Abstract
Tulane virus (TV) is a newly reported calicivirus that was isolated from stool samples of captive rhesus macaques from the Tulane National Primate Research Center (TNPRC). The virus has been cultivated successfully in LLC-MK2 rhesus monkey kidney cells. Its complete genomic sequence suggests that TV represents a new genus and is evolutionarily more closely related to Norovirus than to any other genus of Caliciviridae. In this study, we demonstrated that RNA transcripts made in vitro from the full-length genomic cDNA of TV were infectious upon transfection into permissive LLC-MK2 cells. The recombinant virus exhibited plaque morphologies and growth kinetics similar to those of the wild-type virus in this cell line. Capping was required for TV RNA infectivity. Although a subgenomic RNA has been detected in TV-transfected cells, a separate subgenomic RNA transcript was not required for the initial transfection to establish the replication. Transfection of truncated RNA lacking open reading frame 2 (ORF2) and ORF3 or TV-norovirus chimeric RNA resulted in abortive replication without the production of infectious progeny viruses, indicating that both ORFs are essential for the replication of TV. A heterologous insertion at the 5′ end of the genome also hampered viral replication, suggesting that an authentic 5′ end of the genome is critical for replication. The availability of the complete genomic sequence and the reverse genetics system described herein make TV a valuable model for studying calicivirus pathogenesis and replication.
Caliciviruses (CVs) are important human and animal pathogens causing a wide variety of diseases, including respiratory infections, vesicular lesions, gastroenteritis, and hemorrhagic disease. Caliciviridae consists of four well-defined genera, Norovirus, Sapovirus, Lagovirus, and Vesivirus (9, 10), and a newly proposed genus, “Becovirus,” or “Nabovirus,” represented by bovine enteropathogenic CVs (Newbury agent 1 and Nebraska) (17, 18, 20). The Tulane virus (TV) that was recently discovered by our group represents a putative sixth genus of Caliciviridae, with a proposed name of “Recovirus” (7).
The CV genome is a positive-sense, single-stranded RNA with a poly(A) tail. Virions encapsidate a genomic RNA (∼7.5 kb) and a subgenomic RNA (∼2.3 kb). The genomic RNA of noroviruses (NVs), vesiviruses, and TV contains three open reading frames (ORFs). ORF1 encodes the nonstructural (NS) polyprotein, which is autocleaved to at least six functional units, including the N-terminal protein, an NTPase, a small protein with yet unknown function, VPg, the viral protease, and the RNA-dependent RNA polymerase (RdRp). ORF2 encodes a viral capsid protein, whereas ORF3 encodes a minor structural protein. In the genome of sapoviruses, lagoviruses, and “becoviruses,” the NS polyprotein- and capsid-encoding genes are fused into ORF1.
NVs and sapoviruses, commonly referred to as human CVs, are important etiologic agents of acute gastroenteritis of humans in both developed and developing countries. Both NVs and sapoviruses are genetically and antigenically diverse. Based on phylogenetic analysis, NVs are classified into five genogroups that can be divided further into over 30 genetic clusters or genotypes (27). Enteric CVs that are genetically and antigenically closely related to human CVs have also been isolated from animals (5, 14, 16, 23), suggesting a possible zoonotic nature of CV gastroenteritis.
Rhesus macaques are susceptible to a number of infectious agents (19) and thus can potentially serve as carriers for human diseases. Jiang et al. reported the presence of NV-specific antibodies in several nonhuman primate species, including rhesus macaques (11). In our laboratory, reverse transcription PCR (RT-PCR) screening of stool samples of rhesus macaques housed at the Tulane National Primate Research Center (TNPRC) led to the identification of a novel CV, Tulane virus (GenBank accession number EU391643) (7). TV has been cultured successfully in LLC-MK2 rhesus monkey kidney cells, where it caused typical cytopathic effects (CPE). Phylogenetic analysis of the TV genome indicated that TV represents a new genus within the family of Caliciviridae that is more closely related to Norovirus than to any of the other genera (7). Whether gastroenteritis or other disease associated with TV infection in rhesus macaques remains to be studied.
The reverse genetics system is a powerful tool for studying virus replication, viral protein function, and virus-host interactions. Several such systems for human and animal CVs have been established, with various levels of success (1-4, 13, 15, 21, 25, 26). Since TV was isolated from stool samples of animals and a permissive cell line for its replication in vitro is readily available, TV could potentially serve as an attractive model system for human CVs. To this end, we confirmed the completeness of the TV genomic clone and established a reverse genetics system for TV. In vitro-transcribed, capped, full-length TV RNA was infectious in LLC-MK2 monkey kidney cells. This system could be valuable for studying CVs.
MATERIALS AND METHODS
Cell culture.
LLC-MK2 rhesus monkey kidney cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The cells were cultured with media 199 (Mediatech, Manassas, VA) supplemented with 5% heat-inactivated fetal bovine serum, 200 U/ml penicillin, 200 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (Invitrogen, Carlsbad, CA). The cell culture was maintained at 37°C in the presence of 5% CO2.
Viral RNA isolation, full-length TV cDNA cloning, and sequencing.
