Abstract
The two ends of RSV linear DNA are independently inserted into host DNA by integrase in vivo. We previously showed that the range of U3 sequences that are acceptable substrates for integrase appeared to be greater than the range of acceptable U5 sequences in vivo. We have done additional experiments to determine which U3 sequences are good integrase substrates. On the U3 end, there does not appear to be a stringent requirement for the canonical CA, integrase can efficiently remove three nucleotides, and six nucleotides are sufficient to allow integration with reasonable, albeit reduced, efficiency.
During retrovirus reverse transcription, the polypurine tract (PPT) primer is generated by specific RNase H cleavages of the RNA genome adjacent to U3. The PPT primer is then used to prime plus-strand DNA synthesis, and the subsequent removal of this PPT primer by RNase H defines the left (U3) end of the linear viral DNA. Several studies have shown that the PPT sequence is important for the proper generation and removal of the PPT primer by RNase H (7, 8, 14, 15). We previously reported that alternate PPTs affected the specific cleavages that remove the PPT in a Rous sarcoma virus (RSV)-derived vector, RSVP(A)Z (3, 4). One of the alternate PPTs, the duck hepatitis B virus PPT in the reverse orientation (DuckHepBFlipPPT), was preferentially cleaved by the RNase H of RSV reverse transcriptase in a way that added 5′-TACAT to the end of U3 (because it is the minus strand on the U3 end that is joined to host DNA by integrase [IN], the minus-strand sequence is shown). This 5′-TACAT sequence is an exact match to positions +2 to +6 of U3 (U3+2 to U3+6) (Fig. 1A and B). Although about 40% of the proviruses generated in an infection with the DuckHepBFlipPPT virus were properly integrated, the CA in the 3′ end of the U3 at the virus/host junction in these proviruses was not the canonical CA, but rather the CA in the duplicated 5′-TACAT. RSV IN appears to recognize the duplicated 5′-TACAT at the U3 end, removes a single nucleotide (T), and joins the CA to host DNA (16).
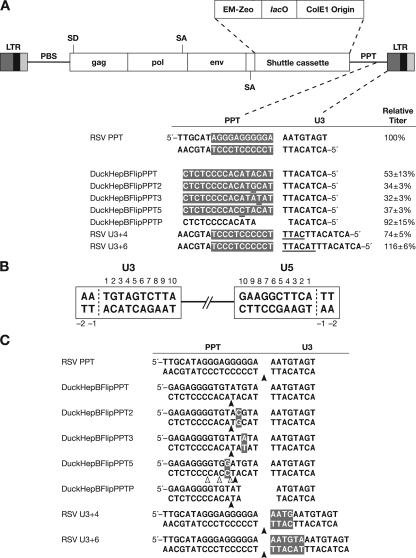
FIG. 1.
Structure of the RSVP vectors. (A) The PPT (white letters with a shaded background) and U3 sequences of wild-type RSV and the various mutants are shown. Mutations are underlined. The complete descriptions of the titers for each mutant are given in reference 4. (B) Schematic numbering of the nucleotide positions in the ends of the provirus. The last nucleotide on each end of a normal provirus is number 1. (C) Cleavages of the mutant PPTs by the RNase H of RSV reverse transcriptase. (Data are from reference 4.) The positions of the cleavages are inferred from the sequences of 2-LTR circle junctions isolated from cells infected with the viruses carrying the mutant PPTs. The arrows depict the positions of the predominant RNase H cleavages. Mutated sequences are shown in white letters with a shaded background.
Both the C and the A in the CA dinucleotide in the 5′-TACAT duplication were individually mutated by changing either the A to a G (DuckHepBFlipPPT2) or the C to a T (DuckHepBFlipPPT3). The position of the RNase H cleavage was not affected by the mutations in the CA dinucleotide in the 5′-TACAT, generating either 5′-TACGT or 5′-TATAT on the U3 end of the linear viral DNA from DuckHepBFlipPPT2 or DuckHepBFlipPPT3, respectively (Fig. 1C) (4). However, when the A residue one nucleotide downstream of the 5′-TACAT sequence was changed to a C (DuckHepBFlipPPT5), RSV RNase H no longer cleaved at the position (U3+5) favored in the parental DuckHepBFlipPPT virus. Although there was preferential cleavage at position U3+4, RNase H cleaved the PPT sequences at various positions, including U3+5, U3+7, and U3+9 (Fig. 1C) (4). We wanted to test how aberrant U3 ends are treated by RSV IN and to ask whether a CA was required for the efficient integration of the U3 end. Full-length integrated viral DNAs were recovered from cells infected with these mutant viruses as described previously (18). Recovered plasmids were sequenced, and the chicken genomic sequences were analyzed by BLAT searches (http://genome.ucsc.edu/cgi-bin/hgBlat).
