of our σA structure and a model of canonical A-form dsRNA of the sequence used. σA molecules were placed in the center of the RNA. The partial specific volume for σA (0.728 ml/g) was calculated from its amino acid composition automatically by HYDROPRO. For dsRNA a value of 0.55 ml/g was used. For the different complexes, weight-averaged values were employed, giving 0.669, 0.692, and 0.792 for the putative 1:1, 2:1, and 3:1 σA-dsRNA complexes, respectively. As recommended in the HYDROPRO manual, values of 3.2, 2.8, and 3.0 were used for the effective atomic radii of protein, dsRNA, and protein/dsRNA samples, respectively.
For electron microscopy, σA alone at 0.05 mg/ml and mixtures of σA with dsRNA (0.05 mg/ml σA; 1.8 μM 36-bp dsRNA; molar ratio, 15:1) in PBS buffer plus 2 mM magnesium chloride were applied to the clean side of carbon on mica (carbon/mica interface) and negatively stained with 1% (wt/vol) sodium silicotungstate (pH 7.0). Micrographs were taken under low-dose conditions with a JEOL 120 EX II microscope at 100 kV and a nominal magnification of ×40,000.
Figures 1A and B, 2A, and 4 were prepared using Pymol (PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA). Figure 3A was prepared using the program CCP4 mg (31).
FIG. 1.
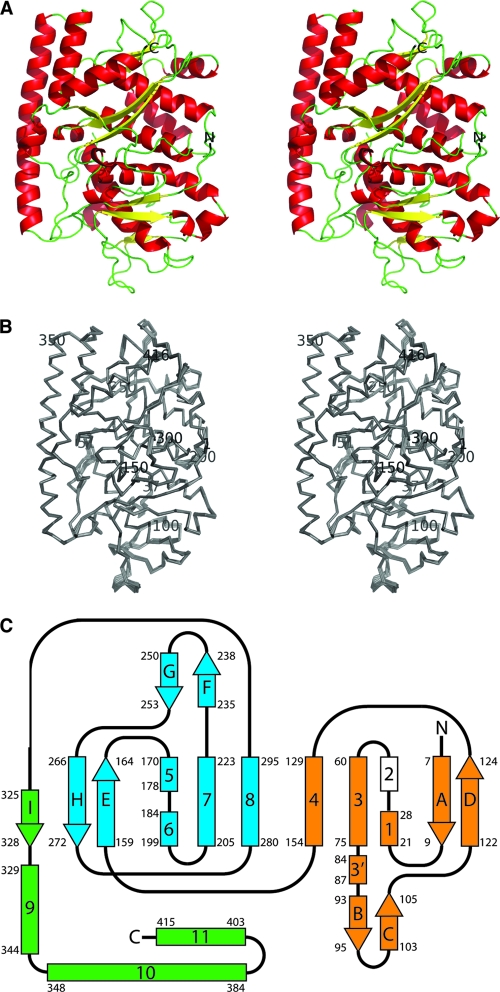
Structure of the avian reovirus σA protein. (A) Stereo view from the top of the σA pyramid (ribbon representation). The monomer with the lowest average temperature factor, G, was used to make this figure. The amino and carboxy termini of the protein are labeled. (B) Stereo view of main chain superimposition of all 12 σA copies in the asymmetric unit of the crystal. The amino-terminal Met, carboxy-terminal Ala, and every 50th residue are labeled, as are the amino acids flanking the disordered loop (Gln37 and Ile61). (C) Topology of the σA protein chain. Alpha-helices are shown as rectangles labeled with numbers, and beta-strands are shown as arrows labeled with letters. The two structurally homologous domains 1 and 2 are shown in orange and blue, and the carboxy-terminal domain 3 is shown in green. Helix 2 (in white) is not observed in our structure but is inferred from the structure of mammalian reovirus σ2 (see text). Starting and ending residues of the secondary structure elements are labeled.
FIG. 2.
