Abstract
Viral emergence can result from the adaptation of endemic pathogens to new or altered host environments, a process that is strongly influenced by the underlying sequence diversity. To determine the extent and structure of intrahost genetic diversity in a recently emerged single-stranded DNA virus, we analyzed viral population structures during natural infections of animals with canine parvovirus (CPV) or its ancestor, feline panleukopenia virus (FPV). We compared infections that occurred shortly after CPV emerged with more recent infections and examined the population structure of CPV after experimental cross-species transmission to cats. Infections with CPV and FPV showed limited genetic diversity regardless of the analyzed host tissue or year of isolation. Coinfections with genetically distinct viral strains were detected in some cases, and rearranged genomes were seen in both FPV and CPV. The sporadic presence of some sequences with multiple mutations suggested the occurrence of either particularly error-prone viral replication or coinfection by more distantly related strains. Finally, some potentially organ-specific host effects were seen during experimental cross-species transmission, with many of the mutations located in the nonstructural protein NS2. These included residues with evidence of positive selection at the population level, which is compatible with a role of this protein in host adaptation.
Emerging viruses that gain new host ranges are major threats to human and animal health. However, the evolutionary mechanisms that allow viruses to infect new hosts and establish self-sustaining transmission chains are complex and far from understood. In most cases studied to date, several mutations together determine the new host range, but where, when, and how these mutations arise have remained largely enigmatic. Many cases of viral emergence probably represent multistage adaptations to an altered host environment, where the initial emerging virus is poorly adapted to the recipient host and causes only inefficient transmission. Further mutations are then required for complete adaptation to the new host. The success of the new pathogen is therefore likely to be highly influenced by the genetic diversity of the viral population, with more variable viral populations being particularly prone to generating successful emerging infections.
The extent of genetic diversity present within viral populations is determined largely by a balance between erroneous replication (measured as the mutation rate) and purifying selection (reflected in the population substitution rate, defined as the number of fixed mutations/nucleotide site/year), with particularly high error rates in RNA viruses that replicate using RNA-dependent RNA polymerases (15), or, in the case of retroviruses, reverse transcriptases. Although large double-stranded DNA viruses possess mutation rates far lower than those seen in RNA viruses, some small single-stranded DNA (ssDNA) viruses appear to both mutate (14, 39) and have substitution rates closer to those of RNA viruses than to those of double-stranded DNA viruses (reviewed in reference 16). The parvoviruses represent a particularly well-studied case, with the mean substitution rate for the canine parvovirus (CPV) capsid protein gene at ∼1 × 10−4 substitutions/nucleotide/year (38, 42). However, the extent and structure of within-host variability in ssDNA viruses, including coinfection as a potential source of genetic diversity, have rarely been analyzed.
Various phenomena such as recombination, nonrandom codon usage, and host effects such as hypermutation mediated by APOBEC3G (apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3B) might also affect standing genetic diversity, although their role in the evolution of ssDNA viruses is largely unclear. In addition, some rapidly evolving viruses such as human immunodeficiency virus undergo tissue-specific variation (24, 47). Tissue-specific effects may also occur during infections with parvoviruses, but this has not been analyzed previously.
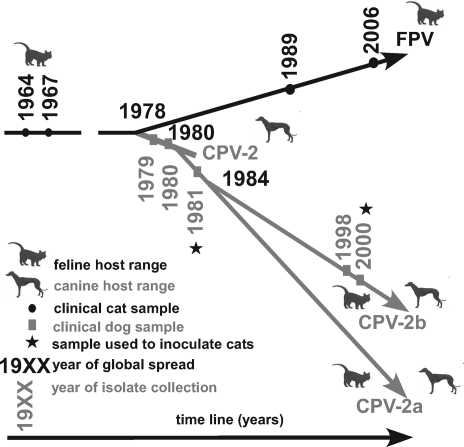
CPV emerged in the 1970s as a new virus of dogs, derived from either feline panleukopenia virus (FPV) or a very closely related virus of another host (Fig. 1). After circulating undetected in dogs in Europe or Eurasia for a few years, the virus spread globally in 1978 (35). This virus is referred to as CPV type 2 (CPV-2) to distinguish it from the distantly related minute virus of canines. In 1979, a variant strain of CPV-2 (CPV-2a) emerged and replaced CPV-2 globally by the end of 1980 (37). The emerging strain differed antigenically and readily infected cats, while CPV-2 did not replicate in felines (44). In 1984, an antigenic variant of CPV-2a with apparently identical host range, which is referred to as CPV-2b, arose. CPV-2a and CPV-2b are currently cocirculating in the global dog population, but their relative frequencies appear to vary among geographic regions and are potentially also subject to temporal fluctuation (reviewed in reference 20). Natural infections of cats and wild felines with CPV have been reported (4, 18, 31), but FPV has remained the more prevalent parvovirus causing disease in cats.
FIG. 1.
Overview of natural virus samples analyzed. The host from which the sample was collected, the virus type, and year of collection are indicated. The likely natural host range and year of global spread are indicated. Viruses used for inoculation of kittens are marked.
Both FPV and CPV cause acute infections of young animals, resulting in the development of long-lived protective immunity (7). Infection occurs through the oronasal route, and initial replication takes place in the lymphatic tissues of the oropharynx (see Fig. 4A). After the primary viremia, the virus begins to replicate in the thymus, spleen, and bone marrow 2 to 3 days after infection. The virus then becomes disseminated throughout the animal, and replication is seen in the gut-associated lymphoid tissues, particularly the Peyer's patches. The virus subsequently replicates in the rapidly dividing epithelial cells within the crypts of the small intestinal villi and is shed at high levels in the feces 4 to 7 days postinfection (6, 7, 29, 33). The virus appears to be cleared less than 2 weeks postinfection, with no residual replicating virus remaining (reviewed in reference 33).
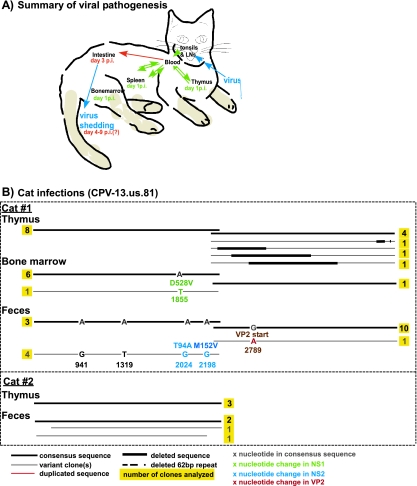
FIG. 4.
