Abstract
Rotavirus nonstructural protein NSP3 interacts specifically with the 3′ end of viral mRNAs, with the eukaryotic translation initiation factor eIF4G, and with RoXaN, a cellular protein of yet-unknown function. By evicting cytoplasmic poly(A) binding protein (PABP-C1) from translation initiation complexes, NSP3 shuts off the translation of cellular polyadenylated mRNAs. We show here that PABP-C1 evicted from eIF4G by NSP3 accumulates in the nucleus of rotavirus-infected cells. Through modeling of the NSP3-RoXaN complex, we have identified mutations in NSP3 predicted to interrupt its interaction with RoXaN without disturbing the NSP3 interaction with eIF4G. Using these NSP3 mutants and a deletion mutant unable to associate with eIF4G, we show that the nuclear localization of PABP-C1 not only is dependent on the capacity of NSP3 to interact with eIF4G but also requires the interaction of NSP3 with a specific region in RoXaN, the leucine- and aspartic acid-rich (LD) domain. Furthermore, we show that the RoXaN LD domain functions as a nuclear export signal and that RoXaN tethers PABP-C1 with RNA. This work identifies RoXaN as a cellular partner of NSP3 involved in the nucleocytoplasmic localization of PABP-C1.
The cytoplasmic poly(A)-binding protein (PABP-C1) is considered a bona fide translation initiation factor which enhances translation by binding the 3′ poly(A) tail of the cellular mRNAs and simultaneously interacting with eukaryotic translation initiation factor 4G (eIF4G) (27, 29). eIF4G is a scaffold protein that allows mRNA circularization by providing sites of interaction for PABP-C1 and eIF4E, the protein that binds the 5′ end of capped mRNAs. eIF4G then coordinates the assembly of several other translation initiation factors, such as eIF4A, eIF3, and the small ribosomal subunit (37). In synergy with the cap structure present at the 5′ end of most mRNAs, PABP-C1 stimulates the translation of cellular polyadenylated mRNAs by enhancing 40S ribosome subunit recruitment and 60S subunit joining (20). Furthermore, PABP-C1 binding to eIF4G increases the affinity of eIF4E for the cap structure (6, 28) by lowering its dissociation rate. Thus, PABP-C1 enhances translation by promoting the binding of mRNA to eIF4G and by lowering dissociation of the 5′ cap structure from the eIF4G/eIF4E complex. Normally evenly dispersed throughout the cytoplasm, PABP-C1 is redistributed into stress granules (SGs) under conditions of stress, such as oxidative stress or heat shock (22). SGs are cytoplasmic foci formed by the condensation of mRNAs stalled during translation and bound by the related RNA-binding proteins TIA-1 and TIA-R. SGs are not translationally competent, but rather serve as local storage and protection compartments for mRNAs under translational arrest during cellular stress. Although it is primarily cytoplasmic, PABP-C1 has been detected nevertheless in the nucleus of several mammalian cells (1, 17, 50, 51) associated with nuclear pre-mRNP (17). PABP-C1 is thus regarded as a shuttling protein that participates in mRNA maturation and nuclear export.
Rotaviruses are the major cause of gastroenteritis worldwide (34) with an estimated death toll of 600,000 children each year, mainly in developing countries. In developed countries, the rotavirus burden is measured by the number of outpatient visits and hospitalizations (in Europe, 1 out of 50 children will be hospitalized for rotavirus gastroenteritis by 5 years of age), by the direct medical costs ($250 million/year in the United States), and by the number of nosocomial infections (2). During rotavirus infection, dimers of the nonstructural protein NSP3 specifically bind the 3′ nonpolyadenylated end of viral mRNAs and to the translation initiation factor eIF4G (39, 40). In this context, NSP3 mimics PABP-C1, circularizing the viral mRNA and thus increasing the translation initiation efficiency (48). During rotavirus infection, NSP3 displaces PABP-C1 from eIF4G, promoting the translation of viral mRNAs while inhibiting the translation of cellular poly(A) mRNAs (33, 39, 40).
Another cellular protein, a binding partner of NSP3, has been identified and called RoXaN (49). RoXaN (ZC3H7B, FLJ13787, KIAA1031) contains several different protein-protein interaction domains: tetratricopeptide repeats (TPR) and a leucine- and aspartic acid-rich (LD) domain are present at the amino terminus, and several repeats of RNA-binding zinc fingers constitute the carboxy-terminal half. The role RoXaN plays in uninfected cells and during rotavirus infection is still not known.
We examined the subcellular localization of cytoplasmic PABP-C1 during rotavirus infection and during the expression of NSP3 in the absence of viral infection. We show that rotavirus NSP3 expression redirects cytoplasmic PABP-C1 to the cell nucleus and that this localization requires the interaction of NSP3 with eIF4G and with the LD domain of RoXaN.
MATERIALS AND METHODS
Cells and viruses.
Embryonic monkey kidney MA104, COS7, and embryonic human kidney HEK293 cells were maintained in Eagle's minimum essential medium (BioWhittaker, Emerainville, France) supplemented with 10% fetal bovine serum (HyClone, SouthLogan, UT), 100 IU/ml penicillin (BioValley, Marne la Vallée, France), and 100 μg/ml streptomycin (BioValley).
The bovine RF strain of group A rotavirus was used to infect MA104 cells. Viral infectivity was determined by plaque assays using MA104 cells, as described previously (40). Infections were performed at a multiplicity of infection (MOI) of 10 PFU/cell or 1 PFU/cell in Eagle's minimum essential medium in the presence of trypsin (0.44 μg/ml; Sigma, St. Quentin Fallavier, France) and antibiotics, but without serum.
Antibodies.
A rabbit polyclonal antibody (PAb) specific for the N-terminal part of NSP3 was obtained by immunization of rabbit with the recombinant His-tagged protein (amino acids [aa] 3 to 178) expressed in Escherichia coli and purified by using a nickel chelate column as described previously (38). The specificity of the anti-NSP3 immunoglobulin G (IgG) has been checked by Western blot, immunoprecipitation, and immunofluorescence assays of noninfected and rotavirus-infected MA104 cells.
A rabbit PAb specific for the amino terminus of RoXaN was obtained by repeated injections of a peptide consisting of aa 1 to 17 of RoXaN coupled to bovine serum albumin (Eurogentec, Belgium). IgGs were purified by using a protein A column (Melon gel purification kit; Pierce, Roquefort, IL) and RoXaN-specific IgGs were affinity purified with the peptide (aa 1 to 17) coupled to Sephadex beads. Purified IgGs were diluted 1/100 for immunofluorescence.
Mouse monoclonal antibodies (MAb) specific for rotavirus RF nonstructural proteins NSP5 and NSP3 have been described previously (3, 41). 10E10 MAb specific for PABP-C1 (14) and polyclonal goat TIA1-specific antiserum were purchased from Abcam (Cambridge, United Kingdom) and Santa Cruz (Santa Cruz, CA), respectively. Rabbit beta-galactosidase-specific polyclonal antiserum was obtained from Promega (Madison, WI). Green fluorescent protein (GFP)- and Flag M5-specific mAbs were purchased from Sigma-Aldrich, and the T7 epitope tag (T7tag)-specific MAb was purchased from Novagen (Darmstadt, Germany).
Secondary antibodies conjugated to horseradish peroxidase, goat anti-mouse IgG, and donkey anti-goat IgG were from Jackson Laboratories, and goat anti-rabbit IgG was from Sigma-Aldrich. AlexaFluor 488, 546, or 647 goat anti-mouse IgG; AlexaFluor 488, 546, or 647 goat anti-rabbit IgG; and Alexa Fluor 488, 546, or 647 goat anti-guinea pig IgG (Molecular Probes, Inc., Eugene, OR) were used as described below. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI).
Transfections, immunoprecipitations, and RNA binding assays.
HEK293 cells (approximately 2 × 106 per 60-mm poly-l-lysine-coated plate [BDBiocoat, Erembodegem, Belgium]) and MA104 cells (approximately 1 × 104 per 12-mm glass coverslip) were plated 24 to 36 h prior to transfection. Transfections were performed using Lipofectamine 2000 (InvitroGene, Cergy Pontoise, France) according the manufacturer's instructions. Cells were washed and processed for immunofluorescence or coimmunoprecipitation analysis 36 to 48 h after transfection.
For immunoprecipitations, cells were washed once with 1 ml of cold (4°C) phosphate-buffered saline (PBS) supplemented with Complete protease inhibitor cocktail (one tablet per 50 ml; Roche, Meylan, France) and lysed in 1 ml of TMGK buffer (20 mM Tris-HCl [pH 8], 20 mM MgCl2, 110 mM KCl, 1% Triton X-100). Cell debris was removed by centrifugation. Immunoprecipitation was performed by adding 1 μl of MAb or 1 μl of PAb to the whole-cell lysate supernatant and incubating overnight at 4°C with end-over-end rotation. Fifty microliters of a 50% suspension of protein A-Sepharose beads in TMGK buffer was then added, and the incubation was continued for 1 h at 4°C. Protein A-Sepharose beads were then subjected to centrifugation (1,300 × g, 30 s), and washed three times with 400 μl TMGK buffer.