Plaque-purified TV that had been passed 25 times in LLC-MK2 cells was used for cloning the full-length TV cDNA (7). The viruses released in the culture media were concentrated by polyethylene glycol precipitation followed by cesium chloride (CsCl) density gradient purification, as described previously (7). Viral RNA was isolated by using Trizol reagent (Invitrogen) or a QIAamp viral RNA mini kit (Qiagen, Valencia, CA).
The first-strand cDNA of TV was synthesized from the viral RNA by use of a modified lock-docking oligo(dT) primer, (T)25VN-3′ (where N is A, C, G, or T and V is A, G, or C) (Clontech, Mountain View, CA). Two overlapping TV cDNA fragments that covered the full-length genome were amplified by use of first-strand cDNA as a template with primers p1094/p1079 and p1069/p1083 (Table 1). A bacteriophage T7 promoter was also incorporated into the end of the 5′ untranslated region (5′ UTR) of the upstream fragment (see Fig. 2A). The cDNA fragments were then assembled into a full-length TV cDNA by sequential cloning in a modified pBlueScript II vector (Stratagene, La Jolla, CA), in which an MluI restriction digestion site upstream of the original multiple cloning sites was introduced by mutagenesis (Quick Change kit; Stratagene, La Jolla, CA). The cloned full-length TV cDNAs derived from the cell culture-adapted virus were sequenced using a standard procedure with a series of gene-specific primers designed from TV sequences determined from TV in stool samples (7).
TABLE 1.
Primers used in the experiments
| Primer name | Primer sequenced | TV nt positions | Orientation |
|---|---|---|---|
| p1094 | 5′-TTGACGCGTAATACGACTCACTATAGTGACTAGAGCTATGGATACG-3′ | 3-23 | Forward |
| p1079 | 5′-ATAGTCGACTCACAAGAATCCAGAACAACC-3′ | 2926-2906 | Reverse |
| p1069 | 5′-CGCGGATCCCCCAGTGATGATTATTACGAC-3′ | 2396-2417 | Forward |
| p1083a | 5′-GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT-3′ | Reverse | |
| p1205 | 5′-GTTGTCTTTAGATAGGCTGGCGCTGCGTT-3′ | 5970-6000 | Forward |
| p1206 | 5′-AACGCAGCGCCAGCCTATCTAAAGACAAC-3′ | 6000-5970 | Reverse |
| p978 | 5′-ATGGAAAACAGCAAAACTGAACAAG-3′ | 4380-4404 | Forward |
| p1275R | 5′-CTGTGGGTGGCACCAAGAATGTGAACTTAAAATCAGG-3′ | 4976-4959 | Reverse |
| p1275Fb | 5′-CCTGATTTTAAGTTCACATTCTTGGTGCCACCCACAG- 3′ | 4959-4976 | Forward |
| p1276Rb | 5′-CAAACGCAGCGCCAGCCATTATAATGCACGTCTGCGCC-3′ | 6002-5984 | Reverse |
| p1276F | 5′-GGCGCAGACGTGCATTATAATGGCTGGCGCTGCGTTTG-3′ | 5984-6002 | Forward |
| p1158c | 5′-TTGACGCGTAATACGACTCACTATAGGGTGACTAGAGCTGCCA CCATGGTGAGCAAGGGCGAG-3′ | 1-14 | Forward |
| p1159c | 5′-CACAGAATCTATGGACGTATCCATGGATCCCTTGTACAGCTC GTCCATG-3′ | 38-15 | Reverse |
| p1081 | 5′-GACTAGAGCTATGGATACGTCC-3′ | 3-24 | Forward |
| p1096 | 5′-ATAGTCGACTCAGGTAGAAACATCATCACC-3′ | 1019-1001 | Reverse |
| p1082 | 5′-GCCCCAGGGATGGGTCAGGCCATTGAGACGG-3′ | 272-241 | Reverse |
| p1091 | 5′-CGTTTCCAATCATGCCTCCTG-3′ | 233-213 | Reverse |
| p1189 | 5′-GGATGGACACAACAACTCCTAGAGATCGTGTACC-3′ | 491-525 | Forward |
| p1190 | 5′-GGTACACGATCTCTAGGAGTTGTTGTGTCCATCC-3′ | 525-491 | Reverse |
| p1072 | 5′-CCAAGGGGGTATTG TGCCTGTTATTGTC-3′ | 920-893 | Reverse |
| p1076 | 5′-AATGGATCCCGGCTCCAAAGGTTGAGC-3′ | 1002-1019 | Forward |
| p1077 | 5′-ATAGTCGACTCAGGAGCATCCCAACTGGAG-3′ | 1791-1773 | Reverse |
| p1078 | 5′-AATGGATCCCCGCCGCTTGTAGGAACTAAAGAC-3′ | 3036-3060 | Forward |
Oligo(dT) primer.
Primer for NV cDNA amplification.
Primer for eGFP cDNA amplification.
Underlining represents the TV nucleotide sequences; italic font represents the T7 promoter sequence.