Analysis of the sequence recovered from experimental CPV-13.us.81 infection of cats. (A) Summary of the expected pathogenesis of FPV in cats, including the viral location at various days postinfection (p.i.) and organs infected. (B) Characterization of viral sequences recovered from different organs, including the type of nucleotide substitution and the attained protein (for nonsynonymous changes) of each infected cats.
The CPV genome (depicted in Fig. 2, below the consensus sequences, and Fig. 3, top) contains two open reading frames (ORFs), one of which encodes the two nonstructural proteins NS1 and NS2 and the other of which encodes the two structural proteins VP1 and VP2 (40). The amino-terminal ends of NS1 and NS2 overlap and are identical in sequence, but the carboxy-terminal domain of NS2 is derived by differential splicing, fusing it to a different reading frame, which overlaps that of NS1. The sequence of VP2 is completely contained within that of VP1, which has an additional 143-amino-acid-long N-terminal sequence. VP2 makes up 90% of the viral capsid, which is the major determinant of canine and feline host ranges (8, 22, 32). Since the feline host range of CPV-2a was not due to back mutations to the FPV sequences, some of the newly arisen mutations that characterized CPV-2a were likely of a compensatory nature (43, 45).
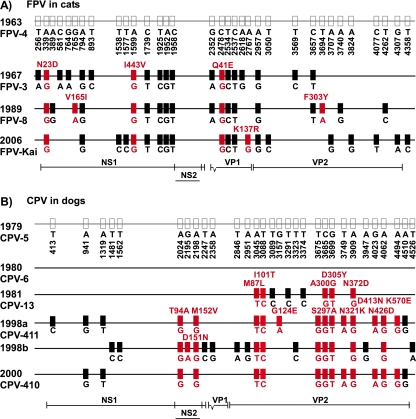
FIG. 2.
Mutations in the consensus sequences. The isolate consensus sequences from clinical samples of FPV (A) and CPV (B) infections were compared by using the oldest FPV and CPV samples as reference sequences in each case. The nucleotide position is indicated at the top, and the character state of individual nucleotides is indicated below each sequence. Changes in amino acid sequence are characterized above the sequence, where appropriate. Synonymous changes are indicated by black boxes, and nonsynonymous changes are indicated by red boxes, while the location in the viral genome can be inferred by referring to the genome map shown below the sequences.
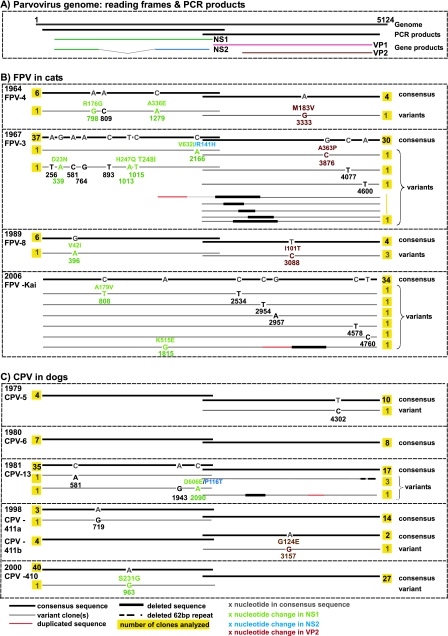
FIG. 3.
Intrahost diversity in FPV and CPV samples. (A) The gene regions covered by PCR amplification and a corresponding translation map of the parvovirus genome are indicated. (B and C) Divergent viral sequences detected in animals naturally infected with FPV (B) or CPV (C) are shown. The location of mutations in the parvovirus genome and the type of nucleotide substitution are indicated for each divergent sequence, the number of individual clones analyzed for each sample and the gene product affected by nonsynonymous mutations are indicated, and the year of isolation is identified. For comparison, in the case of the CPV-infected cat described previously by Battilani et al. (4), 10 out of 14 clones analyzed for the VP2 gene harbored one or more mutations each, which distinguished the clone from the consensus sequence.
Here, we analyze the DNA sequence diversity and population structures of CPV and FPV within samples collected during natural and experimental infections. We define both the sequence variation generated de novo and that which likely results from coinfection. FPV samples from various tissues or feces were collected over a 43-year period, with the oldest sample originating from 1963. The earliest CPV sample was collected in 1979, the second year of the pandemic in dogs. We inoculated cats with CPV collected from naturally infected dogs and analyzed the developing population structures of the virus in different feline organs, comparing the sequence diversity in these populations to that present in the inoculum.
MATERIALS AND METHODS
Virus samples.
The viruses examined are listed in Table 1. All samples were collected as original clinical specimens (feces or tissues) from infected animals and were stored at −80°C. Diluted samples were either added directly to PCRs without any prior preparation or purified using the QIAamp DNA Mini kit (Qiagen, Valencia, CA) according to the manufacturer's recommendations before amplification.
TABLE 1.
Characterization of FPV and CPV isolates from virus samples analyzed from natural infectionsa
| Virus | GenBank accession no. | Host | Yr of collection | Sample tissue | No. of clones
|
Rearranged genome region (nt) (VP2)
|
||
|---|---|---|---|---|---|---|---|---|
| NS1 | VP2 | Deleted | Inserted | |||||
| FPV-4.us.64 | EU659112 | Cat | 1964 | Spleen | 7 | 5 | ||
| FPV-3.us.67 | EU659111 | Cat | 1967 | Kidney | 39 | 38 | 1981-3010 | 1549-1980 |
| 2525-2889 | ||||||||
| 2710-3090 | ||||||||
| 2689-3336 | ||||||||
| 2843-3311 | ||||||||
| CPV-5.us.79 | EU659116 | Dog | 1979 | Spleen | 4 | 11 | ||
| CPV-6.us.80 | EU659117 | Dog | 1980 | Feces | 7 | 8 | ||
| CPV-13.us.81 | EU659118 | Dog | 1981 | Feces | 37 | 21 | 2383-3587 | 3702-3990 |
| FPV-8.us.89ab | EU659113 | Mountain lion | 1989 | Tissue | 7 | 7 | ||
| FPV-8.us.89bb | EU659114 | |||||||
| CPV-411.us.98ab | EU659120 | Dog | 1998 | Feces | 8 | 17 | ||
| CPV-411.us.98bb | EU659121 | |||||||
| CPV-410.us.100 | EU659119 | Dog | 2000 | Feces | 41 | 27 | ||
| FPV-kai.us.106 | EU659115 | Cat | 2006 | Small intestine | 40 | 40 | 3153-3420 | 2764-3154 |
The host species, year of isolation, and tissue type are indicated, and the numbers of clones analyzed for each gene fragment are shown. GenBank accession numbers for the respective consensus sequences are provided, and rearranged genomes are described, where applicable.