RNA binding assays were performed by incubating transfected cell lysates with 50 μl of a 50% suspension (in TMGK buffer with 200 mM KCl) of oligoribonucleotides coupled to Sepharose or to cross-linked acrylamide beads (Sigma) for 2 h at ambient temperature. Mouse IgGs coupled to Sepharose beads were used as the nonspecific binding control. After three washes with TMGK buffer, proteins bound to the beads were recovered in loading buffer, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and identified by immunoblotting.
Immunoblots.
Cells or beads were recovered in loading buffer (10 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 150 mM 2-ß-mercaptoethanol), and proteins were resolved by SDS-PAGE in Laemmli buffer and then transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, France) by transverse electrophoresis in 10 mM CAPS (Sigma) (pH 11)-10% ethanol buffer. Next, the membrane was incubated for 1 h at room temperature in PBS containing 0.1% Tween 20-5% dry milk or in SuperBlock (Pierce) blocking reagent solution and then washed three times with PBS-0.1% Tween 20 and incubated overnight at 4°C with primary antibody (1:1,000 for PABP-C1; 1:5,000 for beta-galactosidase-, T7tag-, and Flag M5-specific antibodies; 1:4,000 for NSP3-specific polyclonal rabbit antiserum; and 1:2,500 for enhanced GFP [eGFP]-specific MAb). The membrane was washed three times in PBS containing 0.1% Tween 20 and incubated for 1 h at room temperature with secondary antibody (1:10,000 dilution for goat mouse-specific IgG and 1:20,000 for goat rabbit-specific IgG or donkey goat-specific IgG; Jackson Laboratories, Bar Harbor, ME) conjugated to horseradish peroxidase and then washed again three times in PBS containing 0.1% Tween 20, incubated with peroxidase substrate (SuperSignal; Pierce) and, finally, exposed to film for various amounts of time.
Immunofluorescence staining and confocal microscopy.
MA104 cells (1.5 × 104) seeded on 12-mm glass coverslips were infected with rotavirus RF at an MOI of 1 or 10 PFU/cell in 200 μl of incomplete media supplemented with trypsin. After adsorption at 37°C for 1 h, 800 μl incomplete medium was added to each well and cells were incubated at 37°C for various times. Cells were fixed with 4% paraformaldehyde at room temperature. Fixed cells were washed twice in PBS and permeabilized with 1% Triton X-100 (Bio-Rad, Hercules, CA) in PBS for 20 min at room temperature with shaking. Nonspecific binding of antibody was blocked by the incubation of cells for 15 min in PBS containing 5% boiled horse serum (Sigma), 0.5% Triton X-100 (Bio-Rad), and 0.2% fish gelatin (Sigma). Subsequent washes and dilutions were performed with this wash-dilution buffer, but with 1% boiled horse serum, unless otherwise indicated. Cells were incubated with the primary antibody for 1 h and then washed three times. Cells were incubated with AlexaFluor 488, 546, 633, or 647 goat anti-mouse IgG; AlexaFluor 488, 546, 633, or 647 goat anti-rabbit IgG; and Alexa Fluor 488, 546, 633, or 647 conjugated goat-, mouse-, rabbit-, or guinea pig-specific IgG (Molecular Probes, Inc., Eugene, OR) at a dilution of 1:1,000 for 1 to 1.5 h. Cells were washed three times for 20 min with wash-dilution buffer and twice for 10 min with PBS. Coverslips were immediately mounted or the cells were incubated with DAPI for 10 min and then mounted on glass slides with ProLong Antifade (Molecular Probes). Cells were visualized with a Leica SP2 confocal fluorescence microscope. Coverslips containing mock-infected or -transfected cells were included in every experiment and processed in parallel with infected cells. Mock-infected or -transfected cells were examined first and used to set the background on the confocal microscope to black before images of infected or transfected cells were obtained. Images were processed with Adobe Photoshop (Adobe Systems, Inc., San Jose, CA) and ImageJ freeware (http://rsb.info.nih.gov/ij/index.html).
Oxidative stress with arsenite and leptomycin B treatment.
Cells plated on coverslips for 24 h were treated for 30 min with a final concentration of 100 mM sodium arsenite. Cells transfected for 24 h were treated with leptomycin B (LMB, 1 ng/ml, Sigma) for 90 min. Cells were then fixed and processed for immunofluorescence as described above.
Quantification of nuclear and cytoplasmic PABP.
Pictures of randomly selected microscopic fields of MA104 cells transfected with wild-type (wt) NSP3, NSP3 R187E, or NSP3 K191E, where NSP3 was stained red, PABP was stained green, and nuclei were stained blue, were taken. Coded pictures were given to two observers who were blinded to the experimental conditions, and cells expressing NSP3 and cells with clearly nuclear PABP were counted. Statistical analysis (χ2 test) did not detect significant differences between each observer but did detect significant differences (P < 0.01) between the percentage of nuclear PABP in wt NSP3-transfected cells (72%, n = 137) versus mutated NSP3-transfected cells (35% with NSP3K191E, n = 137; and 32% with NSP3R187E, n = 187).
Plasmid construction, site-directed mutagenesis, and yeast two-hybrid assay.
NSP3 cDNA RF strain (GenBank accession number Z21639) has been described previously (40). For mammalian cell expression, NSP3 cDNA was amplified by PCR and cloned into the pTEJ4 (48) mammalian vector under the control of the ubiquitin promoter. A fragment encoding NSP3 150 to 313 was amplified by PCR and cloned in frame with the eGFP gene into the peGFP-N1 vector (Invitrogen). Human PABP-C1 cDNA (a kind gift of T. Grange, Paris VI University, Paris, France) amplified by PCR was cloned into the pcDNA 3.1/Hygro(−) vector (Invitrogen) in frame with the Flag epitope (49). eIF4G and RoXaN cDNAs cloned into pcDNA 3.1/Hygro(−) Flag or T7tag have been described previously (7, 49).
The plasmid pCMV-NLS-NES-gal containing the simian virus 40 T antigen nuclear localization signal (NLS) and the rabies virus P protein (aa 6 to 60) nuclear export signal (NES) fused in frame to the bacterial beta-galactosidase gene cloned into the pCMV plasmid, has been described elsewhere (35) and was a generous gift from D. Blondel (VMS, Gif/Yvette, France). A fragment of RoXaN cDNA encoding aa 258 to 297 and encompassing the RoXaN LD domain (aa 264 to 272) was amplified by PCR with appropriate primers with appended BglII sites and cloned in frame into the BglII site of pCMV-NLS-gal, giving the plasmid pCMV-NLS-LD-gal. A similar construct was obtained by amplification of the same region from a RoXaN cDNA bearing mutations at aa 267 to 269, which impair NSP3-RoXaN interaction (49), giving pCMV-NLS-LDmut-gal.
PCR-based site-directed mutagenesis with complementary oligonucleotides (QuikChange; Stratagene, Massy, France) was used to introduce point mutations into NSP3 cDNA. The NSP3 Δ4G (with a stop codon at position 239) construct has been described previously (48). The carboxy-terminal deletion of RoXaN was obtained by introducing a stop codon into the coding sequence of RoXaN cloned in pCDNA3.1-T7tag Hygro(−) by site-directed mutagenesis. Amino-terminal deletions were obtained by the introduction of a second NotI restriction site by site-directed mutagenesis, and then NotI digestion followed by religation removed the 5′ fragment of RoXaN cDNA between the T7tag and the new NotI site and put the T7tag in frame with the shortened RoXaN open reading frame. The accuracy of the nucleotide sequences obtained was checked by DNA sequencing using the Taq dye terminator technique and an ABI 3100 sequencer (Foster City, CA).
NSP3, eIF4G, and RoXaN cDNAs cloned into pGAD and pGBT9 vectors have been described previously (1, 14, 31, 49-51) and were used in the yeast two-hybrid assays with the AH109 (Clontech, Saint-Germain-en-Laye, France) strain of Saccharomyces cerevisiae (38).
Homology modeling of the NSP3 coiled-coil region.
The crystal structure of the human vimentin coil 2B fragment (Protein Data Bank entry 1GK4) was used as a template. The sequence of the coiled-coil region of NSP3 was projected onto chains A and B of 1GK4, using the programs ViTO (8) and MODELLER (13). The model was manually inspected using COOT (12) and subjected to mild minimization with REFMAC (32).
RESULTS
PABP-C1 is localized to the nucleus of rotavirus-infected cells but forms cytoplasmic stress granules upon oxidative stress.
In normal cells, poly(A) binding protein (PABP-C1) is localized primarily to the cytoplasm (1, 14, 31, 50, 51), most likely complexed with the poly(A) tail of cellular mRNAs, with translation initiation complex eIF4F and other cellular proteins, such as Paip1, Paip2, or eRF3 (11, 23, 47, 50). Since rotavirus infection evicts PABP-C1 from the translation initiation complex eIF4F (39), we examined the subcellular localization of PABP-C1 in rotavirus-infected cells. In addition, a recent report (31) indicated that PABP-C1 was present in the nuclei of cells infected with the rotavirus RRV strain.
