Abstract
MicroRNAs have emerged as important players in tissue-specific mammalian gene regulation and have also been exploited in experimental targeting of gene expression. We have constructed a recombinant adenovirus that contains sequences complementary to the liver-specific microRNA 122 (miR122) in the 3′ untranslated region of the E1A gene. In Huh7 cells, which resemble normal hepatocytes in expressing high levels of miR122, this feature resulted in strongly reduced levels of E1A mRNA and protein. This property allowed us to generate a novel recombinant adenovirus that was severely attenuated in cells of hepatic origin but replicated normally in other cells. This strategy may be useful in circumventing liver toxicity associated with the systemic delivery of oncolytic adenoviruses. These data provide the first example of exploiting differential microRNA expression patterns to alter the natural tropism of a DNA virus. In addition, these results suggest that other microRNAs expressed in a tissue- or transformation-specific manner may also be used for the targeting of adenoviral replication and that the same principle may be applied to other viruses that have shown promise as oncolytic or gene delivery platforms.
Although discovered just over a decade ago, microRNAs (miRNAs) are now considered to be key regulators of gene expression in eukaryotes (4, 27, 35). The human genome encodes hundreds of miRNAs, and more than 10% of human genes have been estimated to be under miRNA regulation (23, 27). miRNAs are expressed in tissue- and differentiation state-specific patterns and are often differentially expressed or deleted in various human cancers (11, 22, 45, 46). After transcription and processing, miRNAs are incorporated into a protein complex dubbed RISC (RNA-induced silencing complex) to suppress the expression of target genes via several mechanisms, such as translational inhibition or mRNA degradation (5, 13).
The miRNA system has recently been successfully exploited to regulate transgene expression in genetically modified mice (33) and in cultured cells transduced with gene delivery vectors (9). These studies have established the endogenous miRNA machinery as a versatile and efficient system for the experimental targeting of gene expression to certain cell types according to tissue, cell lineage, and differentiation state.
We have previously shown that the introduction of a target site for the ubiquitously expressed let-7 miRNA into the positive-strand RNA genome of poliovirus can be used to divert the cellular RNA interference machinery to efficiently suppress the replication of an animal virus (17). Recently, Edge and colleagues showed that ectopic let-7 target sites can also be used to develop an attenuated vesicular stomatitis virus (14). Interestingly, the levels of let-7 expression in many cancer cells are low, thus allowing the let-7 target-modified vesicular stomatitis virus to preferentially replicate in these cells, a feature that could be exploited in oncolytic virotherapy (14).
Conditionally replicating adenoviruses (CRAds) have emerged as a possible modality for the treatment of cancer (3, 25, 34). Two types of approaches to achieve tumor-selective viral replication have been reported. One is to introduce loss-of-function mutations into the viral regulatory protein E1A or E1B that compromise viral replication in normal but not in transformed cells that typically have defects in the Rb/p16 and p53/p14ARF signaling pathways (7, 16, 19, 37). The other approach is the use of heterologous regulatory elements to achieve cancer cell-specific expression of E1A (reviewed in reference 41). For example, Rodriguez et al. inserted a prostate-specific antigen gene enhancer element upstream of the E1A gene to restrict viral replication to prostate cancer cells (40). Recently, regulatory elements from prostaglandin-endoperoxide synthase-2 (2) and fibroblast growth factor-2 (43) have been employed to favor the stability and translation, respectively, of E1A mRNA in certain types of cancer cells.
Despite these advances, additional means for better cancer- and tissue-specific targeting of adenoviral replication are needed. Of note, both in humans and in nonhuman primates systemic administration of replication-competent as well as replication-deficient adenoviruses has been associated with significant infection of hepatocytes (18, 36, 38), which may lead to liver toxicity, posing a major complication for the systemic use of oncolytic adenoviruses.
In this paper, we describe the construction of a novel type of CRAd in which the expression of the E1A gene and, consequently, viral replication in hepatic cells are specifically suppressed by the liver-specific miRNA 122 (miR122). These data show that tissue-specific miRNA expression patterns can be exploited also to engineer the tropism of DNA virus replication and may help to overcome liver toxicity associated with the systemic delivery of oncolytic adenoviruses.
MATERIALS AND METHODS
Reporter plasmids and luciferase assays.
To introduce various numbers of miR122 target sites into a reporter plasmid, oligonucleotides 5′-CGCGTGGAGTGTGACAATGGTGTTTGTACCGGT-3′ and 5′-CGCGACCGGTACAAACACCATTGTCACACTCCA-3′ were annealed and ligated into Mlu I sites introduced 5′ or 3′ of firefly luciferase (FFluc) cDNA in the pcDNA3 (Invitrogen)-derived plasmid psiRNALUC. A549 or Huh7 cells in 12-well plates were transfected with 5 ng of empty or miR122 target site-containing psiRNALUC, 5 ng of pcDNA-renilla, 1.5 μg of unrelated empty carrier plasmid, and when indicated, 100 nM miRIDIAN miR122 inhibitor (Dharmacon) by using Lipofectamine 2000 (Invitrogen). Twenty-four to 48 h posttransfection, luciferase activity was measured using a dual-luciferase assay system (Promega).