Two distinct virus sequences were isolated, referred to as sequences “a” and “b.”
PCR, cloning, and sequencing.
PCR analysis was performed using high-fidelity Phusion Hot Start DNA polymerase (New England Biolabs, Beverly, MA) and primer pairs that either flanked the region between nucleotides (nt) 118 and 4837 in the viral genome (compared to the CPV-2 reference sequence under GenBank accession number M38245.1) or amplified that region as two overlapping fragments (nt 118 to 2419 and nt 2033 to 4837 [reverse primer 1] or nt 2033 and 4680 [reverse primer 2] [all primer sequences are available from the authors upon request]). PCR products were purified by agarose gel purification using the QIAquick gel extraction kit (Qiagen) and were either prepared for cloning into the pSMART GC HK vector according to the manufacturer's recommendations (Lucigen Corporation, Middleton, WI) or used without further preparation for cloning into the pJET1.2 vector (Fermentas, Glen Burnie, MD) using the manufacturer's blunt-end cloning protocol. Inserts were sequenced using primer sets that covered the entire insert in both directions (primer sequences are available from the authors upon request).
qPCR analysis.
A TaqMan real-time PCR assay for the quantification of parvovirus genomes was established, targeting a conserved 60-nt region located between nt 1039 and 1114 of the CPV genome (quantitative PCR [qPCR] forward primer AAATGAAACCAGAAACCGTTGAA and qPCR reverse primer TCCCGCGCTTTGTTTCC). A TaqMan minor-groove binding probe (ABI) was designed to bind to the amplified region (TaqMan minor-groove binding probe ACAGTGACGACAGCAC). The linearized sequence of a CPV-2b genome, cloned into a plasmid, was used as an external standard for the quantification of copy numbers.
Sequence analysis.
Isolate consensus sequences were published previously (19) and are available under GenBank accession numbers EU659111 to EU659121. Mutations detected in the course of this study were compared to all publicly available FPV and CPV sequences covering the respective genome regions (alignments were identical to those published previously) (19).
Some nucleotide sequences contained deletions or insertions, which were further analyzed using BLAST (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). These sequences were aligned separately to the consensus sequence. Gene rearrangements were confirmed only if the exact boundaries of the deletion or duplication event could be inferred from the sequence information. To confirm that those rearranged sequences were present in the original samples before PCR amplification, we chose one of the gene rearrangements, detected in the FPV-kai.us.106 sample, and designed PCR primer pairs for which one primer each binds within the duplicated gene region (the sequence is available from the authors upon request).
Experimental cat infections.
All animal infections were approved by the Cornell University Animal Care and Use Committee. The original virus-containing specimens (not tissue culture passaged) were resuspended in phosphate-buffered saline (pH 7.2) and sterile filtered through a 0.22-μm filter prior to inoculation. The viral titers in CPV isolates CPV-13.us.81 and CPV-410.us.100 were determined as 50% tissue culture infective doses (TCID50) in Nordon Laboratories feline kidney (NLFK) cells as described previously (44), and the viral genome copy numbers were determined by qPCR. Groups of two ∼5-week-old parvovirus-seronegative kittens (Liberty Research, Waverly, NY) were inoculated through the oronasal route with 5 × 105 TCID50 of CPV-13.us.81 or with 3 × 105 TCID50 of CPV-410.us.100. Those inocula contained 5.5 × 1014 or 3.4 × 1014 viral copy numbers, respectively, as determined by qPCR. The kittens were monitored daily for clinical symptoms, and viral shedding was assessed by PCR analysis of rectal swabs. On days 0 and 6 postinoculation, serum samples were collected to confirm the absence of antiviral antibodies in hemagglutination inhibition tests as described elsewhere previously (34). On day 6 (for the CPV13 virus-challenged kittens) or 8 (for the CPV410 virus-challenged kittens) after inoculation, the kittens were euthanized, and thymus, bone marrow, and fecal samples were then collected and examined for viral DNA by PCR. PCR products (where these were generated) were cloned and sequenced as described above.
RESULTS
Overall, we obtained approximately 900,000 nt of sequence, including 190 clones covering the NS1/NS2 gene region and 172 clones covering the VP1/VP2 gene region (Table 1).
Analysis of natural FPV infections.
We analyzed viral genetic diversity in clinical FPV samples collected over a 43-year period (Fig. 1), representing a variety of host tissues (Table 1). Most mutations that distinguished isolate consensus sequences were synonymous (Fig. 2A). No mutations were detected in the NS1 carboxy terminus where NS2 is encoded in a different reading frame, and only 1 nonsynonymous mutation was observed among the 12 mutations in the VP2 protein gene (8.3%).
The population structures within individual FPV samples were very homogenous and comparable across samples (Fig. 3A). Eleven of 90 sequences (12.2%) that covered the capsid protein gene harbored a mutation, with 3 of those being nonsynonymous. The VP2 Ile 101-to-Thr change was present in three of seven sequences from the same sample. Six of 93 (6.5%) NS1-covering sequences harbored one or more mutations each, 64% of which were nonsynonymous. One mutation at nt 2166 resulted in nonsynonymous mutations in both the NS1 and C-terminal NS2 reading frames (Fig. 3A).