To investigate the localization of PABP-C1 in rotavirus (strain RF)-infected cells, MA104 cells infected at an MOI of 1 were fixed 18 h postinfection, and PABP-C1 localization was examined with MAb 10E10 (Fig. 1). To distinguish between infected cells and uninfected cells, cells were also stained with a NSP3-specific polyclonal antibody (Fig. 1A; see also Fig. 3B and H and Fig. 10D for the uninfected cell control). Surprisingly, in rotavirus-infected cells (NSP3 positive), PABP-C1 was not found in the cytoplasm, but it formed speckles in the nucleus (Fig. 1B and C) as described for cells overexpressing PABP-C1 (1). Thus, during rotavirus infection, PABP-C1 is relocalized to the nucleus of MA104 cells.
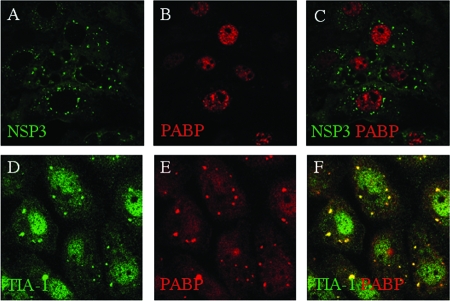
FIG. 1.
Localization of PABP-C1 in rotavirus-infected and uninfected stressed cells. (A to C) MA104 cells were infected with rotavirus, fixed, and incubated with NSP3- and PABP-C1-specific antibodies. Secondary antibodies coupled to Alexa fluorophores stained NSP3 green and PABP-C1 red. Panel C is a merged image of panels A and B. (D to F) MA104 cells, stressed by sodium arsenite treatment, were fixed and incubated with TIA-1- and PABP-C1-specific antibodies. Secondary antibodies coupled to Alexa fluorophores stained TIA-1 green and PABP-C1 red. Panel F is a merged image of panels D and E.
FIG. 3.
Localization of PABP-C1 in cells expressing wt NSP3 or NSP3Δ4G. MA104 cells were transfected with pTEJ4 wt NSP3 or pTEJ4 NSP3Δ4G, fixed with PAF 36 h posttransfection, and incubated with NSP3- and PABP-C1-specific antibodies. Secondary antibodies coupled to Alexa fluorophores stained NSP3 green (A and G) and PABP-C1 red (B and H). Nuclei were stained blue with DAPI (C). Panel D is a merged image of panels A and B. Panel E is a merged image of panels A and C. Panel F is a merged image of panels A to C. Panel I is a merged image of panels G and H. Note that the two cells with nuclear PABP-C1 in panel H do not express NSP3Δ4G (G and I).
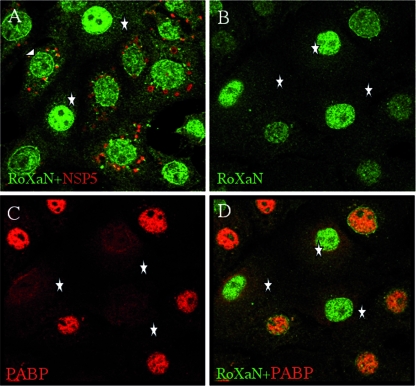
FIG. 10.
Localization of RoXaN in rotavirus-infected cells. MA104 cells, infected with rotavirus RF strain at an MOI of 1 were fixed 18 h postinfection and incubated with RoXaN- and NSP5-specific antibodies (A) or with RoXaN- and PABP-C1-specific antibodies (C and D). Secondary antibodies coupled to Alexa fluorophores stained PABP-C1 red, RoXaN green, and NSP5 red. Panels D is a merged image of panels B and C. Stars indicate noninfected cells, with nuclear RoXaN and without viroplasms in panel A with nuclear RoXaN and cytoplasmic PABP in panels B to D. The triangle indicates RoXaN forming a rim in an infected cell.
It is possible that the nuclear localization of PABP-C1 during rotavirus infection is a feature of rotavirus infection or is peculiar to MA104 cells, as is, for example, an inability to form stress granules. Therefore, we examined whether PABP-C1 localized to stress granules upon oxidative stress in MA104 cells as has been described for numerous other cell lines (21). The subcellular localization of TIA-1 and PABP-C1 was determined by confocal microscopy after treatment of MA104 with sodium arsenite, a strong inducer of stress granules. As illustrated in Fig. 1D to F, following arsenite-induced oxidative stress, TIA-1 formed stress granules in MA104 cells. Under these conditions, PABP-C1 localized in part to cytoplasmic foci with TIA-1 and in part into the nucleus (Fig. 1D), whereas in untreated cells, PABP-C1 was diffusely distributed in the cytoplasm (see Fig. 10D) and TIA-1 was present in the nucleus (data not shown). Therefore, MA104 cells are able to form stress granules upon arsenite treatment, and the nuclear localization of PABP-C1 observed above is the result of rotavirus infection and not a particularity of the cell line used in our study.
Overexpression of PABP-C1 in MA104 cells results in nuclear localization of PABP-C1, but rotavirus infection does not induce overexpression of PABP-C1.
Nuclear localization of PABP-C1 has been observed upon overexpression (1) in mammalian cells by transfection of a PABP-C1-GFP-expressing construct. However, the nuclear localization of PABP-C1 when overexpressed by gene transfection seems to depend on the cell type and on the level of expression of the protein. In NIH 3T3, HeLa JW36, or CV1 cells, transfection of a hemagglutinin-tagged PABP-C1 does not lead to the nuclear accumulation of PABP-C1 (14, 44, 50). In HeLa cells stably expressing a GFP-PABP-C1 fusion protein at a level equivalent to 30% of the endogenous PABP-C1, the fusion protein remains in the cytoplasm. However, transient expression of the same fusion protein results in nuclear localization of PABP-C1 (1).
Whereas rotavirus infection inhibits cellular mRNA translation, the expression of some proteins, such as GRP94 and GRP78, increases during rotavirus infection (52). The translation of PABP-C1 is regulated by a negative feedback loop; PABP-C1 binds the 5′ untranslated region of its own mRNA and inhibits its translation (36). Since rotavirus NSP3 interferes with PABP-C1-dependent translation (30), it is conceivable that rotavirus infection could induce PABP-C1 overexpression by deregulating this negative feedback and, consequently, inducing PABP-C1 nuclear localization. We first determined the localization of PABP-C1 overexpressed in MA104 cells after transfection and then checked whether levels of PABP-C1 were higher in infected cells.
To determine the subcellular localization of PABP-C1 in MA104 overexpressing PABP-C1, cells were transfected with a plasmid construct that allows the expression of Flag-tagged PABP-C1, fixed, and then incubated with a MAb specific for the Flag epitope tag. As illustrated in Fig. 2A to C, when overexpressed in MA104 cells, Flag-PABP-C1 was seen in both the nucleus and the cytoplasm of transfected cells. Thus, PABP-C1 can accumulate in the nucleus when overexpressed in MA104 cells.
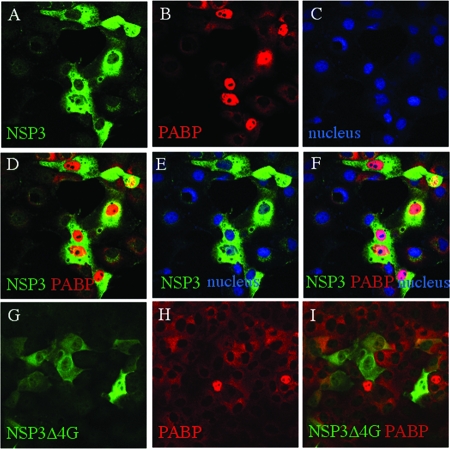
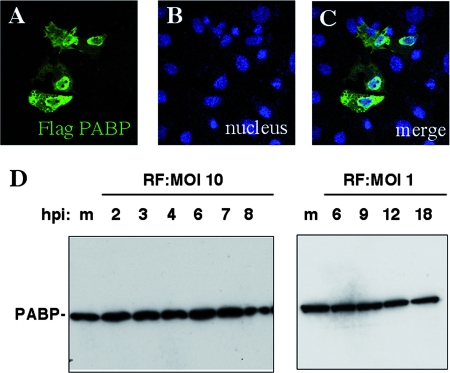
FIG. 2.
Localization of PABP-C1 overexpressed in MA104 cells and expression of PABP-C1 in rotavirus-infected cells. (A to C) MA104 cells were transfected with pcDNA-Flag PABP-C1 for 36 h, fixed, and incubated with Flag-specific antibody. Secondary antibody coupled to Alexa fluorophore stained PABP-C1 green (A). Nuclei were stained blue with DAPI (B). Panel C is a merged image of panels A and B. (D) MA104 cells were infected with rotavirus at a high (10) or low (1) MOI. At the indicated time postinfection (hpi), cell lysates were prepared and protein was separated by SDS-PAGE. PABP-C1 was detected by immunoblotting. m indicates mock-infected cell lysate.
