Abstract
The major immediate-early (IE) region of human cytomegalovirus encodes two IE proteins, IE1 72 and IE2 86, that are translated from alternatively spliced transcripts that differ in their 3′ ends. Two other proteins that correspond to the C-terminal region of IE2 86, IE2 60 and IE2 40, are expressed at late times. In this study, we used IE2 mutant viruses to examine the mechanism by which IE2 86, IE2 60, and IE2 40 affect the expression of a viral DNA replication factor, UL84. Deletion of amino acids (aa) 136 to 290 of IE2 86 results in a significant decrease in UL84 protein during the infection. This loss of UL84 is both proteasome and calpain independent, and the stability of the protein in the context of infection with the mutant remains unaffected. The RNA for UL84 is expressed to normal levels in the mutant virus-infected cells, as are the RNAs for two other proteins encoded by this region, UL85 and UL86. Moreover, nuclear-to-cytoplasmic transport and the distribution of the UL84 mRNA on polysomes are unaffected. A region between aa 290 and 369 of IE2 86 contributes to the UL84-IE2 86 interaction in vivo and in vitro. IE2 86, IE2 60, and IE2 40 are each able to interact with UL84 in the mutant-infected cells, suggesting that these interactions may be important for the roles of UL84 and the IE2 proteins. Thus, these data have defined the contribution of IE2 86, IE2 60, and IE2 40 to the efficient expression of UL84 throughout the infection.
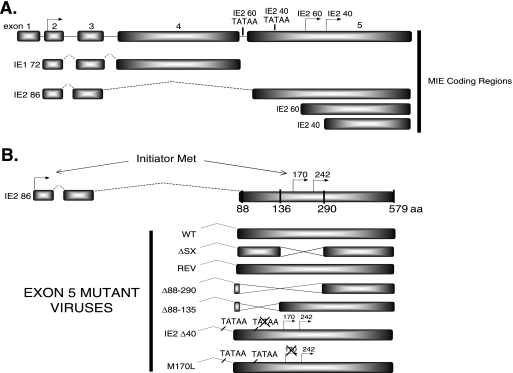
Human cytomegalovirus (HCMV) is the major viral cause of birth defects and poses a severe threat to immunocompromised and transplant patients (for review, see reference 33). Gene expression has been classified into three major groups, referred to as the immediate-early (IE), early, and late genes, which are temporally regulated throughout the infection. The two major IE (MIE) genes, IE1 72 and IE2 86 (encoded by UL122 and UL123), are of particular interest for understanding the various regulatory mechanisms that govern a productive viral infection. They can transactivate viral early promoters, serve in viral promoter repression, and alter the expression of many host cellular genes in order to make the environment favorable for viral replication (for reviews, see references 14 and 33). Both MIE proteins arise from a single transcript that consists of five exons that are differentially spliced to produce IE1 72 (exons 1 to 4) and IE2 86 (exons 1 to 3 and 5) (54-56). While IE1 72 is dispensable for infection at a high multiplicity of infection (MOI), IE2 86 is essential (16, 18, 31, 32, 36, 37, 59).
At late times in infection, transcripts that arise from within exon 5 of the UL122 gene encode the late IE2 60 and IE2 40 proteins, which correspond to the C-terminal region of IE2 86 (41). The IE2 60 protein is expressed from an initiator methionine at amino acid (aa) 170, with the putative TATAA region occurring in the intron between exons 4 and 5. The IE2 40 protein is expressed from a 1.5-kb RNA, with translation initiating at methionine 242, and a putative TATAA box just upstream of the IE2 60 translation initiation site. It has been proposed that a small amount of IE2 60 protein is also expressed from the RNA encoding IE2 40 (60). These proteins have been shown to have a role in transactivation as well as repression of both MIE genes (23, 40, 41, 52). Our laboratory has determined that the IE2 60 and IE2 40 proteins also have a role in the expression of the two early-late viral proteins pp65 (UL83) and UL84; UL83 is a tegument protein, while UL84 is an essential DNA replication factor. Although IE2 60 and IE2 40 are dispensable for the infection, they are required for efficient replication at later stages in the viral life cycle (60).
Many of the functions of both IE1 72 and IE2 86 have been studied in transient-transfection assays and, more recently, in the context of infection using bacterial artificial chromosomes (BACs) with defined mutations in the genes (31, 36, 37, 42, 45, 59-61). One viral mutant of particular interest lacks the region between aa 136 and 290 of IE2 86 (termed IE2 86ΔSX) (45, 60). Since the initiating methionines for IE2 60 and IE2 40 (aa 170 and 242, respectively) are deleted, this virus also does not express IE2 60 and IE2 40. Previous studies of the IE2 86ΔSX virus revealed that it grows slowly and is significantly debilitated in its ability to produce infectious virus. Furthermore, there is a severe lag in IE2 86 protein expression, although IE1 72 expression remains normal. Early genes do not seem to be affected, while expression of many late proteins is greatly reduced. In particular, UL83 and UL84 show significant decreases in protein expression during the later stages of the infection process, and these defects can be partially overcome in complementing cell lines that express IE2 86 and IE2 40 (45, 46, 60).
The only viral protein that IE2 86 has been shown to interact with in the infection is UL84 (44, 51). UL84 is present in low levels at early times in infection and accumulates to high levels after the onset of viral DNA replication (20). In transient assays, it is required for ori-Lyt-dependent replication (35, 47, 63, 65) and through its interaction with IE2 86 appears to be important for the activation of the bidirectional promoter within ori-Lyt (12, 62). However, it also downregulates the ability of IE2 86 to activate some early promoters in transient assays (17). Other properties of UL84 are that it interacts with an RNA stem-loop sequence within the RNA/DNA hybrid region of ori-Lyt, displays UTPase activity, and shows some homology to the DExD/H box family of helicases (11, 13). However, many of its specific functions during the course of the infection remain to be determined.