In most FPV samples, one mutation was detected every 4 × 10−5 to 6 × 10−5 nt, and the mutations were somewhat more frequently located in the NS1 than in the VP2 coding region (see Table 3). About half of the mutations detected in viral genomes from infected feline hosts can also be found in FPV GenBank sequences, such as 10 of 20 mutations in regions with sufficient sequence data available in GenBank. Of the 12 nonsynonymous mutations, 4 (33%) were also present in GenBank FPV sequences (Table 2), indicating a likely relatively high associated fitness of these mutations. Moreover, in the cases of FPV-8.us.89 and CPV-411.us.98, the possibility of coinfections is underlined by the presence of these respective mutations in the global FPV population.
TABLE 3.
Analysis of heterogeneity in the viral populationa
| Expt and sample | No. of mutations within animal | Nonsynonymous fraction | Average no. of mutations/nt | Mutations in NS1 and NS2
|
Mutations in VP1 and VP2
|
||
|---|---|---|---|---|---|---|---|
| No. | Fraction | No. | Fraction | ||||
| Disregarding mutations that distinguish putatively coinfecting strains | |||||||
| FPV-4.us.64 | 4 | 0.75 | 1.4 × 10−4 | 3 | 0.75 | 1 | 0.25 |
| FPV-3.us.67 | 11 | 0.45 | 5.8 × 10−5 | 8 | 0.73 | 3 | 0.27 |
| FPV-8.us.89a | 1 | 1.00 | 3.8 × 10−5 | 1 | 1.0 | ||
| FPV-8.us.89b | 0 | ||||||
| FPV-kai.us.06 | 7 | 0.29 | 3.7 × 10−5 | 2 | 0.29 | 5 | 0.71 |
| CPV-5.us.79 | 1 | 0 | 2.6 × 10−5 | 0 | 0 | 1 | 1.00 |
| CPV-6.us.80 | 0 | ||||||
| CPV-13.us.81 | 3 | 0.33 | 2.2 × 10−5 | 3 | 1.00 | 0 | 0 |
| CPV-411.us.98a | 1 | 0 | 2.3 × 10−5 | 1 | 1.00 | 0 | 0 |
| CPV-411.us.98b | 1 | 1.00 | 3.6 × 10−5 | 0 | 0 | 1 | 1.00 |
| CPV-410.us.00 | 1 | 1.00 | 6.1 × 10−6 | 1 | 1.00 | 0 | 0 |
| Kitten 1 | 18 | 0.56 | 1.8 × 10−4 | 17 | 0.94 | 1 | 0.06 |
| Kitten 2 | 0 | ||||||
| Accounting also for mutations that distinguish putatively coinfecting strains | |||||||
| FPV-8.us.89b | 4 | 1.00 | 1.2 × 10−4 | 1 | 0.25 | 3 | 0.75 |
| CPV-411.us98b | 20 | 0.3 | 3.2 × 10−4 | 7 | 0.35 | 13 | 0.65 |
| Comparison to data from previously published studies | |||||||
| CPV catc | 19 | 0.63 | 7.8 × 10−4 | ||||
The total number of mutations detected in each sample is indicated. In cases where coinfection was suspected (i.e., samples FPV-8.us.89 and CPV-411.us.98), the data were analyzed both by looking at the two viral populations separately and disregarding mutations distinguishing the potential subpopulations and by disregarding the potential of coinfection and counting all mutations detected within an infected animal. Comparable estimates were then calculated from data from a previously published study of heterogeneity in a CPV-infected cat (4). The number of mutations within an animal equals the total number of mutations detected, thus counting mutations that were present in more than one sequence multiple times. The average number of mutations/nucleotide equals the total number of mutations detected in the respective sample divided by the number of nucleotides analyzed.
Counting the total number of mutations that distinguish sequences recovered from the respective samples.
From data published in reference 4.
TABLE 2.
Characterization of within-host mutations detected in individual FPV or CPV sequencesa
| Virus | Virus type | Yr of isolation | nt position | Protein region | Amino acid change | Observed in nature | Functional implication of nonsynonymous change |
|---|---|---|---|---|---|---|---|
| FPV-4 | FPV | 1964 | 798 | NS1 | R176G | No | DNA bindingd |
| 809 | NS1 | —b | No | —b | |||
| 1279 | NS1 | A336E | No | Helicased | |||
| 3333 | VP2 | M183V | No | Unclear | |||
| FPV-3 | 1967 | 256 | NS1 | —b | —c | —b | |
| 339 | NS1 | D23N | Yes | DNA bindingd | |||
| 581 | NS1 | —b | Yes | —b | |||
| 764 | NS1 | —b | Yes | —b | |||
| 893 | NS1 | —b | Yes | —b | |||
| 1013 | NS1 | H247Q | Yes | DNA bindingd | |||
| 1015 | NS1 | T248I | Yes | DNA bindingd | |||
| 2166 | NS1/NS2 | V632I (NS1) R141H (NS2) | No | VP2 phosphorylationd | |||
| 3876 | VP2 | A363P | No | Helicased | |||
| 4077 | VP2 | —b | Yes | —b | |||
| 4600 | VP2 | —b | —c | —b | |||
| FPV-8 | FPV | 1989 | 396 | NS1 | V42L | No | DNA bindingd |
| 3088 | VP2 | I101T | Yes | TfR bindingf | |||
| FPV-Kai | FPV | 2006 | 808 | NS1 | A179V | No | DNA bindingd |
| 1815 | NS1 | K515E | No | VP2 phosphorylationd | |||
| 2534 | VP1 | —b | Yes | —b | |||
| 2954 | VP2 | —b | No | —b | |||
| 2957 | VP2 | —b | Yes | —b | |||
| 4578 | VP2 | —b | —c | —b | |||
| 4760 | VP2 | —b | —c | —b | |||
| CPV-5 | CPV-2 | 1979 | 4302 | VP2 | —b | No | Unclear |
| CPV-6 | CPV-2 | 1980 | —b | —b | —b | —b | —b |
| CPV-13 | CPV-2a | 581 | NS1 | —b | Yes | —b | |
| 1981 | 1943 | NS1 | —b | No | —b | ||
| 2090 | NS1/NS2 | D606E (NS1)/P116T (NS2) | No | Intracellular traffickingd | |||
| CPV-411ae | CPV-2b | 1998 | 719 | NS1 | —b | No | —b |
| CPV-411be | CPV-2b | 1998 | 3157 | VP2 | G124E | No | —b |
| CPV-410 | CPV-2b | 2000 | 963 | NS1 | S231G | No | Unclear |
| CPV-411 | CPV-2b | 1998 | 413 | NS1 | —b | Yes | —b |
| 1998 | 941 | NS1 | —b | Yes | —b | ||
| 1319 | NS1 | —b | Yes | —b | |||
| 1481 | NS1 | —b | Yes | —b | |||
| 1562 | NS1 | —b | Yes | —b | |||
| 2195 | NS1/NS2 | D151N (NS2) | Yes | VP2 phosphorylationd | |||
| 2247 | VP1 | —b | Yes | —b | |||
| 2358 | VP2 | —b | Yes | —b | |||
| 2846 | VP2 | —b | Yes | ||||
| 2951 | VP2 | —b | Yes | —b | |||
| 3157 | VP2 | G124E | No | Unclear | |||
| 3323 | VP2 | —b | Yes | —b | |||
| 3749 | VP2 | N321K | Yes | Possibly TfR binding | |||
| 3947 | VP2 | —b | Yes | —b | |||
| 4023 | VP2 | —b | Yes | —b | |||
| 4494 | VP2 | K570E | Yes | Unclear | |||
| 4510 | VP2 | —b | Yes | —b | |||
| 4526 | VP2 | —b | —c | —b |
Shown are the locations of single mutations detected in FPV or CPV samples. Nonsynonymous mutations are further characterized, and mutations present in other GenBank sequences are indicated by boldface type.