Construction and production of adenoviruses.
pShuttle122 was made by inserting three target sites for miR122 immediately after the stop codon in pSEMII as described above for the luciferase reporter plasmids. pSEMII was derived from pSE1 containing a subgenomic adenovirus type 5 (Ad5) fragment including a wild-type E1A gene (6) but engineered to carry a wild-type E1A enhancer/promoter plus a novel MluI site in the 3′ untranslated region (UTR) of the E1A gene. pSEMII and pShuttle122 were further modified to create pShuttleK and pShuttleK-122 by inserting annealed oligonucleotides 5′-CGCGCACCATGGTGTCGACG-3′ and 5′-CGCGCGTCGACACCATGGTG-3′ into the 5′ UTR of the E1A gene 23 bp upstream of the endogenous E1A codon ATG to create a novel out-of-frame translational start site. Homologous recombination between the modified subgenomic adenoviral genomes and pAd5/3-Δ24 (26) and the generation of infectious adenovirus stocks were carried out using standard procedures. The titers of the purified viruses were as follows: 1.6 × 109 PFU/ml for Ad5/3-Δ24, 1.6 × 107 PFU/ml for Ad5/3-122, and 4 × 108 PFU/ml for both Ad5/3K and Ad5/3K-122.
Western blot analysis.
Cells were lysed with 1% NP-40 lysis buffer (150 mM NaCl; 50 mM Tris-HCl, pH 7.9; 1% Nonidet P-40) in the presence of protease inhibitors. Protein concentrations were determined by the Bio-Rad protein assay, and 50 μg of each sample was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then blotted onto a nitrocellulose membrane (Bio-Rad). After being blocked overnight at 4°C with a solution of phosphate-buffered saline and 0.05% Tween 20 containing 5% nonfat dry milk, the membranes were treated with mouse monoclonal antibody specific to adenoviral E1A proteins (sc-58653; Santa Cruz Biotechnology) or adenovirus-infected patient serum (a kind gift from Jukka Suni, HUSLAB, Helsinki) as a primary antibody and then with horseradish peroxidase-conjugated secondary antibodies against mouse or human immunoglobulins (DakoCytomation). An enhanced chemiluminescence detection system (Millipore) was used for detection.
Quantitative RT-PCR.
Total RNA was extracted from infected cells by using a GenElute mammalian total RNA miniprep kit (Sigma). RevertAid Moloney murine leukemia virus reverse transcriptase (RT; Fermentas) was used for cDNA synthesis. Oligonucleotide sequences for the amplification of the E1A S13 gene were as follows: forward primer, 5′-TCCGGAGCCGCCTCACCTTTC-3′, and reverse primer, 5′-GGCTCAGGTTCAGACACAGGACTGTAG-3′. The human housekeeping GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene was used as an internal control and amplified with the following primers: forward, 5′-GAGTCAACGGATTTGGTCGT-3′, and reverse, 5′-TTGATTTTGGAGGGATCTCG-3′. PCR was done using a LightCycler instrument (Roche Molecular Biochemicals) with a DyNAmo Capillary Sybr green quantitative PCR kit (Finnzymes). After an initial incubation at 95°C for 10 min, 35 amplification cycles were conducted as follows: denaturation at 95°C for 10 s, annealing at 55°C for 10 s, and extension at 72°C for 12 s. To quantify the differences in E1A mRNA levels in Huh7 cells, the LightCycler datum point curves were compared to similar analyses generated using a dilution standard and were graphed as percentages relative to the level in the specimen with the highest level of expression.
Virus replication and cell viability assays.
Virus titers were determined by using a 50% tissue culture infective dose. Permissive 293FT cells were seeded into 96-well plates (104 cells/well), and the next day, eight serial 10-fold dilutions of freeze-thawed lysates of infected cells were added to the 96-well plates. After 10 days of incubation, the wells with observable cytopathic effects (CPE) in each dilution were counted and the virus titers were calculated according to the standard Kärber method. Cell viability was measured using the CellTiter 96 AQueous One solution cell proliferation assay (Promega) and a multiwell plate reader (Multiskan EX; ThermoLabsystems) to determine the optical densities of the reaction mixtures at 490 nm.
RESULTS
miR122 is expressed at high levels in liver but not in other tissues (12, 29). To test the potential of miR122 as a cell type-specific suppressor of foreign genes introduced into cells, we constructed a series of FFluc expression vectors containing one or more target sites for miR122 in the 5′ or 3′ UTR (Fig. 1A). Due to the perfect complementarity of these target sites to miR122, we hypothesized that they could mediate the destruction of luciferase mRNA in liver cells. As a liver cell model, we used Huh7 cells representing a relatively well-differentiated hepatocellular carcinoma cell line, which differs from most other hepatocellular carcinoma lines and instead resembles normal hepatocytes in expressing high levels of miR122 (12). As examples of nonhepatic cells not expressing miR122, we used carcinoma cell lines derived from human lung (A549), prostate (PC3), and cervix (HeLa) and the embryonic kidney-derived cell line 293FT.