Rotavirus-infected cell lysates recovered at different times postinfection were probed with MAb 10E10 specific for PABP-C1. Cell lysates were also prepared at different times postinfection and from infection at different MOIs to be able to detect even a transient enhancement of PABP overexpression. As illustrated in Fig. 2D, none of the conditions tested showed any increase in PABP-C1 expression compared to that seen with mock-infected cells. Thus, the nuclear localization of PABP-C1 during rotavirus infection is not due to an overexpression of PABP-C1 induced by rotavirus infection.
PABP-C1 nuclear localization requires interaction of NSP3 with eIF4G.
Rotavirus nonstructural protein NSP3 evicts PABP-C1 from translation initiation complexes because it interacts with eIF4G at the same position as PABP-C1 but with a hundredfold-higher affinity (15). It is possible that during rotavirus infection, PABP-C1 freed from eIF4G is redirected to the nucleus. To determine whether the nuclear localization of PABP-C1 was a consequence of NSP3 interaction with eIF4G, NSP3 was transiently expressed in MA104 cells in the absence of rotavirus infection. Transfected cells expressing NSP3 were identified by immunofluorescence with a rabbit NSP3-specific PAb (Fig. 3A), PABP-C1 was visualized using MAb 10E10 (Fig. 3B), and cell nuclei were labeled with DAPI (Fig. 3C). The merged PABP-C1, NSP3, and nucleus images (Fig. 3D to F) indicate that PABP-C1 was indeed present in the nuclei of cells expressing high level of NSP3, whereas in untransfected cells or in cells expressing low level of NSP3, PABP-C1 was diffused into the cytoplasm. Quantification showed that 74% of the cells expressing NSP3 versus less than 2% of normal cells have a nuclear PABP.
Nuclear localization of PABP-C1 was not observed when a mutant of NSP3 (NSP3Δ4G) unable to bind eIF4G and to evict PABP-C1 (38) was expressed in MA104 cells (Fig. 3G to I). Note that this deletion mutant is in part localized in the nucleus, while full-length NSP3 is exclusively cytoplasmic. Therefore, nuclear localization of PABP-C1 during rotavirus infection is due to the interaction of NSP3 with eIF4G and does not require other viral factors.
NSP3 expression does not impair CRM1-dependent export.
As PABP-C1 is a shuttling protein, its localization in the nucleus of NSP3-expressing cells could result from increased import of cytoplasmic PABP-C1 freed from eIF4G into the nucleus or a failure of PABP-C1 to exit from the nucleus. We have shown that NSP3 is involved in the first mechanism through its interaction with eIF4G, but the possibility that it also perturbs PABP-C1 exit from the nucleus cannot be excluded.
PABP-C1 export from the nucleus depends on a leptomycin-sensitive, CRM1-dependent pathway (51). To determine whether NSP3 perturbs this pathway, we transfected cells with a plasmid carrying the bacterial beta-galactosidase coding sequence fused to the simian virus 40 NLS and to the rabies P protein NES (pNLS-NES-gal). It has been shown that the product of this chimera is mainly cytoplasmic due to a continuous NLS-dependent nuclear import and a potent CRM1-dependent nuclear export (see Fig. 5 in reference 35). Thus, if NSP3 is capable of blocking CRM1-dependent nuclear export, the localization of the NLS-NES-gal protein would be primarily nuclear. Figure 4A to C shows that after transfection of pNLS-NES-gal into MA104 cells, the chimeric protein is, as expected, localized to the cytoplasm and that coexpression of wt NSP3 (Fig. 4D to F) does not modify this localization. Therefore, the nuclear localization of PABP-C1 observed during rotavirus infection or NSP3 expression is not due to a general impairment of nuclear export through the CRM1 export pathway.
FIG. 5.
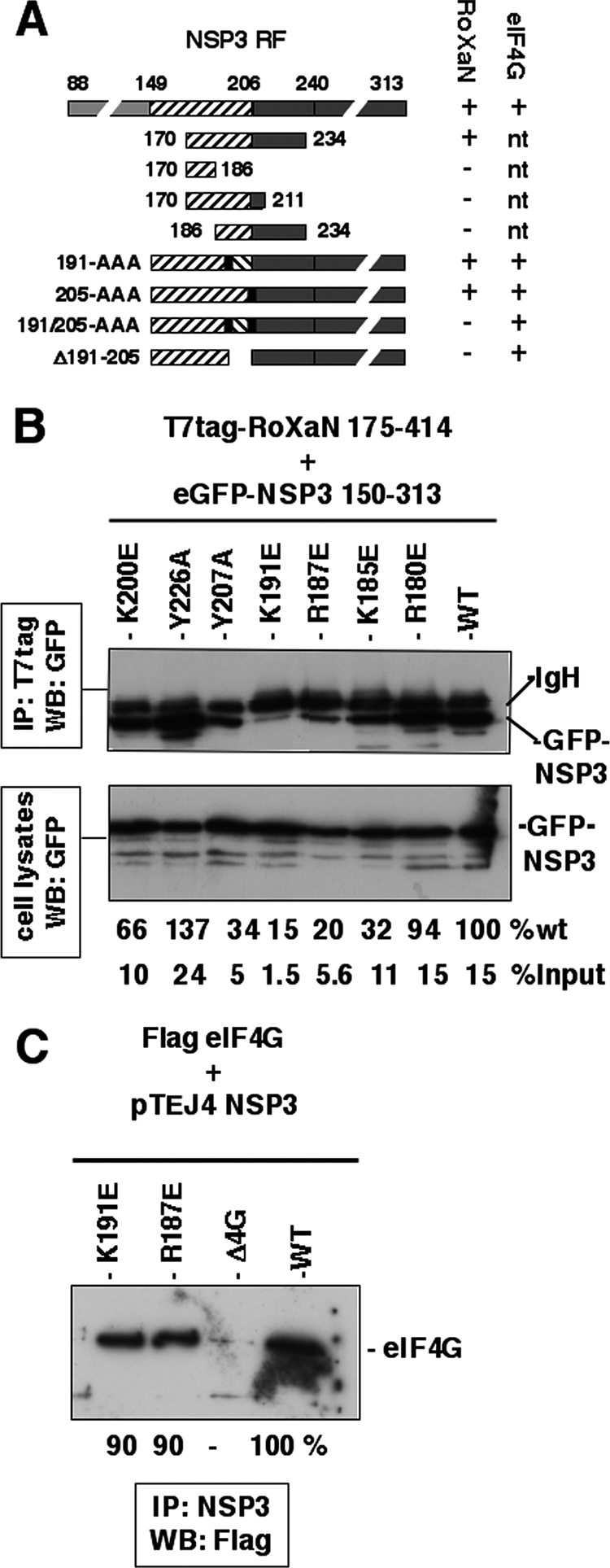
Fine mapping of the NSP3 RoXaN interaction domain in NSP3. (A) Fine mapping of the NSP3 RoXaN interaction domain in NSP3 by yeast two-hybrid assay. A schematic representation of NSP3 structural and functional domains is shown. The RNA-binding domain (gray box), the dimerization (striped box) and the eIF4G-binding domain (black box) of NSP3 are indicated along their positions. NSP3 fragments cloned in pGBT9 used in the two-hybrid assay to delineate the interaction domain of NSP3 with RoXaN are schematically represented. Black vertical bars represent the insertion of an alanine triplet at position 191 or 205. Interactions with RoXaN (1 to 977) or eIF4G (160 to 965) (position according to the MN_198241.1 variant of eIF4G1) were determined by the capacity of yeast to grow (+) or not (−) on medium lacking leucine as described previously (38, 49). nt, not tested. (B) Interaction of NSP3 punctual mutants with RoXaN tested by coimmunoprecipitation. Plasmids encoding NSP3 150 to 313 fused to eGFP and mutated at the indicated positions were cotransfected with a plasmid encoding T7tag-RoXaN 175 to 414. After immunoprecipitation (IP) of RoXaN, eGFP-NSP3 chimeras were revealed by Western blotting (WB) with an eGFP-specific MAb. The immunoblot of the unprocessed cell lysates is shown as the control (lower panel). %wt indicates the intensity of each eGFP-NSP3 band in the immuno- precipitation (measured with ImageJ software) relative to that of wt NSP3. %input indicates the intensity of each eGFP-NSP3 band in the immunoprecipitation (measured with ImageJ software) relative to that of the eGFP-NSP3 band in the cell lysates. (C) Interaction of mutants K187 and K191 with eIF4G in vivo. Plasmids encoding wt, Δ4G, K187E, and K191E NSP3 were transfected with plasmid encoding full-length eIF4G variant 8/1 (accession number AF002815) (7) fused to the Flag epitope. eIF4G coimmunoprecipitated with NSP3 was revealed by Western blotting with a Flag-specific MAb.
FIG. 4.
