Abstract
Human immunodeficiency virus (HIV) infections in sub-Saharan Africa represent about 56% of global infections. Study of active-site mutations (the V82A single mutation and the V82F I84V double mutation) in the less-studied South African HIV type 1 subtype C (C-SA) protease indicated that neither mutation had a significant impact on the proteolytic functioning of the protease. However, the binding affinities of, and inhibition by, saquinavir, ritonavir, indinavir, and nelfinavir were weaker for each variant than for the wild-type protease, with the double mutant exhibiting the most dramatic change. Therefore, our results show that the C-SA V82F I84V double mutation decreased the binding affinities of protease inhibitors to levels significantly lower than that required for effective inhibition.
AIDS remains a serious infectious disease challenge in developing countries (12), with Africa home to more than 70% of people infected with human immunodeficiency virus (HIV) worldwide (6). Although a low adult prevalence rate exists in Western countries, the overall figure is greater in sub-Saharan Africa (12), and South Africa, in particular, is at the epicenter of the epidemic. Of the 10 subtypes of HIV type 1 (HIV-1) within the major group of the virus, subtype C (C-SA) accounts for ∼95% of infections in South Africa (7). Because antiretroviral drugs have been developed and tested against subtype B, an important question relates to the effectiveness of protease (PR) inhibitors against proteases from non-B subtypes.
Our work addressed the effects of active-site mutations (V82A and the V82F I84V double mutation) in the wild-type C-SA HIV-1 PR (containing eight polymorphic sites) in relation to the wild-type subtype B HIV-1 PR (Fig. 1). These polymorphisms do not affect viral fitness but do influence thermodynamic inhibitor binding and can amplify the effects of drug-resistant mutations (2-5, 9, 13, 21, 28-30). Sequence data from the National Institute for Communicable Diseases (NICD, South Africa) have revealed the presence of the V82A mutation in PR inhibitor-treated patients. The V82F I84V double mutation used in this study has not been observed in clinical isolates, although it was previously identified by passaging HIV-1 in the presence of increasing concentrations of HIV-1 PR inhibitors and is known to confer multidrug cross-resistance (4, 15, 20). The polymorphic substitutions and active-site mutations (V82A and V82F I84V) in the C-SA PR molecule were used to determine (i) PR catalytic efficiency and biochemical fitness and (ii) acetyl-pepstatin and drug (saquinavir, ritonavir, indinavir, and nelfinavir) binding energetics.
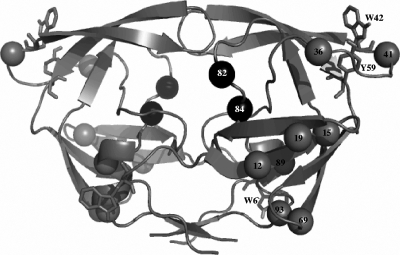
FIG. 1.
Ribbon representation of the homodimeric structure of HIV-1 PR, indicating the topographical positions occupied by the eight consensus amino acid residues in the South African subtype C: T12S, I15V, L19I, M36I, R41K, H69K, L89M, and I93L (gray spheres). Spheres representing the positions occupied by amino acids 82 and 84 are shown in black. Tryptophan and tyrosine residues are represented in stick format. The PDB code used for this subtype B HIV-1 PR structure is 1HXW. This figure was created with PyMol.
Source of sequence data and verification of the HIV-1 PR expression plasmid.
PR sequence data were obtained from the AIDS Virus Research Unit (NICD, South Africa). The pET-11b expression plasmid was a gift from J. Tang (University of Oklahoma Health Science Center, Oklahoma City) (11). The wild-type C-SA PR (T12S I15V L19I M36I R41K H69K L89M I93L) and the V82A and V82F I84V mutants were generated using a QuikChange kit (Stratagene). The coding region for each PR was confirmed by DNA sequencing.
Overexpression and purification of the HIV-1 PR.