FIG. 1.
Characterization of miR122 target sequences as cell type-specific suppressors of reporter gene expression. (A) Schematic illustration of the FFluc and Renilla luciferase constructs used and sequence of the miR122 target site inserted in different copy numbers into FFluc constructs. (B) Effects of the indicated combinations of miR122 target sequences in the 5′ and/or 3′ UTR of FFluc constructs on the ratio of FFluc versus Renilla luciferase activity in cotransfected A549 (striped bars) or Huh7 (black bars) cells. The ratio for cells transfected with an unmodified FFluc construct (control) was set to 100%, and the other values for the same cell type are expressed relative to this reference. The numbers of miR122 target sequences and their positioning are indicated under the corresponding bars. (C) Suppression of the negative effect of miR122 target sites by an miR122 inhibitor. Huh7 cells were transfected with a Renilla luciferase construct and an unmodified or miR122 target site-containing FFluc construct (3x3′, corresponding to the 3′ three-target-site construct indicated in panel B), together with (+) or without (−) a synthetic antagomir for miR122. The FFluc/Renilla luciferase activity ratio obtained with unmodified FFluc in the absence of the antagomir was set to 100%, and the other values are expressed relative to this reference.
As shown in Fig. 1B, the presence of a single target site in the 5′ or 3′ UTR of FFluc mRNA resulted in the specific suppression of FFluc expression in Huh7 cells but not in A549 cells or in the other non-liver-derived cell lines tested, including 293FT, PC3, and HeLa (data not shown). Cotransfection with a synthetic miR122 inhibitor (antagomir-122) potently suppressed the negative effect of miR122 target sites in Huh7 cells (Fig. 1C), confirming that the inhibitory effect of miR122 target sites was indeed mediated by miR122.
To test the ability of miR122 to suppress E1A expression in the context of the infection of liver-derived cells with a replication-competent adenovirus, we constructed a virus designated Ad5/3-122, which is a derivative of Ad5/3-Δ24 (26) containing three copies of the miR122 target site in the 3′ UTR of the E1A gene. In good agreement with the reporter gene data, E1A protein levels in Ad5/3-122-infected Huh7 cells were dramatically lower than those in Ad5/3-Δ24-infected Huh7 cells, whereas in A549 cells, the viruses expressed similar amounts of E1A protein (Fig. 2A). Yet the replication of the modified Ad5/3-122 virus in Huh7 cells was attenuated only moderately compared to that of Ad5/3-Δ24, as judged by the development of CPE in colorimetric cell viability assays (data not shown). Accordingly, like lysates from A549 cells, lysates from Ad5/3-Δ24- and Ad5/3-122-infected Huh7 cells showed similar patterns and levels of expression of total adenoviral proteins (Fig. 2A). These results established that adenoviral E1A expression could be efficiently inhibited in a cell type-specific manner by this strategy but also indicated that the degree of E1A suppression was not sufficient to have a significant impact on virus replication in this study system.
FIG. 2.
Inhibitory effects of engineered miR122 target sites on E1A protein (A) and mRNA (B) expression in infected liver cells. (A) Huh7 and A549 cells were infected with Ad5/3-Δ24 (Ad5/3) or Ad5/3-122 (122) at a MOI of 0.05 or left uninfected (−). Three days later, the cells were harvested and analyzed by Western blotting for E1A protein expression (top panels) and total immunoreactive adenoviral protein expression (bottom panels). α-E1A, anti-E1A antibody; α-Ad, antiadenovirus antibody. (B) Real-time RT-PCR was used to quantify E1A mRNA from total RNA extracted from Huh7 cells infected with Ad5/3-Δ24 or Ad5/3-122 at a MOI of 0.05. The relative expression of E1A mRNA at 1, 2, and 3 days (d) postinfection with these two viruses was determined based on an E1A mRNA dilution standard and the parallel quantification of GAPDH mRNA from the same specimens.
To test if the reduced levels of E1A protein in Ad5/3-122-infected Huh7 cells were due to reduced E1A mRNA (as hypothesized) rather than reduced E1A translation (the normal mode of miRNA action), we extracted RNA from Huh7 and A549 cell cultures infected for 1, 2, or 3 days with the Ad5/3-Δ24 or Ad5/3-122 virus and measured E1A mRNA expression by quantitative RT-PCR. The level of E1A mRNA produced by the Ad5/3-122 virus was significantly reduced compared to that produced by the control virus in Huh7 cells but not in A549 cells, whereas similar amounts of control (GAPDH) mRNA were detected in all the infected cultures (data not shown). A quantitative analysis of the raw PCR data revealed that in Huh7 cells, Ad5/3-122 expressed less than 10% of the level of E1A mRNA produced by Ad5/3-Δ24 (Fig. 2B). This finding indicated that the cell type-specific negative regulation indeed occurred mainly via mRNA destruction.