NSP3 expression does not impair CRM1-dependent export. MA104 cells were transfected with pCMV-NLS-NES-gal alone (A to C) or with pTEJ4 wt NSP3 (D to F), fixed with PAF 36 h posttransfection, and incubated with beta-galactosidase- and NSP3-specific antibodies. Secondary antibodies coupled to Alexa fluorophores stained beta-galactosidase green (A and D) and NSP3 red (E). Nuclei were stained with DAPI (B). Panel C is a merged image of panels A and B. Panel F is a merged image of panels D and E.
NSP3 mutants unable to interact with RoXaN.
We previously described another cellular protein partner of NSP3 called RoXaN (49). The functional role of this cellular protein is still unknown. RoXaN (977 aa) contains TPR at its amino terminus (positions 37 to 149) followed by an LD domain (264 to 272) and four pairs of zinc fingers (see Fig. 1B in reference 49). RoXaN interaction with NSP3 involves the LD domain of RoXaN and the coiled-coil domain of NSP3 between aa 149 and 240 (49). The cellular protein paxillin, which contains several LD domains (46), has been shown to modulate the nuclear localization of PABP-C1 (50, 51), so we investigated whether RoXaN could play a similar role.
To study a possible role of NSP3-RoXaN interaction on PABP-C1 localization, we looked for mutations in NSP3 that could abolish its interaction with RoXaN without affecting its binding to eIF4G. The use of the yeast two-hybrid assay and deletions in the 149 to 240 region allowed us to delineate the NSP3-RoXaN interaction domain as amino acids 170 to 234 of NSP3 (Fig. 5A). Further deletions at the carboxy (stop codon at position 186 or 211) or amino (start codon at 186) terminus or internal deletion (deletion of 191 to 205) of this region totally abolished the interaction (Fig. 5A). Insertions of three alanines at position 191 or 205, to modify the pitch of the coiled-coil, did not abolish the interaction of NSP3 with RoXaN, but the same insertions in combination did abolish the interaction (Fig. 5A). This result underlines the importance of the tertiary structure of the 170-to-234 region of NSP3 for its interaction with RoXaN and the importance of using only limited modification of the 170-to-234 coiled-coil domain in order to study the role of NSP3-RoXaN interaction on PABP-C1 localization. To introduce point mutations in NSP3 that could abolish RoXaN-NSP3 interaction without affecting eIF4G binding, we first built a model of the three-dimensional structure of the 163-to-239 coiled-coil region of NSP3 and compared it to the three-dimensional structure of the focal adhesion kinase targeting domain of the focal adhesion kinase bound to the LD2 domain of paxillin (4, 16). According to crystallographic studies, LD motifs contact a hydrophobic space between two helices, engaging charge-charge interactions with basic residues surrounding a hydrophobic patch. A NSP3-parallel coiled-coil between positions 170 and 234 could offer such a binding surface. Mutations that change positively a charged residue (K or R at positions 180, 185, 187, 191, and 200) to a negatively charged glutamic acid residue were then introduced into the NSP3 150-to-313 coding sequence fused to eGFP. Mutations that changed one of the two large amino acid residues, Y207 and Y226, to a small amino acid (alanine) were also introduced in the same eGFP-NSP3 150-to-313 chimera. All these mutations were then tested for RoXaN interaction by coimmunoprecipitation after cotransfection of eGFP-NSP3 150-to-313 mutants and T7-tagged RoXaN 178 to 414 into HEK293 cells (Fig. 5B). None of the point mutations totally abolished the interaction of NSP3 with RoXaN, indicating that the RoXaN-NSP3 interaction probably occurs on a large surface of NSP3, but mutants R187E and K191E clearly showed a reduced interaction with RoXaN, with a low recovery level compared to that of the wt and that of the input material (20 and 15% of the wild-type level and only 1.5 and 5.6% of the input level, respectively). It should be noted that, although none of the insertions of three alanines were able to disrupt NSP3-RoXaN interaction when tested by the two-hybrid assay (Fig. 5A), NSP3-RoXaN interaction was indeed abolished when these insertions were introduced in the full-length NSP3 protein and were tested by coimmunoprecipitation (data not shown). This observation confirms that coimmunoprecipitation is an accurate method to study NSP3-RoXaN interactions, whereas the two-hybrid assay cannot be used to quantify such interactions. When introduced in the full-length NSP3, the R187E and K191E mutations did not modify interaction of NSP3 with eIF4G, as illustrated in Fig. 5C. Hence, the R187E and K191E mutations in NSP3 satisfy the prerequisite of a much lower interaction with RoXaN without affecting eIF4G binding and, hence, could be used to test the role of NSP3-RoXaN interaction on PABP-C1 localization.
NSP3 mutants unable to interact with RoXaN do not induce nuclear localization of PABP-C1.
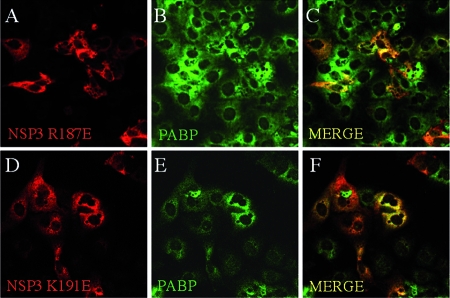
To study the effect of the R187E and K191E mutations of NSP3 on the localization of PABP-C1, the R187E and K191E mutations were introduced into the pTEJ4-RF07 construct and the plasmids were transfected into MA104 cells. PABP-C1 and NSP3 R187E or K191E were then examined by indirect immunofluorescence. Under these conditions (Fig. 6), few cells present nuclear PABP-C1 when NSP3 R187E or K191E mutants were expressed compared to results with wt NSP3 (Fig. 3). Quantification of cells positive for NSP3 staining and for nuclear PABP-C1 indicated a significant difference (P < 0.01, χ2 test) in PABP nuclear localization between cells expressing wt NSP3, where PABP-C1 is nuclear in 74% of the cells (n = 137) (compared to less than 1% in nontransfected cells), and cells expressing NSP3 K191E and R187E, where only 35% (n = 137) and 32% (n = 187) of the cells exhibit nuclear PABP-C1, respectively. The difference between the two NSP3 mutants is not statistically significant. Thus, diminishing the strength of NSP3-RoXaN interaction dramatically reduces the efficiency of PABP-C1 nuclear localization.
FIG. 6.
NSP3 mutants unable to interact with RoXaN do not induce nuclear localization of PABP-C1. MA104 cells transfected with plasmids encoding NSP3 mutants K187E and K191E (panels A to C and D to F, respectively) were fixed and incubated with NSP3- and PABP-C1-specific antibodies. Secondary antibodies coupled to Alexa fluorophores stained NSP3 red and PABP-C1 green. Panels C and F are merged images of panels A and B and panels D and E, respectively.
PABP-C1 interaction with RoXaN.
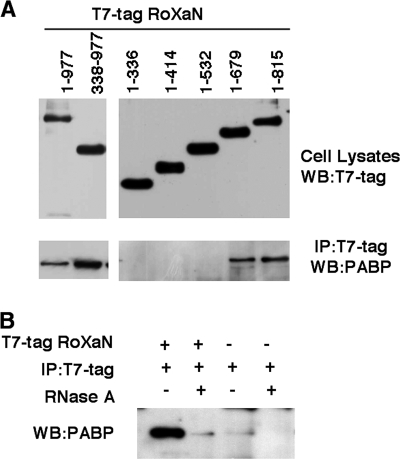
The observations described above indicate that, in addition to the NSP3-eIF4G interaction, a NSP3-RoXaN interaction is essential for substantial PABP-C1 nuclear localization. It can be envisioned that in normal cells, RoXaN interacts with PABP-C1 and sequesters PABP-C1 into the cytoplasm or that RoXaN is involved in the nuclear export of PABP-C1. Furthermore, interaction of PABP with the LD domain of paxillin has been detected in vitro and paxillin is involved in the subcellular localization of PABP (51). In order to detect an interaction of PABP-C1 with RoXaN, plasmids encoding the full-length RoXaN (1 to 977) or different combinations of the TPR, LD, and zinc finger domains of RoXaN tagged with T7tag were transfected in HEK293 cells, and the proteins were immunoprecipitated with a T7tag-specific antibody. PABP-C1 was detected among the proteins coimmunoprecipitated with RoXaN by immunoblotting with MAb 10E10 following SDS-PAGE (Fig. 7A). The full-length RoXaN (1 to 977) and the fragment 338 to 977 encompassing all the zinc fingers were both able to coimmunoprecipitate PABP-C1. Inversely, PABP-C1 was not coimmunoprecipitated with RoXaN fragment 1 to 336, which encompasses the TPR and LD domains. Overlapping carboxy-terminal deletions of RoXaN (Fig. 7A) show that the two zinc fingers between aa 546 and 637 are required to coimmunoprecipitate PABP-C1 with RoXaN.
FIG. 7.