Our previous work showing that the IE2 86ΔSX virus exhibits a severe dysregulation of UL84 at late times postinfection (60) prompted us to examine the interactions and regulatory mechanisms that govern the expression of the UL84 and IE2 proteins at both early and late times postinfection. Here, we show that infection with the IE2 86ΔSX virus results in a marked decrease in the protein expression of UL84 throughout the infection. In contrast, the transcription of UL84 and the mRNAs encoding UL85 and UL86, which are 3′ coterminal with the UL84 RNA (43), remains normal in this mutant infection. The defect in accumulation of the UL84 protein is after the initiation of translation, as nuclear export of the UL84 RNA, as well as the distribution of the mRNA on polyribosomes (polysomes), is unaffected in the IE2 86ΔSX infection. Immunoprecipitation analyses of lysates from cells infected with IE2 mutant viruses and in vitro glutathione-S-transferase (GST) binding assays reveal that a domain that plays a prominent role in the formation of a complex of IE2 86 and UL84 lies between aa 290 and 369 of IE2 86, although there may be some contribution from other regions within the protein. Consistent with these results is the finding that UL84 is found in a complex with IE2 60 and IE2 40 in virus-infected cells. Taken together, these data suggest that an interaction of UL84 with IE2 86 alone may play a role in the expression of UL84, but this interaction is not sufficient for accumulation of normal levels of the UL84 and IE2 86 proteins. Moreover, an interaction of IE2 60 and IE2 40 with UL84 is likely necessary for proper expression of the UL84 protein and may help regulate the overall progress of the viral infection at late times.
MATERIALS AND METHODS
Construction of viruses.
The WT-EGFP, IE2 86ΔSX-EGFP, and Rev-EGFP viruses have been previously described (45). Other versions of the wild-type (WT) and IE2 86ΔSX viruses were previously described in the paper by Sanders et al. (46) or were produced from the AD169 BAC in the same fashion as the enhanced green fluorescent protein (EGFP) viruses, except that EGFP was not present in the recombinant BAC. Briefly, the IE2 86ΔSX C-F virus was constructed from the IE2 86ΔSX BAC. This virus contains the Cre and FLP recombinases, which can be used in inducible cell lines to produce either IE2 86 using the Cre induction or IE2 40 or IE2 60 using the FLP induction. These inductions are temporally regulated in that IE2 86 is expressed at the IE times of the infection (driven by the MIE promoter), while IE2 60 and IE2 40 are expressed at early to late times in the infection (driven by the HCMV 1.2-kb promoter). A WT version of this virus was also constructed (HB5 C-F) for comparison (46). All versions of the IE2 86ΔSX virus have been extensively studied to ensure that each behaves identically to the original IE2 86ΔSX-EGFP virus. The IE2 86Δ88-290 and IE2 86Δ88-135 mutants were created using the QuikChange (Stratagene) protocol as described in the manufacturer's instructions, except that separate PCRs were carried out for the forward and reverse reactions initially (five cycles). The forward and reverse reaction mixtures were then pooled, and the QuikChange PCR was allowed to continue for 18 cycles. The following primers were used to create the mutations within the IE2 86 cDNA in the pSG5-J(BglII-StuI)WT vector: IE2 86Δ88-290 sense, 5′-CACCATCAGGTGACAGCCACCATGGGCGC-3′, and antisense, 5′-GCGCCCATGGTGGCTGTCACCTGATGGTG-3′; IE2 86Δ88-135 sense, 5′-CACCATCAGGTGACGGGGCATCCGCTACTCC-3′, and antisense, 5′-GGAGTAGCGGATGCCCCGTCACCTGATGGTG-3′.
Following mutagenesis, these mutations were placed into the UL122-123 coding region contained in the WT HCMV AD169 BAC pHB5 (gift from M. Messerle) (7) using a counterselection BAC modification kit (Gene Bridges) to create an HCMV BAC containing the desired mutations, as previously described (61).
The IE2 Δ40 mutant has been previously described (60). The M170L mutant virus was constructed as described above. The primers used to create the mutation at the start site of IE2 60 were sense, 5′-CTCCCGCGCCTATCCTCCTGCCCCTCATCA-3′, and antisense, 5′-TGATGAGGGGCAGGAGGATAGGCGCGGGAG-3′.
All mutations were confirmed by restriction digest and sequencing (Eton Biosciences). The recombinant BACs were digested with restriction endonucleases and subjected to field inversion gel electrophoresis to ensure that there were no rearrangements of the genome. All viruses were reconstituted, titers were determined, and viruses were propagated from the BACs as previously described (59, 61).
Cell culture and infections.
Human foreskin fibroblasts (HFFs) were obtained from the University of California San Diego Medical Center and cultured in Earle's minimal essential medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen), 1.5 μg/ml amphotericin B (Invitrogen), 2 mM l-glutamine (Invitrogen), 100 U/ml penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen). Cells were incubated at 37°C with 7% CO2 and allowed to grow 3 days past confluence before infection for G0 synchronization. At the time of infection, the HFFs (passage numbers 12 to 23) were trypsinized, replated, and infected at the MOI indicated. At various times postinfection, cells were washed with phosphate-buffered saline (PBS), trypsinized, and processed accordingly.
Western blotting.
Infected or mock-infected cells were harvested at various times postinfection. Cells were lysed in reducing sample buffer containing 50 mM Tris (pH 6.8), 2% sodium dodecyl sulfate (SDS), 10% glycerol, 5% 2-mercaptoethanol, 50 mM leupeptin, 100 mM pepstatin A, 50 mM sodium fluoride, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM sodium orthovanadate, 5 mM β-glycerolphosphate. Samples then were sonicated to shear DNA and aid cellular lysis and then assayed for protein content. Samples were heated to 95°C for 5 min, and equivalent protein amounts were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose. Membranes were stained with amido black to assess protein loading, blocked in 5% nonfat milk in Tris-buffered saline that included 0.05% Tween 20, and then incubated with the antibodies CH16.0 mouse antibody (MAb; 1:10,000), UL84 MAb (1:2,000 to 1:10,000), UL85 MAb (1:200), UL86 MAb (1:50), monoclonal IE2 86 MAb 8140 (1:1,000), or β-actin MAb Ac-15 (1:5,000 to 1:10,000), washed, and then incubated with horseradish peroxidase-immunoglobulin G (IgG)-coupled anti-mouse antibody (1:5,000 to 1:10,000). CH16.0 was purchased from Virusys. Anti-UL84 antibodies were kind gifts from G. Pari and E. S. Huang. Anti-UL85 and anti-UL86 were kind gifts from B. Britt. Anti-IE2 86 was purchased from Chemicon, and anti-β-actin was purchased from Sigma-Aldrich. Horseradish peroxidase-coupled anti-mouse IgG antibody was obtained from Calbiochem. After incubation and washes, proteins were detected with SuperSignal chemiluminescent substrate (Pierce Biotechnology) according to the manufacturer's instructions.