—, not applicable (synonymous change).
—, sequence coverage in GenBank was insufficient to assess the presence or absence from the population.
Functions represent those reported for the related minute virus of mice and have not been analyzed for FPV or CPV.
Since CPV-411.us.98 most likely contained two coinfecting virus strains, we analyzed the two virus populations separately and then compared the two consensus sequences present in sample CPV-411.
Transferrin receptor.
Finally, genomic rearrangements were detected within the capsid protein gene regions of FPV-3.us.67 and FPV-kai.us.106 (Table 1) (with the FPV-kai.us.106 clones spanning the complete viral coding region between nt 140 and 4815) and were verified as described above (data not shown).
Analysis of natural CPV infections.
Clinical CPV samples were isolated over a 22-year period, starting the year after CPV spread worldwide (Fig. 1). Most samples were in feces, except for CPV-5.us.79, which was a sample from the spleen (Table 1). The sequences were assigned to the previously defined CPV-2 (CPV-5.us.79 and CPV-6.us.80), CPV-2a (CPV-13.us.81), and CPV-2b (CPV-410.us. 00 and CPV-411.is.98) antigenic types (Fig. 1) (35, 37). Most mutations that distinguished the isolate consensus sequences clustered in the capsid protein gene (Fig. 2B). Eleven of 20 (55%) mutations in this gene were nonsynonymous, while genome-wide, 14 nonsynonymous mutations were found among the 30 mutations (46.7%). Three mutations were located in the carboxy-terminal region of NS1 where NS2 is encoded in a different reading frame, changing residue 641 of NS1 or residues 94 and 152 of NS2.
The CPV-411.us.98 sample, collected from a puppy infected during a shelter outbreak, harbored two clearly distinct viral populations (Fig. 3B). Both strains were of the CPV-2b type, and all but one of the distinguishing mutations were also detected in GenBank sequences (Table 2), indicating that this likely represented a coinfection by two different viruses. The sample also contained a single capsid protein gene clone that was of the CPV-2 type, possibly a remnant of vaccine virus. However, since the possibility of contamination in this case could not be ruled out, the sequence was excluded from further analyses.
In most CPV samples, one mutation was detected every 2 × 10−5 to 4 × 10−5 nt, and mutations again appeared to be somewhat more frequent in the NS1 than VP2 ORF (Table 3). Four of the 97 NS1 clones (4.1%) and two of the 83 VP2 clones (2.4%) harbored mutations from the consensus sequences, with individual clones harboring one or two mutations. Only one of the seven mutations detected within CPV-infected dogs (14.3%) was also detected in sequences from GenBank (Table 2). We found evidence of one gene duplication and deletion in the VP2 gene fragment of CPV-13.us.81 (Table 1), and 3 of 21 CPV-13.us.81 sequences differed in the frequencies of a repeated 62-nt sequence located in the 3′ end of the viral genome (Fig. 3B), a phenomenon described previously for CPVs and FPVs (36).
Analysis of experimental cat infections.
Only low levels of viral replication and little evidence of clinical disease were seen in the two kittens inoculated with CPV-13.us.81. Kitten 1 developed mild clinical signs on day 4 after inoculation and shed small amounts of parvovirus in the feces the following day, while kitten 2 showed mild clinical signs and viral shedding on day 6 postinoculation. Neither of the kittens inoculated with CPV-410.us.100 showed clinical signs, and no clear evidence of viral fecal shedding was detected by conventional PCR or qPCR (data not shown).
We analyzed a total of 29 NS1- and 22 VP2-spanning clones isolated from different tissues of the two CPV-13.us.81 inoculated animals (Fig. 4B). In general, all viral populations were highly homogeneous. Mutations were detected in the bone marrow (VP2 Asp528 to Val) and feces of kitten 1, with the later affecting the start codon of VP2. Two distinct sequences were recovered from the feces of kitten 1, with four of the eight clones being identical to the challenge virus (Fig. 3B). The other four clones harbored the same set of four mutations in the NS1 region, two synonymous changes in the NS1 carboxy-terminal sequence and nonsynonymous in the NS2 ORF (Thr94 to Ala and Met152 to Val) and two synonymous changes located in the NS1 amino terminus. All mutations except the one in the VP2 gene of kitten 1 were also present in GenBank sequences (Table 4). Gene rearrangements were detected in sequences from the thymus of kitten 1 (nt 2134 to 2875, 2474 to 3342, and 2676 to 3770), but such rearrangements were not detected in the challenge virus. Both forms of 62-nt repeat arrangements were present in the challenge virus and were also detected after experimental passage in cats.
