Abstract
The genomes of herpesviruses, including human cytomegalovirus (CMV), are double-stranded DNA molecules maintained as episomes during infection. The viral DNA lacks histones when encapsidated in the virion. However, it has been found histone associated inside infected cells, implying unidentified chromatin assembly mechanisms. Our results indicate that components of the host cell nucleosome deposition machinery target intranuclear CMV DNA, resulting in stepwise viral-chromatin assembly. CMV genomes undergo limited histone association and nucleosome assembly as early as 30 min after infection via DNA replication-independent mechanisms. Low average viral-genome chromatinization is maintained throughout the early stages of infection. The late phase of infection is characterized by a striking increase in average histone occupancy coupled with the process of viral-DNA replication. While the initial chromatinization affected all analyzed parts of the CMV chromosome, a subset of viral genomic regions, including the major immediate-early promoter, proved to be largely resistant to replication-dependent histone deposition. Finally, our results predict the likely requirement for an unanticipated chromatin disassembly process that enables packaging of histone-free DNA into progeny capsids.
The DNA within our cells exists in the form of chromatin. The basic building block of eukaryotic chromatin is the nucleosome, which contains 146 bp of DNA wrapped nearly twice around an octamer of the four core histone proteins H2A, H2B, H3, and H4 (30). DNA replication-coupled nucleosome assembly normally occurs during S phase of the cell cycle. At the replication fork, parental nucleosomes are distributed between the two nascent chromatids, and the remaining gaps are filled by de novo histone deposition (21). Nucleosomes are assembled in a two-step process in which a tetramer of H3 and H4 or two H3-H4 dimers are first deposited onto DNA and then two dimers of H2A-H2B are added (6, 52). Disassembly of a nucleosome likely reverses these steps (14). H3-H4 complexes are initially positioned by the chromatin assembly factor 1 (CAF1) multisubunit chaperone, which is tethered to the replication processivity clamp (proliferating cell nuclear antigen [PCNA]). Other histone chaperones, including antisilencing factor 1 (ASF1), synergize with CAF1 to promote nucleosome assembly during DNA replication (44, 47). Although the replication-coupled pathway accounts for the bulk of normal cellular nucleosome assembly, a fraction occurs outside S phase and has been termed replication-independent nucleosome assembly (1).
The herpesvirus family encompasses a group of large, ubiquitous DNA viruses with eight human-pathogenic members. Human cytomegalovirus (CMV), also known as human herpesvirus 5, establishes invariably lifelong infections in the majority of people worldwide. Primary CMV infections or viral reactivations can cause life-threatening diseases in immunologically immature or compromised individuals, including AIDS patients, transplant recipients, and congenitally infected neonates (34). Completion of one productive infectious cycle takes about 48 to 96 h in primary human fibroblasts, which represent the standard system for the study of lytic CMV infection. Following host cell entry, viral capsids are transported to the nuclear pores, where they release the ∼230,000-bp linear double-stranded DNA genomes into the nucleus. After that, the CMV genomes are rapidly circularized and serve as templates for transcription and replication. Viral transcription and genome duplication proceed within discrete nuclear inclusions that develop from small sites known as promyelocytic leukemia bodies or nuclear domain 10 (8) and take over large parts of the nuclear space at late times postinfection. During productive infection, roughly 200 CMV genes are expressed in a triphase cascade pattern (34). Immediate-early (IE) gene products are transcribed within the first hours postinfection in the absence of de novo viral-gene expression and protein synthesis. They include the major IE proteins IE1-72kDa and IE2-86kDa, which antagonize innate and intrinsic immune responses and activate expression of early gene products (9, 32, 42, 53, 54). The early proteins are typically involved in the process of viral-DNA replication, which starts around 24 h post-CMV infection in primary human fibroblasts. It has been proposed that the initial steps in DNA replication of herpesviruses take place by a theta-like mechanism, followed by a rolling-circle mechanism at later stages of the lytic infection cycle (34). Activation of late gene expression requires viral-DNA synthesis and produces the regulatory and structural proteins involved in progeny virion assembly and release.
From the earliest work, the genomes of herpesviruses were believed to be completely free of histones when encapsidated in virions (3, 4, 12, 16, 27, 43). This view has been confirmed by more recent work, including Western blotting experiments with purified herpes simplex virus type 1 (HSV-1) capsids (41) and mass spectrometry-based proteome analyses of Epstein-Barr virus (EBV), Kaposi's sarcoma-associated herpesvirus, and CMV particles (33). A series of studies also provided evidence for an exclusively, or at least predominantly, nonnucleosomal structure of intranuclear herpesvirus genomes in productively infected cells (27, 28, 36, 37, 43, 50). Furthermore, it has been reported that HSV-1 DNA accumulates in replication compartments that exclude histones (35, 49), and no evidence for replication-coupled viral-chromatin assembly was found in these infections (41). Nonetheless, recent chromatin immunoprecipitation (ChIP) experiments have revealed that histones H3 and H4, as well as posttranslationally modified core histone forms, are associated with HSV-1 and CMV genomes inside lytically infected cells (18, 20, 23, 38, 40, 41, 46). Dynamic changes in histone occupancy likely have major impacts on viral DNA metabolism, including transcription, replication, and repair.
Here, we systematically analyzed the dynamics of histone and nucleosome occupancy on CMV episomes during the temporal course of a productive infection in resting primary human fibroblasts. Our results define discrete, consecutive stages of CMV genome chromatinization in these cells, including DNA replication-independent and -dependent mechanisms. The data also reveal that viral genomes become more extensively chromatinized than has been anticipated during the lytic cycle of herpesvirus infection.
MATERIALS AND METHODS
Cell culture and virus infections.
Human fetal diploid lung fibroblasts (MRC-5) were obtained from the European Collection of Cell Cultures and grown in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. The cells were cultured at 37°C in a humidified 5% CO2 atmosphere, and growth-arrested early-passage cells (15 to 25 population doublings before senescence) were used in all experiments. To arrest fibroblasts by contact inhibition, cultures were kept in a fully confluent state for 10 days with a single medium change after 3 days. Virus was grown on MRC-5 cells, and the titers were determined by standard plaque assay on growth-arrested fibroblasts. All infections were carried out at a multiplicity of 5 PFU per cell with the Towne strain of CMV (32). The DNA polymerase inhibitors aphidicolin and phosphonoacetic acid (PAA) were purchased from Sigma-Aldrich.
MNase accessibility assay.
Micrococcal-nuclease (MNase) digestion of chromatin in permeabilized cells, followed by purification and characterization of digested DNA, was done according to the method of Zaret (58). MRC-5 cells were grown and infected in six-well dishes. At the desired time points, the cells were washed with 1 ml permeabilization solution I (150 mM sucrose, 80 mM KCl, 35 mM HEPES [pH 7.4], 5 mM K2HPO4, 5 mM MgCl2, 0.5 mM CaCl2). After the solution was aspirated, 360 μl of 0.1% lysolecithin (prewarmed to 37°C) was added, and the solution was incubated for 2 min at 37°C. After another washing step with 1 ml permeabilization solution I, 360 μl of permeabilization solution II (150 mM sucrose, 50 mM Tris [pH 7.5], 50 mM NaCl, 2 mM CaCl2) containing between 24 U and 96 U of MNase (Roche) was added. The samples were incubated for 1.5 to 30 min at room temperature or 37°C with gentle agitation. The solution was aspirated, and cell lysis was performed by adding 360 μl of 2× TNESK buffer (20 mM Tris [pH 7.4], 200 mM NaCl, 2 mM EDTA, 2% sodium dodecyl sulfate [SDS], 70 μg proteinase K [Roche]). Once lysis had occurred, 360 μl of lysis dilution buffer (150 mM NaCl, 5 mM EDTA) was added, and after the remaining cells were scraped off, the suspension was transferred to a reaction tube and incubated for 15 h at 37°C for complete lysis. DNA extraction was performed twice with phenol-chloroform/isoamyl alcohol (25:24:1) and once with chloroform in Phase Lock Gel Heavy tubes (Eppendorf). Exactly 600 μl of the final aqueous phase was incubated with 0.25 μg of DNase-free RNase (Roche) for 1 h at 37°C. DNA precipitation was performed by addition of 60 μl 3 M sodium acetate (pH 5.2), 20 μg glycogen (Roche), and 1.65 ml ethanol. As soon as a white stringy precipitate was visible, it was spooled out with a glass microcapillary tube, washed briefly in 70% ethanol, air dried for a few seconds, and dissolved in 60 μl of TE buffer (10 mM Tris [pH 8.1], 1 mM EDTA). The rest of the sample was incubated overnight at −20°C to precipitate shorter DNA fragments. After centrifugation (30 min; 4°C; 16,000 × g), the DNA pellet was washed with 70% ethanol, air dried, and dissolved in 60 μl TE buffer containing the spooled-out DNA.