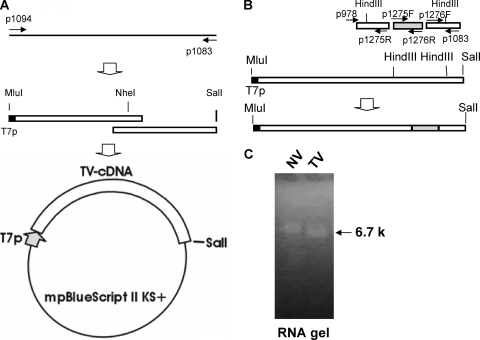
FIG. 2.
Generation of full-length TV cDNA, TV-NV chimeric cDNA, and RNA transcripts. (A) Cloning of TV recombinant construct. The black bar represents TV first-strand cDNA, the thin arrows show primer binding sites, the white box represents TV cDNA, and the black box (or the thick gray arrow) represents the T7 promoter (T7p). (B) Generation of TV-NV chimeric cDNA. The gray box represents a portion of NV cDNA. (C) RNA transcripts of full-length TV and NV. The arrow indicates the size of RNA transcripts, based on the size of the RNA control.
Generation of deletion, knockout, and chimeric TV cDNAs.
TV ORF1 cDNA (with deletion of ORF2 and -3) was constructed by amplification of the entire ORF1 region with primers p1094 and p1079, followed by cloning into the modified pBlueScript II vector. An ORF3 knockout mutation was generated by deletion of 2 nucleotides (A and T at nucleotide [nt] positions 5984 and 5985, respectively, resulting in a frameshift) at the start codon of ORF3, using the full-length TV cDNA as a template and primers p1205 and p1206. To generate a TV-NV chimeric cDNA construct, primers p978, p1275R, p1275F, p1276R, p1276F, and p1083 were used to amplify a fusion coding region for the TV capsid S domain, NV VA387 capsid P domain (GenBank accession number AY038600), and TV VP2 and 3′ UTR (see Fig. 2B). The fragment was digested with HindIII and placed into the corresponding coding region of the full-length TV cDNA construct. To generate an eGFP-TV fusion cDNA construct, primers p1158 and p1159 were used to amplify the eGFP coding sequence, and primers p1081 and p1096 were used to amplify the TV N-terminal protein coding sequence. An overlapping PCR was performed to amplify enhanced green fluorescent protein (eGFP)- and N-terminal protein-coding cDNA. The cDNA was placed into the full-length TV cDNA construct at MluI and ClaI digestion sites.
cDNA library screening.
TV RNA isolated from cell culture-adapted viruses and TV-specific reverse primer p1082 were used for the first-strand-cDNA synthesis. A TV 5′ cDNA library was generated using a Universal RiboClone cDNA synthesis system (Promega, Madison, WI). TV cDNA clones from the library were screened by PCR with primer M13F or M13R and TV-specific primer p1091. The clones with expected inserts were sequenced with the M13F or M13R primer.
Generation of TV cDNA containing a genetic marker.
A pair of primers, p1189 and p1190, was used to introduce a C-to-A silent mutation at nt 512 of TV cDNA by site-directed mutagenesis (Quick Change kit; Stratagene, La Jolla, CA). This mutation abolished an XhoI restriction digestion site (ΔTV) present in both the wild-type and cell culture-adapted TV cDNA.
In vitro transcription and RNA transfection.
A linearized recombinant plasmid (1 μg) that contained the full-length TV cDNA with a T7 promoter was used as a template for in vitro transcription. A plasmid that contained the full-length Norwalk virus cDNA (GenBank accession number NC_001959) with a T7 promoter was used as a control for in vitro transcription. Uncapped TV RNA and Norwalk virus RNA were generated using a MEGAscript T7 kit (Ambion, Austin, TX), whereas capped TV RNA was generated using a mMESSAGE mMACHINE T7 kit (Ambion, Austin, TX), according to the manufacturer's instructions. Reaction mixtures were incubated at 37°C for 3 h, followed by digestion with 3 U of DNase I (Ambion) for 15 min at 37°C. RNA was quantified by absorbance at 260 nm and diluted in nuclease-free water to 0.2 μg/μl prior to storage at −80°C. RNA integrity was determined by agarose gel electrophoresis and visualization by UV light illumination with ethidium bromide staining.
RNA transfection in LLC-MK2 cells was performed using a TransIT mRNA transfection kit (Mirus, Madison, WI) according to the manufacturer's instructions. Briefly, 0.1 to 0.4 μg in vitro-transcribed TV RNA was incubated with 1 to 2 μl mRNA boost reagent and 1.5 to 3 μl TransIT mRNA reagent in 50 μl serum-free media for 5 min and then was transferred to 75%-confluent LLC-MK2 cells in a 24-well plate containing 0.5 ml complete medium. The cells were examined daily for morphological changes by light microscopy. Cell-free medium (50 μl) was passed to fresh cultures in 24-well plates at 48 to 72 h posttransfection.
To identify the presence of the genetic marker in the recovered ΔTV virus, viral RNA obtained from the fifth passage with the LLC-MK2 cells was amplified with primers p1081 and p1072, and the RT-PCR product was analyzed by agarose gel electrophoresis following XhoI restriction enzyme digestion. The RT-PCR products were also sequenced using the RT-PCR primers.
