Abstract
Repertoire composition, quantity, and qualitative functional ability are the parameters that define virus-specific T-cell responses and are linked with their potential to control infection. We took advantage of the segregation of different hepatitis B virus (HBV) genotypes in geographically and genetically distinct host populations to directly analyze the impact that host and virus variables exert on these virus-specific T-cell parameters. T-cell responses against the entire HBV proteome were analyzed in a total of 109 HBV-infected subjects of distinct ethnicities (47 of Chinese origin and 62 of Caucasian origin). We demonstrate that HBV-specific T-cell quantity is determined by the virological and clinical profiles of the patients, which outweigh any influence of race or viral diversity. In contrast, HBV-specific T-cell repertoires are divergent in the two ethnic groups, with T-cell epitopes frequently found in Caucasian patients seldom detected in Chinese patients. In conclusion, we provide a direct biological evaluation of the impact that host and virus variables exert on virus-specific T-cell responses. The discordance between HBV-specific CD8 T-cell repertoires present in Caucasian and Chinese subjects shows the ability of HLA micropolymorphisms to diversify T-cell responses and has implications for the rational development of therapeutic and prophylactic vaccines for worldwide use.
Virus-specific CD8+ T cells recognize virus-encoded peptides associated with major histocompatibility complex (MHC) class I molecules displayed on the surfaces of the infected cells. Virally infected cells can produce thousands of potentially immunogenic peptides, but CD8+ T cells are usually directed against only a few peptides, and CD8+ T cells specific for different viral determinants can possess different antiviral activities (47). Information regarding virus-specific T-cell repertoires and the potential antiviral efficacies of CD8+ T cells with differing antigen specificities is essential to understand viral pathogenesis and develop vaccines. Such information is limited in the great majority of viral and bacterial infections due to cumbersome methods that are required for the detection and characterization of new MHC class I-restricted epitopes (47). In addition, the identification of the T-cell repertoire against viruses infecting different ethnic populations with distinct HLA class I alleles and haplotype frequencies is particularly complex because different ethnic groups are often infected by different viral strains, which are likely to have coevolved in these populations (11, 16, 29).
The influence that virus heterogeneity and the distinct HLA profiles of the infected subjects has on the repertoire and hierarchy of T-cell responses is difficult to predict. The existence of T-cell responses against conserved regions of different virus strains (13, 46) and the reported degeneracy in HLA-peptide binding, with identical peptides able to bind multiple HLA class I types (6, 14, 35, 37, 42), support the idea of a substantial overlap between the virus-specific T-cell repertoires of subjects of different ethnicities expressing closely related but distinct HLA class I molecules. On the other hand, viral heterogeneity might affect the generation of certain epitopes as strain-specific variations within the epitopes (3) or, in flanking regions, might impair their processing and presentation (23, 34). Even subtle differences in closely conserved HLA class I molecules (28, 37) may severely affect the presentation of specific epitopes (2, 40) or change their conformation (44) sufficiently so that individuals of different ethnicities may focus the response toward different T-cell epitopes.
Given the global distribution of hepatitis B virus (HBV), understanding the commonality or divergence of virus-specific T-cell responses present in HBV-infected patients with different ethnicities is necessary. HBV, a strictly hepatotropic virus, induces a functionally efficient, multispecific CD8+ T-cell response in subjects who resolve the infection, while HBV-specific CD8+ T cells are not present or functionally impaired in chronically infected patients (5). A comprehensive knowledge of HBV-specific CD8+ T-cell specificities is lacking, and with rare exceptions (7, 9, 43), CD8+ T-cell responses have been analyzed using preselected peptides able to bind to common HLA class I molecules (HLA-A2, -A3, -A24, -A11, and -B7) (19, 20, 26, 31, 32, 38, 43, 45). Attempts to define immunodominant regions in the HBV proteome were based on the use of HLA-A2-restricted epitopes (45) and on samples from HBV genotype A (HBVgenA)- or HBVgenD-infected individuals of Caucasian decent. However, 75% of the population of chronically infected patients live in Asia (27), and Asian patients are infected, mostly at birth, by HBVgenB or HBVgenC, which differ by nearly 8% in amino acid composition compared to genotypes A and D (21).
The HLA class I profiles of the two populations differ not only in the frequency of the major HLA class I alleles (i.e., HLA-A11 is present in 51.7% of Chinese and 14% of Caucasians; HLA-B40 is present in 31.5% of Chinese and 14.7% of Caucasians [28]) but are also characterized by substantial differences in allele subtypes. The HLA-A2 molecule, present in nearly 50% of both Caucasians and Chinese, is subdivided into HLA-A2 subtypes, which are differentially expressed in the two ethnic groups (22). More than 95% of HLA-A2+ Caucasians are HLA-A0201+, whereas subtypes HLA-A0203, -A0206, and -A0207 are, respectively, present in 23%, 10%, and 45% of HLA-A2+ individuals of Chinese origin (22). Therefore, we performed the first direct comprehensive analysis of HBV-specific T-cell responses present in patients of different ethnicities (Chinese versus Caucasian) infected by different HBV genotypes (HBVgenB versus HBVgenD) to understand whether and, if so, to what degree host and virus variables influence the virus-specific T-cell response.
MATERIALS AND METHODS
Patient selection.
A total of 76 HBV-infected patients of Chinese Han origin and 78 Caucasians of European descent were enrolled at the Unit of Infectious Diseases and Hepatology of the Azienda Ospedaliero-Universitaria of Parma, Italy; the University Hospital S. Orsola-Malpighi of Bologna, Italy; or the National University of Singapore Hospital. Twenty patients (8 Chinese and 12 Caucasians) had clinical, biochemical, and virological evidence of acute HBV infection (alanine aminotransferase [ALT] levels of >10 times the upper limit of normal, detection of HBsAg and serum anti-HBc immunoglobulin M [IgM], and HBsAg clearance within 2 months from the clinical onset of hepatitis). A total of 64 HBV-infected patients of Chinese origin and 66 of Caucasian origin displayed clinical, biochemical, and virological evidence of chronic HBV infection (HBsAg and anti-HBc positive for at least 6 months) and displayed various ALT and HBV DNA levels. All patients were not undergoing antiviral or immunomodulatory treatment during the study and 6 months before enrollment, and all tested negative for HCV, HDV, and human immunodeficiency virus type 1 (HIV-1).
HBV genotype characterization was performed on all patients enrolled. Chinese chronic HBV patients infected with HBVgenC (n = 29) and 7 undetermined patients were excluded from HBV-specific T-cell analysis, while 32 Chinese chronic HBVgenB patients as well as 8 Chinese acute HBVgenB patients were selected for HBV-specific T-cell analysis. Sixteen Caucasian HBVgenA patients were also excluded and 62 HBVgenD-infected Caucasians were selected for further analysis. HLA-A2 subtypes of HLA-A2 patients (selected by a low-resolution genetic approach) were determined by high-resolution sequencing of the A2 locus (direct sequencing of the alpha 1 and alpha 2 chains).
This study was approved by the ethical committees of the Azienda Ospedaliero-Universitaria of Parma, the University Hospital of Bologna, and the National University of Singapore Hospital, and all subjects gave written informed consent.
Virological assessment.
HBsAg, HBeAg, anti-HBs, anti-HBc IgG and IgM, anti-HBe, anti-HDV, anti-HCV, and anti-HIV were determined by commercial enzyme immunoassay kits (Abbott Labs, Abbott Park, IL; Ortho Clinical Diagnostic, Johnson & Johnson, Raritan, NJ; DiaSorin, Vercelli, Italy). Serum HBV DNA was quantified by PCR (Cobas Amplicor test; Roche Diagnostics, Basel, Switzerland). HBV genotyping was performed by restriction fragment length polymorphism analysis of a pre-S amplicon previously described by Lindh et al. (25).
Isolation of PBMC and in vitro expansion of HBV-specific CD8+ cells.