MNase-digested and untreated control samples (13 μl) were loaded on 1.2% agarose gels and run for 3 h at 120 V in TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA). The gel was stained with 0.5 μg/ml ethidium bromide in double-distilled water for 30 min, photographed, and washed twice for 30 min in ample denaturation solution (3 M NaCl, 400 mM NaOH). Following 15 min of incubation in transfer solution (3 M NaCl, 8 mM NaOH), the transfer stack was assembled using a TurboBlotter unit with a Nytran SuperCharge membrane (Whatman) that had been equilibrated in transfer solution. Downward capillary transfer occurred overnight, after which the positively charged nylon membrane was incubated for 5 min in neutralizing buffer (200 mM sodium phosphate [pH 6.8]). DNA was cross-linked in a Stratalinker 1800 UV cross-linker from Stratagene (240 mJ/cm2). Prehybridization was performed at 68°C for at least 4 h in hybridization buffer (6× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt's solution, 0.5% SDS, 100 μg/ml denatured salmon sperm DNA). For preparation of the hybridization probe, 90 ng of RsaI-digested Towne bacterial artificial chromosome DNA was denatured for 3 min at 95°C in a volume of 45 μl, cooled quickly in ice water, mixed with 50 μCi (5 μl) [32P]dCTP (GE Healthcare) and an aliquot of All-In-One Random Prime DNA Labeling Mix (without dCTP) from Sigma, and incubated for 30 min at 37°C. Purification of the labeled probe was done using Sephadex G-50 columns (Roche). Hybridization was carried out overnight with fresh hybridization buffer containing the denatured 32P-labeled probe. Afterward, the membrane was washed twice for 15 min in 2× SSC containing 0.1% SDS, twice for 20 min in 0.1× SSC with 0.1% SDS, and briefly in 0.1× SSC. All washing steps were performed at 68°C. The damp membrane was placed in a development folder and exposed for an appropriate time to a Bio-Rad Imaging Screen K, which was finally scanned in a Molecular Imager FX from the same company.
FAIRE.
Formaldehyde-assisted isolation of regulatory elements (FAIRE) analysis was done essentially following a published protocol (11). Growth-arrested MRC-5 cells (6 × 107) were infected with CMV for 2, 16, or 48 h. One half of the cells were cross-linked for 10 min at 37°C with 1% formaldehyde added directly to the culture medium, followed by a 5-min incubation with 125 mM glycine at room temperature to stop the reaction. The cells were washed twice with ice-cold phosphate-buffered saline without calcium and magnesium ions (PBS), scraped into ice-cold serum-free culture medium, and collected by low-speed centrifugation (800 × g; 10 min; 4°C). Subsequently, the supernatant was removed and the cell pellet was snap-frozen in liquid nitrogen. The second half of the cells were prepared in the same way but without formaldehyde and glycine treatment. After thawing in ice water, the pellet was carefully resuspended in 1 ml SDS lysis buffer (50 mM Tris [pH 8.1], 10 mM EDTA, 1% SDS, 1% protease inhibitor cocktail III [Calbiochem]), and the lysate was incubated on ice for 10 min. Chromatin was sheared into fragments of predominantly 300 to 500 bp by sonication (Branson Sonifier 450; seven 15-s pulses at setting 2.5). After centrifugation (10 min; 16,000 × g; 4°C), a 200-μl aliquot of the supernatant was diluted with 200 μl TE buffer and extracted twice with phenol-chloroform/isoamyl alcohol (25:24:1) and once with chloroform in Phase Lock Gel Heavy tubes. DNA from 300 μl of the aqueous phase was subsequently precipitated by the addition of 30 μl 3 M sodium acetate (pH 5.2), 20 μg glycogen, and 830 μl ethanol. This step was followed by an overnight incubation at −20°C. After that, the precipitate was collected by centrifugation for 30 min at 16,000 × g and 4°C. The pellet was washed with 70% ethanol, air dried, and resuspended in 40 μl double-distilled water. Quantification of purified DNA (0.05 to 5%, depending on the cross-linking state, locus, and postinfection time point analyzed) was carried out by real-time PCR using the LightCycler Fast Start DNA MasterPLUS Sybr green I kit from Roche according to the manufacturer's instructions. The primer sequences and PCR conditions are listed in Table 1. The identities of the PCR products were verified by melting-curve analysis. DNA levels were calculated using the efficiency-corrected relative-quantification strategy described in Roche Applied Science Technical Note no. LC 13/2001.
TABLE 1.
Oligonucleotides used in this study
| Name or locus | Orientationa | Sequence (5′→3′) | Temp (°C)b | Time (s)c | PCR product (bp) | Efficiencyd |
|---|---|---|---|---|---|---|
| MIE-P | Fw | CTTACGGGACTTTCCTACTTG | 58 | 15 | 284 | 1.87 |
| Rv | CGATCTGACGGTTCACTAA | |||||
| MIE-T | Fw | CCTAGTGTGGATGACCTA | 58 | 8 | 112 | 1.88 |
| Rv | GTGACACCAGAGAATCAG | |||||
| UL54-P | Fw | CACCAAAGACACGTCGTT | 58 | 8 | 71 | 1.94 |
| Rv | GTCCTTTGCGACCAGAAT | |||||
| UL54-T | Fw | GTGTGCAACTACGAGGTA | 58 | 8 | 113 | 1.96 |
| Rv | GACAGCACGTTGGTTACA | |||||
| UL32-P | Fw | ACTGGTACTGCGGTTCTA | 58 | 8 | 150 | 1.88 |
| Rv | TCCACAACACCACGGTGA | |||||
| UL32-T | Fw | CCGATTACAACGACGTCA | 66-56e | 8 | 106 | 1.77 |
| Rv | GTGGATGTCGTCGTCATT | |||||
| oriLyt | Fw | GAATACAGCGATCCCTAG | 58 | 10 | 171 | 1.86 |
| Rv | GGGTTCCACCTATCTGAA | |||||
| GAPDH | Fw | CCTCACAGTTGCCATGTA | 66-56e | 8 | 71 | 1.97 |
| Rv | GATGGTACATGACAAGGTG |
Fw, forward primer; Rv, reverse primer.
Annealing temperature used for PCR.
Polymerization time used for PCR.
PCR amplification efficiency determined as described in Roche Applied Science Technical Note no. LC 13/2001.
“Touch-down” conditions were used.
ChIP assay.
Approximately 3 × 107 growth-arrested MRC-5 cells were infected with CMV for 30 min to 96 h. Formaldehyde was added to the medium at a final concentration of 1% to cross-link protein-DNA complexes, and the culture dishes were incubated for 10 min at 37°C. Subsequently, glycine was added to a final concentration of 125 mM to stop the cross-linking reaction, and the cells were incubated for 5 min at room temperature. After aspiration of the supernatant, the cells were scraped into ice-cold culture medium and collected by centrifugation (800 × g; 10 min) at 4°C. After that, the cell pellet was washed twice in ice-cold PBS, frozen in liquid nitrogen, and stored at −80°C. After thawing in ice water, the pellet was carefully resuspended in 1 ml lysis buffer (50 mM Tris [pH 8.1], 10 mM EDTA, 1% SDS, 1% protease inhibitor cocktail III) and incubated on ice for 10 min. Chromatin was sheared into fragments of predominantly 300 to 500 bp by applying seven 15-s pulses at setting 2.5 using a Branson Sonifier 450 probe tip. Samples were allowed to cool in ice water for 2 min between the sonication steps. After repeated centrifugation (10 min; 16,000 × g; 4°C), the clarified supernatant was separated into five 200-μl aliquots and an input sample. Each 200-μl aliquot was then diluted 10-fold in lysis dilution buffer (16.7 mM Tris [pH 8.1], 167 mM NaCl, 1.2 mM EDTA, 1.1% Triton X-100, 0.01% SDS, 1% protease inhibitor cocktail III) and rotated for 30 min with 75 μl protein A agarose/salmon sperm DNA slurry (Millipore) at 4°C prior to centrifugation (1 min; 100 × g; 4°C). The supernatant was incubated with the appropriate antibody (listed in Table 2) at 4°C overnight with gentle rotation. Ten micrograms of normal rabbit immunoglobulin G (IgG) served as a negative control. Antigen-antibody complexes were precipitated by incubation with 60 μl protein A agarose/salmon sperm DNA slurry at 4°C with gentle rotation, followed by centrifugation. The supernatant was aspirated, and the pellet was washed consecutively with 1 ml each of low-salt buffer (20 mM Tris [pH 8.1], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), high-salt buffer (20 mM Tris [pH 8.1], 0.5 M NaCl, 2 mM EDTA, 1% Triton X-100, 0.1% SDS), lithium chloride (LiCl) buffer (10 mM Tris [pH 8.1], 0.25 M LiCl, 1 mM EDTA, 1% Igepal-CA630, 1% deoxycholic acid), and twice with TE buffer. Elution of the chromatin-antibody complexes was carried out by incubation with 250 μl freshly prepared elution buffer (100 mM NaHCO3, 1% SDS) at 65°C for 15 min, followed by 15 min of rapid rotation at room temperature. After centrifugation, 200 μl of the supernatant was removed, and the elution procedure was repeated with another 250 μl elution buffer. This time, 250 μl of the supernatant was removed and pooled with the first eluate. NaCl was added to a final concentration of 170 mM, and each sample was incubated for 5 h at 65°C to reverse the chemical cross-linking. Five microliters of the input chromatin sample was diluted 20-fold in lysis dilution buffer and reverse cross-linked, as well. Then, EDTA and Tris (pH 6.5) were added to a final concentration of 9 mM and 36 mM, respectively. After incubation at 37°C for 1 h in the presence of 20 μg proteinase K, the DNA was extracted twice with phenol-chloroform/isoamyl alcohol (25:24:1) and once with chloroform using Phase Lock Gel Heavy tubes. To 400 μl of each ChIP sample and 90 μl of each input sample, sodium acetate was added to a final concentration of 270 mM. Then, the input and output samples were mixed with 20 μg glycogen and precipitated with 2.5 volumes of ethanol overnight at −20°C. Subsequently, the DNA was collected by centrifugation (30 min; 16,000 × g; 4°C), washed once with 70% ethanol at room temperature, air dried, and resuspended in 40 μl double-distilled water. Quantification of DNA was done by real-time PCR as described above. Between 0.05 and 5% of the final DNA material was used per PCR, depending on the type of sample, locus, and postinfection time point analyzed.