The observed wild-type-like replication of Ad5/3-122 in Huh7 cells despite strongly reduced levels of E1A may be explained by the results of previous studies that have reported the capacity of adenovirus to replicate normally despite strongly reduced levels of E1A (20). To overcome this problem, we designed a strategy to uniformly reduce E1A protein levels in all cell types and hypothesized that such a lowered baseline level of E1A would still support normal adenovirus replication but that, in this setting, the suppression of E1A mRNA by miR122 may be sufficient to prevent viral replication in liver cells. To achieve this reduction, we inserted an ectopic consensus site for translational initiation (ACCAUGG) (28) into the 5′ UTR of E1A mRNA in the wrong reading frame relative to the E1A coding region and named the resulting virus Ad5/3K. As shown in Fig. 3A, E1A protein expression was indeed reduced in the expected uniform manner in A549 cells as well as in Huh7 cells infected with the Ad5/3K virus. As also expected, these reduced E1A protein levels were not associated with a notably lower replicative capacity as judged by the development of CPE in the infected cultures and colorimetric viability assays (data not shown) or by Western blot analysis of total adenoviral protein expression (Fig. 3A).
FIG. 3.
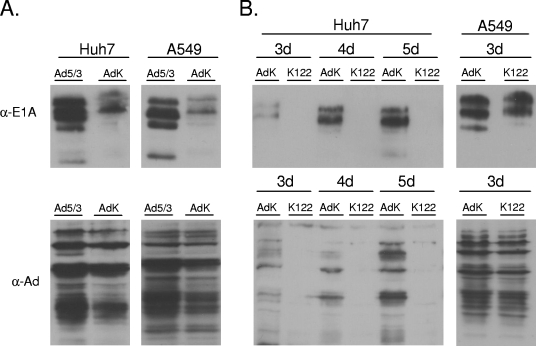
Abolishment of total adenoviral protein expression in liver cells by miR122 target sites. (A) Placing of an out-of-frame translational initiation site upstream of the E1A open reading frame results in a cell type-independent reduction in E1A translation without reducing overall adenoviral protein expression. Huh7 and A549 cells were infected with Ad5/3-Δ24 (Ad5/3) or Ad5/3K (AdK) at a MOI of 0.05. Cells were analyzed 3 days postinfection for adenoviral protein expression as described in the legend to Fig. 2. α-E1A, anti-E1A antibody; α-Ad, antiadenovirus antibody. (B) Liver-specific loss of detectable adenoviral protein expression can be achieved by the introduction of miR122 target sites into the Ad5/3K virus background. Huh7 and A549 cells were infected with Ad5/3K or its derivative Ad5/3K-122 (K122) at a MOI of 0.05 and analyzed 3 days (3d) later for adenoviral protein expression as described in the legend to Fig. 2.
Based on these results, we generated Ad5/3K-122, which is a derivative of Ad5/3K containing three tandem copies of the miR122 target sequence in its E1A gene 3′ UTR. The infection of Huh7 cells with Ad5/3K-122 or Ad5/3K at a multiplicity of infection (MOI) of 0.05 now revealed a dramatic effect of miR122 target sites on viral replication. In addition to having undetectable levels of E1A, Huh7 cells infected with Ad5/3K-122 failed to express other adenoviral proteins at levels detectable by Western blot analysis. In contrast, Huh7 cells infected in parallel with Ad5/3K expressed increasing amounts of E1A and other viral proteins. No differences in the expression of E1A or other viral proteins between A549 cells infected with Ad5/3K-122 and those infected with Ad5/3K were observed (Fig. 3B).
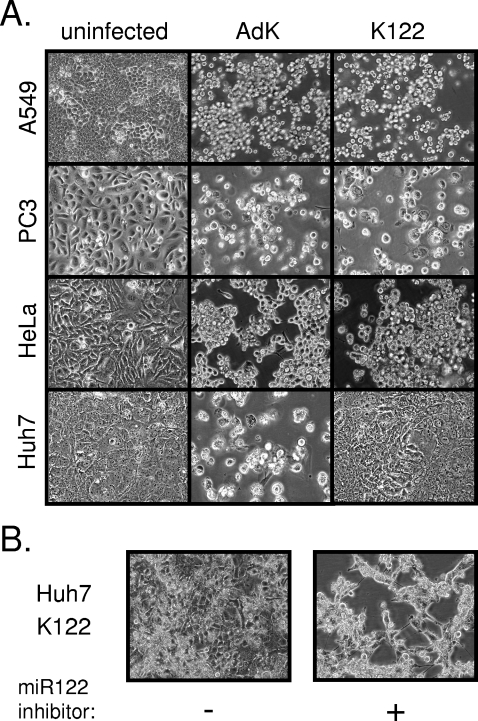
The lack of a noticeable CPE in Huh7 cells infected with Ad5/3K-122 supported the idea that this virus was unable to productively replicate in liver-derived cells (Fig. 4A). Similar to the examples from 6 days postinfection shown in Fig. 4A, these Huh7 cultures showed no evidence of a CPE even upon prolonged observation. In contrast, indistinguishable CPE in other cell types (A549, PC3, and HeLa) infected with these two viruses were observed. Strikingly, a marked CPE in Huh7 cells infected with Ad5/3K-122 also developed in the same period of time if the cells were transfected with antagomir-122 prior to infection (Fig. 4B), confirming that the differential capacity of Ad5/3K-122 to replicate in these cell lines was indeed determined by miR122.