PABP-C1 RoXaN interaction mediated by RNA. (A) PABP and RoXaN coimmunoprecipitation. Lysates of HEK293 cells transfected with plasmids expressing complete T7tag-RoXaN (1 to 977) or deletion mutants, as indicated, were immunoprecipitated with a T7tag-specific MAb. PABP-C1 coimmunoprecipitated with RoXaN was revealed by immunoblotting (lower panels). The expression of the T7tag-RoXaN mutants was controlled by immunoblotting of the unprocessed cell lysates with a T7tag-specific MAb (upper panels). (B) RoXaN tethers PABP with RNA. Lysates of HEK293 cells mock transfected or transfected with a plasmid expressing complete T7tag-RoXaN (1 to 977) were immunoprecipitated with a T7tag-specific MAb. Half of each immunoprecipitate was treated with RNase A, and PABP-C1 that coimmunoprecipitated with RoXaN was revealed by immunoblotting. IP, immunoprecipitation; WB, Western blotting.
The requirement of a zinc finger for coimmunoprecipitation of PABP-C1 suggested that the PABP-C1-RoXaN interaction could be mediated by an RNA intermediate bound to PABP-C1 RNA recognition motifs (RRMs) and to RoXaN zinc fingers. Indeed, treatment of the cell lysate with RNase A after immunoprecipitation but prior to SDS-PAGE abolished the coimmunoprecipitation of PABP-C1 (Fig. 7B) and the RRM region of PABP-C1 (aa 10 to 366) was coimmunoprecipitated with RoXaN only if the RNase treatment was omitted (data not shown).
To definitively establish that RoXaN is an RNA-binding protein, an RNA pull-down assay was used. Oligoribonucleotides linked to Sepharose beads were used to precipitate full-length T7tag-RoXaN or T7tag-RoXaN fragment 1 to 336 (as a negative control). As illustrated in Fig. 8A, only the full-length RoXaN could be recovered with poly(U) oligoribonucleotides and, to a much lesser extent, with poly(A). RoXaN fragment 1 to 336, which neither contains zinc fingers nor coprecipitates PABP-C1 (Fig. 7B), was not retained by any oligoribonucleotides (Fig. 8B). Interestingly, under the same conditions, Flag-PABP-C1 showed a marked preference for poly(A) sequence over poly(U) sequence (Fig. 8C).
FIG. 8.
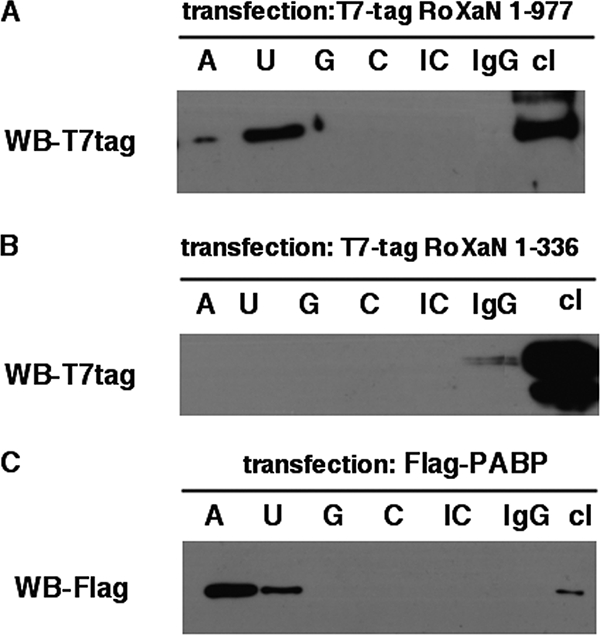
RoXaN is an RNA-binding protein. Lysates of HEK cells transfected with a plasmid expressing T7tag-RoXaN (1 to 977) or T7tag-RoXaN 1 to 336 (Fig. 7A) or Flag-PABP-C1 were incubated with poly(A), poly(U), poly(C), or poly(G) single-stranded RNA or poly(I:C) double-stranded RNA or mouse IgG coupled to Sepharose beads. Beads were washed three times, and proteins bound on beads were analyzed by immunoblotting with T7tag-specific (A and B) or Flag-specific (C) antibodies. Transfection and immunoblot controls are provided by unprocessed cell lysates (cl). WB, Western blotting.
Together, these results indicate that PABP-C1 and RoXaN are two RNA-binding proteins with different sequence specificity; RoXaN can thus tether PABP-C1 through an RNA intermediate.
RoXaN LD domain functions as a NES.
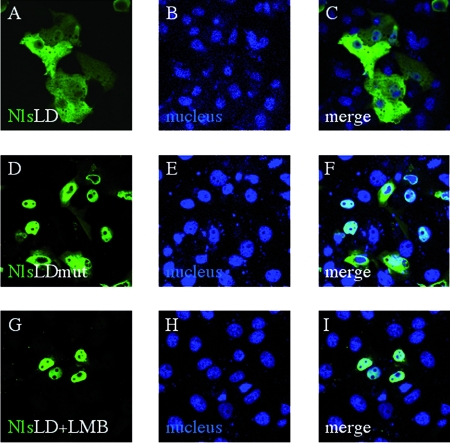
PABP-C1 has been shown to exit the nucleus using a CRM1-dependent pathway (1). The CRM1-dependent export pathway relies on the interaction of CRM1 with a NES (18). NESs are defined by a succession of hydrophobic amino acids matching the pattern ΦX2-3ΦX2-3ΦXΦ, where Φ represents any hydrophobic residue, most frequently leucine, and X represents any amino acid. As the LD domain of RoXaN matches the NES consensus sequence (Table 1), we examined whether RoXaN LD domain (aa 264 to 271) could function as a NES. To this aim, aa 257 to 293 of RoXaN were fused to a NLS-beta-galactosidase chimera, and the fusion protein was localized by indirect immunofluorescence in MA104-transfected cells. An additional control was provided by the fusion of the same region of RoXaN but amplified from a mutant (49) in which the amino acids at positions 267 to 269 of the LD domain have been mutated to alanines (Table 1). Figure 9 provides a comparison of the subcellular localization of the two fusion proteins. It is clear that the presence of an intact, wt LD domain with the NLS-beta-galactosidase protein leads to the accumulation of the chimeric protein in the cytoplasm (Fig. 9A to C) but that the mutated chimera is present primarily in the nucleus (Fig. 9D to F), as is the parental NLS-beta-galactosidase chimera (reference 35 and data not shown). Moreover, treating cells transfected with the NLS-LD-beta-galactosidase construct with LMB led to nuclear accumulation of the chimeric protein (Fig. 9G to I), demonstrating that nuclear export of NLS-LD-beta-galactosidase requires a functional CRM1-export pathway. These results show that the RoXaN LD domain can fulfill the function of a NES.
TABLE 1.
LD and NES sequencesa
| Description | Sequence |
|---|---|
| LD consensus | LDxLLxxL |
| LD RoXaN | LDtLLdsLsL |
| NES consensus | Φx2-3Φx2-3ΦxΦ |
| LD mut RoXaN | LDtaaasLsL |
| Paxillin LD2 | lseLDrLLleL |
Comparison of the LD domain consensus sequence and the NES consensus sequence with the LD domain of RoXaN and paxillin LD2 (45). Amino acids that match the NES consensus sequence are in bold letters. Amino acids that match the LD consensus sequence are in capital letters. Mutations introduced in the RoXaN LD domain (LD mut RoXaN) are underlined. Φ represents L, I, V, F, or M, and x refers to any amino acid.
FIG. 9.
The RoXaN LD domain is a NES. MA104 cells transfected with pCMV-NLS-LD-beta-galactosidase (A to C and G to I) or the pCMV-NSL-LD-mut-beta-galactosidase (D to F) for 36 h were treated with leptomycin B for 90 min (G to I) or left untreated (A to F) and then fixed and incubated with beta-galactosidase-specific antibody. Secondary antibody coupled to Alexa fluorophore stains beta-galactosidase green. Nuclei are stained blue with DAPI.
Localization of RoXaN and PABP-C1 in uninfected and rotavirus-infected cells.
We have shown that RoXaN and PABP can be linked on the same RNA, that the LD domain of RoXaN can function as a NES, and that PABP is accumulated in the nuclei of rotavirus-infected cells. We next sought to determine the localization of RoXaN in infected and uninfected cells. To determine the subcellular localization of RoXaN and PABP in uninfected and rotavirus-infected cells, an antibody raised against the amino-terminal 17 aa of RoXaN was used (Fig. 10). MA104 cells were infected at a low MOI such that not all the cells were infected; thus, infected and uninfected cells were simultaneously present on the same microscopic field and could easily be compared with each other. In uninfected cells (NSP5 negative), RoXaN was present mainly in the nucleus, while in infected cells (NSP5 positive), the nucleus was notably depleted of RoXaN. In some infected cells (Fig. 10A), RoXaN forms a ring around the nucleus reminiscent of proteins found in nuclear pores (5). In the same experiment, we also looked at the localization of RoXaN and PABP-C1 in rotavirus-infected cells (Fig. 10B to D). Infected cells are recognizable because, as described above, PABP-C1 accumulates in the nucleus. In these infected cells, RoXaN is less intensely stained in the nucleus than it is in uninfected cells. Conversely, uninfected cells (Fig. 10) show an even distribution of PABP-C1 in the cytoplasm, whereas RoXaN is concentrated in the nucleus.