Peripheral blood mononuclear cells (PBMC) were isolated from fresh heparinized blood by Ficoll-Hypaque density gradient centrifugation and resuspended in AIM-V medium (Invitrogen, Carlsbad, CA) with 2% pooled human AB serum. For the in vitro assays, the cells were used either directly ex vivo or after a 10-day antigen-specific in vitro stimulation. For the latter, 20% of the PBMC was first stimulated with 10 μg/ml of all the overlapping peptides from the respective HBV genotypes for 1 h at 37°C, then washed at 3.0 × 106 cells/ml before coculturing with the remaining PBMC in AIM-V medium with 2% pooled human AB serum supplemented with interleukin-2 (IL-2; R&D Systems, Abingdon, United Kingdom) (20 IU/ml) and seeded at 1 ml/well in 24-well plates. The immunological assays were performed on day 10 of the expansion.
Synthetic peptides and antibodies.
Two panels of 313 15-mer peptides overlapping by 10 residues were used to test HBV-specific T-cell responses. The peptides covered the overall sequence of HBVgenD (GenBank accession number AF121241) and HBVgenB (GenBank accession number AF121243) and were purchased from Chiron Mimotopes (Victoria, Australia) or synthesized at the peptide synthesis facility of Massachusetts General Hospital using 9-fluorenylmethoxy carbonyl chemistry. The purity of the peptides was above 80%, and their composition was confirmed by mass spectrometry analysis. The designed peptides presented at least 95% similarity with those encoded by HBV genomes sequenced from five Chinese and five Caucasian patients studied. The 15-mer core and X peptides were pooled in a 9 by 8 matrix, containing eight or nine peptides/pool, respectively, using a concept similar to what was previously described for HIV (1). Envelope peptides were pooled in a 9 by 9 matrix containing nine peptides/pool, while polymerase peptides were pooled in a 14 by 12 matrix containing 12 or 14 peptides/pool, respectively. All peptides were first diluted at 40 mg/ml in dimethyl sulfoxide and then further diluted in RPMI medium at a working dilution (between 1 mg/ml and 1 ng/ml).
Optimally defined HLA-A2-restricted cytotoxic T lymphocyte (CTL) epitopes (Core18-27, Env183-91, Env335-43, Env338-47, Env370-79, and Pol455-63 [see Table 1]) of HBV genotypes A, B, C, and D were purchased from Proimmune (Oxford, United Kingdom) and from GenScript (Piscataway, NJ). The peptide sequences were based on genotype-specific sequences of 24 GenBank entries (6 HBVgenA, 8 HBVgenB, 6 HBVgenC, and 4 HBVgenD). Furthermore, a viral amino acid sequence analysis of the Core18-27 and Env183-91 regions of the Chinese and Caucasian HLA-A2+ patients studied confirmed the genotype-specific sequence of the infecting viral strain. Anti-CD8 (phycoerythrin [PE]-Cy7), anti-CD3 (peridinin chlorophyll protein-Cy5.5), and anti-CD107a (fluorescein isothiocyanate) antibodies were purchased from Becton Dickinson Pharmingen (San Jose, CA). Anti-gamma interferon (anti-IFN-γ; PE) was purchased from R&D Systems (Minneapolis, MN).
TABLE 1.
Tabulated summary of CD8+ T-cell responses against known HLA-A2-restricted epitopes in HLA-A2+ Chinese and Caucasian patientsa
| Peptide | Amino acid sequence
|
No. of positive patients/total no. with:
|
||||||
|---|---|---|---|---|---|---|---|---|
| Acute/resolved disease
|
Chronic disease
|
|||||||
| <107 HBV DNA copies
|
>107 HBV DNA copies
|
|||||||
| Genotype B | Genotype D | Chinese | Caucasian | Chinese | Caucasian | Chinese | Caucasian | |
| Envelope | ||||||||
| Env183-191b | FLLTKILTI | FLLTRILTI | 0/5 | 7/8 | 2/8 | 7/8 | 0/6 | 2/9 |
| Env335-343b | WLSLLVPFV | WLSLLVPFV | 1/5 | 4/6 | 0/8 | 1/8 | 0/6 | 0/9 |
| Env338-347 | LLVPFVQWFV | LLVPFVQWFV | 2/5 | 0/6 | 1/8 | 1/8 | 0/6 | 1/9 |
| Env348-357 | GLSPTVWLSV | GLSPTVWLSV | 1/5 | 4/7 | 0/8 | 3/8 | 0/6 | 1/9 |
| Polymerase | ||||||||
| Pol455-463b | GLSRYVARL | GLSRYVARL | 1/5 | 5/7 | 0/8 | 1/8 | 0/6 | 0/9 |
| Core | ||||||||
| Core18-27b | FLPSDFFPSI | FLPSDFFPSV | 2/5 | 7/8 | 1/8 | 6/8 | 0/6 | 0/9 |
PBMC from HLA-A2+ Caucasian and Chinese acute or chronic patients (segregated by HBV DNA levels) were expanded in vitro for 10 days and tested against known HLA-A2-restricted HBV epitopes. The sequences of the Core18-27 and Env183-91 peptides differ by one amino acid (as indicated in bold) between the B and D genotypes.
HLA-A2 supertype binding capacity.
Intracellular cytokine staining (ICS) and degranulation assays.
In vitro-expanded PBMC were incubated in medium alone (control) or with viral peptides (5 μg/ml) for 5 h in the presence of brefeldin A (10 μg/ml). After a washing, the cells were stained with anti-CD8 PE-Cy7 and anti-CD3 peridinin chlorophyll protein-Cy5.5 monoclonal antibody (MAb) for 30 min at 4°C and then fixed and permeabilized using Cytofix/Cytoperm fixation/permeabilization solution (BD Biosciences, San Jose, CA), according to the manufacturer's instructions. The cells were stained with anti-IFN-γ PE for 30 min on ice, washed, and analyzed by flow cytometry. To assess degranulation activity, CD107a PE antibody (BD Pharmingen, San Diego, CA) was added to all wells at the beginning of the 5-h incubation with T cells. Following the incubation, the cells were washed and labeled with anti-CD8 PE-Cy7.
CTL clones and EBV-transformed B-cell lines.
HBV Core18-27-specific CD8+ T-cell clones were generated from HLA-A2+ HBV patients with acute hepatitis B as previously described (17). Epstein-Barr virus (EBV) B-cell lines with known HLA-A2 subtypes (kindly provided by Chan Soh Ha, Department of Microbiology, National University of Singapore) were grown and maintained in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum, 20 mM HEPES, 0.5 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin, MeM amino acids with l-glutamine, MeM nonessential amino acids (Invitrogen, Carlsbad, CA), and 5 μg/ml Plasmocin (InvivoGen, San Diego, CA) to prevent mycoplasma contamination.
IFN-γ ELISPOT assay.
IFN-γ enzyme-linked immunospot (ELISPOT) assays were performed as previously described (7) using a panel of 313 overlapping peptides covering the entire peptide sequence of HBVgenB or HBVgenD pooled in the described mixtures and used in patients infected with the respective HBV genotype. HBV-specific T-cell responses were analyzed in IFN-γ ELISPOT assays either ex vivo using fresh or frozen PBMC or after short-term peptide-specific polyclonal T-cell expansion (10 days). Briefly, 96-well plates (Multiscreen-HTS; Millipore, Billerica, MA) were coated overnight at 4°C as recommended by the manufacturer with 5 μg/ml capture mouse anti-human IFN-γ MAb (1DIK; Mabtech, Sweden). The plates were then washed five times with phosphate-buffered saline and blocked with AIM-V supplemented with 10% heat-inactivated fetal calf serum for 30 min at room temperature. A total of 1 × 105 PBMC or 5 × 104 cells from short-term polyclonal T-cell lines were seeded per well, in duplicate for each individual peptide mixture. The plates were incubated for 18 h at 37°C in the presence or absence of peptides (at a final concentration of 5 μg/ml). After five washes with phosphate-buffered saline, 100 μl of 0.5 μg/ml biotinylated anti-human IFN-γ MAb (7B6-1; Mabtech, Sweden) was added; plates were incubated for 2 h at room temperature and thereafter washed five times, 100 μl of streptavidin-alkaline phosphatase (1:2,000 dilution) (Mabtech, Sweden) was added to each well for 1 h at room temperature, and plates were incubated in the dark. The plates were again washed five times, and 50 μl of alkaline phosphatase substrate (5-bromo-4-chloro-3-indolyl phosphate-nitro blue tetrazolium chloride [BCIP-NBT]; KPL, Gaithersburg, MD) was added. After 10 to 15 min, the colorimetric reaction was stopped by washing with distilled water. The plates were air-dried, and spots were counted using an automated ELISPOT reader (ImmunoSpot; CTL, Cleveland, OH). The number of IFN-γ-producing cells was expressed in spot-forming units (SFU) per 1 × 105 cells. The number of specific IFN-γ-secreting cells was calculated by subtracting the nonstimulated control value from the stimulated sample. Positive controls consisted of PBMC stimulated with staphylococcal enterotoxin B (SEB) or phytohemagglutinin. In the direct ex vivo assays, a well was considered positive when the number of SFU was more than 5 and at least three times above the mean of the unstimulated control wells (3 wells/patient). The responses to peptide mixtures were also analyzed directly ex vivo in nine healthy subjects, and only a single individual showed the presence of a positive response. The positivity criteria for in vitro ELISPOT assays is less stringent, including wells that have SFU of more than 5 and at least two times above the mean of the unstimulated control wells. However, ICS was applied to every positive sample to reconfirm the response and to determine the T-cell subset (CD8 or CD4) responsible for IFN-γ production.