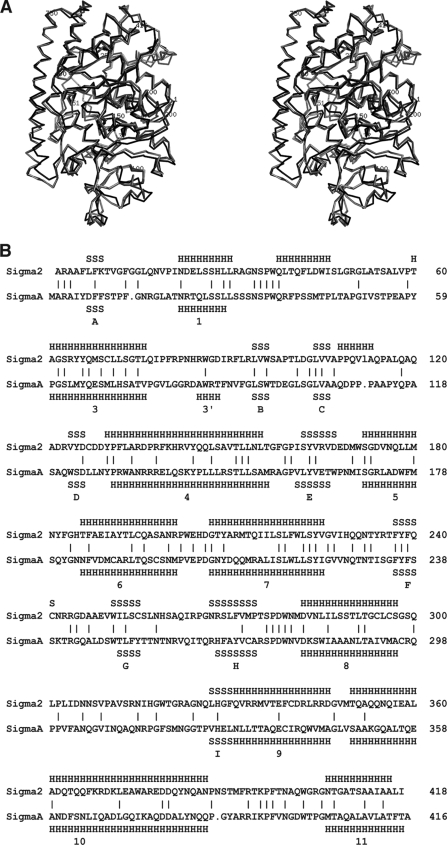
Comparison of avian reovirus σA structure with mammalian reovirus σ2. (A) Superposition of σA onto σ2. σA chain G was superposed on σ2 chains D and E (PDB entry 1EJ6 [33]), leading to an overall RMSD of 1.6 Å for 390 or 392 superimposed C-alpha atoms, respectively (20). σA is shown in black, σ2 chain D is shown in dark gray, and σ2 chain E is shown in light gray. (B) Structure-based alignment of σA and σ2. Secondary structure elements in σA are indicated (S for strand, H for helix) and numbered.
FIG. 4.
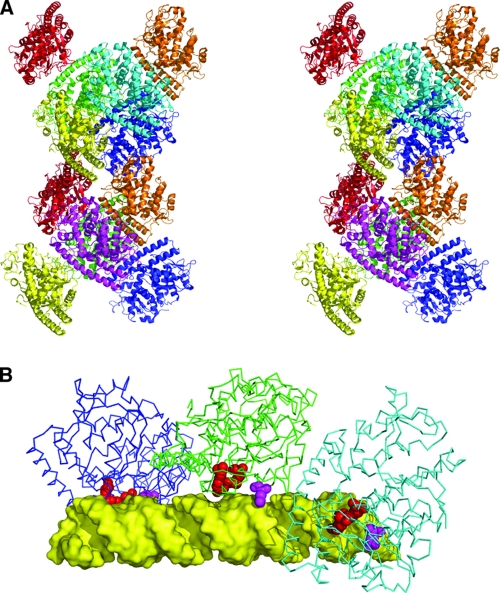
Assembly of σA molecules in the crystal and model of dsRNA binding. (A) Stereogram of the dodecameric assembly of σA in the crystallographic asymmetric unit. Monomers are shown in cartoon format and colored differently (two yellow, two orange, two red, two green, two blue, one magenta, and one cyan). (B) Model of a σA trimer interacting with a 36-bp canonical A-form dsRNA (yellow). Residues implicated in RNA binding, Arg155 and Arg273, are shown in red; Gln305 is shown in magenta.
Protein structure accession number.
The coordinates have been deposited in the protein structure database (http://www.rcsb.org) under accession code 2VAK; the structure factors are also available.
RESULTS
Bacterial expression, purification, and crystallization of the σA protein have been described previously (15). Molecular replacement yielded a solution with 12 copies of a model based on the mammalian reovirus core protein σ2. The σA molecules occupy 38% of the unit cell (Matthews coefficient 3.2 [26]). The final refinement was performed without using noncrystallographic symmetry restraints; statistics are shown in Table 1. The final model contains amino acids Met1-Gln37 and Ile51-Ala416 of σA for each of the 12 chains, 12 sulfate ions, and 3,003 water molecules. Some monomers also contain a vestige of the expression linker segment remaining after factor Xa protease cleavage (Ile-Ser-Glu-Phe-Gly-Ser-Thr), whereas in others this segment is partially or completely disordered. The conformation of the expression tail will not be further discussed here and is not included in the analyses described below.
TABLE 1.
Refinement statistics
| Parameter | Valuea |
|---|---|
| Space group | P1 |
| Cell parameters (a, b, c, α, β, γ) | 103.2 Å, 129.9 Å, 144.0 Å, 93.8°, 105.1°, 98.2° |
| Resolution range | 30.0-2.34 Å (2.47-2.34 Å) |
| No. of reflections used in refinementb | 281,587 (32,494) |
| No. of reflections used for R-free | 1,385 (119) |
| R-factorc | 0.210 (0.26) |
| R-free | 0.271 (0.34) |
| No. of protein/sulfate/water atoms | 38,161/60/3,003 |
| Avg B protein/sulfate/water | 37.2 Å2/60.4 Å2/37.2 Å2 |
| Ramachandran statisticsd | 91.0%/8.5%/0.4%/0.1% |
| RMSDe (bonds/angles) | 0.014 Å/1.4° |
Values in parentheses are for the highest-resolution bin, where applicable.