Infections with the DuckHepBFlipPPT2, DuckHepBFlipPPT3, and DuckHepBFlipPPT5 mutants gave rise to several proviruses in which IN used sequences at the end of U3 other than the CA dinucleotide that is 1 base pair (bp) from the end of U3, to generate otherwise normal integration products (Table 1). The viral sequences joined to host DNA include 5′-CG (position −6) for DuckHepBFlipPPT2, 5′-TA (positions −4 and −6) and 5′-AT (position −5) for DuckHepBFlipPPT3, and 5′-CA (position −6) and 5′-TA (position −4) for DuckHepBFlipPPT5 (Table 1). However, the canonical CA dinucleotide in U3 was not used for integration in any of the proviruses isolated from these mutants. Infections with the mutant viruses also produced aberrant proviruses. In most cases, the terminus of the U3 long terminal repeat (LTR) was deleted, and more rarely, there was an insertion of the PPT and flanking sequences. In addition, there were duplications and deletions of the host sequences at the target site, ranging in size from a few to thousands of nucleotides (Table 1) (see Fig. S1 in the supplemental material).
TABLE 1.
Recovery of full-length integrated viral DNA
| Mutant | U3 (sequence)a | U5a | Size of duplication (bp) | No. of cases |
|---|---|---|---|---|
| DuckHepBFlipPPT2 | −6 | 1 | 6 | 3 |
| Complicatedb (tcaagc-5′) | 1 | 6 | 1 | |
| Variable | 1 | Variable | 11 | |
| DuckHepBFlipPPT3 | −6 | 1 | 6 | 4 |
| −4 | 1 | 6 | 3 | |
| −5 | 1 | 5 | 1 | |
| 5144 (CTtttt-5′) | 1 | 6 | 1 | |
| 6917 (Acccgg-5′) | 1 | 6 | 1 | |
| Variable | 1 | Variable | 14 | |
| DuckHepBFlipPPT5 | −6 | 1 | 6 | 7 |
| −4 | 1 | 6 | 2 | |
| 6956 (acatca-5′) | 1 | 6 | 1 | |
| Complicatedc (aagacc-5′) | 1 | 5 | 1 | |
| −4 | 22 | 190 | 1 | |
| Variable | 1 | Variable | 5 | |
| DuckHepBFlipPPTP | 1 | 1 | 6 | 15 |
| 1 | 1 | 5 | 7 | |
| 1 | 1 | 7 | 1 | |
| Variable | 1 | Variable | 2 | |
| RSV U3+4 | −4 | 1 | 6 | 12 |
| −4 | 1 | 5 | 2 | |
| 329 (accact-5′) | 1 | 6 | 1 | |
| Variable | 1 | Variable | 3 | |
| RSV U3+6 | −6 | 1 | 6 | 16 |
| −6 | 1 | 5 | 3 | |
| Variable | 1 | Variable | 5 |
The last viral nucleotide at each end of the proviruses is indicated by a number, using the numbering system shown in Fig. 1B. Bold capital letters indicate sequences that are part of a microhomology with the host DNA.
For the detailed structure, see D10 in Fig. S1 in the supplemental material.
For the detailed structure, see D8 in Fig. S1 in the supplemental material.
In case of DuckHepBFlipPPTP virus, in which the last three nucleotides (5′-TAC) of the PPT and the terminal nucleotide (T) of U3 were deleted, RNase H cleaves the PPT in a way that leaves three nucleotides on the U3 end beyond the canonical CA dinucleotide in U3 (Fig. 1C). We previously changed the sequence (TT) between the primer binding site at the U5 end to GGT in the mutant virus RSVP(HIV2). In infections with RSVP(HIV2), RSV IN removed three nucleotides beyond the CA and properly integrated the processed U3 end, although the titer of this virus titer was only 3.5% of the wild-type level (16). However, surprisingly, the titer of DuckHepBFlipPPTP virus was comparable to that of the wild-type virus (93%) (4). Most of the proviruses derived from infections with the DuckHepBFlipPPTP virus were integrated normally and flanked by a 5- or 6-bp duplication at the target site; in one provirus, there was a 7-bp duplication. Only 2 of 25 DuckHepBFlipPPTP proviruses were integrated aberrantly. This suggests that RSV IN can efficiently remove three nucleotides beyond the CA from the U3 end of DuckHepBFlipPPTP virus DNA.