Image analysis.
A series 3B ImmunoSpot image analyzer (Cellular Technology) specifically designed for the ELISPOT assay was used. The digitized images were analyzed for the presence of areas in which the color density exceeded the background by an amount set on the basis of the comparison of wells with peptide and wells without peptide. After background and noise subtraction, custom software is used to analyze spot morphology for circularity and density distribution to identify and separate touching and overlapping spots. Objects that meet these criteria are recognized as spots and counted. The measurement of the mean spot size is a built-in function of the software.
Statistical analysis.
An unpaired t test was used in two instances: (i) to determine the significance of the difference in the mean percentage of positive mixtures between acute and chronic patients and (ii) to determine the significance of the difference in the ELISPOT assay-derived mean spot sizes between acute and chronic patients stimulated with HBV peptides or SEB/phytohemagglutinin. Differences with a P value less than 0.05 were considered statistically significant.
RESULTS
Comprehensive analysis of HBV-specific T-cell responses in HBVgenB- and HBVgenD-infected patients.
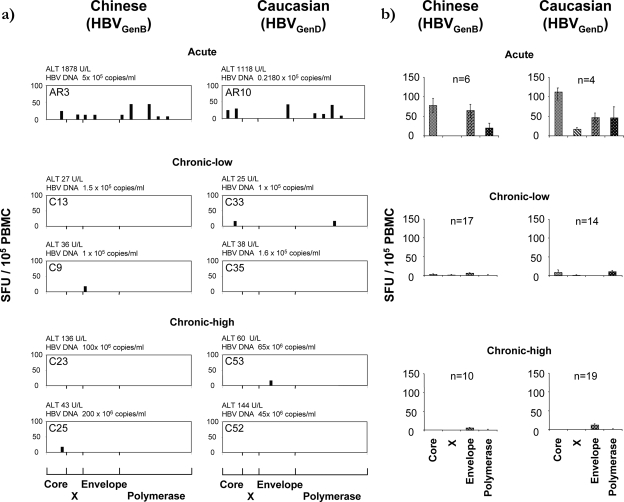
To identify similarities/differences in the breadth and magnitude of HBV-specific T-cell responses between Caucasian and Chinese patients, HBV-specific IFN-γ-producing T cells were tested by direct ex vivo ELISPOT assays in 34 patients of Chinese origin (6 acute and 27 chronic HBVgenB infected) and in 37 Caucasian subjects (4 acute and 33 chronic HBVgenD infected) (the full clinical profiles of all patients tested are shown in Fig. S1 in the supplemental material). PBMC of Chinese patients were stimulated with the HBVgenB peptide panel, while Caucasian patients were stimulated with HBVgenD peptides. Consistent with previous data, obtained mainly for Caucasian patients (7, 9), HBV-specific T-cell responses were detected ex vivo only in patients with acute HBV infection (6/6 Chinese and 4/4 Caucasian subjects). Responses in chronic patients (18% [5/27] of Chinese and 15% [5/33] of all Caucasian individuals) were rarely observed ex vivo (Fig. 1a and b), indicating that clinical status was a stronger predictor of detectable responses than ethnicity or infecting genotype. In line with previous results (45), the envelope-specific T-cell response appears to be the only weak response detectable ex vivo in chronic HBV patients with a high HBV load (Fig. 1b).
FIG. 1.
Ex vivo quantitative profile of HBV-specific T cells in Chinese and Caucasian HBV patients. (a) Overlapping peptide pools were used to stimulate PBMC directly ex vivo in the IFN-γ ELISPOT assay. Results from selected acute and chronic Chinese and Caucasian patients, segregated based on HBV DNA and ALT levels (shown above the graphs), are displayed. Each bar represents the response to an individual peptide mixture. (b) Mean ex vivo frequency of IFN-γ-producing cells in patients of different ethnicities. Each bar represents the response to the mixtures covering the indicated HBV protein, with error bars indicating the standard error.
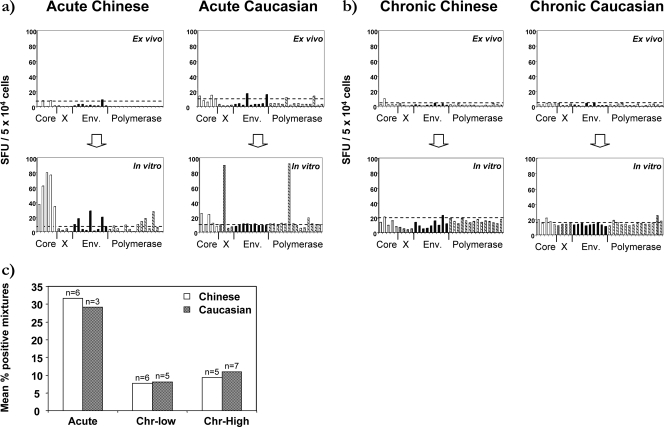
HBV-specific T-cell responses were also compared after in vitro expansion in 17 HBVgenB-infected Chinese and 15 HBVgenD-infected Caucasian subjects. HBV-specific T-cell frequency was generally low directly ex vivo but became clearly detectable in all acute patients after in vitro expansion (Fig. 2a), while such expansion was defective in chronic patients irrespective of ethnicity and HBV genotype (Fig. 2b). The data for all patients are summarized in Fig. 2c, which provides a comparison of the numbers of peptide mixtures able to elicit ELISPOT assay responses in acute versus chronic HBV patients. We observed the same general pattern of efficient expansion and multispecific T-cell responses in acute patients compared to weak and narrower T-cell responses in chronic patients, irrespective of ethnicities and HBV genotypes. Furthermore, previous reports suggested that the magnitude of the HBV-specific T-cell response is inversely correlated with the serum HBV DNA level (7, 45); however, this relationship was not observed in our data, and there was no correlation between HBV-specific T-cell expansion and HBeAg/anti-HBe status, which is probably attributable to the limited sample size (data not shown).
FIG. 2.
Quantification of HBV-specific T cells after in vitro expansion. (a and b) ELISPOT analysis was performed on PBMC directly ex vivo (top panels) and after 10 days of in vitro expansion (bottom panels) using the same pools of peptides. Results from two selected acute (a) and chronic (b) patients are displayed. Each bar represents the response to an individual peptide mixture. The threshold of positivity (above 5 SFU and two times above the mean SFU of the unstimulated control) is indicated as a dashed line. Env., envelope. (c) Each bar represents the percentage of positive mixtures observed from the ELISPOT assays after in vitro expansion in the different categories (n indicates the number of patients tested in each category). Acute and chronic Chinese and Caucasian patients were segregated based on HBV DNA levels (Chr-low, <107 HBV DNA copies/ml; Chr-high, >107 HBV DNA copies/ml).
Functional defects of HBV-specific T cells from chronic patients.