Overexpression and purification of the wild-type and variant PRs were similar. Briefly, plasmid DNA encoding each PR was transformed into Escherichia coli BL21(DE3)/pLysS cells. Proteases were overexpressed as inclusion bodies and purified as previously described (28). After refolding, the PR was estimated to be ≥99% pure, with a monomeric size of 11 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (18), and an apparent oligomeric molecular mass of 22 kDa by using size exclusion high-performance liquid chromatography. Protein concentrations were determined spectrophotometrically using an extinction coefficient (E1%) of 11.8 at 280 nm (23).
Active PR concentration determination and peptide binding energetics.
Active PR concentrations were determined calorimetrically using a VP-ITC calorimeter (MicroCal Inc., Northampton, MA). Briefly, an enzyme solution (20 μM) was titrated with acetyl-pepstatin (200 to 300 μM) dissolved in 10 mM sodium acetate, pH 5.0. Raw data were analyzed using Origin5 software. The stoichiometry was used to determine the concentration of active PR. Binding of acetyl-pepstatin to each PR was characterized by successive endothermic heats (positive ΔH). Titration data for wild-type C-SA PR yielded a Gibbs free energy (ΔG) of −8.7 kcal/mol, a binding enthalpy (ΔH) of 7.7 kcal/mol, a binding entropy (−TΔS) of −16.4 kcal/mol, and a dissociation constant (Kd) of 400 nM. Acetyl-pepstatin binding to the V82A and V82F I84V variant PRs was also characterized by a favorable −TΔS. The binding affinity for acetyl-pepstatin was not significantly affected for the V82A and V82F I84V variant PRs.
Spectroscopic methods.
Far-UV (250 to 190 nm) circular dichroism was performed using 15 μM protein in 10 mM sodium acetate buffer, pH 5.0. The spectra of all the PRs exhibited minima at 216 nm, typical of predominantly β-sheeted protein (7, 19).
Intrinsic fluorescence emission spectra (emission maximum, 355 nm) were obtained by selectively exciting tryptophan and tyrosine residues. Wavelength emission maxima indicated that tryptophan residues were highly solvent exposed, because the emission spectrum of tryptophan in water is between 350 and 355 nm (17). However, the fluorescence intensity of the double mutant was significantly enhanced (40 to 60%) relative to that of the wild-type and V82A HIV-1 PRs, suggestive of tertiary structural changes.
Steady-state kinetics.
PR hydrolytic activity was measured by monitoring the relative decrease in absorbance of a chromogenic peptide substrate at 300 nm. An extinction coefficient of 1,800 M−1 cm−1 at 300 nm was used to convert the absorbance change to reaction rates. No major differences in enzyme function were observed among all the PRs (Table 1). The kcat/Km and kcat values for substrate cleavage were 3.6-fold lower for the V82A mutant and 6-fold lower for the V82F I84V mutant than for the wild-type C-SA PR, consistent with the results obtained for the subtype B PR (16, 26).
TABLE 1.
Kinetic parameters for the wild-type C-SA, V82A, and V82F I84V HIV-1 PRs
| Protease | Sp act (μmol/min/mg) | Km (μM) | kcat (1/s) | kcat/Km (1/μM·s) |
|---|---|---|---|---|
| Wild-type C-SA | 0.91 | 76 | 2.74 | 0.036 |
| V82A mutant | 2.75 | 71 | 0.71 | 0.010 |
| V82F I84V mutant | 0.95 | 81 | 0.49 | 0.006 |
Inhibition studies.
Fifty percent inhibitory concentrations (IC50) and Ki values were determined by using increasing inhibitor concentrations (Table 2). Inhibition constants for the wild-type and V82A PRs were in the nanomolar range and were comparable to previous data (16). Against the V82F I84V variant, however, inhibition constants for all inhibitors were drastically weaker; the ratios of the IC50 and Ki values for this variant to those for the wild type ranged from 47 to 497 (Table 2). Saquinavir was the least affected PR inhibitor, with a 47-fold increase in the Ki and a 100-fold increase in the IC50, whereas indinavir showed the greatest increases (447- and 497-fold, respectively).
TABLE 2.