TABLE 2.
Antibodies used in this study
| Antigen | Name or no. | Source | Typea | IFb | ChIPc | WBd |
|---|---|---|---|---|---|---|
| ASF1A | 87 | 59 | R | 1:500 | NA | 1:4,000 |
| CAF1 p48 | ab1765 | Abcam | R | 1:500-1:2000 | NA | 1:2,000 |
| CMV IE2 | 3A9 | Unpublishede | M | 1:20 | NA | NA |
| CMV ppUL44 | GTX26501 | GeneTex | M | 1:1,000 | NA | 1:4,000 |
| GAPDH | ab9485 | Abcam | R | NA | NA | 1:2,000 |
| H2A | ab18255 | Abcam | R | 1:200 | 20 | NA |
| H2B | ab1790 | Abcam | R | 1:500 | 10 | NA |
| H3 | 9715 | Cell Signaling | R | 1:100 | NA | NA |
| H3 | ab1791 | Abcam | R | NA | 10 | NA |
| H3K9me2 | 07-441 | Upstate | R | 1:250 | NA | NA |
| H4 | ab10158 | Abcam | R | 1:100 | 10 | NA |
| PCNA | ab18197 | Abcam | R | 1:2,000 | NA | 1:2,000 |
R, rabbit polyclonal; M, mouse monoclonal.
Dilution in PBS-T with 0.02% human IgG. IF, immunofluorescence. NA, not applicable.
Amount used for one sample (microliters). NA, not applicable.
Dilution in PBS. WB, Western blotting. NA, not applicable.
A. Marchini, H. Zhu, and T. Shenk, unpublished data.
Immunofluorescence microscopy.
MRC-5 fibroblasts on no. 1 coverslips (18 by 18 mm) were mock infected or infected with CMV. At 8 to 72 h postinfection, the cells were rinsed briefly in PBS with 0.05% Tween 20 (PBS-T), fixed with paraformaldehyde (2% in PBS) for 15 min at room temperature, washed three times for 5 min each time in PBS-T, and permeabilized for 15 min with Triton X-100 (0.1% in PBS). This was followed by three 5-min washes in PBS-T. As an alternative to Triton X-100, cells were treated with methanol for 10 min at −20°C, followed by two short wash steps in PBS-T. After fixation and permeabilization, samples were incubated for 1 h at room temperature with 100 μl blocking solution (2% bovine serum albumin in PBS-T). To prevent nonspecific binding of rabbit IgG to CMV-encoded Fcγ receptors (2), IgG from human serum (Sigma-Aldrich) was added to the blocking solution at a final concentration of 0.02%. After a wash step in PBS-T for 5 min, each sample was reacted with 75 μl primary antibody solution in PBS-T with 0.02% human IgG containing a combination of a CMV IE2- or ppUL44-specific mouse monoclonal antibody and one of several polyclonal rabbit sera directed against human histone or chromatin assembly proteins (Table 2). After a 1-h incubation at room temperature with the primary antibodies, samples were washed three times for 5 min each time in PBS-T and incubated under light protection for 1 h with 75 μl PBS-T containing the following two antibody conjugates, each at a 1:1,000 dilution (2 μg/ml): Alexa Fluor 594 goat anti-mouse IgG (H+L) (highly cross-adsorbed) and Alexa Fluor 488 goat anti-rabbit IgG (H+L) (highly cross-adsorbed) (Invitrogen). In a subset of experiments 4′,6-diamidino-2-phenylindol (DAPI) (Roche) was added to the secondary-antibody solution at a final concentration of 0.2 μg/ml. After that, coverslips were washed three times in PBS-T and once in PBS and mounted on glass slides using ProLong Gold or SlowFade Gold antifade reagent (Invitrogen). Where DAPI was not used, DRAQ5 (5 mM; Alexis) was diluted 1:3,000 in SlowFade Gold mounting medium. Slides were analyzed and images acquired using a Leica DMRX epifluorescence microscope equipped with a digital camera system (Retiga; Q-Imaging) or a Zeiss LSM 510 Meta confocal microscope. Images were cropped and processed using Image-Pro Plus 6.2 (Q-Imaging), Zeiss LSM 510, and/or Adobe Photoshop CS (version 8.0) software.
Western blot analysis.
Whole-cell extracts were prepared by sonication in lysis buffer (50 mM Tris [pH 8.0], 50 mM NaCl, 0.1% SDS, 1% Igepal CA-630, 0.5% sodium deoxycholate), and proteins were analyzed as previously described (42). For a list of antibodies, see Table 2.
RESULTS
Nucleosomal arrangement of CMV genomes during productive infection.
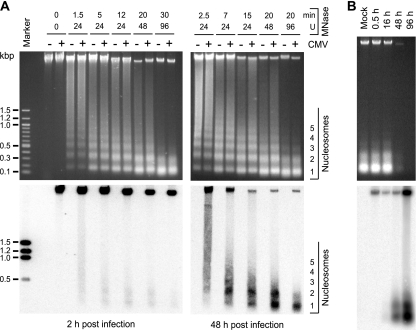
Previous reports have shown core histone association of intracellular CMV genomes, but to our knowledge, a nucleosomal arrangement of DNA from the virus has not been unequivocally demonstrated. MNase digestion was used to determine whether the CMV genome becomes associated with nucleosomes during productive infection. MRC-5 cells were mock infected or infected with the CMV Towne strain, and permeabilized cells were treated with MNase at early (2-h) and late (48-h) postinfection time points. Different amounts of enzyme and various incubation times were used to generate partial to almost complete digestion of cellular and viral genomic DNA. DNA fragments were visualized in stained agarose gels (Fig. 1A, top). DNA laddering was detectable only in samples of digested cells and not in control genomic DNA that was either not treated with MNase (0 U; 0 min) (Fig. 1A) or purified free of protein prior to digestion (data not shown).
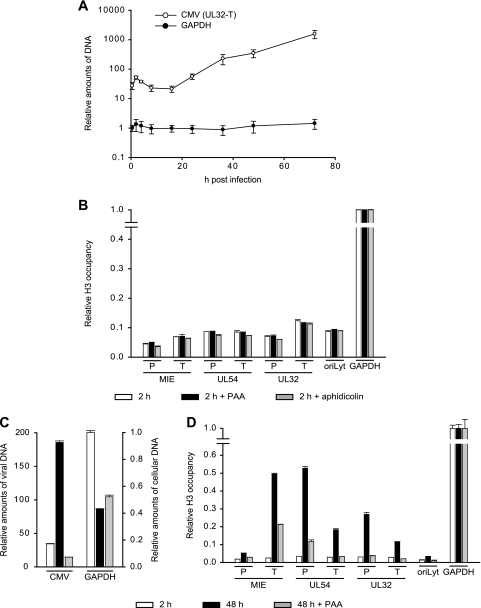
FIG. 1.