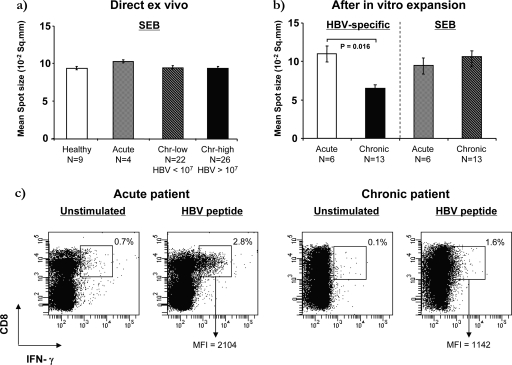
The functional alteration of IFN-γ production by T cells has been reported in both Chinese (10) and Caucasian (7) chronic HBV patients. While such defects were shown to be HBV specific in Caucasians, a recent report suggested that T-cell defects present in Chinese chronic HBV patients were pervasive and caused by programmed death 1 ligand (PD-L1) upregulation on dendritic cells (10). To determine if there was an impairment of IFN-γ production in chronic HBV patients, we examined the relative amount of IFN-γ secreted by HBV-specific and non-HBV-specific T cells from healthy and HBV-infected subjects. Spot sizes (18) obtained by direct ex vivo ELISPOT assays from a total of 48 Chinese chronic HBV patients (22 with HBV DNA loads of <106 and 26 with HBV DNA loads of >106), 4 patients with acute HBV infection, and 9 healthy control subjects were measured. The average spot size of the non-HBV-specific T cells (SEB stimulated) was similar across all groups and was not influenced by HBV DNA load (Fig. 3a). Identical results were obtained with in vitro-expanded cells (Fig. 3b), confirming that non-HBV-specific T-cell function was not affected by HBV status. However, the average spot size of HBV-specific T cells was reduced nearly 50% in cells from chronic patients compared to individuals with resolved infection (P = 0.016) (Fig. 3b), demonstrating that the defect in IFN-γ production was selectively detectable in HBV-specific T-cell populations of chronic patients.
FIG. 3.
Quantification of IFN-γ production in acute and chronic Chinese patients. The mean spot size of the positive wells in the ELISPOT assay was used as an estimate of the quantity of IFN-γ produced by single cells after stimulation. (a) Direct ex vivo quantification of unicellular IFN-γ production in healthy, acute, and chronic Chinese patients after SEB stimulation. Each bar represents the mean spot size with error bars indicating the standard error of the mean. Chr, chronic. (b) Quantification of unicellular IFN-γ production in acute and chronic Chinese patients after 10 days of in vitro expansion. Only significant differences are shown (HBV-specific acute patients versus chronic patients [mean ± standard error of the mean], 10.9 ± 0.7 versus 6.9 ± 1.0 10−2 mm2). (c) IFN-γ production by HBV-specific CD8 T cells determined by ICS. Representative acute and chronic HBV-infected Chinese patients are shown. The percentages of IFN-γ+ cells out of the CD8+ cells after gating on the CD3+ fraction of PBMC and the MFI of the double-positive population are indicated. Cells incubated without the HBV polymerase peptide pool were used as the unstimulated negative control in which the IFN-γ+ gate was set.
These results were validated by measuring the fluorescence intensity of IFN-γ+, HBV-specific T cells. Representative results from one acute and one chronic patient are shown in Fig. 3c. The mean fluorescence intensity (MFI) of IFN-γ+ HBV-specific cells detected in the acute and chronic patients differed significantly. IFN-γ+ HBV-specific cells from acute patients had a high MFI (MFI = 2,104), while IFN-γ+ HBV-specific T cells from chronic patients had a low MFI and were often difficult to distinguish from unstimulated cells. Taken together, these results show that Chinese patients with chronic hepatitis B, like Caucasians (7), seem to harbor a functional T-cell defect in IFN-γ production restricted to virus-specific cells.
Ethnicity and HBV genotype influence the HBV-specific CD8+ T-cell repertoire of HBV-infected patients.
The above results show that neither race nor HBV genotype significantly influences the general quantitative and qualitative profile of the HBV-specific T-cell response; however, these variables could still impact the diversity of the HBV-specific CD8+ T-cell repertoire. Therefore, we determined if ethnicity and HBV genotype can influence CD8+ T-cell responses against six HLA-A2-restricted epitopes that have previously been shown to be promiscuously presented by multiple HLA-A2 subtypes (Table 1) (6). High-resolution HLA-A2 typing was performed on the subjects to determine their HLA-A2 subtypes. All HLA-A2+ Caucasians displayed HLA-A0201, while HLA-A2+ Chinese patients displayed different HLA-A2 subtypes (A0201, n = 3; A0203, n = 5; A0206, n = 4; A0207, n = 8). Patient PBMC were stimulated for 10 days with the peptides corresponding to the sequence of the infecting HBV genotype, and the frequency of the HBV-specific CD8+ T cells was analyzed by ICS for IFN-γ production.
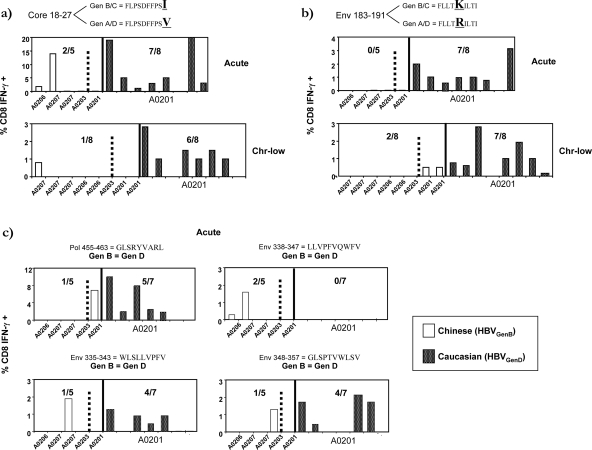
Figure 4a and b show the results of the CD8+ T-cell responses against the Core18-27 and Env183-91 peptides, which differ by one amino acid between HBVgenB/C and HBVgenA/D and frequently stimulate virus-specific CD8+ T cells in HLA-A0201+ Caucasian HBV patients (4, 6, 30, 45). As expected, 13/16 HLA-A0201+ Caucasian patients responded to the Core18-27(V) epitope. In contrast, CD8+ T cells specific to Core18-27(I) were present in only 3/13 Chinese patients (Fig. 4A). Interestingly, none of the A201+ Chinese patients responded to HBVgenB/C Core18-27(I), and specific responses were only detectable in Chinese patients expressing HLA-A0207 (n = 2) and HLA-A0206 (n = 1).
FIG. 4.
Induction of CD8+ T-cell response against known A2-restricted epitopes in HLA-A2+ Chinese and Caucasian patients. Bars represent the frequency of Core18-27 (a)- or Env183-91 (b)-specific CD8+ T cells in individual patients with the indicated HLA-A2 subtypes. PBMC of Chinese patients were expanded with HBVgenB/C Core18-27(I) or Env183-91(K) peptide, while PBMC of Caucasian patients were expanded with HBVgenA/D Core18-27(V) peptide or Env183-191(R) (all at 1 μM) for 10 days, before restimulating the lines with the corresponding stimulatory peptide and analyzing the frequency of the CD8+ cells producing IFN-γ with ICS. Sequencing of the Core18-27 region of the HBV infecting the chronic Chinese and Caucasian patients confirmed the presence of the Core18-27(I) and Core18-27(V) sequences in the Chinese and Caucasian patients, respectively. The hatched line demarcates the A0201-expressing subjects. (c) Frequency of CD8+ T-cell response against conserved HBV epitopes (genotype B = genotype D) Pol456-63, Env338-47, Env335-43, and Env348-57 in acute Chinese and Caucasian HBV patients.
CD8+ T-cell responses to the Env183-91 epitope were highly influenced by HLA-A2 micropolymorphisms: 14/16 HLA-A0201+ Caucasian HBV patients responded to the Env183-91(R) epitope from HBVgenA/D, and 2/3 HLA-A0201+ Chinese patients (2/2 chronic patients) were capable of responding to the Env183-91(K) epitope from HBVgenB/C. In contrast, all 11 Chinese patients expressing the A0203, A0206, or A0207 HLA-A2 subtype were unable to respond to the Env183-91(K) peptide (Fig. 4b).