Inhibition characteristics and thermodynamic parameters for the binding of inhibitors to the wild-type subtype B, wild-type C-SA, and V82A and V82F I84V mutant HIV-1 PRs
| HIV-1 PR and inhibitor | Inhibition characteristic (nM)a
|
Thermodynamic parameter
|
||||
|---|---|---|---|---|---|---|
| IC50 | Ki | Kd (nM) | ▵G (kcal/mol) | ▵H (kcal/mol) | −T▵S (kcal/mol) | |
| Wild-type subtype Bb | ||||||
| Saquinavir | 0.40 | −12.80 | 1.90 | −14.70 | ||
| Ritonavir | 0.03 | −14.40 | −3.70 | −10.70 | ||
| Indinavir | 0.48 | −12.70 | 2.10 | −14.80 | ||
| Nelfinavir | 0.26 | −13.10 | 2.60 | −15.70 | ||
| Wild-type C-SA | ||||||
| Saquinavir | 0.12 (1) | 0.62 (1) | 0.84 | −12.40 | 6.40 | −18.80 |
| Ritonavir | 1.25 (1) | 0.50 (1) | 0.21 | −13.20 | −4.20 | −9.10 |
| Indinavir | 3.02 (1) | 0.49 (1) | 0.95 | −12.30 | 2.70 | −15.00 |
| Nelfinavir | 2.34 (1) | 0.52 (1) | 0.30 | −13.00 | 4.10 | −17.10 |
| V82A mutant | ||||||
| Saquinavir | 0.28 (2.3) | 0.97 (1.6) | 1.85 | −12.00 | 5.50 | −17.50 |
| Ritonavir | 6.26 (5.0) | 1.08 (2.2) | 2.00 | −11.90 | −2.20 | −9.70 |
| Indinavir | 26.01 (8.6) | 1.20 (2.4) | 12.70 | −10.80 | 2.90 | −13.70 |
| Nelfinavir | 2.87 (1.2) | 0.90 (1.7) | 0.66 | −12.50 | 4.30 | −16.90 |
| V82F I84V mutant | ||||||
| Saquinavir | 12 (100) | 29 (47) | 98 | −9.60 | 0.80 | −10.40 |
| Ritonavir | 250 (200) | 97 (194) | 230 | −9.10 | 0.60 | −9.60 |
| Indinavir | 1,500 (497) | 219 (447) | 450 | −8.70 | 3.80 | −12.50 |
| Nelfinavir | 240 (103) | 56 (108) | 110 | −9.50 | 4.00 | −13.50 |
The ratio of the value for the mutant to the value for the wild type is given in parentheses for the variant PRs.
Thermodynamic parameters for the wild-type HIV-1 subtype B PR were taken from Velazquez-Campoy et al. (29) for comparison purposes.
Thermodynamics of clinical inhibitor binding.
Due to the high affinity of clinical inhibitors for the HIV-1 PR, binding thermodynamics were measured in competition experiments with acetyl-pepstatin (14, 24, 27). The clinical inhibitors bound the wild-type C-SA PR with high affinities in an entropically driven process (Table 2). The binding of saquinavir, indinavir, and nelfinavir to the wild-type C-SA PR was enthalpically unfavorable, while the binding of ritonavir was slightly favorable enthalpically (Table 2).
The binding affinity of the V82A PR for saquinavir and nelfinavir was reduced 2.2-fold; for ritonavir and indinavir it was reduced 10- and 13-fold, respectively. Consequently, this mutation lowered the ΔG toward all inhibitors relative to that of the wild-type C-SA PR (Table 2). The effect of the V82A mutation on ΔH amounted to 2 kcal/mol for ritonavir and 0.2 kcal/mol for indinavir and nelfinavir, with a gain in ΔH of 0.9 kcal/mol for saquinavir. Entropic changes amounted to 1.3 kcal/mol for saquinavir and indinavir and 0.2 kcal/mol for nelfinavir, with a slight gain of 0.6 kcal/mol for ritonavir. The V82A mutation compensates for the loss in binding entropy by a small gain in binding enthalpy for saquinavir and nelfinavir. This suggests that some functional characteristics of the compounds render them less susceptible to binding site distortions or changes associated with this mutation (10, 25). On the other hand, indinavir and ritonavir lose enthalpic contributions to the binding energy, resulting in a higher binding affinity loss.