Nucleosomal arrangement of intracellular CMV DNA during productive infection. (A) MRC-5 cells were mock infected or infected with CMV. At 2 or 48 h postinfection, permeabilized cells were reacted with increasing amounts of MNase for the indicated incubation times at room temperature (24 and 48 U) or 37°C (0 and 96 U). DNA was prepared and separated in ethidium bromide-stained 1.2% agarose gels (top). The same samples were transferred to nylon membranes and hybridized to a 32P-labeled probe derived from a bacterial artificial chromosome clone of the complete CMV (Towne) genome (32) (bottom). To ensure comparable signal intensities despite varying amounts of viral DNA, a shorter film exposure was chosen for the 48-h than for the 2-h samples. Marker, 100-bp DNA ladder (New England Biolabs). (B) At the indicated postinfection time points, permeabilized cells were incubated for 30 min with 96 U MNase at 37°C, and samples were treated as described for panel A. For the 48- and 96-h samples, only 1/10 or 1/50 of the DNA amounts used in the mock and 0.5- and 16-h lanes were loaded, respectively.
The DNA isolated from MNase-treated cells and separated in agarose gels was subjected to Southern blotting and hybridized to a radioactively labeled DNA probe encompassing the entire CMV genome (Fig. 1A, bottom). No signals were detected in samples from mock-infected cells, while bands of full viral-genome-length size and specific digestion products were evident in all lanes corresponding to CMV-infected cells. At 2 h after infection, the viral genomic DNA proved to be largely resistant to MNase digestion, even after extended incubation with high enzyme concentrations. This observation suggests that most CMV DNA molecules were still present within capsids at this postinfection time point. However, a small proportion of the total DNA was detected in mono- and dinucleosome size fragments under intermediate MNase reaction conditions. This observation shows that at least a fraction and/or subgenomic parts of the input viral DNA molecules were already assembled in nucleosomes by 2 hours after the initial virus-cell contact. In the late phase of infection (48 h), a substantially (>10-fold) larger proportion of total viral DNA was found in nucleosomal fragments compared to the early time point. Again, mainly mono- and dinucleosome size bands were observed. Improved detection of nucleosomal fragments at 48 versus 2 h postinfection may have resulted from more extensive CMV genome chromatinization. Alternatively, it may simply reflect the differences in global accessibility of viral DNA to nuclease digestion. To discriminate between these possibilities, viral chromatin was examined at additional postinfection time points by MNase assay (Fig. 1B). Comparison between samples from 16 and 48 h postinfection clearly demonstrates an increase in nucleosome assembly at similar overall accessibilities of viral DNA.
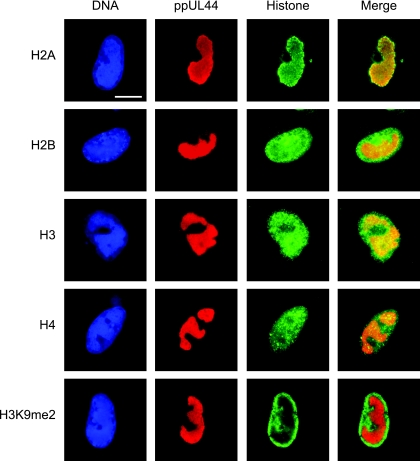
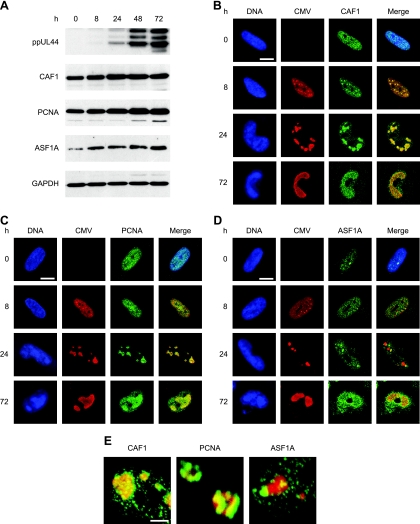
The presence of core histones in the subnuclear compartments of viral-DNA accumulation was confirmed by indirect immunofluorescence microscopy using primary antibodies specific for total H2A, H2B, H3, or H4 protein. The viral replication compartments were simultaneously stained with an antibody directed against the CMV DNA polymerase accessory subunit ppUL44 (Fig. 2). Singly stained histone patterns were indistinguishable from double-staining results, and none of the four histone-specific antibodies detected cross-reacting bands on Western blots of CMV-infected or uninfected cell lysates (data not shown). In uninfected cells, each of the four human histone species displayed a relatively even, diffuse, or micropunctate nuclear distribution (data not shown). At late times after CMV infection, all core histones were detected, both at the centers of viral replication and in the nuclear space outside these structures. However, differences were observed in the extents to which individual histones colocalized with the viral nuclear compartments. H2A was strongly enriched in the periphery and, to a lesser extent, within the viral structures. While H3 was also found to codistribute with viral replication compartments, this was less evident for H4 and H2B. In fact, H2B appeared to be more abundant outside the viral structures. A modified histone form (H3 dimethylated at lysine 9) known to be underrepresented in CMV chromatin (20; A. Nitzsche, C. Paulus, and M. Nevels, unpublished data) was largely excluded from the viral replication compartments, thus serving as a negative control. In addition, ChIP assays revealed that all four human core histone classes were physically associated with the CMV genome at both early and late times postinfection (see Fig. 4 and data not shown).
FIG. 2.
Subnuclear distribution of core histones in relation to late viral replication compartments. MRC-5 cells were infected with CMV for 48 h, fixed with paraformaldehyde/methanol, and stained with a mouse monoclonal antibody specifically detecting the CMV ppUL44 DNA polymerase accessory protein and rabbit polyclonal antibodies directed against the C-terminal histone fold domain of H2A, H2B, H3, or H4. A rabbit antiserum specific for histone H3 dimethylated at lysine 9 (H3K9me2) was used as a negative control. Samples were subsequently stained with DAPI, a mouse-specific Alexa Fluor 594, and a rabbit-specific Alexa Fluor 488 conjugate. Representative nuclei showing DAPI, ppUL44, and the respective histone staining are shown. Additionally, merge images of ppUL44 and histone signals are presented. Scale bar, 10 μm.
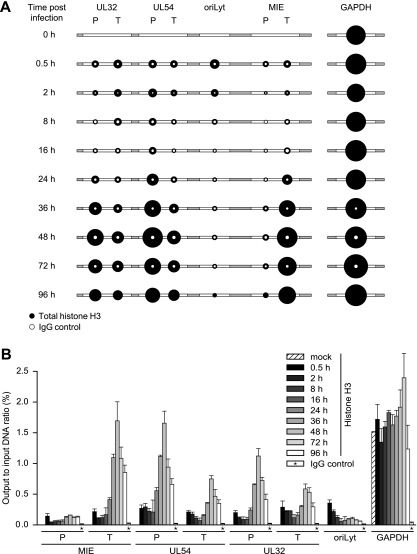
FIG. 4.
Temporal patterns of histone H3 occupancy in selected regions of the CMV genome. MRC-5 cells were infected with CMV, and cell extracts were subjected to ChIP using an antibody specifically directed against the C-terminal domain of H3 at the indicated times (0.5 to 96 h) postinfection. Normal rabbit IgG was used to control for nonspecific precipitation. Quantitative PCR was performed on input and coprecipitated (output) DNAs with primers specific for the indicated viral genomic regions and human GAPDH. (A) The circular areas represent the mean output-to-input DNA ratios determined from at least two independent ChIP assays, each quantified in duplicate. Where white circles are missing, no specific PCR products were detected. (B) The data set used for the schematic representation in panel A is shown as bars (mean values) with standard deviations. For the IgG controls, average values from all 10 time points are shown.
These results indicate that during productive infection of human fibroblasts, CMV genomes become extensively occupied with host-derived core histones, most likely forming canonical nucleosomes. These data also present the first indication that nucleosome association of the viral genome increases between the early and late stages of infection.
FAIRE analysis confirms differential early and late states of CMV chromatin.
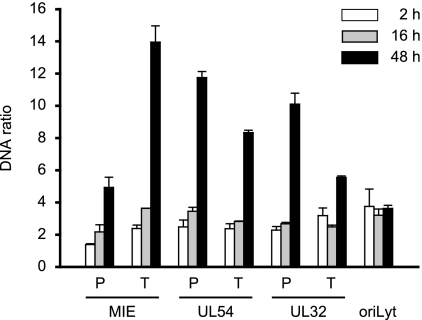
To further examine the possibility that CMV genomes exist in distinct early and late states that differ in the extents of chromatinization, we employed a recently developed method known as FAIRE (11, 39). The FAIRE procedure provides a means of enriching nucleosome-depleted DNA fragments from total chromatin. This DNA can then be quantified by real-time PCR at specific regions of interest. The reciprocal values from the PCR results are indicative of the extents of nucleosome occupancy at the respective sequences. We applied FAIRE to seven carefully selected viral indicator regions. The indicator loci were spaced throughout the unique long (UL) region of the viral genome and represented the three kinetic classes of herpesvirus genes and a marker for viral-DNA replication: the major IE (MIE) transcription unit, the UL54 early gene, the UL32 late gene, and the core element from the origin of lytic viral-DNA replication (oriLyt) (34). Concerning the coding genes, we analyzed both promoter sequences surrounding the transcription start sites (MIE-P, UL54-P, and UL32-P), as well as sequences at the 3′ ends of the open reading frames (MIE-T, UL54-T, and UL32-T).