TABLE 4.
Mutations and rearrangements detected in individual CPV sequences after experimental passage in catsa
| Area of isolation | Gene region | nt position | Amino acid change | Observed in nature | Functional implication |
|---|---|---|---|---|---|
| Feces | NS1 | 941 | — | Yes | — |
| Feces | NS1 | 1319 | — | Yes | — |
| Feces | NS1/NS2 | 2024 | T94A (NS2) | Yes | Intracellular traffickingb |
| Feces | NS1/NS2 | 2198 | M152V (NS2) | Yes | Interaction with members of the 14-3-3 protein familyb |
| Feces | VP2 | 2798 | Start codon | No | VP2 start codon |
| Bone marrow | NS1/NS2 | 1855 | D528V | Yes | VP2 phosphorylationb |
Samples from cat 1 were used. Single mutations in samples collected after experimental infection with CPV-13.us.81 are shown. Mutations also observed in GenBank sequences are indicated by boldface type. —, not applicable.
Functions represent comparable functions seen in the related minute virus of mice. The respective protein function has not been analyzed for FPV or CPV.
DISCUSSION
The emergence of CPV provides a valuable opportunity to examine the mechanisms of cross-species transmission, allowing a comparison of essentially the same viruses under endemic and emerging conditions. Here, we define the levels of variation among the viral sequences in different hosts, comparing the long-adapted FPV in cats, CPV in dogs at various times after its global spread in 1978, and CPV sequences during single experimental passages in cats. We compared the degree of sequence diversity in naturally infected dogs and cats, distinguishing between newly arising variation and heterogeneity due to coinfection, and analyzed the effect of host switching on the degree of sequence heterogeneity detected.
Intrahost sequence diversity.
In general, we observed low levels of sequence variation during natural infections by either FPV or CPV and also during the experimental cross-species transfer of CPV to cats. Only 6.3% of viral sequences cloned from natural infections harbored any mutations. This contrasts with most previous studies of intrahost population structure during parvovirus infections but has also been suggested by one other recent study of CPV in dogs (3). Battilani et al. analyzed individual virus genomes isolated from a single CPV-infected cat and detected high levels of sequence diversity within a 1,745-nt fragment of the VP2 gene (4), with 10 distinct sequences observed among 14 analyzed viral clones (71%). Two antigenically distinct CPV variants (CPV-2a and a variant with the VP2 D426E substitution, referred to as CPV-2c) were isolated from this single animal, indicating that at least part of the observed variation was likely due to superinfection rather than newly arising mutations.
Evidence for coinfections.
Notably, our study provided strong evidence for multiple infections. In particular, three of seven clones analyzed for FPV-8.us.89 contained the same change (VP2 residue Ile101 to Thr) in the capsid protein gene while being otherwise identical to the consensus sequence. This particular mutation is circulating in the FPV (Table 2) and CPV (19) populations, thereby making a coinfection event a plausible explanation. The CPV-411.us.98 isolate presented particularly strong evidence of coinfection. This sample contained two genetically distinct CPV-2b viruses (CPV-411a.us.98 and CPV-411b.us.98), which are distinguished by 6 and 12 mutations in the NS1 and VP1 genes, respectively. All but one of the mutations differentiating those two viral strains are present in GenBank sequences, again supporting the idea of coinfection since the mutations are clearly circulating in the global CPV population. If we assumed that coinfection was the source of some heterogeneity, the average number of mutations per nucleotide was in the range of 2 × 10−5 to 6 × 10−6 in all but one of the analyzed samples (the estimate for kitten 1 was excluded here, as the artificial host switching might impact the developing population structure, and the estimates might therefore not be comparable). Conversely, the estimates were considerably higher for the questionable samples if it was assumed that all variation arose by de novo mutation. These higher estimates were in the range estimated based on a previous report by Battilani et al. (3), so similar reasoning might explain the high diversity reported by those authors. Coinfections have been described for vaccine and field strains of CPV (13) and for several related parvovirus family members, including human parvovirus B19 (5). Coinfections with multiple parvovirus strains may thus occur frequently, potentially facilitating recombination (41).
Gene rearrangements.
We detected several gene duplication and deletion events among the FPV and CPV genomes examined. Such rearranged sequences have long been recognized among the parvoviruses (1, 10, 21) and likely result from template switching of the polymerase during replication. This process, which is also thought to be responsible for nonhomologous and homologous recombination among RNA viruses (23, 28), may be facilitated by the complex secondary structures of the viral ssDNA genome. Recombination among CPV genomes was suggested to explain the origins of genomes containing various combinations of mutations after extended tissue culture passage (2).
Effect of host species, organ, or sampling time after CPV emergence.
The viral population structures in all isolates were generally characterized by low levels of sequence diversity regardless of their host species, organ, time of isolation, or specific viral strain. On average, one mutation was detected every 104 to 105 nt, which is comparable to the annual substitution rate estimates of these viruses obtained on a population level (19, 43). Since variant CPV sequences arise very readily during tissue culture passage (2), strong purifying selection appears to quickly purge most arising mutants during natural infections, resulting in the low degree of sequence diversity within infected hosts. We detected no observable differences in intrahost population structure between CPV isolates collected during the first wave of spread (from 1979 and 1980) and those collected after the virus had been circulating in dogs for longer times, suggesting that the dynamics of intrahost mutation and selection did not change markedly during this time period.