FIG. 4.
Cell type- and miR122-dependent CPE of an adenovirus carrying miR122 target sites. (A) The indicated cell lines were infected with Ad5/3K (AdK) or Ad5/3K-122 (K122) at a MOI of 0.05. Cells were photographed 4 days later (or 6 days later in the case of Huh7 cells) to document the appearance of a CPE. Uninfected control cell cultures are shown for comparison. (B) Huh7 cells infected with Ad5/3K-122 as described in panel A following pretreatment with the miRIDIAN miR122 inhibitor. +, present; −, absent.
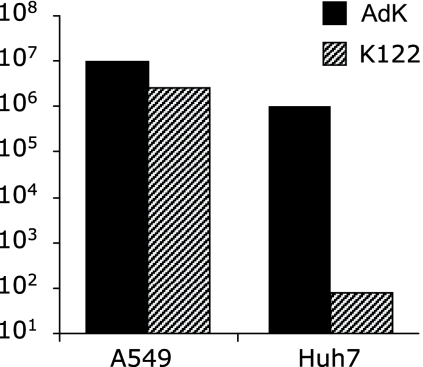
To directly quantify the suppression of replication by miR122, we compared the amounts of cell-associated infectious virus produced by Huh7 and A549 cells infected with Ad5/3K-122 or Ad5/3K. Five days postinfection with these viruses at a MOI of 0.05, almost no infectious virus (79 PFU/ml) could be recovered from Ad5/3K-122-infected Huh7 cells whereas the virus titer in Ad5/3K-infected Huh7 cells was more than 10,000-fold higher (1.0 × 106 PFU/ml) and no difference in the rates of replication of these two viruses in A549 cells was seen (Fig. 5).
FIG. 5.
Effect of miR122 target sites on viral replication in liver-derived and control cells. Huh7 and A549 cells were infected with Ad5/3K (AdK) or Ad5/3K-122 (K122) at a MOI of 0.05, and the titers of cell-associated infectious virus at three (A549) or five (Huh7) days postinfection were determined by a standard 50% tissue culture infective dose limiting dilution method. Values are expressed as PFU per milliliter.
Undetectable levels of adenoviral proteins, the lack of a CPE, and the decreasing amounts of infectious virus recovered from Huh7 cells infected with Ad5/3K-122 indicated that this virus was severely attenuated if not completely unable to replicate in these cells. In contrast, as judged by the same parameters, the Ad5/3K virus, which lacked the target sites for miR122 but was otherwise identical, did not show any evidence of attenuation in Huh7 cells. These data establish the introduction of miR122 target sites as a powerful strategy for suppressing adenovirus replication in liver cells without compromising the capacity to replicate in other cell types.
DISCUSSION
In this study, we have shown that cell type-specific miRNA expression can be exploited to create a CRAd that fails to productively replicate in liver cells. This strategy adds a valuable new approach to the current strategies for generating oncolytic adenoviruses. Moreover, our results provide the first example of hijacking the cellular RNA interference machinery to suppress the replication of a DNA virus in animal cells.
Because adenovirus replication can proceed normally despite low E1A protein levels (20), additional nonspecific measures (decreasing the translational capacity of E1A mRNA) were needed to complement liver-specific miRNA-mediated E1A mRNA suppression in order to bring E1A protein levels below a threshold at which they no longer supported viral replication in the Huh7 hepatoma cells. It is possible, however, that in normal liver this threshold may be less stringent and that the degree of miR122-mediated suppression of E1A mRNA may be sufficient to prevent the replication of an adenovirus expressing E1A transcripts that are translated normally.
It has been shown previously in a nonhuman primate model that acute hepatotoxicity of systemically administered adenoviral vectors can develop without viral gene expression in hepatocytes (10). However, extensive infection of hepatocytes is known to occur upon high-dose systemic infection of monkeys or humans with replication-competent adenoviruses, such as ONYX-015 (see, e.g., reference 18) or adenoviral vectors (see, e.g., reference 36). Thus, the availability of an adenovirus that cannot replicate in hepatocytes should allow the development of better systemic therapies involving replication-competent adenoviruses. Moreover, in the context of nonreplicating therapeutic gene delivery vectors, the same principle may also be applied to limit their gene expression in the liver.
A special instance in which placing an oncolytic adenovirus under negative control by miR122 may be particularly useful may be the virotherapy of metastatic disease of the liver. Intravascular injection of ONYX-015 into the liver has been tested as a therapy for metastatic colorectal cancer, with modestly promising results (18, 36). However, despite the failure to express E1B, which should bias replication of this virus into transformed cells, PCR and immunostaining studies of a patient who died shortly after ONYX-015 treatment revealed viral antigens also in normal liver cells and amounts of viral DNA that were even greater than in the metastatic lesions (18). Thus, additional means for preventing adenoviral expression and replication in normal hepatocytes are clearly needed for the development of better CRAd-based therapies for metastatic liver disease.