Antibody generation and immunofluorescence analysis.
TV cDNA coding regions for helicase (nt 1002 to 1790) and RdRp (nt 3036 to 4358) were determined by homologous comparison with those of several CVs. The helicase coding region was amplified using primers p1076 and p1077, while the RdRp coding region was amplified using primers p1078 and p1079. cDNAs were cloned into pQE30 vector (Qiagen) and expressed in Escherichia coli M15 (Qiagen). His-tagged TV helicase and TV RdRp proteins were purified using Talon resin (Clontech). Mouse polyclonal anti-TV helicase and anti-TV RdRp antibodies were generated using standard protocols.
Indirect immunofluorescence was performed using LLC-MK2 cells that were grown on poly-d-lysine-coated coverslips (BD Biosciences, San Jose, CA) and transfected with in vitro-generated TV. At 24 or 48 h posttransfection, cells were treated with RSB buffer (10 mM Tris [pH 7.4], 10 mM NaCl, and 5 mM MgCl2), fixed with 3.7% formaldehyde for 10 min, and then permeabilized with 0.5% IGEPAL [tert-octylphenoxy poly(oxyethylene)ethanol] CA-630 (NP-40) for 20 min. Expression of TV helicase and TV RdRp was detected with mouse polyclonal anti-TV helicase and anti-TV RdRp antibodies and fluorescein isothiocyanate-conjugated secondary antibody (Jackson ImmunoResearch, West Grove, PA). Nuclei were stained with 0.2 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) (Sigma, St. Louis, MO).
Single-cycle growth curve and plaque assay.
For construction of a single-cycle growth curve, LLC-MK2 cells (2 × 105 cells/well) in 12-well tissue culture plates were incubated with TV (wild type or recombinant) at a multiplicity of infection of 0.025 for 1 h on a rotary shaker at 37°C in a CO2 incubator. Each of the plates had three wells with culture. After three washes with 1 ml of serum- and antibiotic-free M199 medium, plates were filled with 1 ml/well complete M199 culture medium and incubated at 37°C in a CO2 incubator. At each time point, one plate was sealed with Parafilm and stored at −70°C; when cultures had been collected at all time points, cultures were freeze-thawed three times, harvested by combining the individual wells from a single time point, cleared from cell debris, serially diluted 10-fold, and titrated.
A plaque assay was performed with 12-well tissue culture plates. Each dilution was inoculated into three wells. Cultures were incubated with 0.5 ml of serially diluted virus for 1 h on a rotary shaker at 37°C in a CO2 incubator, washed once with M199 medium, and overlaid with 2 ml of 1.5% methylcellulose-M199 medium. Plates were incubated at 37°C in a CO2 incubator. Plaques were counted under a light microscope and after crystal violet staining (Sigma-Aldrich, St. Louis, MO) on day 5 pi. Endpoint titers (PFU/0.5 ml) were calculated as the means from the three individual wells at the highest dilution with plaque formation.
RESULTS
Determination of the 5′ sequence of the TV genome.
As described in our previous study, due to the limited availability of the TV-positive stool specimens, the 5′ end of the TV genome was determined by the use of a tissue culture-adapted TV clone (7). The TV genome is 6,714 nt long, which is the shortest genome among all known CVs and about 1,000 nt shorter than most of the NV genomes (12). Sequence alignment of TV and selected NVs showed that most of the predicted structural and nonstructural regions of the two genera have comparable sizes, except for the 5′ end. The NV 5′ end encodes an N-terminal protein of 38 to 45 kDa, while the predicted N-terminal protein of TV has a molecular mass of 26 kDa (7).
An intact 5′ end of the cDNA is critical for the development of an infectious clone. To further determine the precise 5′-end sequence of TV, we performed a conventional cDNA library screening, targeting the 5′ cDNA of cell culture-adapted TV. The screening of this library revealed identical sequences in several clones but with variable numbers of G's (three to eight G's) at the 5′ end. This result was consistent with that from a previous 5′ rapid amplification of cDNA ends with dGTP tailing carried out using the first-strand cDNA. The fact that the TV genome starts with three G's was later clarified by 5′ rapid amplification of cDNA ends with dATP tailing. The variation in the number of G's at the 5′ end of the TV genome among various cDNA library clones was most likely due to the terminal transferase activity of the reverse transcriptase (Powerscript) used in the first-strand-cDNA synthesis (according to the manufacturer's manual). This result has also been confirmed by sequencing of cDNA transcribed from the end-to-end-ligated TV genomic RNA (data not shown). Thus, we concluded that, as originally predicted, the TV genome contains 6,714 nt, with three G's at the 5′ end and an additional poly(A) tail at the 3′ terminus (7). This conclusion was also confirmed by the transfection experiments in which infectious viruses were recovered from RNA transcribed in vitro.
Construction of full-length TV genomic cDNA.