Analysis of CD8+ T-cell responses against epitopes conserved between HBVgenB/C and HBVgenA/D (out of 24 full HBV genome entries in GenBank [see Materials and Methods and Table 1]) confirmed the impact of HLA-A2 subtypes on the HBV-specific T-cell repertoire. While patients expressing HLA-A0201 were capable of responding to the Pol455-63, Env335-43, and Env348-57 epitopes (Fig. 4c), only sporadic promiscuous responses could be detected in the other HLA-A2 subtypes: one HLA-A0206+ patient (Env338-47), two HLA-A0207+ patients (Env338-47 and Env335-43), and one HLA-A0203+ patient (Env348-57). Responses to the Pol455-63 epitope, which was targeted by 6/8 HLA-A0201+ subjects, were not seen at all in HLA-A0201-negative individuals (Fig. 4c). Conversely, responses to Env338-47, a peptide with only predicted HLA-A2-binding ability (30), were present in HLA-A0206+ and HLA-A0207+ acute subjects but undetectable in all HLA-A0201+ subjects tested.
Hierarchy of HBV-specific CD8 T cells in HLA-A0206+ and HLA-0203+ patients.
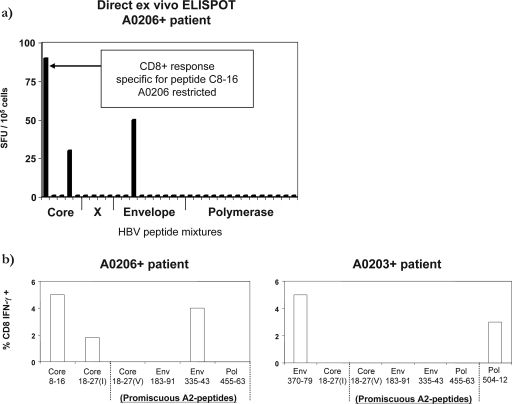
We then investigated whether the HBV-specific CD8+ T-cell repertoire of Chinese patients expressing different HLA-A2 subtypes focused on specificities which differ from those previously defined in A201+ Caucasian patients. To avoid the bias associated with focusing on previously identified epitopes, HBV-specific T-cell responses were assessed using 15-mer overlapping peptides covering the entire HBV proteome, followed by characterization of fine specificity and HLA restrictions of detectable responses. Given the extensive cell requirements for such comprehensive analyses, we were able to perform such thorough analysis in one HLA-A0206+ and one HLA-A0203+ patient with acute HBV infection. In the HLA-A0206+ acute HBV patient, three responses were found directly ex vivo, with a dominant response targeting a region of the core protein (Fig. 5a). Detailed analysis of the phenotype, fine specificity, and HLA restriction of the responsive T cells revealed that this response recognized a novel HLA-A0206-restricted Core8-16 (EFGASVELL) epitope (data not shown). The two additional responses present at lower frequencies were directed against a second region in the core and one in the envelope which did not overlap with the previously known A2 epitopes (Fig. 5a).
FIG. 5.
Hierarchy of HBV-specific CD8+ T-cell responses in a HLA-A0206+ patient and a HLA-A0203+ Chinese acute HBV patient. (a) Direct ex vivo ELISPOT frequency of HBV-specific T cells in the PBMC of an HLA-A0206+ acute patient. Triplicate wells of unstimulated cells gave a mean of three spots/well of background. (b) Frequency of CD8+ T cells specific for the indicated peptides in the HLA-A0206+ and HLA-A0203+ patients tested after in vitro expansion. Bars represent the frequency of peptide-specific CD8+ T cells after in vitro expansion.
Responses against “promiscuous” HLA-A2 epitopes were also analyzed but were not detected directly ex vivo (data not shown). However, after in vitro stimulation using the corresponding optimal HLA-A2 peptides, the A0206+ acute patient demonstrated the presence of Core18-27(I) (HBVgenB/C) and Env335-43 CD8+ responses, which were subdominant in terms of frequency compared to the Core8-16-specific CD8+ response (Fig. 5b).
The A2-restricted CD8+ T-cell repertoire in the HLA-A0203+ patient differed significantly from that of the HLA-A0206+ patient. Analysis of the HBV-specific CD8+ T-cell repertoire was performed only after in vitro expansion, and it revealed the presence of an HLA-A0203-restricted response to two HBV epitopes rarely recognized in Caucasian subjects (Env370-79 and Pol504-12; Fig. 5b). Responses against all other known HLA-A2-restricted HBV epitopes were undetectable, both directly ex vivo and after in vitro expansion (data not shown and Fig. 5b). These results confirm that the HLA-A2 polymorphisms in the Chinese subjects lead to a different focus of the HLA-A2-restricted responses compared to the response patterns seen in the HLA-A0201+ Caucasian population.
Effect of HBV genotype and HLA-A2 subtypes on CTL recognition.
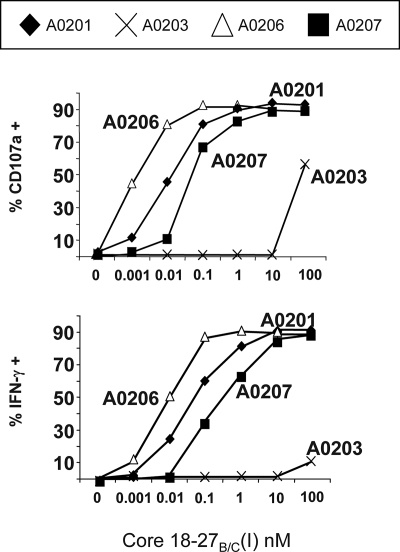
To further dissect how the combination of amino acid differences in HBV genotypes and HLA-A2 subtypes influence CD8+ T-cell response patterns, we assessed the recognition of genotype-specific epitope variants in the context of several HLA-A2 subtypes. Core18-27-specific CD8+ T-cell clones generated from an A201+ HBV-infected patient can efficiently recognize the Core18-27(I) peptide variant when presented by different A2 subtypes (Fig. 6). Peptide recognition was effective in the context of HLA-A0206-, HLA-A0207-, and HLA-A0201-positive EBV-transformed B-cell lines, which trigger CD8+ T-cell activation when pulsed with low (10 pM) concentrations of Core18-27(I) peptide. In contrast, only minimal CD8+ T-cell activation was observed after stimulation with peptide-loaded HLA-A0203+ target cells, even at the highest peptide concentration (100 nM) (Fig. 6).
FIG. 6.
Functional presentation of Core18-27 by HLA subtypes common in the Chinese population. Frequency of CD107a+ or IFN-γ+ CTLs after stimulation with EBV B-cell lines, expressing the various HLA-A2 subtype molecules (HLA-A0201, -A0206, -A0207, and -A0203), pulsed with graded concentrations of HBVgenB/C Core18-27(I). The results are representative of two repeated experiments.
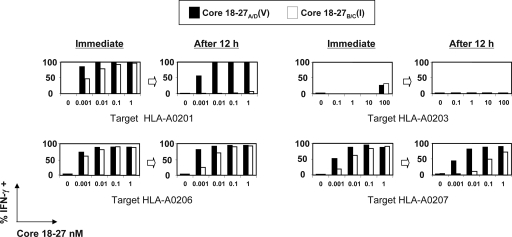
We then analyzed, in parallel, the ability of HLA-A2 subtypes to present the two genotype-specific Core18-27 epitope variants as well as the stability of the peptide/HLA-A2 complex, a parameter associated with peptide immunogenicity (36). Antigen-presenting cells with defined HLA-A2 subtypes were pulsed with increasing concentrations of Core18-27(V) or Core18-27(I) peptides and then used to stimulate Core18-27(V)-specific CD8+ T-cell clones. Effector cells were added immediately or after a 12-h period. Both the Core18-27(V) as well as its 27(I) variant peptide were presented effectively by HLA-A0201, -A0206, and -A0207 but not by HLA-A0203 if effector cells were added immediately. However, when the loaded peptides were allowed to dissociate from the antigen-presenting cells before the addition of the effector T cells (after a 12-h period), the Core18-27(I) variant was in all cases less stimulatory than the Core18-27(V) peptide. In the context of HLA-A0201, the genotype B Core18-27(I) peptide lost essentially all immunogenicity, whereas immunogenicity was retained when the Core18-27(I) peptide was presented by HLA-A0206. Not surprisingly, the little stimulatory efficiency of HLA-A203+ targets was completely lost after the 12-h dissociation period (Fig. 7). It is interesting to note that the peptide presentation data recapitulate the frequency of Core18-27-specific CD8+ T cells detected in HBV-infected patients. The hierarchy of Core18-27(I) presentation (A0206 > A0207 > A0201 > A0203) described by the presentation experiments in Fig. 7 is in line with the presence of Core18-27(I)-specific CD8+ T-cell responses found in some HLA-A0206+ and -A0207+ HBV patients and their absence in HLA-A0201+ and -A0203+ patients (Fig. 4a).