Interestingly, the binding affinities of clinical inhibitors for the V82F I84V mutant were sufficiently low to allow thermodynamic measurements by direct calorimetric titrations. Clinical inhibitor binding was characterized by unfavorable enthalpic contributions (Table 2). The ΔG was −9.6 kcal/mol for saquinavir, −9.1 kcal/mol for ritonavir, −8.7 kcal/mol for indinavir, and −9.5 kcal/mol for nelfinavir. The −TΔS parameter, therefore, contributed between −10 kcal/mol and −14 kcal/mol to the ΔG and compensated for the unfavorable ΔH, thereby providing favorable binding affinities for saquinavir, ritonavir, indinavir, and nelfinavir. However, the double mutation reduced the binding affinities of clinical inhibitors 117- to 1,095-fold. Kd values for clinical inhibitors were in agreement with inhibition constants measured in enzyme inhibition experiments. Against the V82F I84V variant, saquinavir, indinavir, and nelfinavir lose significant entropic contributions relative to both wild-type and V82A C-SA PRs (Table 2). Binding entropy loss is partially compensated for by a slight gain in binding enthalpy for saquinavir and nelfinavir, while indinavir and ritonavir lose enthalpic contributions to the binding energy and consequently suffer higher affinity losses. Molecular dynamic simulations have shown that the subtype B V82F I84V variant induced rapid, frequent flap tip curling, suggesting a more open conformation (1, 22). This is consistent with low binding affinities for most inhibitors in clinical use, primarily as a consequence of a larger energetic penalty for the burial of larger exposed hydrophobic surface areas.
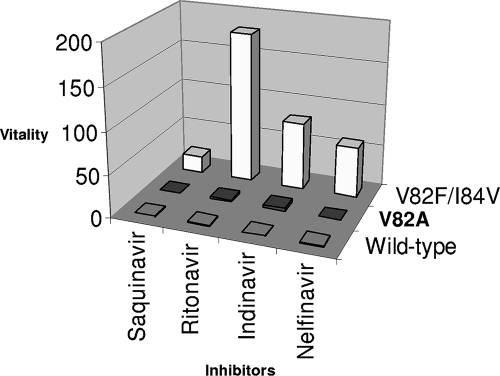
Relative vitality.
The vitality/viral fitness of variant PRs in the presence of an inhibitor was calculated from reference 8 as follows: [(Kd × kcat/Km)mutant]/[(Kd × kcat/Km)wild type].
The data in Fig. 2 indicate that the V82A mutation is less likely to be selected in the presence of saquinavir and nelfinavir. However, there was an increase (2.6- or 3.7-fold) in the vitality of the V82A variant in the presence of indinavir or ritonavir, consistent with the results obtained for subtype B PR (8). The double mutant displayed increased vitality against all inhibitors. This mutant is particularly selective for indinavir and ritonavir, with vitality values of 80 and 180, respectively, in agreement with inhibition and thermodynamic studies.
FIG. 2.
Vitality values for the C-SA HIV-1 proteases with saquinavir, ritonavir, indinavir, and nelfinavir.
Implications for drug resistance.
In subtype B, the V82F I84V mutation lowers the binding affinities for saquinavir and nelfinavir ∼21-fold, for indinavir 68-fold, and for ritonavir 368-fold (29). However, when the binding energetics of clinical inhibitors for the wild-type subtype B PR are compared to those for each subtype C PR (this study), it is clear that inhibitory potency is lowered for the V82A variant but much more for the V82F I84V variant. Within the C-SA subtype, the V82F I84V mutation lowers the affinity of inhibitors by factors ranging from ∼245 to 7,667.
In conclusion, although the clinical inhibitors used in this study exhibit high affinities of binding to the C-SA wild-type and V82A enzymes, the affinity of inhibitors for the V82F I84V mutant is weakened far below the level required for effective inhibition. This can, therefore, have serious implications for the long-term viability of PR inhibition therapy for the clade C-SA virus.
Acknowledgments
This work was supported by the National Research Foundation (Focus Area grant 2069111), the Carnegie Corporation of New York, a University of the Witwatersrand AIDS Institute grant, the Andrew Mellon Foundation, and the Medical Research Council.