Because of the limited quantity of TV-positive stool samples, viral RNA extracted from tissue culture-propagated TV was used for the generation of the full-length TV genomic cDNA. In addition, by using RNA transcripts based on tissue culture-adapted viruses, we had a better chance of success in developing the infectious clone in the permissive cell line LLC-MK2. The full-length genomic TV cDNA was sequenced and compared with the published wild-type sequence. The full-length wild-type TV sequence was generated by direct sequencing of five overlapping regions that were reamplified from an RNA template obtained from a TV-positive stool sample (7), whereas the cDNAs of the tissue culture-adapted TVs were generated by ligation of two overlapping cDNAs covering the entire TV genome obtained from plaque-purified, tissue culture-adapted TV. The two corresponding sets of sequences confirmed each other, including the primer binding sites and junction regions.
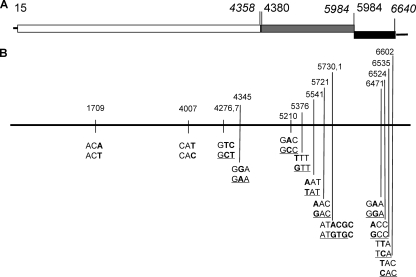
However, sequence variations between the two sets of sequences have also been observed, with a significant number of mutations in the protein-encoding regions, mainly in the P domain of the capsid protein and the C terminus of the minor structural protein VP2 (Fig. 1). These sequence variations could be due to differences between a highly homogeneous RNA population following the plaque purification (and cloning) and a potential quasi-species nature of the viruses in vivo. On the other hand, however, the tissue culture-adapted viruses were plaque purified and passed in the LLC-MK2 cells ∼25 times; thus, these variations were likely true mutations. The significantly high number of mutations in the capsid P domain, particularly in the P2 subdomain compared to other regions, indicates potential adaptive mutations of TV in the LLC-MK2 cells. The finding of an even higher mutation rate in the VP2 gene may suggest that this gene is essential for replication and at the same time is under selection pressure by a host factor(s).
FIG. 1.
TV genome organization and mutations identified in the genome of cell culture-adapted TV. (A) Organization of TV genome. The open box represents ORF1, the gray box represents ORF2, and the black box represents ORF3. The initial nucleotide for each ORF is in lightface font, and the end nucleotide for each ORF is in italics. (B) Mutation identified in the genome of cell culture-adapted TV. The black bar represents the TV genome. The numbers above the bar correspond to mutation sites. Mutated nucleotides are highlighted in bold, whereas mutations resulting in amino acid substitutions are underlined.
Recovery of infectious TV by transfection of TV RNA.
A full-length TV genomic cDNA was constructed with an upstream bacteriophage T7 promoter and a downstream SalI restriction digestion site by use of a modified pBlueScript II vector (Stratagene, La Jolla, CA) (Fig. 2A). In vitro transcription using the linearized cDNA construct as a template generated RNA transcripts of an expected size (Fig. 2C). Both capped and uncapped RNAs were made in the presence and absence of the cap analogue m7G(5′)ppp(5′)G, respectively, and transfection of both the capped and uncapped TV RNAs into LLC-MK2 cells resulted in cell death within 48 h, while the mock-transfected cells were unaffected.
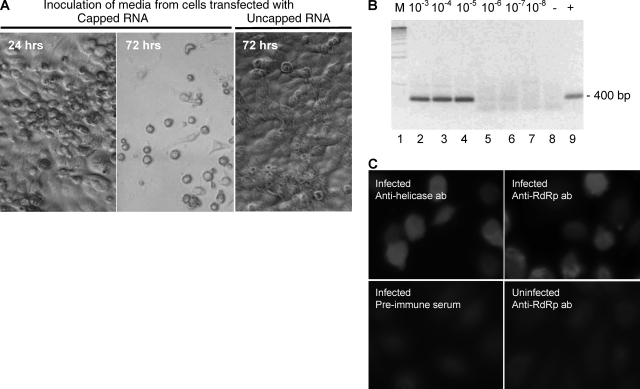
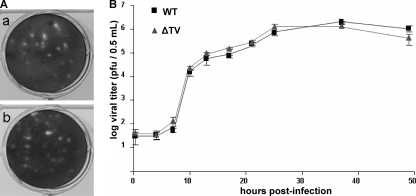
To determine if cell death upon transfection was due to virus replication and if infectious virus could be recovered from the RNA transfection, fresh LLC-MK2 cells were inoculated with media harvested 48 h posttransfection. Typical CPE of cell rounding and detaching were observed within 24 to 48 h in wells inoculated with media from the capped TV RNA transfection cultures but not in wells inoculated with media from the uncapped RNA transfection cultures (Fig. 3A). Plaque formation was observed 2 to 3 days postinfection, and within 4 to 6 days all cells became infected. The recovered virus remained infectious through subsequent passages. The plaque morphologies and growth kinetics of recombinant virus and wild-type virus are similar in LLC-MK2 cells (Fig. 4). We later repeated the transfection experiments with smaller amounts of the RNAs (0.2 μg/transfection) and transfection reagents (1 μl boost reagent and 1.5 μl transfection reagent/transfection) and found that the CPE previously observed in the culture transfected with the uncapped RNA was nonspecific, attributed to RNA and reagent overdose.
FIG. 3.