No sigma cutoff or other restrictions were used for inclusion of observed reflections.
R = Σ‖Fobs(hkl)| − |Fcalc(hkl)‖/Σ|Fobs(hkl)|.
According to the program PROCHECK (24). The percentages are indicative of residues in the most favored, additionally allowed, generously allowed, and disallowed regions of the Ramachandran plot, respectively.
Estimates provided by the program REFMAC (27).
Structure and topology of σA.
The overall topology of the σA monomer is roughly tetrahedral in form with an approximately rectangular base (Fig. 1). The base is relatively planar, 65 to 70 Å long and 45 to 50 Å wide, and encompasses the proposed site of interaction with the main core protein λA (see below). The distance from the base to the apex, which is expected to form the interaction site with μB in the mature virion, is around 45 Å. The 12 crystallographically independent copies in the unit cell are virtually identical (root mean square deviation [RMSD] of C-alpha atoms after superimposition of 0.2 to 0.7 Å; Fig. 1B). Each σA monomer contains 12 alpha-helices, one mixed three-stranded beta-sheet, and three antiparallel two-stranded beta-sheets (Fig. 1C).
There is a gap where the protein chain is disordered in all 12 copies present in the crystal between helices 1 and 3. No interpretable electron density was observed for σA residues 38 to 50 in this region. In σ2, the structurally equivalent amino acids are 39 to 52 (Fig. 2). In the previously solved mammalian reovirus core structure, this region contains an alpha-helix in one of the σ2 copies (residues 39 to 46) which is melted in the other two (33). This region contacts λ1 in the mammalian reovirus core (33), so it is conceivable that the σA alpha-helix becomes ordered only upon binding to λA. We have designated the putative equivalent helix in σA as helix 2. The fact that this loop is flexible and can adopt various conformations suggests that it may be used by the protein to adapt to the different binding environments that the λA shell presents.
The N and C termini are both oriented toward the apex of the molecule and are solvent accessible in this context (although in the virion they are likely to be buried in the interface with μB). The first secondary structure element in the N-terminal region, beta-strand A, however, is buried in the core of the molecule. Phe-8 and Phe-9 in this strand both pack against surrounding hydrophobic and aromatic residues. Helices 1, 3, and 4 (and the putative helix 2 [see below]), together with beta-sheets B-C and A-D, form a discrete subdomain, which is packed against another subdomain with the same topology (helices 5, 6, 7, and 8 and sheets G-F and H-E). These two domains are packed against a third domain formed by helices 9, 10, and 11 and beta-strand I (which lies alongside strand E, forming the H-E-I beta-sheet motif). The apex of the pyramid is largely formed by the loop between helix 8 and strand I.
The regions of σA contacting λA and μB can be predicted from the previously published crystal structure of the mammalian reovirus core (PDB code 1EJ6 [33]), the published pseudoatomic model of the entire mammalian reovirus obtained by fitting known crystal structures into cryo-electron microscopic density (PDB code 2CSE [41]), and cryo-electron microscopic studies of avian reovirus (40). The interaction site with μB mainly comprises residues contained within this third domain, while the proposed λA binding site is almost exclusively composed of residues from domains 1 and 2. Domains 1 and 2 present the same secondary structural elements on the surface at the λA binding site, potentially indicating domain duplication, where the two domains have evolved to interact with different facets of the λA component in the inner shell.
Structural and functional comparison with other proteins.
Analysis of the whole structure, or domains 1, 2, or 3 independently, by three-dimensional secondary structure matching (20) did not reveal any close structural homologues for σA, other than mammalian reovirus σ2, with which it shares 29% sequence identity. The overall topologies of these proteins are very similar; however, there are small differences in some surface residues, which may be related to inherent flexibility in these regions (Fig. 2A). Most of the surface residues that differ between σA and σ2 vary in position between the 12 independent σA monomers in the unit cell (Fig. 1B and 2A). One notable difference between σA and σ2 occurs at the carboxy terminus of helix 10 (residues 374 to 388 in the σA structure), which is markedly distorted in σA.
dsRNA binding of σA.
With regard to dsRNA binding, σA is functionally, though not structurally, analogous to mammalian reovirus σ3. σA and σ3 both bind dsRNA and have been implicated in PKR inhibition (13, 16). As with σA, the ability of σ3 to bind dsRNA has been linked to its capacity to inhibit dsRNA-dependent activation of PKR through competitive binding. It has been proposed that σ3 binds dsRNA as a dimer and that multiple dimers may line up along the RNA molecule with their twofold axes aligned along the RNA local dyads (30).