FAIRE was performed at 2 and 48 h post-CMV infection of MRC-5 cells (Fig. 3). At the early time point, substantially larger amounts of FAIRE-extracted DNA were detected from all seven examined viral genomic regions compared to a GAPDH (glyceraldehyde-3-phosphate dehydrogenase) control, indicating a much lower degree of nucleosome association with the CMV genome than the cellular locus (data not shown). At 48 h, the FAIRE results indicated up to a sixfold-higher degree of chromatinization at the six viral coding genes compared to the 2-h time point. At the same time, we observed only a little (<1.5-fold) change in the oriLyt and GAPDH (Fig. 3 and data not shown). The FAIRE results strongly support the view of distinct early and late states of CMV chromatin that differ markedly in the extents of nucleosome assembly. They also indicate that not all CMV genomic regions are equally affected by the late increase in viral-genome chromatinization.
FIG. 3.
Differential chromatinization of the CMV genome at early versus late times postinfection as analyzed by FAIRE. MRC-5 cells were infected with CMV and collected at 2, 16, or 48 h postinfection. Enrichment for nucleosome-depleted chromatin by FAIRE extraction was performed, and DNA from the aqueous phase was quantified by real-time PCR using primer pairs specific for seven selected viral indicator loci and human GAPDH. The results were normalized to GAPDH and are representative of triplicate experiments with standard deviations. They are presented as the ratio of viral DNA recovered from non-formaldehyde-fixed cells divided by the amounts of the same DNA in the corresponding cross-linked samples. The data therefore reflect the degrees of nucleosome assembly in the respective viral genomic regions.
Temporal pattern of histone occupancy on CMV genomes in productively infected cells.
To assess the dynamics of nucleosome association of the CMV genome in a more specific and comprehensive manner, we performed a series of ChIP assays at the seven previously selected viral indicator loci. The analysis was carried out using a rabbit-derived antibody directed against the globular domain of core histone H3 precipitating total amounts of the protein as a surrogate marker of nucleosomes. As a negative control, we used normal rabbit IgG. MRC-5 cells infected with CMV were subjected to ChIP, followed by quantitative PCR at nine time points between 0.5 and 96 h postinfection. Importantly, both the input and the precipitated (output) DNAs were quantified, and the results are presented as relative output-to-input ratios to account for the varying copy numbers of viral genomes at different stages during infection (Fig. 4). Histone H3 was specifically detected above the normal IgG background in all viral and cellular genomic regions and postinfection time points under investigation. However, striking time- and locus-dependent differences in H3 association of the CMV chromosome were observed, while histone levels at the GAPDH gene showed only minor changes. At 0.5 h postinfection, comparable amounts of H3 protein could be detected at the selected CMV sequences with only ∼2-fold variations between the individual viral loci. This observation supports the view that sequences in at least a subset of CMV genomes become assembled into nucleosomes immediately upon nuclear entry. Importantly, the H3 levels in the CMV DNA were ∼5- to >10-fold lower than the cellular control locus, suggesting a relatively low average degree of viral-genome chromatinization at this very early stage of infection. Following the initial histone deposition, H3 levels in the tested regions of the viral chromosome remained constantly low or even decreased throughout the early phase of infection. For example, at the MIE-P and the oriLyt, the amounts of DNA-associated histone H3 consistently dropped by 2.4- or >6-fold, respectively, in the 0.5-h to 16-h postinfection interval. In contrast, at the MIE-T and the UL54-P, histone levels did not change significantly (<1.3-fold) between the same time points unless the values were normalized to the IgG control. The early phase of low histone occupancy was succeeded by a striking, gradual increase in H3 deposition at the CMV chromosome starting from 24 h and peaking at 48 h postinfection (Fig. 4). At 48 h, the increase in H3 occupancy averaged between ∼5-fold (UL32-T) and ∼12-fold (UL32-P) at five out of the seven tested viral loci compared to the 16-h time point. At the UL54-P and MIE-T sequences, H3 even reached levels comparable to those of the GAPDH gene at this stage. However, the late histone deposition did not affect all parts of the CMV genome to similar extents, as we observed only little change at the MIE-P and the oriLyt (2.7- and 1.8-fold increases, respectively). Finally, between 48 h and 96 h post-CMV infection, a moderate (up to ∼2-fold) reduction in H3 occupancy was measured across all viral loci. For a subset of regions and time points, ChIP experiments were repeated using antibodies directed against other core histones (H2A and H2B) with results comparable to those for H3 (data not shown).
Taken together, our ChIP data confirm the presence of two major, discrete stages of net histone deposition on intracellular CMV genomes: an early stage in which histones become initially deposited and a late stage of much more efficient histone association in a subset of, but not all, viral genomic regions. Both assembly stages are followed by a phase of reduced average histone occupancy indicative of partial chromatin disassembly.
CMV chromatin assembly by DNA replication-independent and replication-coupled mechanisms.
Chromatin assembly in eukaryotic cells can be divided into DNA replication-dependent and -independent mechanisms (44, 47). During infection of primary human fibroblasts, CMV DNA replication is known to start at around 24 h (34). We confirmed this notion by quantitative PCR in which viral-DNA levels did not rise at time points up to 16 h but increased continuously between 24 and 72 h postinfection (Fig. 5A). This strongly suggests that the observed initial histone deposition on the CMV genome (0.5 to 16 h postinfection) is mediated by mechanisms that are independent of DNA synthesis. To exclude the possibility that early viral chromatin assembly occurs in conjunction with low-level DNA replication mediated by viral or cellular DNA polymerases, we performed ChIP assays of cells infected for 2 h in the presence of PAA or aphidicolin. PAA is a specific inhibitor of the CMV DNA polymerase, whereas aphidicolin inhibits both CMV and cellular DNA polymerase alpha activities (7, 19). PAA and aphidicolin were used at concentrations that were confirmed to result in complete inhibition of the respective enzyme activities (Fig. 5C and data not shown). As expected, neither PAA nor aphidicolin had significant effects on the levels of histone H3 deposition regarding all seven indicator regions of the CMV genome at 2 h postinfection (Fig. 5B). These results verify that early CMV chromatin assembly occurs through DNA replication-independent mechanisms.
FIG. 5.
Consecutive stages of replication-independent and -dependent histone H3 deposition on the CMV genome. (A) MRC-5 cells were infected with CMV for 0.5 to 72 h, relative amounts of viral and cellular DNAs were determined at the UL32-T and GAPDH loci by quantitative PCR, and the results were normalized to GAPDH at 0.5 h (set to 1). The data present the mean amounts of DNA from three independent experiments with standard deviations. (B) MRC-5 cells were pretreated with PAA (200 μg/ml) or aphidicolin (2 μM) for 1 h or left untreated prior to infection with CMV. Infection continued for 2 h in the presence of the inhibitors. ChIP was performed using an antibody against the C-terminal domain of histone H3, and the amounts of input and coprecipitated DNA were determined by quantitative PCR with eight specific primer pairs, as indicated. PCR results from two independent experiments, each quantified in duplicate, are presented as mean output-to-input DNA fractions normalized to the output-to-input ratio of GAPDH without drug treatment (set to 1). The error bars indicate standard deviations. (C) MRC-5 cells were infected with CMV and either left untreated or treated with PAA (200 μg/ml) immediately following infection. After 24 h, the culture medium was changed, and fresh drug was added where applicable. Samples were collected at 2 and 48 h postinfection. Relative amounts of viral (means of all seven viral indicator loci) and cellular (GAPDH) DNAs were determined by quantitative PCR and normalized to GAPDH at 2 h postinfection (set to 1). The bars represent mean values with standard deviations from three experiments. (D) The samples from panel C were subjected to ChIP as described for panel B. PCR results from two independent experiments, each quantified in duplicate, are presented as mean output-to-input DNA fractions normalized to the output-to-input ratio of GAPDH at 2 h (set to 1). The error bars indicate standard deviations.