Infectivity of TV. (A) Media from cells transfected with capped RNA or uncapped RNA 48 h posttransfection were used for inoculation of cells. Only capped TV RNA yielded infectious virus. The images were taken at 24 or 72 h after incubation with virus-containing media. (B) Detection of TV RNA by RT-PCR. Lane 1 (M), DNA kilobase marker; lanes 2 to 7, RNA isolated from cells infected with serially diluted viral stocks; lane 8 (−), negative control of RT-PCR, in which the RT reaction step was omitted; lane 9 (+), RT-PCR positive control. (C) Detection of nonstructural proteins in transfected cells. The preimmune serum used as a control in the immunofluorescence analysis was collected from the mouse prior to injection with purified TV RdRp protein. ab, antibody.
FIG. 4.
Plaque morphologies and growth kinetics of wild-type (WT) and in vitro-generated (ΔTV) TV. (A) WT TV (a) and ΔTV (b) exhibit similar plaque morphologies. (B) Growth curves of WT and ΔTV viruses. LLC-MK2 cells were infected with WT or ΔTV at a multiplicity of infection of 0.025.
To further confirm that the observed CPE was associated with TV replication, the levels of TV RNA in the culture from the fifth passage were estimated by semiquantitative RT-PCR (Fig. 3B). The viral RNA titer was found to be comparable to that of the wild-type virus. Furthermore, high levels of expression of TV helicase and RdRp genes in the TV-infected cultures were demonstrated by immunofluorescence with the TV-specific antibodies (Fig. 3C). The expression of TV capsid protein was also detected by indirect immunofluorescence analysis using anti-TV VP1 antibodies (data not shown).
Recovery of genetically modified TV.
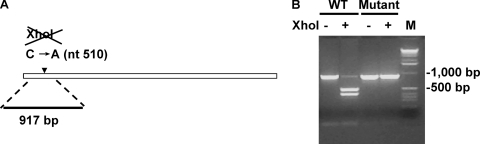
To exclude the possibility that the recovered viruses were a result of contamination with TVs present in the laboratory, a cDNA clone containing a silent mutation that eliminated an authentic XhoI site (ΔTV) present in both the wild-type and the tissue culture-adapted TV was generated (Fig. 5A). Transfection of in vitro-transcribed ΔTV RNA resulted in CPE and infectious viruses. Both restriction enzyme analysis and sequencing of RT-PCR amplicons obtained from these cultures revealed the loss of the XhoI digestion site, confirming that the infectious viruses were recovered from the in vitro-transcribed RNA (Fig. 5B).
FIG. 5.
Construction of a TV genome containing a genetic marker. (A) Schematic representation of a genetically marked TV genome. (B) Restriction digestion analysis of RT-PCR products from genetically marked TV. WT, wild type. −, without XhoI digestion; +, with XhoI digestion; M, 1-kb DNA marker.
Both ORF2 and ORF3 are essential for TV infectivity.
In our studies of developing the infectious TV clone, we observed that a transfection of mutated TV RNA with a deletion of ORF2 and -3 or with an ORF3 knockout mutation caused CPE and cell death within 24 to 48 h posttransfection but that no infectious viruses were generated from the transfected cells (Table 2). This result indicated that the nonstructural gene may be sufficient for initiation of RNA replication but that both ORF2 and ORF3 are essential for generation of progeny viruses.
TABLE 2.
CPE of transfection with various modified TV RNA transcripts
| RNA transcript | CPE at transfectiona | CPE at blind passages (1 to 5)a |
|---|---|---|
| Capped full-length | ++++ | ++++ |
| Uncapped full-length | − | − |
| Capped ORF1 | ++ | − |
| Capped ORF3 knockout | ++ | − |
| Capped TV-NV chimeric | ++ | − |
| Capped eGFP-TV fusion | ++ | − |
| Mock | − | − |
++++, strong CPE; ++, moderate CPE; −, without CPE.
After the success in development of the infectious TV clone, we further studied the roles of these genes in viral replication by construction of TV-NV chimeric clones with a replacement of the TV capsid gene or the P domain with a human NV capsid gene or P domain (VA387, GII-4). Similar CPE and cell death were observed, but unfortunately we did not detect progeny viruses. To confirm the viral RNA replication, we performed immunofluorescence analysis on cells transfected with the chimeric RNA and the ORF3 knockout mutant RNA and detected a low level of the TV RdRp and the NV capsid proteins (data not shown). These results confirmed our previous observation that the nonstructural region of the TV genome is sufficient for initiation of TV RNA replication in LLC-MK2 cells; however, both the viral capsid protein VP1 and the minor structural protein VP2 are essential for generation of progeny viruses (22). These results also indicated that the human NV capsid proteins may not be compatible with TV in assembling progeny viruses.
An insertion of the eGFP gene at the 5′ end of the TV genome impaired the infectivity of TV.
Since the coding region of the TV N terminus is much smaller than that of other CVs, we tested whether the 5′ end of the genome would tolerate an insertion of heterologous sequences. An eGFP-TV construct, with an eGFP gene inserted between the 5′ UTR and the coding region of the N terminus protein of TV, was generated. This construct was expected to result in an eGFP-N terminus fusion protein upon translation and polyprotein processing. However, transfection of LLC-MK2 cells with capped RNA of eGFP-TV resulted in neither GFP expression nor viral infection. The reason for the lack of GFP expression remains unknown, but the apparent interruption of TV RNA replication by the eGFP gene indicates that the authentic 5′ RNA sequence of the virus is critical for TV infectivity.