When the electrostatic potential is superimposed onto the surface of the σA monomer, a positively charged region, containing among other residues Arg155 and Arg273, is notable (Fig. 3A). We mutated these residues independently to alanine and examined the effects on dsRNA binding by gel shift assays (Fig. 3B). Both mutations abolished the dsRNA binding properties of σA (shown for the Arg155-to-Ala mutation), whereas mutation of Arg134, Arg135, Arg389, or Arg390 had no significant effect on dsRNA binding. Both mutant proteins were soluble, and there were no indications that either of the mutations affected the protein stability or had any structural impact. A further indication that this region may play a role in RNA binding comes from the binding of a sulfate ion at or near Gln305, which is close to Arg155 and Arg273. This sulfate is present in each of the 12 monomers in our structure and may mimic the binding of phosphate in the RNA backbone at this site.
In the gel shift assays using native σA, high-molecular-weight complexes were observed that barely enter the stacking gel (Fig. 3B, middle panel). This suggests either that σA and dsRNA may form large complexes or that the resulting complex may be relatively uncharged at the pH employed here (8.0). The latter is in keeping with a model where σA molecules (theoretical pI 8.6) cover the bulk of the surface of the dsRNA, thus masking negative charges arising from the phosphate backbone. The dsRNA-binding experiments also indicate that σA is able to bind RNA down to a minimum length of at least 21 bp (Fig. 3B, left panel). However, as dsRNA size increases, complexes form at lower σA concentrations, which suggests that the binding is cooperative.
To identify the minimal length of dsRNA bound by σA, gel shift assays were performed with 10-bp, 14-bp, and 18-bp dsRNAs (Fig. 3C). This showed that σA can bind 18-bp dsRNA and also 14-bp dsRNA, albeit less efficiently, while 10-bp dsRNA was not bound by σA. In contrast, σ3 has been reported to require a minimum length of 32 to 45 bp for efficient RNA binding (39). We investigated whether the putative dsRNA binding site is structurally related to the dsRNA binding motif (dsRBM) identified in several other proteins (3), including PKR (28, 29). The archetypal dsRBM consists of a contiguous alpha-beta-beta-beta-alpha fold, in which the two helices are arranged on one side, packed against a three-stranded antiparallel beta-sheet. The residues implicated in dsRNA interaction are located in loop 2 (between beta-strands 1 and 2) and loop 4 (between beta-strand 3 and alpha-helix 2). The dsRNA binding site of σA is somewhat similar and is made up from a discontinuous alpha-beta-beta-beta-alpha subdomain, in which helix 11 and helix 4 and/or helix 8 lie on opposite sides of a three-stranded mixed beta-sheet. The arginines implicated in dsRNA interaction are located in the loop between alpha-helix 4 and beta-strand E and in the loop between alpha-helix 8 and beta-strand H. However, these loops are not conserved and have a different connectivity in the dsRBM, suggesting that the structural similarity is superficial, and hence that there is no direct evolutionary relationship between the σA dsRNA-binding region and the dsRBM.
Interactions between σA molecules in the crystal.
σA is monomeric in salt solutions as judged by size-exclusion chromatography (not shown), sedimentation velocity ultracentrifugation, and electron microscopy (see below), which indicated an absence of higher-order aggregates. However, at 4°C, and in the absence of salt, σA precipitates reversibly, indicating that the protein has a tendency to form higher-order aggregates.
In the crystal structure, 12 σA molecules pack together into what at first view appears to be a double-helical assembly (Fig. 4A). When the interactions are analyzed on the basis of buried surface area using the PISA server (21), three types of interactions with potential biological relevance are observed. We have divided these possible interactions into three classes, the “tail-to-tail,” “head-to-tail,” and “head-to-head” interactions. The tail consists of residues from the N-terminal loop, strand A, strand D, the loop to helix 4, the loop between strands G and H, and the loop between helix 8 and strand I, while the “head” consists of amino acids from the loop between putative helix 2 and helix 3, from helix 10, and the loop between helices 10 and 11. The tail-to-tail interaction has a buried surface of 690 Å2, a solvation energy gain of 12.4 kcal/mol, three hydrogen bonds, and a P value of 0.033; the head-to-tail interaction has a buried surface of 467 Å2, a solvation energy gain of 7.4 kcal/mol, six hydrogen bonds, and a P value of 0.054, while the head-to-head interaction buries a surface of 373 Å2 and has a solvation energy gain of 4.7 kcal/mol, two hydrogen bonds, and a calculated P value of 0.26. None of the interactions observed among σA molecules in our crystal structure are similar to the previously observed interactions among σ2 molecules in the mammalian reovirus core.