The late increase in histone deposition on the CMV chromosome temporally correlates with the onset of viral-DNA replication (compare Fig. 4 and 5A). This observation suggests that late CMV chromatin assembly may be coupled with the process of viral-DNA synthesis. To investigate this possibility, we performed ChIP assays of PAA-treated and untreated cells at 48 h post-CMV infection. Again, PAA was used under conditions that completely inhibited viral-DNA synthesis but did not negatively affect cell survival or the levels of GAPDH DNA (Fig. 5C and data not shown). As shown in Fig. 5D, PAA specifically inhibited the late increase in H3 deposition on the viral genome either entirely (UL54-T, UL32-P, UL32-T, and oriLyt) or at least to a large extent (MIE-P, MIE-T, and UL54-P), while histone occupancy at the GAPDH locus was not affected. From these results, we conclude that, for the most part, late CMV chromatin assembly depends on and is most likely coupled to the process of viral-DNA replication. The different sensitivities to PAA in the tested viral loci may indicate that in some regions of the CMV genome, DNA replication-dependent and -independent mechanisms of chromatin assembly act side by side in the late phase of infection.
Increased levels and relocalization of human nucleosome assembly proteins during CMV infection.
CMV genome chromatinization likely involves interactions between viral components and proteins of the cellular chromatin assembly machinery. To obtain evidence for this view, we analyzed the accumulation and subcellular localization of three key mediators of human nucleosome assembly in CMV-infected fibroblasts: CAF1 p48, PCNA, and ASF1A. ASF1A and CAF1 are histone chaperones involved in H3-H4 deposition, while PCNA is a DNA polymerase processivity factor that targets CAF1 to chromatin via direct interaction (44, 47). Surprisingly, the steady-state levels of all three cellular proteins increased significantly over the course of infection while the levels of GAPDH did not change (Fig. 6A). In uninfected cells, CAF1, PCNA, and ASF1A were predominantly found in a micropunctate nuclear pattern (Fig. 6B to D). ASF1A was additionally detected in one to several larger intranuclear complexes (Fig. 6D). In CMV-infected cells, both CAF1 and PCNA were strongly enriched at the sites of viral-genome localization and replication in late-stage infections, particularly at the 24-h time point (Fig. 6B, C, and E). Recruitment of PCNA to viral replication compartments was preceded by CAF1, since only the latter accumulated at the viral structures in the early phase (8 h) of infection (Fig. 6B and C). In contrast to CAF1 and PCNA, ASF1A-positive complexes did not exactly colocalize with viral replication compartments, but a subset showed juxtaposed association at 8 and 24 h after CMV infection (Fig. 6D and E). These results show for the first time that CMV profoundly affects the accumulation and subnuclear targeting of critical host cell chromatin assembly proteins. The data also strongly suggest that components of the human nucleosome assembly machinery act on CMV chromosomes throughout the viral replication cycle, contributing to both DNA replication-dependent and -independent viral-genome chromatinization.
FIG. 6.
Increased accumulation of human nucleosome assembly proteins and association with intranuclear viral replication compartments during CMV infection. (A) MRC-5 cells were mock infected (0 h) or infected with CMV for 8 to 72 h, as indicated, and proteins from whole-cell extracts were separated in 10% polyacrylamide-SDS gels. After Western blot transfer, the proteins were reacted with the respective antibodies as shown in Table 2. (B to D) Mock- or CMV-infected MRC-5 cells were fixed and permeabilized with paraformaldehyde/methanol (B and C) or paraformaldehyde/Triton X-100 (D) at the indicated time points and incubated with primary antibodies specifically detecting the CMV IE2 (0 and 8 h) or ppUL44 (24 and 72 h) proteins, together with antibodies directed against CAF1 p48, PCNA, or ASF1A, as indicated. Samples were subsequently stained with an Alexa Fluor 594 conjugate, an Alexa Fluor 488 conjugate, and DRAQ5. Single- and dual-color merge confocal images of representative nuclei are shown. Scale bars, 10 μm. (E) Three-dimensional projections of z stacks showing details of the spatial relationship between CMV replication compartments (ppUL44) (red) and complexes containing the indicated cellular chromatin assembly proteins (green) at 24 h postinfection. The frames were acquired with a step width of 0.38 μM and rendered with the Zeiss LSM510 software. Scale bar, 1 μm.
DISCUSSION
Little is known about the physical properties of herpesvirus chromosomes (25, 29). Studies of HSV-1 and other herpesvirus capsids failed to reveal any abundant protein intimately coating the viral DNA. Moreover, recent proteome analyses of purified CMV, EBV, and Kaposi's sarcoma-associated herpesvirus particles did not identify histones, although a variety of other host cell proteins were detected (33). Consistently, our own results did not yield any evidence for the presence of histones in purified virion preparations of CMV (A. Nitzsche and C. Paulus, unpublished data). Instead, early studies showed that HSV-1 capsids are rich in polyamines and suggested that these, rather than histones or other proteins, serve to neutralize the DNA and enable it to become packed tightly within the capsid (10). Thus, it appears that after the capsid has docked with the nuclear pore, the uncoated herpesvirus genome released into the nucleoplasm is initially “naked” with respect to proteins. Nevertheless, previous work has shown that the DNA of human herpesviruses, such as HSV and EBV, appears to be largely arranged in a nucleosomal structure inside latently infected cells (5, 48). Others have provided evidence for histone association (18, 20, 23, 38, 40, 41, 46) and at least partial nucleosome-like chromatinization of HSV and CMV genomes in productively infected cells (12, 23, 24, 51). These observations imply de novo deposition of histones on intranuclear herpesvirus DNA.
Initial DNA replication-independent CMV chromatin assembly.
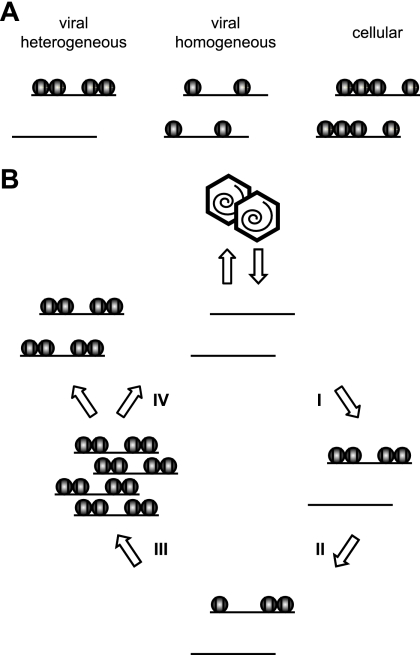
In fact, our results show significant deposition of human histones on CMV genomes by 30 min after high-multiplicity infection of human fibroblasts (Fig. 4), and nucleosomal viral DNA was detected as early as 2 h postinfection under these conditions (Fig. 1A). These observations indicate that at least a subset of CMV genomes become histone associated very quickly upon nuclear entry. However, average histone levels at the CMV chromosome were substantially lower than at a cellular control locus (GAPDH) between 0.5 and 16 h postinfection, as determined by ChIP and FAIRE. In this respect, the incoming viral genome may resemble transfected plasmid DNA. Indeed, it has been recently shown that a transiently transfected DNA template acquires a full complement of core histones but exhibits only intermediate levels of nucleosomal assembly (13). The limited histone deposition in the early stages of CMV infection was shown to be DNA replication independent, because it occurred well before the onset of viral-DNA synthesis (Fig. 5A) and was not affected by chemical inhibitors of viral and cellular DNA polymerases (Fig. 5B). Thus, input DNA episomes of plasmid or herpesvirus origin may generally exist in a rather irregularly chromatinized state, most likely because replication-independent chromatin assembly mechanisms are generally inefficient compared to replication-coupled mechanisms, which account for the bulk of eukaryotic nucleosome assembly. This conclusion would be justified if the episomes formed a largely homogeneous group in terms of nucleosome occupancy (Fig. 7A). However, ChIP and FAIRE assays measure only average histone/nucleosome occupancies. Therefore, our data are also compatible with highly divergent chromatin states between individual DNA molecules resulting in a heterogeneous pool of episomes (Fig. 7A). In this case, a fraction of input CMV genomes may be assembled into nucleosomes while another subset may not become histone associated at all, perhaps because the cellular chromatin assembly machinery does not have access to the latter group. Indeed, by MNase assay, the qualitative patterns of viral DNA present in nucleosomes looked remarkably similar at all tested postinfection time points (Fig. 1), supporting the idea of a rather heterogeneous population of early CMV genomes. The MNase accessibility data may also suggest preferential association of viral DNA with di- and/or mononucleosomes rather than regular nucleosomal arrays, since more extensive laddering of viral chromatin fragments was not observed (Fig. 1). Two critical mediators of nucleosome assembly, CAF1 p48 and ASF1A, were enriched in or close to the intranuclear compartments of CMV genome accumulation before the onset of viral-DNA replication (Fig. 6B to D). These proteins are therefore prime candidates among host cell factors that may be involved in early CMV genome chromatinization.