In the integration step, IN recognizes sequences at the ends of linear viral DNA and usually removes two nucleotides adjacent to the conserved CA dinucleotides on each of the 3′ ends of the linear viral DNA (6, 8, 10, 20). The LTR terminal sequences are involved in specific interactions with IN and are sufficient for proper integration both in vitro and in vivo (1, 2, 9, 13, 22, 23, 24). In human immunodeficiency virus type 1 integration, 7 to 13 bp adjacent to the canonical CA dinucleotide mediate the interaction between the LTR and IN, in vivo and in vitro (11, 12, 13, 19, 21). In the present study, the integration of DNA from mutant viruses that replicated using versions of the DuckHepBFlipPPT suggested that RSV IN interacts with the last six nucleotides from the U3 end of the linear viral DNA. To determine whether the last six nucleotides are all that are required for efficient integration of the U3 end, duplications of the four (RSV U3+4) and six (RSV U3+6) terminal nucleotides of U3 were added to the end of U3. The RSV U3+4 mutation creates a U3 end resembling the normal U5 end (5′-TTCATT), whereas the RSV U3+6 mutation creates a duplicated U3 end (5′-TACATT) (Fig. 1A). In both mutants, the RSV RNase H cleaves at the PPT/U3 junction, producing a linear DNA in which U3 is extended by a duplication of four or six nucleotides (Fig. 1C). However, the percentage of consensus 2-LTR circle junctions was significantly higher than that in an infection with a wild-type virus, and there was a significant decrease in the percentage of large deletions in ends of linear viral DNA compared to that for the wild type (4). Although most of the proviruses that derived from infections with both mutants (RSV U3+4 and RSV U3+6) were integrated normally using the CA dinucleotide in the duplicated insert, about 20% of proviruses were integrated aberrantly. When the aberrant virus/host junctions were examined, there were, in many cases, microhomologies involving one to nine nucleotides between the virus and host sequence (see Table S1 in the supplemental material). We previously showed that, at this type of aberrant junction, there frequently are microhomologies between the host and virus DNA (17, 18). Given that the fraction of 2-LTR circle junctions with large deletions in the U3 sequences (these circles would arise from linear viral DNAs with large deletions in U3) was much lower in both mutants than in the wild type, the fact that infections with both viruses with U3 duplications led to the generation of proviruses in which there was a moderate fraction (20%) with defects at the U3 junction suggests that the last six nucleotides in U3 are sufficient to direct integration with moderate efficiency in vivo but that a larger segment makes contributions to efficient/accurate integration. It was previously shown in vitro that changing the C at position 7 in the U3 terminus of RSV to either an A or a T resulted in a decrease in concerted full-site integration (5). In vivo, both of the mutants we tested (RSV U3+4 [T at position 7] or RSV U3+6 [A at position 7]) caused a modest reduction in the integration of the mutated U3 ends.
We previously showed that about 60% of the proviruses derived from infections with RSVP(U3TCTT), in which the CA of the U3 was mutated to TC, were integrated normally, suggesting that RSV IN can recognize this mutated end sequence, remove the two nucleotides beyond the mutated TC sequence at the terminus of U3, and insert the linear DNA which has both nucleotides in the canonical CA at the mutated U3 terminus (18). Here, we show that RSV IN can process and insert linear viral DNAs that have, in place of the canonical CA, a CG, a TA, and an AT dinucleotide. Because RSV IN can recognize the duplicated 6 bp at the U3 end and properly process and integrate these ends, the terminal 6 bp appear to contain the bulk of the sequence needed for integration of the U3 end in vivo. Based on the behavior of viruses with mutations that give rise to defects in both the U5 and the U3 ends of the linear viral DNA, it appears that RSV IN may recognize the two ends independently, and it also appears that the sequence requirements at the U3 end may be less stringent than those at the U5 end.