The head-to-tail interaction is the only one involving all 12 σA molecules of the asymmetric unit. It joins the 12 molecules into four trimers. Each trimer is related to its corresponding opposite in a double-helical hexamer by a rotation of about 180°. The fit between trimers is very good (RMSD after superposition of 0.4 Å), suggesting that the trimers act as a rigid body. The monomers of the trimer are related by a rotation of approximately 50°, around an axis running through the center of the double helix, which explains the previously reported peak in the self-rotation function at a chi angle of 50° (15). Interestingly, we note that canonical A-form dsRNA also has a helical rotation of around 50°.
The double-helical hexamer is related to the other hexamer in the asymmetric unit by a rotation of around 180°. In this case, the fit between the two hexamers is not so good (RMSD after superposition of 1.6 Å). It is tempting to propose the trimer as a biological unit, arranged around dsRNA as a single-stranded or double-stranded assembly. These models could explain the cooperative binding of σA to dsRNA as well as the large complexes observed in the gel shift assays (Fig. 3B, middle panel) (38). The previously discussed positively charged patch maps to the inside of the helix (Fig. 4B). The width of the cavity inside the double-helical σA assembly varies from 11 to 35 Å, with the cross section being at least 20 Å wide in one dimension, while the diameter of A-form dsRNA is about 10 by 20 Å in cross section. In other words, A-form dsRNA would probably fit, although some minor structural adjustments in the dsRNA or in the σA assembly may be necessary.
The distance between Arg155 and Arg273 in the same molecule is 8.5 Å, while the distances to their counterparts in the next molecule of the trimer are 39 to 40 Å. In a model constructed with canonical A-form RNA and the crystallographic trimer, one arginine side chain could bind a phosphate on one side of the minor groove and the other could bind a phosphate on the other side of the minor groove (distances between phosphates, 10 Å). The next molecule could make the same interaction with phosphates 13 bp further along the RNA chain (distance between phosphates, 38 Å) (Fig. 4B). A structural analysis of the σA-dsRNA complex will be needed to confirm this hypothesis. Mutation of residues involved at the putative protein-protein interface might also be attempted. However, inspection of this interface did not suggest any obvious target residues that, when mutated, would significantly affect protein-protein binding, without having a potential serious structural impact.
Stoichiometry of σA binding to dsRNA.
To obtain independent evidence for our dsRNA binding model and in an attempt to discriminate between our putative single and double σA helices, we performed sedimentation velocity centrifugation experiments on σA-dsRNA complexes and transmission electron microscopy on negatively stained σA-dsRNA samples.
In sedimentation velocity centrifugation experiments, a single peak was obtained in control experiments containing σA or dsRNA alone, indicating that both are monodisperse. The estimated molecular masses were those expected for σA monomers and dsRNA duplexes (Table 2). When excess protein was mixed with different amounts of dsRNA (experiment 1), four reproducible peaks were obtained. Sedimentation coefficients obtained for these peaks are compatible with monomeric σA and three different complexes of σA/dsRNA with stoichiometries of 1:1, 2:1, and 3:1 (Table 2). In experiment 2 (Table 2), excess dsRNA was added to protein. Again, dsRNA of 36 bp gave a monodisperse peak, while in the mixtures two or three peaks were observed, of which one clearly corresponds to uncomplexed dsRNA. The complex peaks have smaller sedimentation coefficients than expected for 1:1 and 2:1 σA/dsRNA complexes, indicating that they are probably in equilibrium with free dsRNA and 1:1 σA/dsRNA complex, respectively. Taken together, the centrifugation experiments suggest a maximum of three σA molecules binding to a 36-bp dsRNA (Fig. 4B), which would be compatible with a single helix of σA molecules covering dsRNA.
TABLE 2.