FIG. 7.
Models of CMV chromatin assembly and disassembly. (A) Schematic of two possible chromatin distributions in early-phase CMV genome populations. By ChIP or FAIRE analysis, both the heterogeneous and the rather homogeneous distributions would be interpreted as reduced histone occupancy relative to a cellular control locus. (B) Hypothetical four-step cycle of CMV chromatin assembly and disassembly (assuming heterogeneous chromatin distributions). Step I, limited initial DNA replication-independent nucleosome assembly on “naked” input viral genomes. Step II, maintenance of low chromatinization levels and partial chromatin disassembly. Step III, DNA replication-dependent nucleosome assembly resulting in chromatinization of most or all newly synthesized viral genomes (note that a subset of viral genomic regions is largely resistant to this step). Step IV, complete chromatin disassembly in a fraction of replicated viral genomes before or during encapsidation.
Maintenance of low average histone occupancy on CMV DNA through the early phase of infection.
Our ChIP assays revealed that average histone levels in the analyzed regions of the CMV genome stayed generally low throughout the entire early phase of infection (Fig. 4). Similar results were obtained using the FAIRE technique (data not shown). Again, this may reflect the inefficiency of cellular DNA replication-independent chromatin assembly mechanisms. Since histone deposition likely poses a major barrier to the process of transcription from the viral genome, it is also conceivable that one or more CMV gene products may antagonize further chromatinization or even promote chromatin disassembly to facilitate viral early gene expression. In support of the latter possibility, the average histone H3 occupancy of viral DNA in several of the analyzed regions dropped between 2 and 16 h postinfection (Fig. 4). Furthermore, there is precedent for a herpesvirus protein reducing the histone association of viral DNA. ChIP assays showed that the VP16 virion transactivator protein of HSV-1 recruits ATP-dependent chromatin-remodeling proteins to viral IE gene promoters. This activity was correlated with strongly reduced histone H3 levels at these promoters in the presence of VP16 (15). It is tempting to speculate that viral tegument or IE transactivators, such as pp71 (ppUL82), IE1-72kDa, or IE2-86kDa, may adopt this function in CMV. In fact, we have previously shown that CMV IE1 and IE2 proteins antagonize histone deacetylation (40), and acetylation of core histone tails has been linked to increased nucleosome mobility (14).
DNA replication-coupled CMV chromatin assembly.
Our ChIP assays clearly show that the average histone association of the CMV chromosome increases by up to 12-fold concurrent with and largely dependent on the process of viral-DNA synthesis (Fig. 4 and 5D). In some regions, histone H3 occupancy appeared to reach levels comparable to those of the cellular reference locus, indicating an extent of viral-genome chromatinization that has not been appreciated in productively herpesvirus-infected cells (25, 29). These results are compatible with our MNase, immunofluorescence, and FAIRE analyses (Fig. 1 to 3). Together, they demonstrate unanticipated DNA replication-coupled assembly mechanisms of CMV chromatin in the late stage of infection that most likely involve the host cell nucleosome assembly machinery. In fact, at least two conserved cellular mediators of replication-dependent nucleosome deposition (CAF1 and PCNA) are recruited to the subnuclear compartments of viral-DNA synthesis during CMV infection (Fig. 6B, C, and E). Moreover, the steady-state levels of all three tested human chromatin assembly proteins specifically increased over the course of the infection cycle, yet the relevance of this observation remains to be determined.
A recent report by Oh and Fraser (41) did not find newly synthesized HSV-1 DNA to be associated with histones, implying a lack of replication-dependent chromatin assembly during productive infection by the virus. This observation suggests that two herpesviruses (HSV-1 and CMV) may have evolved fundamentally different strategies with respect to chromatinization of newly replicated viral DNA. Alternatively, the extent of replication-dependent viral-chromatin assembly may be determined by the host cell environment. Oh and Fraser used an African green monkey kidney (Vero) and a human neuroblastoma cell line for their experiments. In contrast, all experiments in the present work were performed on primary human fibroblasts (MRC-5). Therefore, it is conceivable that differences in available histone pools between cell types may contribute to the observed effects. Normally, DNA replication-dependent chromatin assembly occurs in S phase, where it is coupled to histone synthesis in order to provide material for nucleosomes. However, we used contact-inhibited, resting cells for all our experiments. Moreover, CMV usually arrests cycling cells at the G1/S border upon infection of primary human fibroblasts (55) and inhibits cellular-DNA replication (56). Accordingly, productively CMV-infected cells do not appear to undergo induction of histone synthesis (45) (C. Paulus and M. Bergbauer, unpublished data). Under these conditions, the histone proteins required for chromatinization of CMV DNA have to be derived from available free cellular histone pools or through mobilization from host chromatin. It seems unlikely that the use of resting versus cycling cells accounts for the differences between this study and the report by Oh and Fraser, since the smaller pool of free histones in resting cells is expected to limit their association with newly replicated viral DNA instead of increasing it.
One intriguing finding of this study concerns the fact that not all viral genomic regions under investigation underwent significant DNA replication-coupled chromatin assembly. The MIE-P and the oriLyt proved to be largely resistant to the late increase in histone deposition (Fig. 4). Since both loci are among the most critical cis-regulatory elements in the CMV genome, high-affinity binding by cellular and/or viral regulatory nonhistone proteins may compete with efficient nucleosome deposition in these regions. Indeed, the analyzed regions in both the MIE-P and the oriLyt include target sequences for sequence-specific DNA binding by the CMV IE2-86kDa protein (17, 22, 26, 31, 57).
Complete disassembly of CMV chromatin as a prerequisite for DNA packing into capsids?
At 72 and 96 h postinfection, we observed a decrease in histone H3 occupancy at all CMV genomic regions under investigation (Fig. 4). This effect may reflect either depletion of the available cellular histone pools in the latest stages of infection or active disassembly of CMV chromatin. Since herpesvirus genomes exist as histone-free DNA inside capsids, our results actually imply that the replicated, chromatinized CMV genomes we observe have to undergo complete disassembly before or during packaging or may not be packaged into capsids at all. In the latter case, a subpopulation of replicated viral genomes must exist in late infected cells which somehow escape from DNA replication-coupled chromatin assembly. It is hard to conceive how a significant proportion of newly replicated viral genomes would stay completely histone free in replication compartments that harbor histones and accumulate chromatin assembly factors (Fig. 2 and Fig. 6). Moreover, the analogous levels of average histone occupancy between several viral loci and GAPDH at 48 h postinfection (Fig. 4) indicate that most, if not all, viral genomes carry nucleosomes at this stage. Therefore, we favor a scenario in which CMV chromatin disassembly immediately before or, perhaps more likely, during DNA packaging into capsids is a necessary prerequisite for the formation of infectious virions. Here, the activities of unidentified viral and/or cellular ATP-utilizing chromatin-remodeling factors, which have the ability to disrupt histone-DNA interactions, would almost certainly be required to allow packaging of CMV progeny genomes.
Based on the results of this study and the considerations described above, we propose a general model in which CMV chromatin is assembled and disassembled in four successive steps (Fig. 7B). We believe that these epigenetic events are relevant to all viral-DNA-based processes in CMV and likely other herpesvirus infections, including genome replication, the DNA damage response, and the temporal cascade of transcription. Further investigations into the dynamic structure of viral chromatin and its consequences for the outcome of infection not only may open up new opportunities for antiviral intervention, they may also provide more general information about the fate of naked and/or foreign DNA in the nucleus. In this respect, CMV could serve as a useful model system for the study of chromatin assembly and disassembly processes.
Acknowledgments
We thank Jörg Schröer and Charlotte Reitberger for help with immunofluorescence analyses; Joachim Griesenbeck, Hans-Helmut Niller, and Gernot Längst for helpful discussions and comments on the manuscript; Rugang Zhang and Thomas Shenk for antibodies; and Hans Wolf for continuous support.
This work was supported by grant NE 791/2-1 from the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 10 September 2008.