DISCUSSION
The difficulty of cultivating human NVs in vitro has significantly hampered NV research. In this study, we developed an efficient reverse genetics system for TV which could be a suitable surrogate for human NV. TV has a genome organization typical for CVs. Its nonstructural polyprotein, encoded by ORF1, contains all of the conserved amino acid motifs, and they are in an arrangement similar to that of other CVs. More importantly, although TV may be classified into a new genus of Caliciviridae, it is genetically more closely related to human NVs than to other CV genera by both sequence and phylogenetic analyses. Furthermore, the molecular mass of TV structural proteins encoded by ORF2 and ORF3 is similar to that of NVs. Finally, since TV was originally isolated from stools of rhesus macaques, TV is likely to replicate in the intestine and thus might be a good model for human NVs. It remains unknown whether the virus in infected animals is also associated with clinical illness, such as diarrhea.
We compared the genomic organizations and protein sequence similarities of TV and human NVs in an attempt to identify potential elements unique for TV permissiveness to the host cell. The coding regions for the N-terminal protein, 3A-like protease, and VPg are the most diverse regions between the two genera in terms of size and sequence composition. The N-terminal protein is the most unique to TV because it is only around half the size of the NV N-terminal protein (7). In NVs, the N-terminal protein may play a role in intracellular protein trafficking in host cells (6, 8). In this study, we have demonstrated that an insertion of the eGFP coding region in frame with the coding region of the N-terminal protein impaired the infectivity of TV, indicating that the authentic 5′ end of either the viral RNA or the N-terminal protein is critical for infectivity. In future studies, it will be important to investigate the function of the N-terminal protein and whether the smaller size of this protein plays any role in host permissiveness.
We also noticed similarities and differences between TV and other CVs. For example, the genome of many CVs consists of both genomic and subgenomic RNAs, and both share highly conserved 5′ UTR sequences. We found that even though both genomic and subgenomic RNAs were present during TV replication in the LLC-MK2 cells (unpublished data), the 5′ UTR did not reveal significant homologies with the 5′ region of the putative subgenomic TV RNA. Nevertheless, the 5′ UTR sequence aligned well to a region 21 nt downstream from the ORF2 initiation codon (7). Similarly to feline CV (21), transfection of the genomic TV RNA alone without subgenomic RNA is sufficient to establish infection.
In this report, we demonstrated that the presence of the cap analog m7G(5′)ppp(5′)G is essential in establishing infectious RNA for TV. This is consistent with reports by other groups indicating that VPg or a cap structure is required in generating CV replicon and/or infectious virus (2, 3, 21). In contrast to some CV reverse genetics systems, the TV replication system does not require a helper virus. However, viral replication remains at a low level following the primary transfection, and distinct plaques can be observed only in the subsequent passages, although typical CPE and cell death have been observed following transfection. It remains unknown whether this low replication was due to a low efficiency of RNA capping, a low rate of transfection, or a unique property of TV, as low titers (∼105 PFU/ml) of the tissue culture-adapted TV were usually obtained.
In this study, we also demonstrated that the TV nonstructural gene is sufficient in the initiation of viral RNA replication but that both the major and minor structural genes are required for the production of progeny viruses. Since TV and NVs are genetically closely related, we studied chimeric TV-NV clones by replacing the TV capsid gene with that of human NV in an attempt to recover the progeny chimeric viruses. We also studied a chimeric clone with a replacement of the capsid P domain only, because in a recent study we demonstrated that the NV capsid protein S domain and P domain can be functionally and structurally independent (24). Although neither of the chimeric RNAs resulted in progeny viruses, the successful detection of the viral nonstructural protein (RdRp) and the human NV capsid protein in the transfected cells is encouraging.
Acknowledgments
We thank Xiaoyun Deng and Weiming Zhong for technical support and Ming Tan for reviewing the manuscript.
This study was supported by the National Institutes of Health (R01 AI37093, R01 AI55649, and P01 HD013021) and the Department of Defense (PR033018) (to X.J.) and by a Cincinnati Children's Hospital Research Foundation Trustee grant (to T.F.).
Footnotes
Published ahead of print on 10 September 2008.