Sedimentation velocity centrifugation results and analysis of σA/36-bp dsRNA mixtures
| Component or complex |
s20,w (10−13 s) (%)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Calculated | Expt 1 (excess σA)
|
Expt 2 (excess dsRNA)
|
|||||||||
| σA | 36-mer dsRNA | Mixture
|
36-mer dsRNA | Mixture
|
|||||||
| 1/0.21 | 1/0.41 | 1/0.62 | 1/1.0 | 1/3.1 | 1/9.3 | 1/31 | |||||
| σA | 3.9 | 3.6 (100) | NOa | 3.6 (71) | 3.7 (57) | 3.5 (35) | NO | NO | NO | NO | NO |
| dsRNA | 3.4 | NO | 3.4 (100) | NO | NO | NO | 3.6 (100) | 3.2 (62) | 3.2 (75) | 3.2 (77) | 3.2 (75) |
| Complex | |||||||||||
| 1:1 | 5.6 | NO | NO | 5.8 (7) | 6.0 (11) | 5.4 (16) | NO | 5.2 (21) | 4.4 (19) | 4.3 (23) | 4.2 (25) |
| 2:1 | 7.3 | NO | NO | 7.6 (9) | 8.0 (12) | 7.4 (18) | NO | 6.9 (15) | 5.7 (6) | NO | NO |
| 3:1 | 8.8 | NO | NO | 9.0 (12) | 9.4 (13) | 8.8 (25) | NO | NO | NO | NO | NO |
NO, not observed.
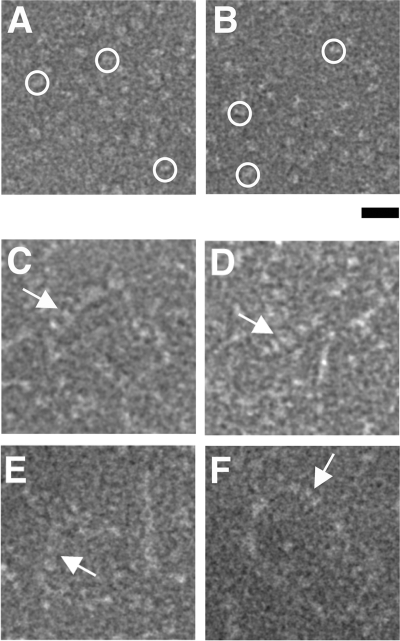
Electron micrographs (Fig. 5) also show multiple σA molecules binding to dsRNA (Fig. 5C to F), while σA alone appears monomeric (Fig. 5A and B). The width of the observed complexes varies from 54 to 98 Å and does not permit distinction between a single helix (expected diameter, 60 Å) or double helices (expected diameter, 100 Å) of σA binding to the dsRNA. In fact, binding may be plastic, with regions of dsRNA covered by single-helical σA and others covered by double-helical σA, depending on the local dsRNA structure. Curiously, the structures formed are clearly longer than the 36-bp dsRNA used, suggesting that multiple dsRNA duplexes covered by σA line up, presumably via σA-σA interactions. Future structural studies by cryo-electron microscopy and/or cocrystallization studies will hopefully shed more light on the non-sequence-specific, cooperative mode of dsRNA binding by RNA.
FIG. 5.
Transmission electron microscopy of σA-dsRNA mixtures. (A and B) σA alone, which appears monomeric. Particles with approximate sizes of monomeric σA are shown by circles. (C to F) Putative σA-dsRNA complexes, indicated by arrows. Bar, 20 nm.
DISCUSSION
Orthoreoviruses can be functionally classified into two subgroups, fusogenic and nonfusogenic, based on their ability to induce cell-cell fusion and form syncytia. The archetypal mammalian orthoreoviruses are nonfusogenic, although they do form specific pores in the membrane, proposed to be part of the membrane penetration pathway during cell entry (1). The remaining members of the genus, including avian orthoreoviruses (2), reptilian orthoreoviruses (10), baboon orthoreoviruses (8), and other mammalian orthoreoviruses like Nelson Bay reovirus (9) and Pulau reovirus (32), are fusogenic. A series of diseases have been attributed to orthoreovirus infection in animals, and recently it has been reported that Melaka reovirus of bat origin, another fusogenic mammalian reovirus, is associated with an acute respiratory disease in humans (5). However, infections with nonfusogenic orthoreoviruses are generally benign, with very rare cases of mild upper respiratory tract illness or enteritis occurring in infants and children.