Footnotes
Published ahead of print on 3 September 2008.
REFERENCES
- 1.Aruksakunwong, O., P. Wolschann, S. Hannongbua, and P. Sompornpisut. 2006. Molecular dynamic and free energy studies of primary resistance mutations in HIV-1 protease-ritonavir complexes. J. Chem. Inf. Model. 462085-2092. [DOI] [PubMed] [Google Scholar]
- 2.Becker-Pergola, G., P. Kataaha, L. Johnston-Dow, S. Fung, J. B. Jackson, and S. H. Eshleman. 2000. Analysis of HIV type 1 protease and reverse transcriptase in antiretroviral drug-naive Ugandan adults. AIDS Res. Hum. Retrovir. 16807-813. [DOI] [PubMed] [Google Scholar]
- 3.Benson, D. A., M. S. Boguski, D. J. Lipman, J. Ostell, and B. F. Ouellette. 1998. GenBank. Nucleic Acids Res. 261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cane, P. A., A. de Ruiter, P. Rice, M. Wiselka, R. Fox, and D. Pillay. 2001. Resistance-associated mutations in the human immunodeficiency virus type 1 subtype C protease gene from treated and untreated patients in the United Kingdom. J. Clin. Microbiol. 392652-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coman, R. M., A. H. Robbins, M. A. Fernandez, C. T. Gilliland, A. A. Sochet, M. M. Goodenow, R. McKenna, and B. M. Dunn. 2008. The contribution of naturally occurring polymorphisms in altering the biochemical and structural characteristics of HIV-1 subtype C protease. Biochemistry 47731-743. [DOI] [PubMed] [Google Scholar]
- 6.Essex, M. 1999. Human immunodeficiency viruses in the developing world. Adv. Virus Res. 5371-88. [DOI] [PubMed] [Google Scholar]
- 7.Foulkes, J. E., M. Prabu-Jeyabalan, D. Cooper, G. J. Henderson, J. Harris, R. Swanstrom, and C. A. Schiffer. 2006. Role of invariant Thr80 in human immunodeficiency virus type 1 protease structure, function, and viral infectivity. J. Virol. 806906-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gulnik, S. V., L. I. Suvorov, B. Liu, B. Yu, B. Anderson, H. Mitsuya, and J. W. Erickson. 1995. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry 349282-9287. [DOI] [PubMed] [Google Scholar]
- 9.Holguín, A., and V. Soriano. 2002. Resistance to antiretroviral agents in individuals with HIV-1 non-B subtypes. HIV Clin. Trials 3403-411. [DOI] [PubMed] [Google Scholar]
- 10.Hong, L., X. C. Zhang, J. A. Hartsuck, and J. Tang. 2000. Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: insights into mechanisms of drug resistance. Protein Sci. 91898-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ido, E., H. P. Han, F. J. Kezdy, and J. Tang. 1991. Kinetic studies of human immunodeficiency virus type 1 protease and its active-site hydrogen bond mutant A28S. J. Biol. Chem. 26624359-24366. [PubMed] [Google Scholar]
- 12.Joint United Nations Programme on HIV/AIDS and World Health Organization. December 2007. AIDS epidemic update. UNAIDS, Geneva, Switzerland. http://data.unaids.org/pub/EPISlides/2007/2007_epiupdate_en.pdf.
- 13.Kantor, R., and D. Katzenstein. 2003. Polymorphism in HIV-1 non-subtype B protease and reverse transcriptase and its potential impact on drug susceptibility and drug resistance evolution. AIDS Rev. 525-35. [PubMed] [Google Scholar]
- 14.King, N. M., M. Prabu-Jeyabalan, E. A. Nalivaika, and C. A. Schiffer. 2004. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 7812012-12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King, R. W., D. L. Winslow, S. Garbe, L. T. Bacheler, S. Stack, and M. J. Otto. 1995. Identification of a clinical isolate of HIV-1 with an isoleucine at position 82 of the protease which retains susceptibility to protease inhibitors. Antivir. Res. 2813-24. [DOI] [PubMed] [Google Scholar]
- 16.Klabe, R. M., L. T. Bacheler, P. J. Ala, S. Erickson-Viitanen, and J. L. Meek. 1998. Resistance to HIV protease inhibitors: a comparison of enzyme inhibition and antiviral potency. Biochemistry 378735-8742. [DOI] [PubMed] [Google Scholar]
- 17.Lacowicz, J. R. 1999. Principles of fluorescence spectroscopy. Plenum Press, New York, NY.