REFERENCES
- 1.Asanaka, M., R. L. Atmar, V. Ruvolo, S. E. Crawford, F. H. Neill, and M. K. Estes. 2005. Replication and packaging of Norwalk virus RNA in cultured mammalian cells. Proc. Natl. Acad. Sci. USA 10210327-10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang, K. O., S. S. Sosnovtsev, G. Belliot, Q. Wang, L. J. Saif, and K. Y. Green. 2005. Reverse genetics system for porcine enteric calicivirus, a prototype sapovirus in the Caliciviridae. J. Virol. 791409-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, K. O., S. V. Sosnovtsev, G. Belliot, A. D. King, and K. Y. Green. 2006. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 353463-473. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhry, Y., M. A. Skinner, and I. G. Goodfellow. 2007. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J. Gen. Virol. 882091-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dastjerdi, A. M., J. Green, C. I. Gallimore, D. W. Brown, and J. C. Bridger. 1999. The bovine Newbury agent-2 is genetically more closely related to human SRSVs than to animal caliciviruses. Virology 2541-5. [DOI] [PubMed] [Google Scholar]
- 6.Ettayebi, K., and M. E. Hardy. 2003. Norwalk virus nonstructural protein p48 forms a complex with the SNARE regulator VAP-A and prevents cell surface expression of vesicular stomatitis virus G protein. J. Virol. 7711790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farkas, T., K. Sestak, C. Wei, and X. Jiang. 2008. Characterization of a rhesus monkey calicivirus representing a new genus of Caliciviridae. J. Virol. 825408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez-Vega, V., S. V. Sosnovtsev, G. Belliot, A. D. King, T. Mitra, A. Gorbalenya, and K. Y. Green. 2004. Norwalk virus N-terminal nonstructural protein is associated with disassembly of the Golgi complex in transfected cells. J. Virol. 784827-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, K., R. Chanock, and A. Kapikian. 2001. Human calicivirus, p. 841-874. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 10.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181(Suppl. 2)S322-S330. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, B., H. M. McClure, R. L. Fankhauser, S. S. Monroe, and R. I. Glass. 2004. Prevalence of rotavirus and norovirus antibodies in non-human primates. J. Med. Primatol. 3330-33. [DOI] [PubMed] [Google Scholar]
- 12.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 19551-61. [DOI] [PubMed] [Google Scholar]
- 13.Katayama, K., G. S. Hansman, T. Oka, S. Ogawa, and N. Takeda. 2006. Investigation of norovirus replication in a human cell line. Arch. Virol. 1511291-1308. [DOI] [PubMed] [Google Scholar]
- 14.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, G., Y. Zhang, Z. Ni, T. Yun, Z. Sheng, H. Liang, J. Hua, S. Li, Q. Du, and J. Chen. 2006. Recovery of infectious rabbit hemorrhagic disease virus from rabbits after direct inoculation with in vitro-transcribed RNA. J. Virol. 806597-6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martella, V., M. Campolo, E. Lorusso, P. Cavicchio, M. Camero, A. L. Bellacicco, N. Decaro, G. Elia, G. Greco, M. Corrente, C. Desario, S. Arista, K. Banyai, M. Koopmans, and C. Buonavoglia. 2007. Norovirus in captive lion cub (Panthera leo). Emerg. Infect. Dis. 131071-1073. http://www.cdc.gov/EID/content/13/7/1071.htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver, S. L., E. Asobayire, A. M. Dastjerdi, and J. C. Bridger. 2006. Genomic characterization of the unclassified bovine enteric virus Newbury agent-1 (Newbury1) endorses a new genus in the family Caliciviridae. Virology 350240-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliver, S. L., A. M. Dastjerdi, S. Wong, L. El-Attar, C. Gallimore, D. W. Brown, J. Green, and J. C. Bridger. 2003. Molecular characterization of bovine enteric caliciviruses: a distinct third genogroup of noroviruses (Norwalk-like viruses) unlikely to be of risk to humans. J. Virol. 772789-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sestak, K., C. K. Merritt, J. Borda, E. Saylor, S. R. Schwamberger, F. Cogswell, E. S. Didier, P. J. Didier, G. Plauche, R. P. Bohm, P. P. Aye, P. Alexa, R. L. Ward, and A. A. Lackner. 2003. Infectious agent and immune response characteristics of chronic enterocolitis in captive rhesus macaques. Infect. Immun. 714079-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smiley, J. R., K. O. Chang, J. Hayes, J. Vinje, and L. J. Saif. 2002. Characterization of an enteropathogenic bovine calicivirus representing a potentially new calicivirus genus. J. Virol. 7610089-10098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sosnovtsev, S., and K. Y. Green. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology 210383-390. [DOI] [PubMed] [Google Scholar]
- 22.Sosnovtsev, S. V., G. Belliot, K. O. Chang, O. Onwudiwe, and K. Y. Green. 2005. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 794012-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugieda, M., H. Nagaoka, Y. Kakishima, T. Ohshita, S. Nakamura, and S. Nakajima. 1998. Detection of Norwalk-like virus genes in the caecum contents of pigs. Arch. Virol. 1431215-1221. [DOI] [PubMed] [Google Scholar]
- 24.Tan, M., P. Huang, J. Meller, W. Zhong, T. Farkas, and X. Jiang. 2003. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: evidence for a binding pocket. J. Virol. 7712562-12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thumfart, J. O., and G. Meyers. 2002. Feline calicivirus: recovery of wild-type and recombinant viruses after transfection of cRNA or cDNA constructs. J. Virol. 766398-6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward, V. K., C. J. McCormick, I. N. Clarke, O. Salim, C. E. Wobus, L. B. Thackray, H. W. Virgin IV, and P. R. Lambden. 2007. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc. Natl. Acad. Sci. USA 10411050-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng, D. P., T. Ando, R. L. Fankhauser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346312-323. [DOI] [PubMed] [Google Scholar]