Sequence analysis and RNA binding assays carried out on the major outer capsid protein, σB (and equivalents) from fusogenic reoviruses, have revealed that this protein, in contrast to its nonfusogenic counterpart σ3, does not bind dsRNA (9, 37, 38). As PKR downregulation in avian reovirus S1133 is caused by σA (13), we analyzed the sequence of the minor inner capsid proteins of fusogenic and nonfusogenic reoviruses to see if the residues implicated in dsRNA binding are conserved among them. This analysis revealed that Arg273 is invariant among fusogenic reoviruses (both avian and mammalian), whereas in nonfusogenic reoviruses it is replaced by Thr. Arg155 is invariant among avian reoviruses but is replaced by Gln in mammalian fusogenic reoviruses and by Gly in mammalian nonfusogenic reovirus. This pattern of conservation suggests that the fusogenic counterparts of the σA protein retain their ability to bind dsRNA (and by extension the potential to downregulate PKR). In nonfusogenic reoviruses, the σ2 proteins, which are structural homologues of σA, do not appear to bind dsRNA. Among this viral subclass RNA binding is linked to the outer capsid protein σ3. These differences in function may profoundly influence host cell interactions and could be related to and/or have evolved together with the syncytial mode of propagation of the fusogenic viruses and are thus intimately tied to their pathogenicity.
In conclusion, the structure, gel shift, sedimentation velocity, and electron microscopy data presented here suggest a cooperative mode of σA binding to dsRNA, which may be of general relevance to all fusogenic reoviruses, thereby contributing to a better understanding of their biology. This improved understanding may in turn lead to better strategies to combat these viruses.
Acknowledgments
This research was sponsored by research grants BFU2005-02974 and BFU2005-24982-E (awarded to M.J.V.) and by a predoctoral FPU fellowship to P.G.-C., all from the Spanish Ministry of Education and Science, and by research grant PGIDIT04BTF203003PR from the Xunta de Galicia (awarded to J.M.-C.). As part of the European Science Foundation EUROCORES Programme EuroSCOPE, the work was also supported by funds from the European Commission under contract ERAS-CT-2003-980409.
We thank Carlos Alfonso and Germán Rivas of the CSIC (Centro de Investigaciones Biológicas, Madrid, Spain) for help with the analytical ultracentrifugation experiments and Patricia Ferraces Casais and Rebeca Menaya Vargas for technical assistance.
Footnotes
Published ahead of print on 17 September 2008.
REFERENCES
- 1.Agosto, M. A., T. Ivanovic, and M. L. Nibert. 2006. Mammalian reovirus, a nonfusogenic nonenveloped virus, forms size-selective pores in a model membrane. Proc. Natl. Acad. Sci. USA 10316496-16501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benavente, J., and J. Martinez-Costas. 2007. Avian reovirus: structure and biology. Virus Res. 123105-119. [DOI] [PubMed] [Google Scholar]
- 3.Chang, K. Y., and A. Ramos. 2005. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2722109-2117. [DOI] [PubMed] [Google Scholar]
- 4.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 313497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chua, K. B., G. Crameri, A. Hyatt, M. Yu, M. R. Tompang, J. Rosli, J. McEachern, S. Crameri, V. Kumarasamy, B. T. Eaton, and L. F. Wang. 2007. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc. Natl. Acad. Sci. USA 10411424-11429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50760-763. [DOI] [PubMed] [Google Scholar]
- 7.Davis, I. W., A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall III, J. Snoeyink, J. S. Richardson, and D. C. Richardson. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35W375-W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawe, S., and R. Duncan. 2002. The S4 genome segment of baboon reovirus is bicistronic and encodes a novel fusion-associated small transmembrane protein. J. Virol. 762131-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan, R. 1999. Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal. Virology 260316-328. [DOI] [PubMed] [Google Scholar]
- 10.Duncan, R., J. Corcoran, J. Shou, and D. Stoltz. 2004. Reptilian reovirus: a new fusogenic orthoreovirus species. Virology 319131-140. [DOI] [PubMed] [Google Scholar]
- 11.Emsley, P., and K. Cowtan. 2004. Model building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 12.Garcia de la Torre, J., M. L. Huertas, and B. B. Carrasco. 2000. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys. J. 78719-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Lopez, C., J. Martinez-Costas, M. Esteban, and J. Benavente. 2003. Evidence that avian reovirus sigmaA protein is an inhibitor of the double-stranded RNA-dependent protein kinase. J. Gen. Virol. 841629-1639. [DOI] [PubMed] [Google Scholar]