REFERENCES
- 1.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 91191-1200. [DOI] [PubMed] [Google Scholar]
- 2.Atalay, R., A. Zimmermann, M. Wagner, E. Borst, C. Benz, M. Messerle, and H. Hengel. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcγ receptor homologs. J. Virol. 768596-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Booy, F. P., W. W. Newcomb, B. L. Trus, J. C. Brown, T. S. Baker, and A. C. Steven. 1991. Liquid-crystalline, phage-like packing of encapsidated DNA in herpes simplex virus. Cell 641007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen, G. H., M. Ponce de Leon, H. Diggelmann, W. C. Lawrence, S. K. Vernon, and R. J. Eisenberg. 1980. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34521-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deshmane, S. L., and N. W. Fraser. 1989. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eickbush, T. H., and E. N. Moudrianakis. 1978. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry 174955-4964. [DOI] [PubMed] [Google Scholar]
- 7.Eriksson, B., B. Oberg, and B. Wahren. 1982. Pyrophosphate analogues as inhibitors of DNA polymerases of cytomegalovirus, herpes simplex virus and cellular origin. Biochim. Biophys. Acta 696115-123. [DOI] [PubMed] [Google Scholar]
- 8.Everett, R. D., and M. K. Chelbi-Alix. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89819-830. [DOI] [PubMed] [Google Scholar]
- 9.Gawn, J. M., and R. F. Greaves. 2002. Absence of IE1 p72 protein function during low-multiplicity infection by human cytomegalovirus results in a broad block to viral delayed-early gene expression. J. Virol. 764441-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson, W., and B. Roizman. 1971. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc. Natl. Acad. Sci. USA 682818-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giresi, P. G., J. Kim, R. M. McDaniell, V. R. Iyer, and J. D. Lieb. 2007. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 17877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall, M. R., N. Aghili, C. Hall, J. Martinez, and S. St Jeor. 1982. Chromosomal organization of the herpes simplex virus type 2 genome. Virology 123344-356. [DOI] [PubMed] [Google Scholar]
- 13.Hebbar, P. B., and T. K. Archer. 2008. Altered histone H1 stoichiometry and an absence of nucleosome positioning on transfected DNA. J. Biol. Chem. 2834595-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henikoff, S. 2008. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 915-26. [DOI] [PubMed] [Google Scholar]
- 15.Herrera, F. J., and S. J. Triezenberg. 2004. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 789689-9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7107-122. [DOI] [PubMed] [Google Scholar]
- 17.Huang, C. H., and J. Y. Chen. 2002. Identification of additional IE2-p86-responsive cis-repressive sequences within the human cytomegalovirus major immediate early gene promoter. J. Biomed. Sci. 9460-470. [DOI] [PubMed] [Google Scholar]
- 18.Huang, J., J. R. Kent, B. Placek, K. A. Whelan, C. M. Hollow, P. Y. Zeng, N. W. Fraser, and S. L. Berger. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 805740-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikegami, S., T. Taguchi, M. Ohashi, M. Oguro, H. Nagano, and Y. Mano. 1978. Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature 275458-460. [DOI] [PubMed] [Google Scholar]
- 20.Ioudinkova, E., M. C. Arcangeletti, A. Rynditch, F. De Conto, F. Motta, S. Covan, F. Pinardi, S. V. Razin, and C. Chezzi. 2006. Control of human cytomegalovirus gene expression by differential histone modifications during lytic and latent infection of a monocytic cell line. Gene 384120-128. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, V., and R. Chalkley. 1985. Histone segregation on replicating chromatin. Biochemistry 246930-6938. [DOI] [PubMed] [Google Scholar]
- 22.Jupp, R., S. Hoffmann, R. M. Stenberg, J. A. Nelson, and P. Ghazal. 1993. Human cytomegalovirus IE86 protein interacts with promoter-bound TATA-binding protein via a specific region distinct from the autorepression domain. J. Virol. 677539-7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kent, J. R., P. Y. Zeng, D. Atanasiu, J. Gardner, N. W. Fraser, and S. L. Berger. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 7810178-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kierszenbaum, A. L., and E. S. Huang. 1978. Chromatin pattern consisting of repeating bipartite structures in WI-38 cells infected with human cytomegalovirus. J. Virol. 28661-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knipe, D. M., and A. Cliffe. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6211-221. [DOI] [PubMed] [Google Scholar]
- 26.Lang, D., and T. Stamminger. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J. Virol. 67323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leinbach, S. S., and W. C. Summers. 1980. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J. Gen. Virol. 5145-59. [DOI] [PubMed] [Google Scholar]
- 28.Lentine, A. F., and S. L. Bachenheimer. 1990. Intracellular organization of herpes simplex virus type 1 DNA assayed by staphylococcal nuclease sensitivity. Virus Res. 16275-292. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman, P. M. 2008. Chromatin organization and virus gene expression. J. Cell. Physiol. 216295-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389251-260. [DOI] [PubMed] [Google Scholar]
- 31.Macias, M. P., and M. F. Stinski. 1993. An in vitro system for human cytomegalovirus immediate early 2 protein (IE2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate early promoter. Proc. Natl. Acad. Sci. USA 90707-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 751870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maxwell, K. L., and L. Frappier. 2007. Viral proteomics. Microbiol. Mol. Biol. Rev. 71398-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2773. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, and M. A. Martin (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia. PA. [Google Scholar]
- 35.Monier, K., J. C. Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2661-665. [DOI] [PubMed] [Google Scholar]
- 36.Mouttet, M. E., D. Guetard, and J. M. Bechet. 1979. Random cleavage of intranuclear herpes simplex virus DNA by micrococcal nuclease. FEBS Lett. 100107-109. [DOI] [PubMed] [Google Scholar]
- 37.Muggeridge, M. I., and N. W. Fraser. 1986. Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system. J. Virol. 59764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 211112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagy, P. L., M. L. Cleary, P. O. Brown, and J. D. Lieb. 2003. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc. Natl. Acad. Sci. USA 1006364-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nevels, M., C. Paulus, and T. Shenk. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA 10117234-17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh, J., and N. W. Fraser. 2007. Temporal association of the herpes simplex virus (HSV) genome with histone proteins during a lytic infection. J. Virol. 823530-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paulus, C., S. Krauss, and M. Nevels. 2006. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc. Natl. Acad. Sci. USA 1033840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pignatti, P. F., and E. Cassai. 1980. Analysis of herpes simplex virus nucleoprotein complexes extracted from infected cells. J. Virol. 36816-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polo, S. E., and G. Almouzni. 2006. Chromatin assembly: a basic recipe with various flavours. Curr. Opin. Genet. Dev. 16104-111. [DOI] [PubMed] [Google Scholar]
- 45.Radsak, K., and B. Schmitz. 1980. Unimpaired histone synthesis in human fibroblasts infected by human cytomegalovirus. Med. Microbiol. Immunol. 16863-72. [DOI] [PubMed] [Google Scholar]
- 46.Reeves, M., J. Murphy, R. Greaves, J. Fairley, A. Brehm, and J. Sinclair. 2006. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J. Virol. 809998-10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz, B. E., and K. Ahmad. 2006. 2. Chromatin assembly with H3 histones: full throttle down multiple pathways. Curr. Top. Dev. Biol. 7431-55. [DOI] [PubMed] [Google Scholar]
- 48.Shaw, J. E., L. F. Levinger, and C. W. Carter, Jr. 1979. Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J. Virol. 29657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simpson-Holley, M., J. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 UL31 and UL34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 785591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sinden, R. R., D. E. Pettijohn, and B. Francke. 1982. Organization of herpes simplex virus type 1 deoxyribonucleic acid during replication probed in living cells with 4,5′,8-trimethylpsoralen. Biochemistry 214484-4490. [DOI] [PubMed] [Google Scholar]
- 51.St Jeor, S., C. Hall, C. McGaw, and M. Hall. 1982. Analysis of human cytomegalovirus nucleoprotein complexes. J. Virol. 41309-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 11651-61. [DOI] [PubMed] [Google Scholar]
- 53.Tavalai, N., P. Papior, S. Rechter, and T. Stamminger. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82126-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor, R. T., and W. A. Bresnahan. 2005. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J. Virol. 793873-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiebusch, L., and C. Hagemeier. 1999. Human cytomegalovirus 86-kilodalton IE2 protein blocks cell cycle progression in G1. J. Virol. 739274- 9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiebusch, L., R. Uecker, and C. Hagemeier. 2003. Human cytomegalovirus prevents replication licensing by inhibiting MCM loading onto chromatin. EMBO Rep. 442-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, Y., S. A. Cei, A. Rodriguez Huete, K. S. Colletti, and G. S. Pari. 2004. Human cytomegalovirus DNA replication requires transcriptional activation via an IE2- and UL84-responsive bidirectional promoter element within oriLyt. J. Virol. 7811664-11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zaret, K. 2005. Micrococcal nuclease analysis of chromatin structure, p. 21.1.1-21.1.17. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 4. John Wiley and Sons, Hoboken, NJ. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, R., M. V. Poustovoitov, X. Ye, H. A. Santos, W. Chen, S. M. Daganzo, J. P. Erzberger, I. G. Serebriiskii, A. A. Canutescu, R. L. Dunbrack, J. R. Pehrson, J. M. Berger, P. D. Kaufman, and P. D. Adams. 2005. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 819-30. [DOI] [PubMed] [Google Scholar]