Northern blot analyses.
HFFs were infected at an MOI of 5 PFU/cell with WT-EGFP, IE2 86ΔSX-EGFP, or rescued IE2 86ΔSX-EGFP (Rev-EGFP) viruses. Approximately 1.5 × 107 cells were harvested at either 24 or 96 h postinfection (p.i.) and mRNA was prepared using the FastTrack 2.0 kit (Invitrogen). Northern blot assays were carried out using the NorthernMax kit (Ambion) according to the manufacturer's instructions. Briefly, 1 μg of mRNA per lane was resolved by agarose gel electrophoresis on a 1% formaldehyde gel and then transferred to a nylon membrane. 32P-labeled probes were synthesized by random priming using the StripEZ DNA kit (Ambion). A 650-bp UL84 probe was generated by restriction digest of the UL84 coding sequence using the vector PTARGET-UL84-HA (gift from G. Pari) with the Eco0901 enzyme. A UL83 probe was generated using the vector pEGFP-pp65 that was digested with BamH1 to remove the entire 1.7-kb UL83 coding region. A β-actin probe was also used as a control for loading. Membranes were hybridized overnight at 42°C and, following washes, were exposed to film for autoradiography.
Immunoprecipitations.
HFFs (3 × 105 to 5 × 105) were used per immunoprecipitation (IP) reaction mixture. Briefly, cells were lysed in radioimmunoprecipitation assay buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) for 30 min and then spun down to remove cellular debris. Lysate was allowed to incubate at 4°C with rotation overnight with protein G-Plus agarose beads (Santa Cruz Biotechnology) coupled with the appropriate antibody. The next day, beads were washed twice in lysis buffer, heated to 95°C for 5 min, and then collected for analysis by SDS-PAGE. An aliquot of the lysate was taken before addition to the beads (“pre”), as well as after incubation with the beads (“post”) to assess protein concentration before and after the IP had been carried out. For the sequential IPs, samples were immunodepleted of all IE1 72 and IE2 86 using the CH16.0 antibody as described above. Following this first IP, the post-binding supernatant was collected and used for a subsequent IP with another antibody that recognizes IE2 86, IE2 60, and IE2 40 (MAb 8140; Chemicon). The second IP therefore contained only IE2 60 and IE2 40 bound to the antibody/bead complex.
Molecular cloning of IE2 86-GST mutants and GST binding assays.
The individual cDNAs of IE2 86 and all of the corresponding deletion mutants provided in the pGEX2TK vector have been previously described (50). Expression and purification of GST fusion proteins were carried out as previously described (24, 50) with the exception that GST fusion protein-bead complexes were resuspended in NETN buffer (20 mM Tris [pH 8.0], 1 mM benzamidine, 1 mM sodium metabisulfate, 0.2 mM phenylmethylsulfonyl fluoride, 0.5% Nonidet P-40, 1 mM dithiothreitol, 1 mM EDTA [pH 8.0], 0.5 mg of bovine serum albumin per ml) containing 200 mM NaCl. After normalized binding of the GST-IE2 proteins, the in vitro-translated protein was added to the mixture and allowed to incubate at room temperature for 1 h with constant rotation. Samples were extensively washed, and bound protein was eluted in reducing sample buffer. Protein was then separated by SDS-PAGE. Following staining (GelCode blue; Pierce Biotechnology) and drying, gels were analyzed for UL84 binding using autoradiography.
In vitro transcription/translation reactions.
In vitro transcription/translation experiments were conducted using the TNT-coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions. The PTARGET-UL84-HA vector (gift from G. Pari) containing the coding region for UL84 under the control of the T7 promoter was used to obtain in vitro-translated protein. Briefly, DNA was incubated with 35S-labeled methionine with the TNT reaction mixture for 90 min. In vitro-translated protein was then either used immediately in the binding reaction or was stored at −80°C for later use.
Drug treatments.
Approximately 1 × 106 cells per time point were infected at an MOI of 5 PFU/cell. Calpeptin (Calbiochem) was added at either 18 h p.i. or 90 h p.i. at a concentration of 50 μM, and cells were incubated for 6 h, or treated with dimethyl sulfoxide (DMSO) for control-treated samples. At 24 and 96 h p.i., cells were washed twice with PBS and then harvested, snap-frozen in liquid nitrogen, and placed at −80°C. Whole-cell lysates were prepared, and protein was analyzed by Western blotting. Lactacystin (Sigma-Aldrich) was added as above to a final concentration of 10 μM.
For the cycloheximide experiments, cells were infected at an MOI of 1 PFU/cell, then at 36 h p.i. were either harvested to assess protein concentration or were washed twice with medium containing 100 μg/ml cycloheximide (Sigma-Aldrich) (or with regular medium for mock-treated samples) and, following washes, were control treated or treated with cycloheximide (100 μg/ml) for 6 h. Cycloheximide was replenished 1 h after treatment. Following drug treatment, cells were harvested for Western blot analysis. Approximately 5 × 105 cells were harvested per time point per virus.
Nuclear/cytoplasmic fractionation.
Following infection at an MOI of 1 to 3 PFU/cell, cells were harvested at 24 h p.i., and the nuclear and cytoplasmic fractions were prepared using the PARIS kit (Ambion) according to the manufacturer's instructions. Briefly, cells were resuspended in ice-cold cell fractionation buffer. Lysate was incubated for 10 min on ice and then centrifuged for 5 min at 500 × g. Supernatant was removed and saved as the cytoplasmic fraction. The pellet following centrifugation was washed in ice-cold cell fractionation buffer and centrifuged again at 500 × g. The pellet was then resuspended in cell disruption buffer and mixed vigorously by pipetting. RNA was then isolated from both fractions using the Nucleospin RNA II kit (Clontech) and was also treated with turbo DNase I (Ambion). RNA was then analyzed by real-time reverse transcription-PCR (RT-PCR) as described previously (60).
Polysome fractionation.