- 14.Guardado-Calvo, P., G. C. Fox, X. L. Hermo-Parrado, A. L. Llamas-Saiz, C. Costas, J. Martinez-Costas, J. Benavente, and M. J. van Raaij. 2005. Structure of the carboxy-terminal receptor-binding domain of avian reovirus fibre sigmaC. J. Mol. Biol. 354137-149. [DOI] [PubMed] [Google Scholar]
- 15.Hermo-Parrado, X. L., P. Guardado-Calvo, A. L. Llamas-Saiz, G. C. Fox, L. Vazquez-Iglesias, J. Martinez-Costas, J. Benavente, and M. J. van Raaij. 2007. Crystallization of the avian reovirus double-stranded RNA-binding and core protein sigmaA. Acta Crystallogr. F 63426-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imani, F., and B. L. Jacobs. 1988. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 sigma 3 protein. Proc. Natl. Acad. Sci. USA 857887-7891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, R. C. 2000. Avian reovirus infections. Rev. Sci. Technol. 19614-625. [DOI] [PubMed] [Google Scholar]
- 18.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47110-119. [DOI] [PubMed] [Google Scholar]
- 19.Kleywegt, G. J., J. Y. Zou, M. Kjeldgaard, and T. A. Jones. 2001. Around O, p. 353-356 and 366-367. In M. G. Rossmann and E. Arnold (ed.), International tables for crystallography, vol. F. Crystallography of biological macromolecules. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 20.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 602256-2268. [DOI] [PubMed] [Google Scholar]
- 21.Krissinel, E., and K. Henrick. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372774-797. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 23.Lamzin, V. S., and K. S. Wilson. 1993. Automated refinement of protein models. Acta Crystallogr. D 49129-149. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26283-291. [Google Scholar]
- 25.Martinez-Costas, J., C. Gonzalez-Lopez, V. N. Vakharia, and J. Benavente. 2000. Possible involvement of the double-stranded RNA-binding core protein σA in the resistance of avian reovirus to interferon. J. Virol. 741124-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews, B. W. 1968. Solvent content of protein crystals. J. Mol. Biol. 33491-497. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 28.Nanduri, S., B. W. Carpick, Y. Yang, B. R. Williams, and J. Qin. 1998. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 175458-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nanduri, S., F. Rahman, B. R. Williams, and J. Qin. 2000. A dynamically tuned double-stranded RNA binding mechanism for the activation of antiviral kinase PKR. EMBO J. 195567-5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olland, A. M., J. Jané-Valbuena, L. A. Schiff, M. L. Nibert, and S. C. Harrison. 2001. Structure of the reovirus outer capsid and dsRNA-binding protein sigma3 at 1.8 Å resolution. EMBO J. 20979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potterton, L., S. McNicholas, E. Krissinel, J. Gruber, K. Cowtan, P. Emsley, G. N. Murshudov, S. Cohen, A. Perrakis, and M. Noble. 2004. Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D 602288-2294. [DOI] [PubMed] [Google Scholar]
- 32.Pritchard, L. I., K. B. Chua, D. Cummins, A. Hyatt, G. Crameri, B. T. Eaton, and L. F. Wang. 2006. Pulau virus; a new member of the Nelson Bay orthoreovirus species isolated from fruit bats in Malaysia. Arch. Virol. 151229-239. [DOI] [PubMed] [Google Scholar]
- 33.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404960-967. [DOI] [PubMed] [Google Scholar]
- 34.Schuck, P. 2000. Size distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 781606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vagin, A., and A. Teplyakov. 2000. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D 561622-1624. [DOI] [PubMed] [Google Scholar]
- 36.Vriend, G. 1990. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 852-56. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Q., J. Bergeron, T. Mabrouk, and G. Lemay. 1996. Site-directed mutagenesis of the double-stranded RNA binding domain of bacterially-expressed sigma 3 reovirus protein. Virus Res. 41141-151. [DOI] [PubMed] [Google Scholar]
- 38.Yin, H. S., J. H. Shien, and L. H. Lee. 2000. Synthesis in Escherichia coli of avian reovirus core protein varsigmaA and its dsRNA-binding activity. Virology 26633-41. [DOI] [PubMed] [Google Scholar]
- 39.Yue, Z., and A. J. Shatkin. 1997. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology 234364-371. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, X., J. Tang, S. B. Walker, D. O'Hara, M. L. Nibert, R. Duncan, and T. S. Baker. 2005. Structure of avian orthoreovirus virion by electron cryomicroscopy and image reconstruction. Virology 34325-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, X., Y. Ji, L. Zhang, S. C. Harrison, D. C. Marinescu, M. L. Nibert, and T. S. Baker. 2005. Features of reovirus outer capsid protein mu1 revealed by electron cryomicroscopy and image reconstruction of the virion at 7.0-Ångstrom resolution. Structure 131545-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]