It is also worth noting that the loss of miR122 expression appears to be correlated with the malignant transformation of hepatocytes, such that Huh7 was unique among the four transformed liver cell lines studied by Chang et al. (12) in maintaining the primary hepatocyte-like expression of miR122. Thus, the suppression of adenovirus replication by miR122 may also be exploited in the targeted virotherapy of primary hepatocellular carcinoma as well as metastatic tumors.
The ability of the miR122 target sequences to specifically inhibit adenovirus replication in a liver-derived cell line but not in cell lines of different origins was striking. Nevertheless, in order to create optimal oncolytic adenoviruses, miRNA-based targeting should probably be combined with one or more of the existing strategies for the targeting of adenoviral entry and replication to the desired tissues and cells. In this regard, it should be emphasized that the introduction of miRNA target sites into the adenoviral genome is readily compatible with any of the previously described approaches to generate CRAds. Because the size of the adenovirus genome cannot be increased by more than 5% and since it may be attractive to include additional therapeutic genes in oncolytic viruses, it is also potentially important that the present miRNA-based approach for altering adenoviral tropism does not require much space.
One issue that cannot be ignored is that the engineered virus may escape miRNA-mediated suppression in liver cells via mutation or deletion of the miR122 target sites. However, during our extended Huh7 cell infection experiments (in which cells were maintained in cultures for up to 2 weeks), no evidence of the emergence of such escape mutants was observed (data not shown), suggesting that it would be a relatively slow process that may not occur before the therapeutic virus would be cleared by the immune system. Moreover, even if such mutants were generated in the infected individuals, the main goal of this approach, i.e., the suppression of viral replication during the initial infection of the liver following systemic administration, would still be achieved. Similarly, it is difficult to rule out that the modified virus would not be able to replicate with extremely slow kinetics (as opposed to being dead) which would not be apparent under the experimental conditions practical for working with in vitro-infected Huh7 cultures. In any case, our results unambiguously show that profound and cell type-specific attenuation of adenoviral replication can be achieved using this strategy.
The rich palette of miRNAs with differential expression patterns (21, 23, 27, 31, 42) and the tendency of certain miRNAs to be lost during carcinogenesis (8, 15, 24, 32, 45) offer great opportunities for developing tumor-specific CRAds. Moreover, the approach described here should be applicable to many other human DNA and RNA viruses, including vesicular stomatitis virus, Sindbis and Semliki Forest viruses, and Newcastle disease virus, as well as herpes and vaccinia viruses, all of which are actively being developed as tools for oncolytic virotherapy (1, 30, 44).
Acknowledgments
We are grateful to Virpi Knuutinen and Virpi Syvälahti for expert technical assistance and Prasanth Viswanathan for assistance in the early phase of this work.
K.S. and A.H. have been supported by grants from the Academy of Finland, the Sigrid Juselius Foundation, Biocentrum Helsinki, and the Helsinki University Central Hospital (HUCH). R.A. has been supported by NIH grants AI36178 and AI064738. A.H. is K. Albin Johansson Research Professor of the Foundation for the Finnish Cancer Institute. E.Y. has been supported by the Helsinki Biomedical Graduate School.
Footnotes
Published ahead of print on 17 September 2008.
REFERENCES
- 1.Aghi, M., and R. L. Martuza. 2005. Oncolytic viral therapies—the clinical experience. Oncogene 247802-7816. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, A., J. Thompson, L. Emiliusen, S. Murphy, R. D. Beauchamp, K. Suzuki, R. Alemany, K. Harrington, and R. G. Vile. 2003. A conditionally replicating adenovirus targeted to tumor cells through activated RAS/P-MAPK-selective mRNA stabilization. Nat. Biotechnol. 21771-777. [DOI] [PubMed] [Google Scholar]
- 3.Alemany, R. 2007. Cancer selective adenoviruses. Mol. Aspects Med. 2842-58. [DOI] [PubMed] [Google Scholar]
- 4.Ambros, V. 2003. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113673-676. [DOI] [PubMed] [Google Scholar]
- 5.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116281-297. [DOI] [PubMed] [Google Scholar]