HFFs (3 × 107 cells per sample) were infected at an MOI of 1 PFU/cell with either the HB5 C-F or IE286ΔSX C-F viruses (46), and then at 48 h p.i., cells were incubated with cycloheximide (100 μg/ml; Sigma) in HFF medium for 30 min. Cells were then washed with PBS containing cycloheximide (100 μg/ml), trypsinized, and resuspended in polysome extraction buffer (140 mM KCl, 5 mM MgCl2, 20 mM Tris-HCl, 0.5 mM dithiothreitol, and 0.1 mg/ml cycloheximide) with 1% Triton X-100 on ice. Lysates were incubated for 30 min on ice, and then the lysate was centrifuged for 10 min at 10,000 × g at 4°C. The supernatant was then either control treated or treated with EDTA (50 mM) for 20 min on ice. Sucrose gradients (10 to 50%) were poured using a Hoefer SB15 gradient maker (Amersham Biosciences). Supernatants were then layered gently onto the gradient and centrifuged in a Beckman L7 ultracentrifuge in an SW-55 swing bucket rotor at 39,000 rpm for 90 min at 4°C. Samples were then fractioned from the heaviest to the lightest fractions (16 fractions total) and dripped into SDS to achieve a final concentration of 1%. Absorbance was measured with a UV spectrophotometer throughout the fractionation. Samples were treated with proteinase K (0.2 mg/ml) and incubated at 37°C for 90 min and then extracted by phenol-chloroform treatment, followed by an ethanol precipitation at −20°C overnight. Samples were spun down the next day at 14,000 rpm for 30 min at 4°C, washed with 70% ethanol, and then spun again for 15 min. The supernatant was removed and pellets were allowed to dry. RNA was then resuspended in Tris-EDTA. The appropriate fractions were pooled according to the polysome (absorbance) profile (six pooled fractions total), and then each sample was treated with DNase by using the Turbo DNase I kit according to the manufacturer's instructions (Ambion). RNA was then analyzed by real-time RT-PCR as described previously (60). A cellular housekeeping gene, glyceraldehyde-6-phosphate dehydrogenase (G6PD) was also assessed. The primers and probe for UL84, IE2 86, and G6PD have been previously described (60). The primers and probe for the UL44 gene were FWD, 5′-GCTTTCGCGCACAATGTCT-3′, and REV, 5′-GCCCGATTTCAATATGGAGTTC-3′, and the probe sequence was 5′-5,6-carboxyfluorescein-CGTGCACGCAGGCCGAGC-3BHQ1-3′.
The quantified RNA from the real-time RT-PCR was then normalized to the total amount of RNA collected in each respective fraction and then normalized again so that the value of the RNA collected in fraction 6 from the WT virus-infected cells was equal to 1, to give relative quantities of RNA in each fraction.
RESULTS
UL84 protein expression is reduced in IE2 86ΔSX virus-infected cells.
The IE2 86 gene is encoded by the UL122 transcript and is alternatively spliced to include exons 1, 2, 3, and 5, while IE1 72 includes exons 1, 2, 3 and 4. Exon 5 also encodes both the IE2 60 and the IE2 40 proteins at late times postinfection (40, 41, 52, 60). These proteins share the C-terminal portion of IE2 86, but they do not have any sequence in common with IE1 72 (Fig. 1A). Figure 1B represents a schematic of the deletions within IE2 86 that were created and subsequently used throughout the course of our studies. The WT-, IE2 86ΔSX-, and Rev-EGFP viruses have been previously characterized (45). A version of the IE2 86ΔSX virus lacking EGFP expression (IE2 86ΔSX) was also constructed to ensure that the defects seen in the mutant virus infection were not due to the addition of EGFP to IE2 86. Another version of IE2 86ΔSX which encodes the Cre and FLP recombinases (IE2 86ΔSX C-F) was also used in these studies and has been previously described (46). This virus can be grown on complementary cell lines expressing IE2 86 and IE2 40 in order to obtain higher-titer virus, thus allowing analysis of this virus in large-scale experiments. The presence of the Cre and FLP recombinases does not affect the growth of the virus on HFFs, however, and only allows generation of higher-titer virus following preparation on complementing cell lines (46). Two other recombinant viruses, IE2 86Δ88-290 and IE2 86Δ88-135, were constructed using the AD169 pHB5 BAC that does not contain EGFP (Fig. 1B) to further assess the role of the N-terminal portion of exon 5 of IE2 86. In addition, mutants lacking expression of either IE2 60 (M170L) or IE2 40 (IE2 Δ40) were analyzed to further define the functional roles of IE2 60 and IE2 40. These viruses contain mutations to disrupt either the putative TATAA box for IE2 Δ40 or the initiator methionine for the M170L virus (Fig. 1B) (60).
FIG. 1.
Schematic representation of the UL122-123 transcripts and deletions created within the IE2 86 coding regions of mutant viruses. (A) UL122-123 transcripts and representation of the MIE genes, IE1 72 and IE2 86, in the wild-type HCMV coding region. Exons 1, 2, 3, and 4 specify IE1 72 mRNA, while exons 1, 2, 3, and 5 specify IE2 86 mRNA. Translation begins at exon 2 for both IE1 72 and IE2 86, while translation of IE2 60 and IE2 40 begins at aa 170 and 242, respectively. (B) Schematic of the mutant viruses used in these analyses to identify the dysregulation of UL84, as well as the important binding domain within IE2 86 for UL84. The WT, IE2 86ΔSX (ΔSX), and a revertant form of the IE2 86ΔSX (REV) are represented in the schematic. Other deletion mutants created, including Δ88-290, Δ88-135, IE2 Δ40, and M170L, are also shown. Diagrams are not to scale.
Previous characterization of the IE2 86ΔSX mutant virus revealed that the kinetics of the infection are slower and the virus grows to reduced titers (45). The mutant virus also expressed markedly lower levels of IE2 86, as well as two early-late proteins, UL83 and UL84. IE1 72 and early gene expression remain normal (45, 60). This mutant is missing the coding region of IE2 86 between aa 136 and 290 of exon 5, which includes the initiator methionines for two smaller IE2 proteins (IE2 60 and IE2 40) that appear later in infection, the sumoylation sites (aa 175 and 180), and the domain that interacts with SUMO-1 (aa 200 to 208) (2, 21, 23, 26, 27, 41, 45, 60). Furthermore, expression of IE2 86 and IE2 40 from inducible cell lines allows recovery of some of these phenotypes, especially at the later stages of infection (46).