FIG. 7.
Time course analysis of Core18-27 epitope presentation by HLA-A2 subtypes. Frequency of IFN-γ+ CTLs tested with EBV B-cell lines, expressing HLA-A0201, -A0206, -A0207, and -A0203, pulsed with graded concentrations of HBVgenB/C Core 18-27(I) or HBVgenA/D Core18-27(V). The different EBV B cells were pulsed with the peptides for 30 min and coincubated with the CTL lines either immediately or after 12 h of in vitro resting. Bars represent the frequency of activated CTLs tested against targets pulsed with Core18-27(V) (black) or Core18-27(I) (white). Similar results were obtained measuring CD107 expression (not shown).
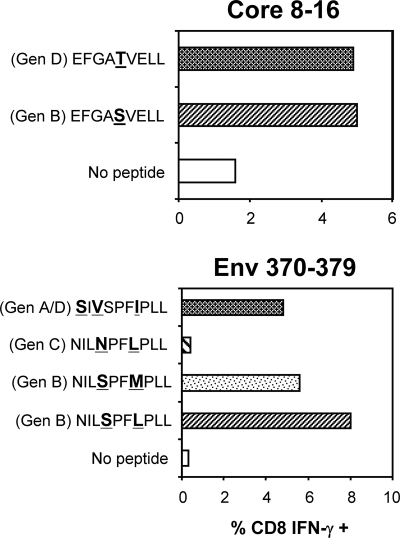
Finally, we tested whether HBVgenB-infected Asian individuals could recognize epitope variants from HBVgenD. T-cell lines specific for different HBVgenB epitopes were generated from Chinese individuals with acute HBVgenB infection and tested for recognition of epitope variants naturally present in genotypes D, A, and C. The single amino acid substitution within the Core8-16 peptide (S12T) did not affect Core8-16-specific CD8+ T-cell recognition (Fig. 8). Similarly, despite the presence of three different amino acids, the peptide corresponding to the sequence Env370-79 of HBVgenA/D was recognized, even though to a lesser degree, by the Env370-79 HBVgenB-specific HLA-A0203-restricted CD8+ T-cell line. In contrast, the single amino acid difference present in the HBVgenC (S373N) variant completely abolished CD8+ T-cell recognition (Fig. 8). Whether such amino acid substitution (S373N) abolished the HLA-A203-binding ability of the peptide or instead altered the T-cell receptor recognition site will need to be properly analyzed.
FIG. 8.
Influence of amino acid variations within different genotypes on T-cell recognition. T-cell lines specific for different HBVgenB-encoded epitopes were expanded from acute patients of Chinese origin (genotype B infected). The T-cell lines indicated were stimulated with peptides corresponding to the different genotypes for 5 h and analyzed by ICS. Each bar represents the percentage of IFN-γ+ CD8 T cells.
DISCUSSION
In this study, we took advantage of the segregation of different HBV genotypes in geographically and genetically distinct host populations to directly analyze the impact that host and virus variables exert on the virus-specific T-cell response. Two important points emerged: (i) HBV-specific T-cell quantity is determined by the virological and clinical profiles of the patients, which outweigh any influence of race or viral diversity, and (ii) HBV-specific T-cell repertoires are divergent in the two ethnic groups with T-cell epitopes frequently found in Caucasian patients and seldom detected in Chinese patients.
In Asia, chronic HBV infection is mainly derived from vertical infection at birth (39), whereas horizontal transmission (household/sexual contact) is the likely route of infection in Caucasians. In addition, infection by different HBV genotypes has also been suggested to influence clinical outcomes or disease severity (15). Furthermore, by inference from HCV infection, host ethnicity may impact clinical parameters and the antiviral CD4+ T-cell response (41). To our knowledge, whether these epidemiological variables might alter the profile of the HBV-specific T-cell response has never been directly tested. Our results show that the epidemiological variables characteristic of the different ethnic populations do not impact the overall virus-specific T-cell quantity. Rather, the level of cellular immunity appears largely defined by the clinical status of the tested patient. HBV-specific T cells were easily detected directly ex vivo in only acute patients, while HBV-specific T-cell responses were very weak in chronic patients. Although different HBV genotypes or transmission routes could trigger different pathogenic mechanisms resulting in viral persistence, our data indicate that the ability to control HBV, or conversely HBV persistence, results in highly similar quantitative profiles of HBV-specific T-cell responses, irrespective of ethnicity and HBV genotype.
We also confirmed, by analyzing IFN-γ production at a single-cell level, that HBV-specific T cells of Chinese chronic patients infected by HBVgenB were defective for IFN-γ production. The IFN-γ defect was found exclusively in HBV-specific T cells while the general T-cell population was unaffected. While this finding is in contrast with recent data (10), which have implicated the apparent upregulation of PD-L1 on myeloid dendritic cells in the suppression of global T-cell function, the data confirm results obtained from anti-HBe+ Caucasian patients, for which the impairment of IFN-γ production, restored by blocking PD-1/PD-L1, was restricted to HBV-specific T cells (7).
HBV genotypes and the individuals' genetic backgrounds instead play an important role in shaping the HBV-specific T-cell repertoire. We found that HLA-A2-restricted CD8+ T-cell epitopes, which frequently induced responses in HLA-A0201 Caucasian patients and possess “HLA class I super-type” binding capacity (6), are scarcely (Core18-27) or completely unable (Env183-91 and Pol455-65) to induce CD8+ T-cell responses in HLA-A0206+, -A0207+, or -A0203+ Chinese patients. Furthermore, a comprehensive analysis of the CD8+ T-cell repertoire performed in one HLA-A0206+ patient and one HLA-A0203+ HBV patient directly confirmed, for naturally infected subjects, how HLA micropolymorphisms (HLA-A0206/A0207 and -A0203 differ from HLA-A0201 by a single amino acid or three amino acids, respectively) and viral amino acid differences impact CD8+ T-cell responses. These two subjects preferentially focused T-cell responses on HLA-A2-restricted epitopes never (Core8-16 and Pol406-12) or scarcely (Env 371-79) (33) detected in HLA-A0201+ subjects, while responses to classical HLA-A2-restricted epitopes were not at all (HLA-A0203+ patient) or rarely (HLA-A0206+ patient) present.
The discordance of the HBV-specific CD8+ T-cell repertoires in the two ethnicities is not surprising if one considers the different mechanisms that might alter the immunogenicity of antigenic peptides. For example, the ability of a peptide epitope to induce CD8+ T cells can be influenced by its bound conformation without necessarily altering the HLA-binding ability. Work on EBV infection has shown that the conformational changes imposed by a single amino acid variation present in two closely related HLA class I molecules (HLA-B3501 and HLA-B3508) are sufficient to alter the ability of an identical peptide to induce a CTL response (44). This scenario might explain our inability to detect the HBV Pol455-63 epitope in HLA-A0203+, -A0206+, and -A0207+ patients, despite its conservation between different genotypes and its reported “promiscuous” ability to bind different HLA-A2 subtypes (6). Furthermore, the ability of HLA-A2 subtypes to preferentially present different sets of peptides (2, 40) is another important contributor to the distinct epitopes targeted in HBV patients expressing different HLA-A2 subtypes. In addition to the alterations imposed by the HLA-A2 polymorphisms, the presence of distinct HBV genotypes in different ethnicities can further increase the potential diversity in the pool of immunogenic peptides. HBV genotypes differ approximately 8% in their amino acid compositions (21), and so it is likely that HBV genotype amino acid variations located within or just outside HBV epitopes might modify epitope processing, presentation, or recognition (23, 34).
Our experiments prove that these amino acid variations can affect T-cell recognition of CD8-specific T-cell epitopes. In particular, we showed for the Core18-27 epitope that the presence of isoleucine instead of valine at position 27 reduced the stability of the peptide/HLA-A2 complex to various degrees in all the HLA-A2 subtypes tested. The Core18-27 isoleucine variation is characteristic of HBVgenB/C and our findings help explain the generally poor immunogenicity of the Core18-27(I) epitope in HLA-A0201+ Chinese subjects. At the same time, the observation that the HBVgenB/C Core18-27(I) peptide is more stably presented by HLA-A0206 and -A0207 (only expressed by Chinese patients) than HLA-A0201 is puzzling. While it could be driven by a possible host-virus coevolution, its evolutionary significance is obscure, as such coevolution would have led to the conservation of a highly avid epitope presented by some of the dominant HLA alleles. Whether this finding can be extrapolated to other epitopes and whether or not these responses are significantly involved in viral control clearly require broader analyses in larger cohorts and more diverse HLA backgrounds.