Supplementary Material
Acknowledgments
We thank Teresa Burdette for help with the preparation of the manuscript.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 3 September 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Balakrishnan, M., and C. B. Jonsson. 1997. Functional identification of nucleotides conferring substrate specificity to retroviral integrase reactions. J. Virol. 711025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bushman, F. D., and R. Craigie. 1991. Activities of human immunodeficiency virus (HIV) integration protein in vitro: specific cleavage and integration of HIV DNA. Proc. Natl. Acad. Sci. USA 881339-1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, K. W., J. G. Julias, W. G. Alvord, J. Oh, and S. H. Hughes. 2005. Alternate polypurine tracts (PPTs) affect the Rous sarcoma virus RNase H cleavage specificity and reveal a preferential cleavage following a GA dinucleotide sequence at the PPT-U3 junction. J. Virol. 7913694-13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, K. W., J. Oh, W. G. Alvord, and S. H. Hughes. 2008. The effects of alternate polypurine tracts (PPTs) and mutations of sequences adjacent to the PPT on viral replication and cleavage specificity of Rous sarcoma virus reverse transcriptase. J. Virol. 828592-8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu, R., and D. P. Grandgenett. 2003. Molecular and genetic determinanats of Rous sarcoma virus integrase for concerted DNA integration. J. Virol. 776482-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craigie, R., T. Fujiwara, and F. Bushman. 1990. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell 62829-837. [DOI] [PubMed] [Google Scholar]
- 7.Dash, C. J., W. Rausch, and S. F. Le Grice. 2004. Using pyrrolo-deoxycytosine to probe RNA/DNA hybrids containing the human immunodeficiency virus type-1 3′ polypurine tract. Nucleic Acids Res. 321539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujiwara, T., and R. Craigie. 1989. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc. Natl. Acad. Sci. USA 863065-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katzman, M., and M. Sudol. 1996. Influence of subterminal viral DNA nucleotides on differential susceptibility to cleavage by human immunodeficiency virus type 1 and visna virus integrases. J. Virol. 709069-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2004. Effects of mutations in the G tract of the human immunodeficiency virus type 1 polypurine tract on virus replication and RNase H cleavage. J. Virol. 7813315-13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaFemina, R. L., P. L. Callahan, and M. G. Cordingley. 1991. Substrate specificity of recombinant human immunodeficiency virus integrase protein. J. Virol. 655624-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leavitt, A. D., R. B. Rose, and H. E. Varmus. 1992. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J. Virol. 662359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda, T., M. J. Kuroda, and S. Harada. 1998. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. 1998. J. Virol. 728396-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McWilliams, M. J., J. G. Julias, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2003. Mutations in the 5′ end of the human immunodeficiency virus type 1 polypurine tract affect RNase H cleavage specificity and virus titer. J. Virol. 7711150-11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powell, M. D., and J. G. Levin. 1996. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J. Virol. 705288-5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh, J., K. W. Chang, and S. H. Hughes. 2006. Mutations in the U5 sequences adjacent to the primer binding site do not affect tRNA cleavage by Rous sarcoma virus RNase H but do cause aberrant integrations in vivo. J. Virol. 80451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh, J., K. W. Chang, W. G. Alvord, and S. H. Hughes. 2006. Alternate polypurine tracts affect Rous sarcoma virus integration in vivo. J. Virol. 8010281-10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oh, J., K. W. Chang, R. Wierzchoslawski, W. G. Alvord, and S. H. Hughes. 2008. Rous sarcoma virus (RSV) integration in vivo: a CA dinucleotide is not required in U3, and RSV linear DNA does not autointegrate. J. Virol. 82503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reicin, A. S., G. Kaplana, S. Paik, S. Marmon, and S. Goff. 1995. Sequences in the human immunodeficiency virus type 1 U3 region required for in vivo and in vitro integration. J. Virol. 695904-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth, M. C., P. L. Schwartzberg, and S. P. Goff. 1989. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell 5847-54. [DOI] [PubMed] [Google Scholar]
- 21.Sherman, P. A., and J. A. Fyfe. 1990. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc. Natl. Acad. Sci. USA 875119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vink, C., D. C. van Gent, Y. Elgersma, and R. H. Plasterk. 1991. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol. 654636-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vora, A., and D. P. Grandgenett. 2001. DNase protection analysis of retrovirus integrase at the viral DNA ends for full-site integration in vitro. 2001. J. Virol. 753556-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshinaga, T., and T. Fujiwara. 1995. Different roles of bases within the integration signal sequence of human immunodeficiency virus type 1 in vitro. J. Virol. 693233-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.