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227680-685. [DOI] [PubMed] [Google Scholar]
- 19.Navia, M. A., P. M. Fitzgerald, B. M. McKeever, C. T. Leu, J. C. Heimbach, W. K. Herber, I. S. Sigal, P. L. Darke, and J. P. Springer. 1989. Three-dimensional structure of aspartyl protease from human immunodeficiency virus HIV-1. Nature 337615-620. [DOI] [PubMed] [Google Scholar]
- 20.Otto, M. J., S. Garber, D. L. Winslow, C. D. Ried, P. Aldrich, P. K. Jadhav, C. E. Patterson, C. N. Hodge, and Y. S. E. Cheng. 1993. In vitro isolation and identification of human immunodeficiency virus (HIV) variants with reduced sensitivity to C-2 symmetrical inhibitors of HIV type 1 protease. Proc. Natl. Acad. Sci. USA 907543-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perno, C. F., A. Cozzi-Lepri, C. Balotta, F. Forbici, M. Violin, A. Bertoli, G. Facchi, P. Pezzotti, G. Cadeo, G. Tositti, S. Pasquinucci, S. Pauluzzi, A. Scalzini, B. Salassa, A. Vincenti, A. N. Phillips, F. Dianzani, A. Appice, G. Angarano, L. Monno, G. Ippolito, M. Moroni, A. d'Arminio Monforte, et al. 2001. Secondary mutations in the protease region of human immunodeficiency virus and virologic failure in drug-naïve patients treated with protease inhibitor-based therapy. J. Infect. Dis. 184983-991. [DOI] [PubMed] [Google Scholar]
- 22.Perryman, A. L., J. H. Lin, and J. A. McCammon. 2004. HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs. Protein Sci. 131108-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polgár, L., Z. Szeltner, and I. Boros. 1994. Substrate-dependent mechanisms in the catalysis of human immunodeficiency virus protease. Biochemistry 339351-9357. [DOI] [PubMed] [Google Scholar]
- 24.Sigurskjold, B. W. 2000. Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Anal. Biochem. 277260-266. [DOI] [PubMed] [Google Scholar]
- 25.Tie, Y., A. Y. Kovalevsky, P. I. Boross, Y.-F. Wang, A. K. Ghosh, J. R. Tozser, W. Harrison, and I. T. Weber. 2007. Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir. Proteins 67232-242. [DOI] [PubMed] [Google Scholar]
- 26.Tie, Y., P. I. Boross, Y.-F. Wang, L. Gaddis, F. Liu, X. Chen, J. Tozser, R. W. Harrison, and I. T. Weber. 2005. Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 Å resolution crystal structures of HIV-1 protease mutants with substrate analogs. FEBS J. 2725265-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Todd, M. J., I. Luque, A. Velazquez-Campoy, and E. Freire. 2000. Thermodynamic basis of resistance to HIV-1 protease inhibition: calorimetric analysis of the V82F/I84V active site resistant mutant. Biochemistry 3911876-11883. [DOI] [PubMed] [Google Scholar]
- 28.Velazquez-Campoy, A., M. J. Todd, S. Vega, and E. Freire. 2001. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc. Natl. Acad. Sci. USA 986062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Velazquez-Campoy, A., S. Vega, and E. Freire. 2002. Amplification of the effects of drug resistance mutations by background polymorphisms in HIV-1 protease from African subtypes. Biochemistry 418613-8619. [DOI] [PubMed] [Google Scholar]
- 30.Velazquez-Campoy, A., S. Vega, E. Fleming, U. Bacha, Y. Sayed, H. W. Dirr, and E. Freire. 2003. Protease inhibition in African subtypes of HIV-1. AIDS Rev. 5165-171. [PubMed] [Google Scholar]