In conclusion, ethnic and viral differences do not alter the vigor of HBV-specific T-cell responses present in patients with HBV infection, but these variables shape distinct HBV-specific T-cell repertoires. The results from our findings show a limited contribution of HLA-A2 “promiscuous” HBV epitopes to the HBV-specific CD8 T-cell repertoire of ethnically different patients and question, at least in the context of the HLA-A2 allele, the ability of the supertype motifs to predict virus-specific CD8+ T-cell responses present in different ethnic groups. From our data, it seems clear that HBV-specific immune monitoring in Asian patients should not rely on the exclusive analysis of epitopes found in Caucasians or conform to algorithms that do not consider the clustering of different viral strains within different ethnic groups. Furthermore, the use of polyepitope vaccines (12), based on peptides selected by HLA class supermotifs or on responses found in Caucasian patients, might divert the response toward incorrect specificities, stimulating nonnatural responses which might be functionally incapable of recognizing the infectious virus, a situation that has already been reported in HIV infections (8, 24). However, it must also be considered that predicted epitopes that might not be naturally immunogenic can be advantageous in breaking immunological tolerance caused by a persistent virus infection, potentially expanding nontolerant, cross-reactive CD8 T cells able to recognize the infecting virus with therapeutic advantage (33). T-cell repertoires elicited by vaccines and their therapeutic effects in patients will have to be carefully analyzed in patients to resolve this conundrum and shed further light on the complex and fascinating diversity of the antiviral T-cell response in humans.
Supplementary Material
Footnotes
Published ahead of print on 17 September 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. R. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 772081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barouch, D., T. Friede, S. Stevanovic, L. Tussey, K. Smith, S. Rowland-Jones, V. Braud, A. McMichael, and H. G. Rammensee. 1995. HLA-A2 subtypes are functionally distinct in peptide binding and presentation. J. Exp. Med. 1821847-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertoletti, A., A. Costanzo, F. V. Chisari, M. Levrero, M. Artini, A. Sette, A. Penna, T. Giuberti, F. Fiaccadori, and C. Ferrari. 1994. Cytotoxic T lymphocyte response to a wild type hepatitis B virus epitope in patients chronically infected by variant viruses carrying substitutions within the epitope. J. Exp. Med. 180933-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertoletti, A., C. Ferrari, F. Fiaccadori, A. Penna, R. Margolskee, H. Schlicht, P. Fowler, S. Guilhot, and F. Chisari. 1991. HLA class I-restricted human cytotoxic T cells recognize endogenously synthesized hepatitis B virus nucleocapsid. Proc. Natl. Acad. Sci. USA 8810445-10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertoletti, A., and A. J. Gehring. 2006. The immune response during hepatitis B virus infection. J. Gen. Virol. 871439-1449. [DOI] [PubMed] [Google Scholar]
- 6.Bertoni, R., J. Sidney, P. Fowler, R. Chesnut, F. Chisari, and A. Sette. 1997. Human histocompatibility leukocyte antigen-binding supermotifs predict broadly cross-reactive cytotoxic T lymphocyte responses in patients with acute hepatitis. J. Clin. Investig. 100503-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boni, C., P. Fisicaro, C. Valdatta, B. Amadei, P. Di Vincenzo, T. Giuberti, D. Laccabue, A. Zerbini, A. Cavalli, G. Missale, A. Bertoletti, and C. Ferrari. 2007. Characterization of hepatitis B virus (HBV)-specific T-cell dysfunction in chronic HBV infection. J. Virol. 814215-4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brander, C., N. Frahm, and B. D. Walker. 2006. The challenges of host and viral diversity in HIV vaccine design. Curr. Opin. Immunol. 18430-437. [DOI] [PubMed] [Google Scholar]
- 9.Chang, J. J., F. Wightman, A. Bartholomeusz, A. Ayres, S. J. Kent, J. Sasadeusz, and S. R. Lewin. 2005. Reduced hepatitis B virus (HBV)-specific CD4+ T-cell responses in human immunodeficiency virus type 1-HBV-coinfected individuals receiving HBV-active antiretroviral therapy. J. Virol. 793038-3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, L., Z. Zhang, W. Chen, Z. Zhang, Y. Li, M. Shi, J. Zhang, L. Chen, S. Wang, and F. S. Wang. 2007. B7-H1 up-regulation on myeloid dendritic cells significantly suppresses T cell immune function in patients with chronic hepatitis B. J. Immunol. 1786634-6641. [DOI] [PubMed] [Google Scholar]
- 11.de Campos-Lima, P. O., R. Gavioli, Q. J. Zhang, L. E. Wallace, R. Dolcetti, M. Rowe, A. B. Rickinson, and M. G. Masucci. 1993. HLA-A11 epitope loss isolates of Epstein-Barr virus from a highly A11+ population. Science 26098-100. [DOI] [PubMed] [Google Scholar]
- 12.Depla, E., A. Van der Aa, B. D. Livingston, C. Crimi, K. Allosery, V. De Brabandere, J. Krakover, S. Murthy, M. Huang, S. Power, L. Babé, C. Dahlberg, D. McKinney, A. Sette, S. Southwood, R. Philip, M. J. Newman, and L. Meheus. 2008. Rational design of a multiepitope vaccine encoding T-lymphocyte epitopes for treatment of chronic hepatitis B virus infections. J. Virol. 82435-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. StJohn, A. Khatri, K. Davis, J. Mullins, P. J. R. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 782187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frahm, N., K. Yusim, T. J. Suscovich, S. Adams, J. Sidney, P. Hraber, H. S. Hewitt, C. H. Linde, D. G. Kavanagh, T. Woodberry, L. M. Henry, K. Faircloth, J. Listgarten, C. Kadie, N. Jojic, K. Sango, N. V. Brown, E. Pae, M. T. Zaman, F. Bihl, A. Khatri, M. John, S. Mallal, F. M. Marincola, B. D. Walker, A. Sette, D. Heckerman, B. T. Korber, and C. Brander. 2007. Extensive HLA class I allele promiscuity among viral CTL epitopes. Eur. J. Immunol. 372419-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fung, S. K., and A. S. Lok. 2004. Hepatitis B virus genotypes: do they play a role in the outcome of HBV infection? Hepatology 40790-792. [DOI] [PubMed] [Google Scholar]
- 16.Gaudieri, S., A. Rauch, L. P. Park, E. Freitas, S. Herrmann, G. Jeffrey, W. Cheng, K. Pfafferott, K. Naidoo, R. Chapman, M. Battegay, R. Weber, A. Telenti, H. Furrer, I. James, M. Lucas, and S. A. Mallal. 2006. Evidence of viral adaptation to HLA class I-restricted immune pressure in chronic hepatitis C virus infection. J. Virol. 8011094-11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gehring, A. J., D. Sun, P. T. Kennedy, E. Nolte-'t Hoen, S. G. Lim, S. Wasser, C. Selden, M. K. Maini, D. M. Davis, M. Nassal, and A. Bertoletti. 2007. The level of viral antigen presented by hepatocytes influences CD8 T-cell function. J. Virol. 812940-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hesse, M. D., A. Y. Karulin, B. O. Boehm, P. V. Lehmann, and M. Tary-Lehmann. 2001. A T cell clone's avidity is a function of its activation state. J. Immunol. 1671353-1361. [DOI] [PubMed] [Google Scholar]
- 19.Hwang, Y. K., N. K. Kim, J. M. Park, K. Lee, W. K. Han, H. I. Kim, and H. S. Cheong. 2002. HLA-A2.1 restricted peptides from the HBx antigen induce specific CTL responses in vitro and in vivo. Vaccine 203770-3777. [DOI] [PubMed] [Google Scholar]