Since IE2 86 has an effect on the regulation of other early-late proteins throughout the infection but is known to interact with only one other viral protein, UL84, we compared the expression of UL84 during the IE2 86ΔSX-EGFP virus infection to that during the WT- and IE2 86ΔSX Rev-EGFP virus infections. To assess the levels of UL84 protein, HFFs were infected at an MOI of 5 PFU/cell and then harvested 12 to 96 h p.i. Lysates were prepared and then analyzed by Western blotting with a monoclonal antibody specific for UL84. Here, we show that throughout the entire infection, there is a significant reduction in the production of the UL84 protein in HFFs infected with the IE2 86ΔSX-EGFP virus, with the effect more pronounced at late times postinfection (Fig. 2A). In the WT-EGFP infection, the level of UL84 steadily increases between 24 and 96 h and can be detected as early as 12 h p.i., whereas in the IE2 86ΔSX-EGFP infection, the protein shows a severe decrease in expression (Fig. 2A). The Rev-EGFP virus fully recovers expression of UL84, indicating that this loss of UL84 expression is due to the deletion within IE2 86 and not due to any other mutation created during mutagenesis of the BAC (Fig. 2A). Furthermore, when assessed by immunofluorescence assays, UL84 in the IE2 86ΔSX infection is still present in the replication centers, where IE2 86 is known to accumulate at late times postinfection, indicating that UL84 may also be functioning in replication processes in infection with the mutant (data not shown). All of the above data were confirmed with the IE2 86ΔSX and IE2 86ΔSX C-F viruses to rule out any possibility that the virus was impaired due to the presence of EGFP on IE2 86 (data not shown). In order to ensure that the WT-, IE2 86ΔSX-, and Rev-EGFP infections were properly matched, other viral proteins that are known to be expressed to normal levels in the IE2 86ΔSX mutant virus-infected cells were assessed and shown to be comparable (data not shown).
FIG. 2.
Analysis of UL84, UL85, UL86, and UL83 RNA and protein at late times postinfection. (A) G0-synchronized HFFs were infected at an MOI of 5 PFU/cell with the WT-EGFP (WT), IE2 86ΔSX-EGFP (SX), or Rev-EGFP (Rev) viruses or were mock infected (M) and harvested at the indicated times postinfection (12 to 96 h). The mock lane was added to indicate that the UL84 antibody was specific for the viral protein. An equal amount of protein from the cell lysates was analyzed by Western blot analyses. β-Actin was used as a loading control. (B) Analysis of the UL84, UL85, and UL86 transcripts in the IE2 86ΔSX-EGFP infection. HFFs were infected or mock infected (M) at an MOI of 5 PFU/cell with the WT-EGFP, IE2 86ΔSX-EGFP, or Rev-EGFP viruses and then harvested at either 24 or 96 h p.i. mRNA was oligo(dT) selected, resolved by agarose gel electrophoresis, and then transferred to a nitrocellulose membrane. 32P-labeled probes were synthesized that recognize UL84, UL85, and UL86 or β-actin. Following hybridization of the probe, membranes were exposed to film for autoradiography. The three transcripts occurring at 2.1, 3.0, and 9.0 kb encode the UL84, UL85, and UL86 RNA transcripts, respectively. Cellular β-actin mRNA served as a loading control. A representative example of three experiments is shown. (C) The same mRNA as for panel B was analyzed by Northern blotting for expression of the UL83 mRNA transcript at 96 h p.i., when UL83 is most abundant. Both the WT-EGFP and IE2 86ΔSX-EGFP virus-infected cells are shown. The UL84 and UL83 protein from this infection is also shown for comparison at 96 h p.i. for the two viruses. (D) Western blot assay results for the UL84, UL85, and UL86 proteins at 72, 96, and 120 h p.i. are shown for the WT-EGFP and IE2 86ΔSX-EGFP viruses. Actin served as a loading control.
Transcription of UL84, UL85, and UL86 is not affected in IE2 86ΔSX-EGFP-infected cells.
Our lab has recently shown that although protein expression is debilitated at late times postinfection, UL84 RNA remains unaltered as analyzed by quantitative real-time RT-PCR. Furthermore, a mutant that does not express either IE2 60 or IE2 40 (IE2 Δ40+60) also shows no loss of the UL84 transcript, even though protein levels are significantly decreased, similar to that of the IE2 86ΔSX infection by 96 h p.i. (60). Since overlapping transcripts are encoded by the UL84 coding region at late times (20, 43), we used Northern blot analyses to assess each of the transcript levels at both early and late times. Briefly, HFFs were infected at an MOI of 5 PFU/cell and then harvested at either 24 or 96 h p.i. mRNA was isolated and then separated by denaturing agarose gel electrophoresis. Levels of mRNA were then analyzed using a probe that recognizes UL84, UL85, and UL86, as well as a probe that recognizes β-actin (as a loading control). Figure 2B shows that at 24 h p.i., a 2.1-kb transcript, which encodes UL84, was expressed in WT-, IE2 86ΔSX-, and Rev-EGFP-infected cells. At 96 h p.i., the 2.1-kb transcript increased in abundance, and transcripts of approximately 3.0 and 9.0 kb, encoding UL85 and UL86, respectively, were also expressed in cells infected with all three viruses. Low levels of other high-molecular-weight transcripts, which likely are read-through RNAs, also appear at late times. At both 24 and 96 h p.i., the levels of each of the transcripts were not significantly affected in cells infected with the WT-, IE2 86ΔSX-, and Rev-EGFP viruses. There was a slight decrease in the levels of the UL84 mRNA, but this difference was minimal and likely does not account for the dramatic loss of UL84 at the protein level. These data confirmed the previous quantitative real-time RT-PCR data (60), providing additional evidence that the reduced levels of UL84 protein in the IE2 86ΔSX infection were not due to reduced levels of RNA. Real-time RT-PCR assays of the samples used for the Northern analyses were also performed in parallel to confirm these results, and no differences were seen between any of the three viruses at either 24 or 96 h p.i. (data not shown).
To confirm that a previously characterized decrease in the UL83 mRNA was occurring, samples from the same infection as in Fig. 2B were analyzed by Northern analyses. At 96 h p.i., the IE2 86ΔSX-EGFP group exhibited a severe defect in the expression of UL83 mRNA (Fig. 2C), which is consistent with previous reports (45). The UL83 protein exhibited a comparable decrease to that of UL84 at this time postinfection (Fig. 2C), further emphasizing the differences in the type of regulation of these two transcripts.