- 6.Bauerschmitz, G. J., K. Guse, A. Kanerva, A. Menzel, I. Herrmann, R. A. Desmond, M. Yamamoto, D. M. Nettelbeck, T. Hakkarainen, P. Dall, D. T. Curiel, and A. Hemminki. 2006. Triple-targeted oncolytic adenoviruses featuring the cox2 promoter, E1A transcomplementation, and serotype chimerism for enhanced selectivity for ovarian cancer cells. Mol. Ther. 14164-174. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff, J. R., D. H. Kirn, A. Williams, C. Heise, S. Horn, M. Muna, L. Ng, J. A. Nye, A. Sampson-Johannes, A. Fattaey, and F. McCormick. 1996. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science 274373-376. [DOI] [PubMed] [Google Scholar]
- 8.Blenkiron, C., and E. A. Miska. 2007. miRNAs in cancer: approaches, aetiology, diagnostics and therapy. Hum. Mol. Genet. 16(R1)R106-R113. [DOI] [PubMed] [Google Scholar]
- 9.Brown, B. D., B. Gentner, A. Cantore, S. Colleoni, M. Amendola, A. Zingale, A. Baccarini, G. Lazzari, C. Galli, and L. Naldini. 2007. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 251457-1467. [DOI] [PubMed] [Google Scholar]
- 10.Brunetti-Pierri, N., D. J. Palmer, A. L. Beaudet, K. D. Carey, M. Finegold, and P. Ng. 2004. Acute toxicity after high-dose systemic injection of helper-dependent adenoviral vectors into nonhuman primates. Hum. Gene Ther. 1535-46. [DOI] [PubMed] [Google Scholar]
- 11.Calin, G. A., and C. M. Croce. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer 6857-866. [DOI] [PubMed] [Google Scholar]
- 12.Chang, J., E. Nicolas, D. Marks, C. Sander, A. Lerro, M. A. Buendia, C. Xu, W. S. Mason, T. Moloshok, R. Bort, K. S. Zaret, and J. M. Taylor. 2004. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 1106-113. [DOI] [PubMed] [Google Scholar]
- 13.Cullen, B. R. 2004. Transcription and processing of human microRNA precursors. Mol. Cell 16861-865. [DOI] [PubMed] [Google Scholar]
- 14.Edge, R. E., T. J. Falls, C. W. Brown, B. D. Lichty, H. Atkins, and J. C. Bell. 2008. A let-7 microRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol. Ther. 161437-1443. [DOI] [PubMed] [Google Scholar]
- 15.Esquela-Kerscher, A., and F. J. Slack. 2006. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 6259-269. [DOI] [PubMed] [Google Scholar]
- 16.Fueyo, J., C. Gomez-Manzano, R. Alemany, P. S. Lee, T. J. McDonnell, P. Mitlianga, Y. X. Shi, V. A. Levin, W. K. Yung, and A. P. Kyritsis. 2000. A mutant oncolytic adenovirus targeting the Rb pathway produces anti-glioma effect in vivo. Oncogene 192-12. [DOI] [PubMed] [Google Scholar]
- 17.Gitlin, L., J. K. Stone, and R. Andino. 2005. Poliovirus escape from RNA interference: short interfering RNA-target recognition and implications for therapeutic approaches. J. Virol. 791027-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamid, O., M. L. Varterasian, S. Wadler, J. R. Hecht, A. Benson III, E. Galanis, M. Uprichard, C. Omer, P. Bycott, R. C. Hackman, and A. F. Shields. 2003. Phase II trial of intravenous CI-1042 in patients with metastatic colorectal cancer. J. Clin. Oncol. 211498-1504. [DOI] [PubMed] [Google Scholar]
- 19.Heise, C., T. Hermiston, L. Johnson, G. Brooks, A. Sampson-Johannes, A. Williams, L. Hawkins, and D. Kirn. 2000. An adenovirus E1A mutant that demonstrates potent and selective systemic anti-tumoral efficacy. Nat. Med. 61134-1139. [DOI] [PubMed] [Google Scholar]
- 20.Hitt, M. M., and F. L. Graham. 1990. Adenovirus E1A under the control of heterologous promoters: wide variation in E1A expression levels has little effect on virus replication. Virology 179667-678. [DOI] [PubMed] [Google Scholar]
- 21.Hsu, P. W., H. D. Huang, S. D. Hsu, L. Z. Lin, A. P. Tsou, C. P. Tseng, P. F. Stadler, S. Washietl, and I. L. Hofacker. 2006. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 34D135-D139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jay, C., J. Nemunaitis, P. Chen, P. Fulgham, and A. W. Tong. 2007. miRNA profiling for diagnosis and prognosis of human cancer. DNA Cell Biol. 26293-300. [DOI] [PubMed] [Google Scholar]
- 23.John, B., A. J. Enright, A. Aravin, T. Tuschl, C. Sander, and D. S. Marks. 2004. Human microRNA targets. PLoS Biol. 2e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John, B., C. Sander, and D. S. Marks. 2006. Prediction of human microRNA targets. Methods Mol. Biol. 342101-113. [DOI] [PubMed] [Google Scholar]
- 25.Kanerva, A., and A. Hemminki. 2004. Modified adenoviruses for cancer gene therapy. Int. J. Cancer 110475-480. [DOI] [PubMed] [Google Scholar]