To better determine the effects of a single cross-species transmission event on the virus, we examined experimental CPV infections of cats, which revealed only limited viral replication in the susceptible cats. The CPV sequences recovered from the cats showed low levels of heterogeneity, but parts of the viral capsid protein gene were deleted in some cases, which is indicative of defective genomes. In addition, one virus isolated from the feces of cat 1 harbored a likely lethal mutation that altered the start codon of VP2. Mutations were detected in the NS1 and NS2 regions of viruses isolated from bone marrow and feces, with approximately 50% of viruses in the feces of cat 1 harboring the same set of four mutations. Two of the changes (at residues 94 and 152 of NS2) mapped to the gene region where NS2 is derived from a different reading frame overlapping the NS1 reading frame and were identified as being potentially positively selected at a population level (19). Since these changes represent two of four apparently linked mutations observed after artificial host switching, a role of some or all of these residues in host adaptation is likely. Little is known about the function of NS2 in CPV, and NS2 knockout mutants showed no obvious differences in replication in cell culture or dogs (46). In rodent parvovirus strains MVM and LuIII, NS2 appears to be required for efficient translation, capsid assembly, and nuclear transport. In these viruses, host-specific effects modulate the functions of NS2 (9, 11, 12, 17, 25, 30). Interestingly, upon infection of severe combined immunodeficient (SCID) mice with MVM in the presence of polyclonal anticapsid antibodies, the viral population harbored nonsynonymous changes in the NS2 C terminus, which likely affected CRM1 binding (27). In this case, nonsynonymous mutations were absent from the capsid protein genes (27), although such mutations arose readily during infections in the absence of polyclonal antibodies (26). The role of NS2 in the host adaptation of CPV therefore merits further study.
Distribution of mutations in the genomes.
The majority of mutations involved in the emergence of the CPV-2 cluster are in the capsid protein ORF, and this region has continued to evolve more rapidly than the nonstructural ORF, at least in CPV (19). Among the FPVs, on the contrary, marked differences in mutation accumulation levels among the genome regions have not been observed (19). Interestingly, the mutations that we detected within individual infected animals showed no clear clustering in any specific genome region. The scarcity of mutations in the capsid protein region after experimental cross-species transmission to cats is also surprising and highlights the weak effect of positive selection within individual hosts.
Acknowledgments
We thank Virginia Scarpino, Wendy Weichert, and Melanie Ho for technical support.
This work was supported by National Institutes of Health grants GM080533 to E.C.H. and AI028385 to C.R.P. K.H. is supported by a graduate assistantship from the College of Veterinary Medicine at Cornell.
Footnotes
Published ahead of print on 3 September 2008.
REFERENCES
- 1.Allen, J. M., D. J. Debelak, T. C. Reynolds, and A. D. Miller. 1997. Identification and elimination of replication-competent adeno-associated virus (AAV) that can arise by nonhomologous recombination during AAV vector production. J. Virol. 716816-6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badgett, M. R., A. Auer, L. E. Carmichael, C. R. Parrish, and J. J. Bull. 2002. Evolutionary dynamics of viral attenuation. J. Virol. 7610524-10529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Battilani, M., L. Gallina, F. Vaccari, and L. Morganti. 2007. Co-infection with multiple variants of canine parvovirus type 2 (CPV-2). Vet. Res. Commun. 31(Suppl. 1)209-212. [DOI] [PubMed] [Google Scholar]
- 4.Battilani, M., A. Scagliarini, S. Ciulli, L. Morganti, and S. Prosperi. 2006. High genetic diversity of the VP2 gene of a canine parvovirus strain detected in a domestic cat. Virology 35222-26. [DOI] [PubMed] [Google Scholar]
- 5.Candotti, D., N. Etiz, A. Parsyan, and J. P. Allain. 2004. Identification and characterization of persistent human erythrovirus infection in blood donor samples. J. Virol. 7812169-12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carman, P. S., and R. C. Povey. 1985. Pathogenesis of canine parvovirus-2 in dogs—histopathology and antigen identification in tissues. Res. Vet. Sci. 38141-150. [PubMed] [Google Scholar]
- 7.Carman, P. S., and R. C. Povey. 1985. Pathogenesis of canine parvovirus-2 in dogs: hematology, serology and virus recovery. Res. Vet. Sci. 38134-140. [PubMed] [Google Scholar]
- 8.Chang, S. F., J. Y. Sgro, and C. R. Parrish. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J. Virol. 666858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, E. Y., A. E. Newman, L. Burger, and D. Pintel. 2005. Replication of minute virus of mice DNA is critically dependent on accumulated levels of NS2. J. Virol. 7912375-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clement, N., B. Avalosse, K. El Bakkouri, T. Velu, and A. Brandenburger. 2001. Cloning and sequencing of defective particles derived from the autonomous parvovirus minute virus of mice for the construction of vectors with minimal cis-acting sequences. J. Virol. 751284-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotmore, S. F., A. M. D'Abramo, Jr., L. F. Carbonell, J. Bratton, and P. Tattersall. 1997. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology 231267-280. [DOI] [PubMed] [Google Scholar]
- 12.D'Abramo, A. M., Jr., A. A. Ali, F. Wang, S. F. Cotmore, and P. Tattersall. 2005. Host range mutants of minute virus of mice with a single VP2 amino acid change require additional silent mutations that regulate NS2 accumulation. Virology 340143-154. [DOI] [PubMed] [Google Scholar]
- 13.Decaro, N., C. Desario, G. Elia, M. Campolo, A. Lorusso, V. Mari, V. Martella, and C. Buonavoglia. 2007. Occurrence of severe gastroenteritis in pups after canine parvovirus vaccine administration: a clinical and laboratory diagnostic dilemma. Vaccine 251161-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drake, J. W. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 887160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drake, J. W. 1999. The distribution of rates of spontaneous mutation over viruses, prokaryotes, and eukaryotes. Ann. N. Y. Acad. Sci. 870100-107. [DOI] [PubMed] [Google Scholar]
- 16.Duffy, S., L. A. Shackelton, and E. C. Holmes. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9267-276. [DOI] [PubMed] [Google Scholar]
- 17.Eichwald, V., L. Daeffler, M. Klein, J. Rommelaere, and N. Salome. 2002. The NS2 proteins of parvovirus minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. J. Virol. 7610307-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gamoh, K., M. Senda, Y. Inoue, and O. Itoh. 2005. Efficacy of an inactivated feline panleucopenia virus vaccine against a canine parvovirus isolated from a domestic cat. Vet. Rec. 157285-287. [DOI] [PubMed] [Google Scholar]
- 19.Hoelzer, K., L. A. Shackelton, C. R. Parrish, and E. C. Holmes. 2008. Phylogenetic analysis reveals the emergence, evolution and dispersal of carnivore parvoviruses. J. Gen. Virol. 892280-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoelzer, K., and C. R. Parrish. 2008. Evolution and variation of the parvoviruses. In E. Domingo, J. Holland, and C. R. Parrish (ed.), Origin and evolution of viruses, 2nd ed. Elsevier, London, United Kingdom.