We also analyzed the protein expression levels of UL85 and UL86 in these studies to determine if they were being regulated in a similar fashion as UL84. Cells were infected at an MOI of 5, and then, at late times postinfection, cells were analyzed by Western blotting for the expression of these proteins. At 72 h p.i., the UL84, UL85, and UL86 proteins exhibited similar decreases in the IE2 86ΔSX virus-infected cells compared to the WT virus-infected cells (Fig. 2D). However, by 96 and 120 h p.i., the UL85 and UL86 protein expression levels had recovered, while the UL84 protein level was still lower than that in the WT virus-infected cells. The decrease in the late UL85 and UL86 proteins is consistent with the growth kinetics of the mutant virus and confirms previous observations that there is a lag in the expression of many late genes (45). These data indicate that UL84 is also regulated in a different fashion than UL85 and UL86.
UL84 RNA in the IE2 86ΔSX virus-infected cells does not show a defect in nuclear export compared to WT.
Given that our analyses of the UL84 RNA revealed no dramatic differences in transcript expression, we next determined whether the RNA was being exported to the cytoplasm appropriately. Briefly, cells were infected at an MOI of 1 to 3 PFU/cell with either the WT or IE2 86ΔSX viruses and harvested at 24 h p.i., and then the nuclear and cytoplasmic fractions were separated by centrifugation. RNA was prepared from each fraction and then analyzed by quantitative real-time RT-PCR. Interestingly, these analyses showed that in the IE2 86ΔSX mutant-infected cells, the RNA was distributed between the two fractions in a comparable fashion as the WT virus-infected cells (Fig. 3). At this time postinfection, the distribution of the UL84 RNA was more cytoplasmic than nuclear in cells infected with both viruses. Distribution of the G6PD RNA and protein was assessed in order to confirm that the fractionation had occurred appropriately (data not shown). These data indicated that the RNA is being appropriately exported out of the nucleus in order to be translated.
FIG. 3.
Nuclear-cytoplasmic distribution of UL84 RNA in WT and IE2 86ΔSX virus infection. At 24 h p.i., cells were harvested and the nuclear and cytoplasmic fractions were separated by centrifugation. RNA was prepared from each fraction and quantitated by quantitative real-time RT-PCR. The percent nuclear versus cytoplasmic was calculated for each sample set. The nuclear fraction (Nuc) and cytoplasmic fraction (Cyto) are shown for both the WT and IE2 86ΔSX (ΔSX) virus-infected cells. Each fraction was normalized to a cellular housekeeping gene (G6PD) to account for the amount of RNA in each reaction mixture. Averages of three experiments are shown, with error bars representing the margins of error between experiments.
There is no defect in the loading or distribution of UL84 or IE2 86 mRNA on polysomes in IE2 86ΔSX virus-infected cells.
To determine whether the UL84 RNA was being appropriately loaded onto polysomes in the IE2 86ΔSX infections, we next assayed the distribution of the RNA in a polysome fractionation assay. Due to the large number of cells necessary for these analyses, we used a stock of both the WT and IE2 86ΔSX viruses that had been prepared on complementary cell lines that expressed both IE2 86 and IE2 40. The characterization of these recombinant viruses (termed HB5 C-F and IE2 86ΔSX C-F) and the complementary cell lines were described in a recent paper by Sanders et al. (46). These viruses have the same growth properties as the WT and IE2 86ΔSX viruses lacking the Cre and FLP recombinases (46). Cells were infected at an MOI of 1 PFU/cell with either the WT C-F or the IE2 86ΔSX C-F viruses and then harvested at 48 h p.i. Immediately following harvest, the cells were lysed and the cytoplasmic fraction was separated from the nuclear fraction. This cytoplasmic fraction was either control treated or treated with EDTA, separated on a sucrose gradient by ultracentrifugation, and then fractioned from the heaviest to the lightest fractions. Treatment of polysome extracts with EDTA has been shown to promote disassembly of free ribosomes into their 40S and 60S subunits, causing a release of the bound mRNA. This results in a shift of the mRNAs into the lighter fractions. The mRNAs bound to large ribonucleoproteins (RNPs), however, should be unaffected by this treatment (6), allowing confirmation that the mRNA measured in the samples lacking EDTA are actually loaded onto polysomes. Absorbance of the polysomes (A254) was measured throughout collection (Fig. 4A). RNA was then prepared from each fraction and analyzed by real-time RT-PCR. At this point in the infection, only the UL84 RNA is present, and thus the 3′-coterminal RNAs expressing UL85 and UL86 do not complicate the real-time RT-PCR analyses (data not shown).
FIG. 4.
Polysome distribution of UL84, IE2 86, UL44, and G6PD RNAs in the WT and IE2 86ΔSX (ΔSX) infections. (A) Absorbance measured during fractionation from the heaviest to the lightest fractions. Fractions were pooled into six separate samples (fractions 1 to 6) and are represented in the figure. Absorbance (A254) was measured throughout the collection. The polysomal fractions are represented in fractions 1, 2, and 3. The monosomal fractions are represented in fractions 4 and 5. Fraction 6 represents the free RNP RNA. (B) The distribution of the RNA and quantification of each fraction is shown for UL84, IE2 86, UL44, and G6PD. Samples were either treated with EDTA (+EDTA) or mock treated (-EDTA). Quantified RNA was measured by real-time RT-PCR and then normalized to the total amount of RNA collected in that fraction. Relative amounts of RNA are shown compared to the total WT RNA present in fraction 6 for each gene, which was set to a value of 1.
Figure 4A shows a representative polysome distribution following fractionation. In the samples without EDTA treatment, the polysome fractions are separated into fractions 1, 2, and 3. The monosome fractions are represented by fractions 4 and 5. Fraction 6 represents the lightest fraction, containing free RNP RNA. In the EDTA-treated samples, the polysome peaks are no longer visible, indicating that the EDTA treatment was efficient in disrupting the polysomes before fractionation. All quantities were normalized to the WT fraction 6 (value equal to 1) to compare the relative amounts of RNA from the two viruses present in each fraction. A representative of three different experiments is shown in Fig. 4A and B.