Thus, in the course of rotavirus infection, although the nucleus appears to be enriched in PABP, it is depleted of RoXaN.
DISCUSSION
In this report, we have shown that, in the absence of PABP-C1 overexpression, rotavirus infection or NSP3 expression lead to the accumulation of PABP-C1 in the nucleus, whereas rotavirus infection depletes the nucleus of RoXaN. Two simultaneous conditions are required to achieve nuclear accumulation of PABP-C1 upon NSP3 expression. First, PABP-C1 in the cytoplasm must be freed from eIF4G, because when NSP3 with a deletion in its eIF4G binding domain is expressed, PABP-C1 remains in the cytoplasm. Second, the LD domain of RoXaN must be trapped by NSP3, because nuclear localization of PABP-C1 is not induced by the expression of NSP3 mutants that have a weakened interaction with RoXaN but that are still able to interact with eIF4G.
The mechanisms of nuclear import and export of PABP-C1 are not well understood, because PABP-C1 primary sequence analysis reveals no canonical NLS or NES. Nevertheless, PABP-C1 is considered to be a nucleocytoplasmic shuttling protein, because it localizes to the nucleus upon treatment with the nuclear export inhibitor leptomycin B (1, 50) and because a GFP-PABP-C1 fusion protein localizes in the nucleus when it is overexpressed (1) after transient transfection. Our results shed some light on these mechanisms. Nuclear accumulation of PABP-C1 was not observed when NSP3Δ4G, which is unable to interact with eIF4G but is still able to interact with RoXaN, was expressed in cells. Hence, in the absence of overexpression of PABP-C1, eviction of PABP-C1 from eIF4G is required for there to be accumulation of PABP-C1 in the nucleus. The two first RRMs of PABP-C1 are required to direct PABP-C1 to the nucleus (1). Interestingly, the same two RRMs are required for full poly(A) binding activity of PABP-C1 and for the binding of PABP-C1 to eIF4G (27) [although eIF4G-PABP-C1 interaction, in mammalian cells, does not depend on the presence of poly(A)] (19, 39). Thus, while PABP-C1 bound to eIF4G appears retained in the cytoplasm, unbound PABP-C1 is targeted to the nucleus, suggesting that eIF4G may compete for a common PABP-C1 binding site with an as-yet-unknown nuclear import factor. Probably the same mechanism leads to nuclear localization of overexpressed PABP-C1: PABP-C1 in excess of eIF4G is directed to the nucleus. When added to the control of PABP-C1 homeostasis via ubiquitination and Paip2A interaction (53), this mechanism would ensure a fine tuning of the quantity of cytoplasmic PABP-C1 in complex with translation initiation complex eIF4F (eIF4G, eIF4A, eIF4E). Taking into account the importance of PABP-C1 in mRNA translation initiation, the control by nuclear localization of the amount of cytoplasmic PABP-C1 is most probably an important way to control mRNA translation. A tight control of mRNA translation could be obtained with a low level of cytoplasmic free PABP-C1 that maintains a strong competition between poly(A) mRNAs for translation initiation factors.
Our results show that NSP3 also interferes with PABP-C1 localization through its interaction with RoXaN. As nuclear localization is the net balance between nuclear import and nuclear export, RoXaN can interfere with one or the other process. A direct role of RoXaN in the nuclear import of PABP-C1 seems very unlikely, because we found that interaction of NSP3 with RoXaN is required for nuclear localization of PABP-C1, but NSP3 is not found in the nucleus of infected cells. Furthermore, we did not detect a direct protein-protein interaction of RoXaN with PABP-C1 and we have no evidence that RoXaN NSP3 interaction modifies the interaction of RoXaN with RNA (our unpublished data). Thus, PABP-C1 being a cargo for RoXaN would require nuclear import of a PABP-C1/RoXaN/RNA ternary complex. But it does not exclude the import of plain RoXaN in the nucleus. Indeed, nuclear import of RoXaN might explain why NSP3D4G is in part present in the nucleus (Fig. 3G and F); in the absence of interaction with eIF4G, this deletion mutant of NSP3 would be hauled by RoXaN to the nucleus.
It is more likely that RoXaN plays a role in nuclear export of PABP-C1. We have shown that RoXaN is a nuclear protein and that its LD domain can function as a NES for the CRM1 pathway. RoXaN and PABP-C1 are both RNA-binding proteins that can bind the same RNA, and it has been shown that PABP-C1 utilizes its RNA binding activity to exit the nucleus (51). Furthermore, PABP-C1 has been involved in the nuclear export of mRNAs and the coupling of mRNA maturation and export (9), and poly(A) sequence regulates nuclear export of mRNA (9). However, while the CRM1 pathway is clearly the PABP-C1 export pathway (1), it does not make a significant contribution to the nuclear export of cellular mRNAs (25). One way to reconcile these observations is to consider that PABP-C1 molecules are exported through the CRM1 pathway with RNA bound to adapter protein. Thus, blocking CRM1 nuclear export with leptomycin B would lead to the accumulation of PABP-C1 in the nucleus but will have little effect on the export of mRNAs from the nucleus. Hence, RoXaN could be such an adapter for the export of a subset of mRNAs through the CRM1 pathway, and its sequestration by NSP3 in the cytoplasm would deplete the nucleus of RoXaN and stop nuclear export of PABP-C1, as we have observed during rotavirus infection (Fig. 10).
It should be noted that paxillin has been involved in the export of PABP-C1 from the nucleus of NIH 3T3 cells via a CRM1-dependent export pathway, and a direct protein-protein interaction has been observed between paxillin and PABP-C1. Furthermore, reducing the level of paxillin by RNA interference leads to the nuclear accumulation of PABP-C1 (50, 51). Paxillin contains five LD domains (45). Treatment of cells with leptomycin B induces a nuclear localization of paxillin, and its second LD domain also fits the leucine-rich NES consensus (Table 1). One might hypothesize that, according to the cell type and physiological conditions, PABP-C1 utilizes different protein adapters containing LD/NES domains to exit the nucleus.
Rotavirus NSP3 has evolved two different interactions with cellular proteins that both lead to the nuclear accumulation of PABP. Interaction with eIF4G evicts PABP from eIF4G and unveils its NLS, and interaction with RoXaN sequesters an element important for nuclear export of PABP-C1. Although the nuclear addressing of PABP-C1 freed from eIF4G can be considered a mere consequence of NSP3-eIF4G interaction, the cytoplasmic recruitment of RoXaN to block PABP export increases the level of complexity of the rotavirus-host cell interactions. What is the advantage for rotavirus interaction with RoXaN? One explanation is that it allows a complete depletion of PABP-C1 from the cell cytoplasm and, thus, it reinforces the shutoff of translation of cellular polyadenylated mRNAs and gives a critical advantage to viral mRNAs to be translated. In addition, PABP-C1 interacts with several other cellular proteins involved in translation termination (10, 47, 50, 51), RNA stability (42, 43), or subcellular localization (51). It is thus possible that by depleting PABP-C1 in the cytoplasm, rotavirus makes some of these factors available in a greater amount for the viral mRNAs that are unable to recruit these factors via PABP-C1 due to the absence of 3′ poly(A) tails.
Identification of the cellular RNAs bound by RoXaN and introduction of the NSP3 mutations, described here, back into the rotavirus genome (24, 26) will allow disentangling the roles of the interactions between NSP3, RoXaN, and PABP-C1.
Acknowledgments
This work has been supported by a grant from Appel d'Offre Virologie from Département Santé Animale, INRA, France. M.M.B. was supported by an International Research Fellowship from the National Science Foundation. M.H. and D.V. were supported by a fellowship from Ministère de l'Education et de la Recherche (France).
This project has benefited from the imaging facilities of the IMAGIF Cell Biology Center of the CNRS Gif campus.
Footnotes
Published ahead of print on 17 September 2008.