- 21.Hogan, A., and E. A. Faust. 1986. Nonhomologous recombination in the parvovirus chromosome: role for a CTATTTCT motif. Mol. Cell. Biol. 63005-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hueffer, K., L. Govindasamy, M. Agbandje-McKenna, and C. R. Parrish. 2003. Combinations of two capsid regions controlling canine host range determine canine transferrin receptor binding by canine and feline parvoviruses. J. Virol. 7710099-10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai, M. M. C. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 5661-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemey, P., A. Rambaut, and O. G. Pybus. 2006. HIV evolutionary dynamics within and among hosts. AIDS Rev. 8125-140. [PubMed] [Google Scholar]
- 25.Li, X., and S. L. Rhode. 1991. Nonstructural protein NS2 of parvovirus H-1 is required for efficient viral protein synthesis and virus production in rat cells in vivo and in vitro. Virology 184117-130. [DOI] [PubMed] [Google Scholar]
- 26.Lopez-Bueno, A., M. G. Mateu, and J. M. Almendral. 2003. High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host. J. Virol. 772701-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Bueno, A., N. Valle, J. M. Gallego, J. Perez, and J. M. Almendral. 2004. Enhanced cytoplasmic sequestration of the nuclear export receptor CRM1 by NS2 mutations developed in the host regulates parvovirus fitness. J. Virol. 7810674-10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, H., R. Xu, H. Cheng, H. S. Kuo, M. During, and R. H. Fang. 2003. Gene transfer into human keloid tissue with adeno-associated virus vector. J. Trauma 54569-573. [DOI] [PubMed] [Google Scholar]
- 29.Meunier, P. C., B. J. Cooper, M. J. G. Appel, M. E. Lanieu, and D. O. Slauson. 1985. Pathogenesis of canine parvovirus enteritis: sequential virus distribution and passive immunization studies. Vet. Pathol. 22617-624. [DOI] [PubMed] [Google Scholar]
- 30.Naeger, L. K., N. Salome, and D. J. Pintel. 1993. NS2 is required for efficient translation of viral mRNA in minute virus of mice-infected murine cells. J. Virol. 671034-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura, M., Y. Tohya, T. Miyazawa, M. Mochizuki, H. T. Phung, N. H. Nguyen, L. M. Huynh, L. T. Nguyen, P. N. Nguyen, P. V. Nguyen, N. P. Nguyen, and H. Akashi. 2004. A novel antigenic variant of canine parvovirus from a Vietnamese dog. Arch. Virol. 1492261-2269. [DOI] [PubMed] [Google Scholar]
- 32.Parrish, C. R. 1999. Host range relationships and the evolution of canine parvovirus. Vet. Microbiol. 6929-40. [DOI] [PubMed] [Google Scholar]
- 33.Parrish, C. R. 1995. Pathogenesis of feline panleukopenia virus and canine parvovirus. Baillieres Clin. Haematol. 857-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parrish, C. R., and L. E. Carmichael. 1983. Antigenic structure and variation of canine parvovirus, feline panleukopenia virus, and mink enteritis virus. Virology 129401-414. [DOI] [PubMed] [Google Scholar]
- 35.Parrish, C. R., C. F. Aquadro, M. L. Strassheim, J. F. Evermann, J. Y. Sgro, and H. O. Mohammed. 1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 656544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parrish, C. R., C. F. Aquadro, and L. E. Carmichael. 1988. Canine host range and a specific epitope map along with variant sequences in the capsid protein gene of canine parvovirus and related feline, mink and raccoon parvoviruses. Virology 166293-307. [DOI] [PubMed] [Google Scholar]
- 37.Parrish, C. R., P. H. O'Connell, J. F. Evermann, and L. E. Carmichael. 1985. Natural variation of canine parvovirus. Science 2301046-1048. [DOI] [PubMed] [Google Scholar]
- 38.Pereira, C. A., E. S. Leal, and E. L. Durigon. 2007. Selective regimen shift and demographic growth increase associated with the emergence of high-fitness variants of canine parvovirus. Infect. Genet. Evol. 7399-409. [DOI] [PubMed] [Google Scholar]
- 39.Raney, J. L., R. R. Delongchamp, and C. R. Valentine. 2004. Spontaneous mutant frequency and mutation spectrum for gene A of phiX174 grown in E. coli. Environ. Mol. Mutagen. 44119-127. [DOI] [PubMed] [Google Scholar]
- 40.Reed, A. P., E. V. Jones, and T. J. Miller. 1988. Nucleotide sequence and genome organization of canine parvovirus. J. Virol. 62266-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shackelton, L. A., K. Hoelzer, C. R. Parrish, and E. C. Holmes. 2007. Comparative analysis reveals frequent recombination in the parvoviruses. J. Gen. Virol. 883294-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shackelton, L. A., C. R. Parrish, U. Truyen, and E. C. Holmes. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA. 102379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Truyen, U., M. Agbandje, and C. R. Parrish. 1994. Characterization of the feline host range and a specific epitope of feline panleukopenia virus. Virology 200494-503. [DOI] [PubMed] [Google Scholar]
- 44.Truyen, U., and C. R. Parrish. 1992. Canine and feline host ranges of canine parvovirus and feline panleukopenia virus: distinct host cell tropisms of each virus in vitro and in vivo. J. Virol. 665399-5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Truyen, U. W. E., J. F. Evermann, E. Vieler, and C. R. Parrish. 1996. Evolution of canine parvovirus involved loss and gain of feline host range. Virology 215186-189. [DOI] [PubMed] [Google Scholar]
- 46.Wang, D., W. Yuan, I. Davis, and C. R. Parrish. 1998. Nonstructural protein-2 and the replication of canine parvovirus. Virology 240273-281. [DOI] [PubMed] [Google Scholar]
- 47.Yamaguchi, Y., and T. Gojobori. 1997. Evolutionary mechanisms and population dynamics of the third variable envelope region of HIV within single hosts. Proc. Natl. Acad. Sci. USA 941264-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]