At 48 h p.i., the distribution of the UL84 RNA was measured for both the WT and IE2 86ΔSX virus-infected cells (Fig. 4B). Interestingly, no major differences were found between the two viruses with regard to the loading or distribution of the UL84 RNA on polysomes. Fractions 2 and 3 (without EDTA) contained the most RNA, were comparable between the two infections, and showed no major shift in the distribution of the RNA. Treating the samples with EDTA shifted the RNA most predominantly to fractions 4 and 5, indicating that the RNA measured in the untreated samples was in fact loaded onto the polysomes. These data indicated that the UL84 RNA is appropriately loaded onto the polysomes in the IE2 86ΔSX virus-infected cells compared to the WT virus-infected cells.
The distributions of the IE2 86, UL44, and G6PD RNAs were also analyzed (Fig. 4B). It is known that the levels of the RNA and protein for UL44 (early protein) remain unaffected in IE2 86ΔSX virus-infected cells (reference 45 and unpublished results). IE2 86 RNA is unaffected in these virus-infected cells, but the IE2 86 protein exhibits decreased expression (45). In each case, the distribution of each of the RNAs was comparable between the two virus-infected cells. Small differences in the amount of RNA in each fraction were noted between experiments, but the differences were not more than twofold. The slight shift in the distribution of the IE2 86 RNA on the polysomes in the IE2 86ΔSX-infected cells is likely due to the smaller size of the transcript as a result of the deletion in exon 5. This result was consistent between experiments. In the untreated samples, the IE2 86 RNA was present most predominantly in the second fraction, as was the UL44 RNA, and each fraction was comparable between the two viruses in both cases. The distribution of G6PD (cellular housekeeping gene) was also assessed and showed no difference in distribution. All samples treated with EDTA also showed that the RNA shifted into the lighter fractions (4 and 5), indicating that the measurements of RNA in the untreated samples were loaded onto the polysomes and that treatment with EDTA shifted the RNA into the monosomal fractions as expected.
Loss of UL84 expression is not due to increased proteasome- or calpain-dependent degradation, and the UL84 protein in the IE2 86ΔSX virus-infected cells is stable.
To determine whether UL84 is being degraded in a proteasome-dependent manner in IE2 86ΔSX infection, a proteasome inhibitor, lactacystin (10 μM), was added at either 18 or 90 h p.i. and then cells were harvested 6 h later at either 24 or 96 h p.i., respectively. The addition of this inhibitor did not result in increased levels of UL84 expression at either early or late times of infection (Fig. 5A). Furthermore, addition of the calpain inhibitor calpeptin (50 μM), which is known to inhibit the Ca2+-stimulated cleavage of p35 to p25 by calpain (57), during the same period was unable to rescue the levels of UL84 (Fig. 5B). As seen at both 24 and 96 h p.i. in the lanes treated with either drug, the levels of UL84 are still much lower than that of the WT-EGFP infection and remain comparable to that of the untreated samples (Fig. 5A and B). These results demonstrate that the low levels of UL84 are not due to enhanced degradation of the protein by either the proteasome or calpain.
FIG. 5.
Analysis of proteasome- and calpain-dependent degradation in IE2 86ΔSX- and WT-infected cells and assessment of the stability of UL84 and IE2 86 proteins in these infections. (A) HFFs were infected at an MOI of 5 PFU/cell with either the WT-EGFP or IE2 86ΔSX-EGFP (SX). Lactacystin (10 μM; Lac, +), or DMSO for mock treatment (-), was added at either 18 or 90 h p.i., and the cells were incubated for 6 h. Cells were harvested at 24 and 96 h p.i. and then analyzed for UL84 protein expression by Western blot analyses. (B) HFFs were infected as above and then treated with calpeptin (50 μM; Cal, +) for 6 h or mock treated with DMSO (-). Cells were harvested and then lysates were analyzed by Western blotting for UL84 protein expression. (C) Cells were infected at an MOI of 1 PFU/cell with the WT or IE2 86ΔSX (SX) and at 36 h p.i. were either harvested to assess protein concentration before drug treatment or were treated with cycloheximide (100 μg/ml; CHX, +) or DMSO (−) as a control for 6 h. Following treatment, cells were harvested at 42 h p.i. to analyze the loss of UL84 and IE2 86 during that time. Two exposures of the IE2 86 blot are shown in order to visualize the small amount of IE2 86 in the IE2 86ΔSX infection (*). β-Actin was analyzed in all of the above experiments in order to assess protein loading.
To assess whether the translation or stability of UL84 was impaired in the IE2 86ΔSX mutant virus, protein levels were analyzed following treatment with cycloheximide. HFFs were infected at an MOI of 1 PFU/cell with the WT and IE2 86ΔSX viruses, and at 36 h p.i. cells were either harvested to analyze the amount of protein present prior to treatment or were control treated or treated with 100 μg/ml cycloheximide. The drug treatment was stopped 6 h later (42 h p.i.), and following washes with PBS, cells were harvested immediately. Our results showed that there was a decrease in the amount of UL84 present in the WT virus-infected cells immediately following drug treatment compared to the untreated sample; however, a portion of the protein still remained (Fig. 5C). There was also a comparable decrease in the amount of UL84 present in the IE2 86ΔSX infection following treatment, indicating that the protein was not turning over more rapidly than in the WT infection (Fig. 5C). These data reveal that the stability of UL84 that is measured by the assay remains the same compared to the WT infection. However, it has not been determined whether the stability of free UL84 is the same as UL84 complexed with other proteins, such as IE2 86.
Since we had previously observed that in IE2 86ΔSX-infected cells IE2 86 levels were severely diminished but RNA levels remained unaffected (45), we also assessed the stability of the IE2 86 protein. IE2 86 appears to be slightly more stable than UL84, but the difference between the cycloheximide-treated and untreated samples was comparable for both the WT virus and the mutant virus (Fig. 5C). As expected, the amount of IE2 86 remains much lower in the IE2 86ΔSX virus-infected cells than that in the WT virus-infected cells. A longer exposure of IE2 86ΔSX is shown in order to more readily assess the levels of IE2 86 present in the IE2 86ΔSX infection.
IE2 86, IE2 60, and IE2 40 are able to interact with UL84 throughout the infection.