REFERENCES
- 1.Afonina, E., R. Stauber, and G. N. Pavlakis. 1998. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 27313015-13021. [DOI] [PubMed] [Google Scholar]
- 2.Angel, J., M. A. Franco, and H. B. Greenberg. 2007. Rotavirus vaccines: recent developments and future considerations. Nat. Rev. Microbiol. 5529-539. [DOI] [PubMed] [Google Scholar]
- 3.Aponte, C., D. Poncet, and J. Cohen. 1996. Recovery and characterization of a replicase complex in rotavirus-infected cells by using a monoclonal antibody against NSP2. J. Virol. 70985-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arold, S. T., M. K. Hoellerer, and M. E. Noble. 2002. The structural basis of localization and signaling by the focal adhesion targeting domain. Structure 10319-327. [DOI] [PubMed] [Google Scholar]
- 5.Boehmer, T., J. Enninga, S. Dales, G. Blobel, and H. Zhong. 2003. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA 100981-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borman, A. M., Y. M. Michel, and K. M. Kean. 2000. Biochemical characterisation of cap-poly(A) synergy in rabbit reticulocyte lysates: the eIF4G-PABP interaction increases the functional affinity of eIF4E for the capped mRNA 5′-end. Nucleic Acids Res. 284068-4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bushell, M., D. Poncet, W. E. Marissen, H. Flotow, R. E. Lloyd, M. J. Clemens, and S. J. Morley. 2000. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis in lymphoma cells: characterisation of an internal fragment generated by caspase-3-mediated cleavage. Cell Death Differ. 7628-636. [DOI] [PubMed] [Google Scholar]
- 8.Catherinot, V., and G. Labesse. 2004. ViTO: tool for refinement of protein sequence-structure alignments. Bioinformatics 203694-3696. [DOI] [PubMed] [Google Scholar]
- 9.Chekanova, J. A., and D. A. Belostotsky. 2003. Evidence that poly(A) binding protein has an evolutionarily conserved function in facilitating mRNA biogenesis and export. RNA 91476-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cosson, B., N. Berkova, A. Couturier, S. Chabelskaya, M. Philippe, and G. Zhouravleva. 2002. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol. Cell 94205-216. [DOI] [PubMed] [Google Scholar]
- 11.Craig, A. W., A. Haghighat, A. T. Yu, and N. Sonenberg. 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392520-523. [DOI] [PubMed] [Google Scholar]
- 12.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 602126-2132. [DOI] [PubMed] [Google Scholar]
- 13.Fiser, A., and A. Sali. 2003. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374461-491. [DOI] [PubMed] [Google Scholar]
- 14.Gorlach, M., C. G. Burd, and G. Dreyfuss. 1994. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp. Cell. Res. 211400-407. [DOI] [PubMed] [Google Scholar]
- 15.Groft, C. M., and S. K. Burley. 2002. Recognition of eIF4G by rotavirus NSP3 reveals a basis for mRNA circularization. Mol. Cell 91273-1283. [DOI] [PubMed] [Google Scholar]
- 16.Hoellerer, M. K., M. E. Noble, G. Labesse, I. D. Campbell, J. M. Werner, and S. T. Arold. 2003. Molecular recognition of paxillin LD motifs by the focal adhesion targeting domain. Structure 111207-1217. [DOI] [PubMed] [Google Scholar]
- 17.Hosoda, N., F. Lejeune, and L. E. Maquat. 2006. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Mol. Cell. Biol. 263085-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutten, S., and R. H. Kehlenbach. 2007. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell. Biol. 17193-201. [DOI] [PubMed] [Google Scholar]
- 19.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 177480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahvejian, A., Y. V. Svitkin, R. Sukarieh, M. N. M'Boutchou, and N. Sonenberg. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 19104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30963-969. [DOI] [PubMed] [Google Scholar]
- 22.Kedersha, N., M. R. Cho, W. Li, P. W. Yacono, S. Chen, N. Gilks, D. E. Golan, and P. Anderson. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 1511257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khaleghpour, K., Y. V. Svitkin, A. W. Craig, C. T. DeMaria, R. C. Deo, S. K. Burley, and N. Sonenberg. 2001. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell 7205-216. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, T., A. A. Antar, K. W. Boehme, P. Danthi, E. A. Eby, K. M. Guglielmi, G. H. Holm, E. M. Johnson, M. S. Maginnis, S. Naik, W. B. Skelton, J. D. Wetzel, G. J. Wilson, J. D. Chappell, and T. S. Dermody. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1147-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohler, A., and E. Hurt. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8761-773. [DOI] [PubMed] [Google Scholar]
- 26.Komoto, S., J. Sasaki, and K. Taniguchi. 2006. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc. Natl. Acad. Sci. USA 1034646-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuhn, U., and E. Wahle. 2004. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 167867-84. [DOI] [PubMed] [Google Scholar]
- 28.Luo, Y., and D. J. Goss. 2001. Homeostasis in mRNA initiation: wheat germ poly(A)-binding protein lowers the activation energy barrier to initiation complex formation. J. Biol. Chem. 27643083-43086. [DOI] [PubMed] [Google Scholar]
- 29.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel, Y. M., D. Poncet, M. Piron, K. M. Kean, and A. M. Borman. 2000. Cap-poly(A) synergy in mammalian cell-free extracts: investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 27532268-32276. [DOI] [PubMed] [Google Scholar]
- 31.Montero, H., M. Rojas, C. F. Arias, and S. Lopez. 2008. Rotavirus infection induces the phosphorylation of eIF2α but prevents the formation of stress granules. J. Virol. 821496-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53240-255. [DOI] [PubMed] [Google Scholar]
- 33.Padilla-Noriega, L., O. Paniagua, and S. Guzman-Leon. 2002. Rotavirus protein NSP3 shuts off host cell protein synthesis. Virology 2981-7. [DOI] [PubMed] [Google Scholar]
- 34.Parashar, U. D., C. J. Gibson, J. S. Bresse, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pasdeloup, D., N. Poisson, H. Raux, Y. Gaudin, R. W. Ruigrok, and D. Blondel. 2005. Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology 334284-293. [DOI] [PubMed] [Google Scholar]
- 36.Patel, G. P., S. Ma, and J. Bag. 2005. The autoregulatory translational control element of poly(A)-binding protein mRNA forms a heteromeric ribonucleoprotein complex. Nucleic Acids Res. 337074-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestova, T. V., V. G. Kolupaeva, I. B. Lomakin, E. V. Pilipenko, I. N. Shatsky, V. I. Agol, and C. U. Hellen. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 987029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piron, M., T. Delaunay, J. Grosclaude, and D. Poncet. 1999. Identification of the RNA-binding, dimerization, and eIF4GI-binding domains of rotavirus nonstructural protein NSP3. J. Virol. 735411-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piron, M., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 175811-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poncet, D., C. Aponte, and J. Cohen. 1993. Rotavirus protein NSP3 (NS34) is bound to the 3′ end consensus sequence of viral mRNAs in infected cells. J. Virol. 673159-3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poncet, D., P. Lindenbaum, R. L'Haridon, and J. Cohen. 1997. In vivo and in vitro phosphorylation of rotavirus NSP5 correlates with its localization in viroplasms. J. Virol. 7134-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddiqui, N., D. A. Mangus, T. C. Chang, J. M. Palermino, A. B. Shyu, and K. Gehring. 2007. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J. Biol. Chem. 28225067-25075. [DOI] [PubMed] [Google Scholar]
- 43.Simon, E., and B. Seraphin. 2007. A specific role for the C-terminal region of the Poly(A)-binding protein in mRNA decay. Nucleic Acids Res. 356017-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tumbarello, D. A., M. C. Brown, and C. E. Turner. 2002. The paxillin LD motifs. FEBS Lett. 513114-118. [DOI] [PubMed] [Google Scholar]
- 46.Turner, C. E. 2000. Paxillin interactions. J. Cell Sci. 1134139-4140. [DOI] [PubMed] [Google Scholar]
- 47.Uchida, N., S. Hoshino, H. Imataka, N. Sonenberg, and T. Katada. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J. Biol. Chem. 27750286-50292. [DOI] [PubMed] [Google Scholar]
- 48.Vende, P., M. Piron, N. Castagne, and D. Poncet. 2000. Efficient translation of rotavirus mRNA requires simultaneous interaction of NSP3 with the eukaryotic translation initiation factor eIF4G and the mRNA 3′ end. J. Virol. 747064-7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitour, D., P. Lindenbaum, P. Vende, M. M. Becker, and D. Poncet. 2004. RoXaN, a novel cellular protein containing TPR, LD, and zinc finger motifs, forms a ternary complex with eukaryotic initiation factor 4G and rotavirus NSP3. J. Virol. 783851-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woods, A. J., T. Kantidakis, H. Sabe, D. R. Critchley, and J. C. Norman. 2005. Interaction of paxillin with poly(A)-binding protein 1 and its role in focal adhesion turnover and cell migration. Mol. Cell. Biol. 253763-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woods, A. J., M. S. Roberts, J. Choudhary, S. T. Barry, Y. Mazaki, H. Sabe, S. J. Morley, D. R. Critchley, and J. C. Norman. 2002. Paxillin associates with poly(A)-binding protein 1 at the dense endoplasmic reticulum and the leading edge of migrating cells. J. Biol. Chem. 2776428-6437. [DOI] [PubMed] [Google Scholar]
- 52.Xu, A., A. R. Bellamy, and J. A. Taylor. 1998. BiP (GRP78) and endoplasmin (GRP94) are induced following rotavirus infection and bind transiently to an endoplasmic reticulum-localized virion component. J. Virol. 729865-9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshida, M., K. Yoshida, G. Kozlov, N. S. Lim, G. De Crescenzo, Z. Pang, J. J. Berlanga, A. Kahvejian, K. Gehring, S. S. Wing, and N. Sonenberg. 2006. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 251934-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]