- 20.Jung, M., B. Hartmann, J. Gerlach, H. Diepolder, R. Gruber, W. Schraut, N. Gruner, R. Zachoval, R. Hoffmann, T. Santantonio, and G. Pape. 1999. Virus-specific lymphokine production differs quantitatively but not qualitatively in acute and chronic hepatitis B infection. Virology 261165-172. [DOI] [PubMed] [Google Scholar]
- 21.Kidd-Ljunggren, K., Y. Miyakawa, and A. H. Kidd. 2002. Genetic variability in hepatitis B viruses. J. Gen. Virol. 831267-1280. [DOI] [PubMed] [Google Scholar]
- 22.Krausa, P., and M. J. Browning. 1996. HLA-A2 polymorphism and immune functions. Eur. J. Immunogenet. 23261-274. [DOI] [PubMed] [Google Scholar]
- 23.Le Gall, S., P. Stamegna, and B. D. Walker. 2007. Portable flanking sequences modulate CTL epitope processing. J. Clin. Investig. 1173563-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leslie, A., D. A. Price, P. Mkhize, K. Bishop, A. Rathod, C. Day, H. Crawford, I. Honeyborne, T. E. Asher, G. Luzzi, A. Edwards, C. M. Rousseau, J. I. Mullins, G. Tudor-Williams, V. Novelli, C. Brander, D. C. Douek, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2006. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J. Immunol. 1774699-4708. [DOI] [PubMed] [Google Scholar]
- 25.Lindh, M., J. E. Gonzalez, G. Norkrans, and P. Horal. 1998. Genotyping of hepatitis B virus by restriction pattern analysis of a pre-S amplicon. J. Virol. Methods 72163-174. [DOI] [PubMed] [Google Scholar]
- 26.Maini, M. K., C. Boni, G. S. Ogg, A. S. King, S. Reignat, C. K. Lee, J. R. Larrubia, G. J. M. Webster, A. J. McMichael, C. Ferrari, R. Williams, D. Vergani, and A. Bertoletti. 1999. Direct ex vivo analysis of hepatitis B virus-specific CD8+ T cells associated with the control of infection. Gastroenterology 1171386-1396. [DOI] [PubMed] [Google Scholar]
- 27.Merican, I., R. Guan, D. Amarapuka, M. J. Alexander, A. Chutaputti, R. N. Chien, S. S. Hasnian, N. Leung, L. Lesmana, P. H. Phiet, H. M. Sjalfoellah Noer, J. Sollano, H. S. Sun, and D. Z. Xu. 2000. Chronic hepatitis B virus infection in Asian countries. J. Gastroenterol. Hepatol. 151356-1361. [DOI] [PubMed] [Google Scholar]
- 28.Middleton, D., L. Menchaca, H. Rood, and R. Komerofsky. 2003. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens 61403-407. [DOI] [PubMed] [Google Scholar]
- 29.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 2961439-1443. [DOI] [PubMed] [Google Scholar]
- 30.Nayersina, R., P. Fowler, S. Guilhot, G. Missale, A. Cerny, H. J. Schlicht, A. Vitiello, R. Chesnut, J. L. Person, A. J. Redeker, and F. V. Chisari. 1993. HLA-A2 restricted cytotoxic T lymphocyte responses to multiple hepatitis B surface antigen epitopes during hepatitis B virus infection. J. Immunol. 1504659-4671. [PubMed] [Google Scholar]
- 31.Penna, A., F. V. Chisari, A. Bertoletti, G. Missale, P. Fowler, T. Giuberti, F. Fiaccadori, and C. Ferrari. 1991. Cytotoxic T lymphocytes recognize an HLA-A2-restricted epitope within the hepatitis B virus nucleocapsid antigen. J. Exp. Med. 1741565-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehermann, B., P. Fowler, J. Sidney, J. Person, A. Redeker, M. Brown, B. Moss, A. Sette, and F. V. Chisari. 1995. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J. Exp. Med. 1811047-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riedl, P., A. Bertoletti, R. Lopes, F. Lemonnier, J. Reimann, and R. Schirmbeck. 2006. Distinct, cross-reactive epitope specificities of CD8 T cell responses are induced by natural hepatitis B surface antigen variants of different hepatitis B virus genotypes. J. Immunol. 1764003-4011. [DOI] [PubMed] [Google Scholar]
- 34.Seifert, U., H. Liermann, V. Racanelli, A. Halenius, M. Wiese, H. Wedemeyer, T. Ruppert, K. Rispeter, P. Henklein, A. Sijts, H. Hengel, P. M. Kloetzel, and B. Rehermann. 2004. Hepatitis C virus mutation affects proteasomal epitope processing. J. Clin. Investig. 114250-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sette, A., and J. Sidney. 1999. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50201-212. [DOI] [PubMed] [Google Scholar]
- 36.Sette, A., A. Vitiello, B. Reherman, P. Fowler, R. Nayersina, W. M. Kast, C. J. Melief, C. Oseroff, L. Yuan, J. Ruppert, et al. 1994. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 1535586-5592. [PubMed] [Google Scholar]
- 37.Sidney, J., B. Peters, N. Frahm, C. Brander, and A. Sette. 2008. HLA class I supertypes: a revised and updated classification. BMC Immunol. 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobao, Y., K. Sugi, H. Tomiyama, S. Saito, S. Fujiyama, M. Morimoto, S. Hasuike, H. Tsubouchi, K. Tanaka, and M. Takiguch. 2001. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*2402, the most common HLA class I allele in East Asia. J. Hepatol. 34922-929. [DOI] [PubMed] [Google Scholar]
- 39.Stevens, C., R. Beasley, and J. Tsu. 1975. Vertical transmission of hepatitis B antigen in Taiwan. N. Engl. J. Med. 292771-774. [DOI] [PubMed] [Google Scholar]
- 40.Sudo, T., N. Kamikawaji, A. Kimura, Y. Date, C. J. Savoie, H. Nakashima, E. Furuichi, S. Kuhara, and T. Sasazuki. 1995. Differences in MHC class I self peptide repertoires among HLA-A2 subtypes. J. Immunol. 1554749-4756. [PubMed] [Google Scholar]
- 41.Sugimoto, K., J. Stadanlick, F. Ikeda, C. Brensinger, E. E. Furth, H. J. Alter, and K. M. Chang. 2003. Influence of ethnicity in the outcome of hepatitis C virus infection and cellular immune response. Hepatology 37590-599. [DOI] [PubMed] [Google Scholar]
- 42.Thimme, R., K.-M. Chang, J. Pemberton, A. Sette, and F. V. Chisari. 2001. Degenerate immunogenicity of an HLA-A2-restricted hepatitis B virus nucleocapsid cytotoxic T-lymphocyte epitope that is also presented by HLA-B51. J. Virol. 753984-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai, S. L., M. H. Chen, C. T. Yeh, C. M. Chu, A. N. Lin, F. H. Chiou, T. H. Chang, and Y. F. Liaw. 1996. Purification and characterization of a naturally processed hepatitis B virus peptide recognized by CD8+ cytotoxic T lymphocytes. J. Clin. Investig. 97577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tynan, F. E., D. Elhassen, A. W. Purcell, J. M. Burrows, N. A. Borg, J. J. Miles, N. A. Williamson, K. J. Green, J. Tellam, L. Kjer-Nielsen, J. McCluskey, J. Rossjohn, and S. R. Burrows. 2005. The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J. Exp. Med. 2021249-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webster, G. J., S. Reignat, D. Brown, G. S. Ogg, L. Jones, S. L. Seneviratne, R. Williams, G. Dusheiko, and A. Bertoletti. 2004. Longitudinal analysis of CD8+ T cells specific for structural and nonstructural hepatitis B virus proteins in patients with chronic hepatitis B: implications for immunotherapy. J. Virol. 785707-5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wentworth, P. A., A. Sette, E. Celis, J. Sidney, S. Southwood, C. Crimi, S. Stitely, E. Keogh, N. C. Wong, B. Livingston, D. Alazard, A. Vitiello, H. M. Grey, F. V. Chisari, R. W. Chesnut, and J. Fikes. 1996. Identification of A2-restricted hepatitis C virus-specific cytotoxic T lymphocyte epitopes from conserved regions of the viral genome. Int. Immunol. 8651-659. [DOI] [PubMed] [Google Scholar]
- 47.Yewdell, J. W. 2006. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25533-543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.