- 26.Kanerva, A., G. V. Mikheeva, V. Krasnykh, C. J. Coolidge, J. T. Lam, P. J. Mahasreshti, S. D. Barker, M. Straughn, M. N. Barnes, R. D. Alvarez, A. Hemminki, and D. T. Curiel. 2002. Targeting adenovirus to the serotype 3 receptor increases gene transfer efficiency to ovarian cancer cells. Clin. Cancer Res. 8275-280. [PubMed] [Google Scholar]
- 27.Kim, V. N., and J. W. Nam. 2006. Genomics of microRNA. Trends Genet. 22165-173. [DOI] [PubMed] [Google Scholar]
- 28.Kozak, M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44283-292. [DOI] [PubMed] [Google Scholar]
- 29.Lagos-Quintana, M., R. Rauhut, A. Yalcin, J. Meyer, W. Lendeckel, and T. Tuschl. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12735-739. [DOI] [PubMed] [Google Scholar]
- 30.Lin, E., and J. Nemunaitis. 2004. Oncolytic viral therapies. Cancer Gene Ther. 11643-664. [DOI] [PubMed] [Google Scholar]
- 31.Liu, C. G., G. A. Calin, B. Meloon, N. Gamliel, C. Sevignani, M. Ferracin, C. D. Dumitru, M. Shimizu, S. Zupo, M. Dono, H. Alder, F. Bullrich, M. Negrini, and C. M. Croce. 2004. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA 1019740-9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu, J., G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz, and T. R. Golub. 2005. MicroRNA expression profiles classify human cancers. Nature 435834-838. [DOI] [PubMed] [Google Scholar]
- 33.Mansfield, J. H., B. D. Harfe, R. Nissen, J. Obenauer, J. Srineel, A. Chaudhuri, R. Farzan-Kashani, M. Zuker, A. E. Pasquinelli, G. Ruvkun, P. A. Sharp, C. J. Tabin, and M. T. McManus. 2004. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 361079-1083. [DOI] [PubMed] [Google Scholar]
- 34.Mathis, J. M., M. A. Stoff-Khalili, and D. T. Curiel. 2005. Oncolytic adenoviruses—selective retargeting to tumor cells. Oncogene 247775-7791. [DOI] [PubMed] [Google Scholar]
- 35.Matranga, C., and P. D. Zamore. 2007. Small silencing RNAs. Curr. Biol. 17R789-R793. [DOI] [PubMed] [Google Scholar]
- 36.Morral, N., W. K. O'Neal, K. Rice, M. M. Leland, P. A. Piedra, E. Aguilar-Cordova, K. D. Carey, A. L. Beaudet, and C. Langston. 2002. Lethal toxicity, severe endothelial injury, and a threshold effect with high doses of an adenoviral vector in baboons. Hum. Gene Ther. 13143-154. [DOI] [PubMed] [Google Scholar]
- 37.O'Shea, C. C., L. Johnson, B. Bagus, S. Choi, C. Nicholas, A. Shen, L. Boyle, K. Pandey, C. Soria, J. Kunich, Y. Shen, G. Habets, D. Ginzinger, and F. McCormick. 2004. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer Cell 6611-623. [DOI] [PubMed] [Google Scholar]
- 38.Raper, S. E., N. Chirmule, F. S. Lee, N. A. Wivel, A. Bagg, G. P. Gao, J. M. Wilson, and M. L. Batshaw. 2003. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 80148-158. [DOI] [PubMed] [Google Scholar]
- 39.Reid, T., E. Galanis, J. Abbruzzese, D. Sze, J. Andrews, L. Romel, M. Hatfield, J. Rubin, and D. Kirn. 2001. Intra-arterial administration of a replication-selective adenovirus (dl1520) in patients with colorectal carcinoma metastatic to the liver: a phase I trial. Gene Ther. 81618-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez, R., E. R. Schuur, H. Y. Lim, G. A. Henderson, J. W. Simons, and D. R. Henderson. 1997. Prostate attenuated replication competent adenovirus (ARCA) CN706: a selective cytotoxic for prostate-specific antigen-positive prostate cancer cells. Cancer Res. 572559-2563. [PubMed] [Google Scholar]
- 41.Saukkonen, K., and A. Hemminki. 2004. Tissue-specific promoters for cancer gene therapy. Expert Opin. Biol. Ther. 4683-696. [DOI] [PubMed] [Google Scholar]
- 42.Sood, P., A. Krek, M. Zavolan, G. Macino, and N. Rajewsky. 2006. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc. Natl. Acad. Sci. USA 1032746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoff-Khalili, M. A., A. A. Rivera, A. Nedeljkovic-Kurepa, A. Debenedetti, X. L. Li, Y. Odaka, J. Podduturi, D. A. Sibley, G. P. Siegal, A. Stoff, S. Young, Z. B. Zhu, D. T. Curiel, and J. M. Mathis. 2008. Cancer-specific targeting of a conditionally replicative adenovirus using mRNA translational control. Breast Cancer Res. Treat. 10843-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vähä-Koskela, M. J., J. E. Heikkilä, and A. E. Hinkkanen. 2007. Oncolytic viruses in cancer therapy. Cancer Lett. 254178-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volinia, S., G. A. Calin, C. G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris, and C. M. Croce. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 1032257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, S. L., H. Y. Chen, P. C. Yang, and J. J. Chen. 2007. Unique microRNA signature and clinical outcome of cancers. DNA Cell Biol. 26283-292. [DOI] [PubMed] [Google Scholar]