The interaction between UL84 and IE2 86 has been shown to be important for inhibiting IE2 86-mediated transactivation of the early and late genes as judged in cotransfection assays (12, 17, 62). However, this interaction has not been extensively characterized in the context of HCMV infection. To further illustrate the contribution of IE2 86, IE2 60, and IE2 40 to UL84 expression, we assessed whether these proteins had the ability to interact with UL84 throughout the entire infection, which might further allude to their involvement in UL84 function. Furthermore, we wanted to address whether IE2 86ΔSX could interact with UL84. We first performed IPs that selectively immunodepleted the lysates of full-length IE2 86 and IE1 72 (using the CH16.0 MAb). This antibody would not immunoprecipitate IE2 40 or IE2 60 unless they had formed heterodimers with IE2 86. In addition, since it is known that UL84 does not interact with IE1 72, any interaction seen in the IP is due to the interaction between IE2 86 and UL84 (17). Subsequent IPs with the monoclonal IE2 86 antibody (mIE2 86) were then used to assess whether IE2 60 and IE2 40 were in complex with UL84 in the absence of the full-length protein (Fig. 6A, schematic).
FIG. 6.
UL84 interacts with IE2 60 and IE2 40 without the contribution of IE2 86. (A) Schematic representation of the experiment, indicating that the post-CH16.0 IP product (step 1) is the same sample as that used for the pre-IP in the mIE2 86 IPs (step 2). The corresponding post and pre samples are denoted with a * in all subsequent parts of the figure. (B) IPs were carried out exactly as described for panel A, and cells were harvested 24 to 96 h p.i. The pre- (lanes 1 to 3) and post- (lanes 7 to 9) IP samples equal approximately 10% of the IP (lanes 4 to 6). (C) The post-binding supernatant (in panel A, this corresponds to Post*, lanes 7 to 9) was subjected to a subsequent IP with an antibody that recognizes IE2 86, IE2 60, and IE2 40 (mIE2 86) to determine if UL84 interacts with IE2 60 and IE2 40 in combination, without the presence of IE2 86. UL84 was analyzed again by Western blotting (lanes 1 to 9). PRE* indicates that this corresponds to the POST* supernatant of the first IP in panel B. The second set of IPs (mIE2 86 IPs) were also analyzed by Western blotting for the presence IE2 86, IE2 60, and IE2 40 using the mIE2 86 antibody (lanes 10 to 18). Only the samples at 72 and 96 h p.i. of the Western blot assay with the mIE2 86 antibody are shown, as IE2 86, IE2 60, and IE2 40 were most abundant at these times. A representative of two experiments is shown.
Briefly, HFFs were infected at an MOI of 1 PFU/cell and then harvested at various times postinfection. Cells were first immunoprecipitated with the CH16.0 antibody that recognizes only full-length IE2 86 and IE1 72 (both the IE2 86ΔSX and WT versions), but not IE2 60 or IE2 40 (Fig. 6A, step 1). However, as noted above, IE2 60 and IE2 40, which are present as a heterodimer with IE2 86, should be immunoprecipitated in step 1, as the dimerization domain is common to all three proteins (1, 10, 15). Following incubation, the bound proteins were eluted from the agarose beads and analyzed by Western blotting for the presence of the UL84 protein. A significant amount of the UL84 in cells infected with WT-, IE2 86ΔSX-, and Rev-EGFP virus was in a complex with IE2 86, as judged by the protein present in the IP lanes (Fig. 6B, lanes 4 to 6). However, a substantial amount of the UL84 protein was not immunoprecipitated in the WT- and Rev-EGFP infections at late times and was present in the post-IP samples (Fig. 6B, lanes 7 to 9). These blots were also probed with the CH16.0 antibody to confirm that IE2 86 had been immunodepleted during the IP (data not shown). Further confirming this, Western blot analysis with the mIE2 86 antibody showed that the full-length protein was efficiently removed from the post-IP supernatant (see below). The pre- and post-binding lanes correspond to approximately 10% of the sample used for the IP (Fig. 6B, lanes 1 to 3 and 7 to 9, respectively). These data show that IE2 86 and IE2 86ΔSX are interacting with UL84 throughout the entire infection but that there is some UL84 that is not complexed with IE2 86 in the WT infection.
To further determine whether the UL84 that was not in the complex with IE2 86 was bound to the IE2 60 and IE2 40 proteins, the post-IP (Fig. 6B, post, lanes 7 to 9) samples from the previous experiment were subjected to an IP with a monoclonal IE2 86 antibody (mIE2 86) that recognizes the C-terminal region included in the full-length IE2 86, IE2 60, and IE2 40 (indicated as step 2 in the schematic shown in Fig. 6A). We confirmed that IE2 60 and IE2 40, but not IE2 86, were present in this IP by Western blotting with the monoclonal IE2 86 antibody (Fig. 6C, lanes 10 to 18), as well as the CH16.0 antibody (data not shown). Western blot analysis with the antibody to UL84 showed that the protein is very abundant in the IP lanes corresponding to the WT and Rev-EGFP infections, but not in the IE2 86ΔSX-EGFP infection (Fig. 6C, lanes 4 to 6). This was as expected, given that IE2 60 and IE2 40 are not produced in the IE2 86ΔSX infection. Furthermore, since no UL84 protein could be detected in the binding reaction product for the IE2 86ΔSX-EGFP infection (Fig. 6C, lane 5), these results also indicate that all full-length IE2 86 was removed in the first binding experiment with the CH16.0 antibody and that any interaction seen was due to the IE2 60 and IE2 40 proteins. Representative Western blotting results with the mIE2 86 antibody are shown at both 72 and 96 h p.i. in Fig. 6C, lanes 10 to 18.
To address whether both IE2 60 and IE2 40 are necessary for a proper interaction or whether each of the proteins interacts with UL84 individually, the same sequential IP reactions were carried out using recombinant viruses that express either the IE2 60 or the IE2 40 protein, but not both. We have recently characterized the viruses that lack expression of either IE2 40 or IE2 40 and IE2 60 combined. Without these two proteins, the levels of UL84 are significantly decreased at 96 h p.i., similar to the decrease in UL84 levels in IE2 86ΔSX virus-infected cells. In addition, the loss of IE2 40 alone results in some loss of UL84 (60).
Since we did not have a mutant that exhibited a total loss of IE2 60, we constructed a new mutant virus that lacked any expression of IE2 60 (Fig. 7A). This mutant, termed M170L, has a mutated initiator methionine (M170) that has been changed to a leucine, thus preventing the expression of IE2 60 (Fig. 7A and B). IE2 40 is still expressed, since its initiator methionine is the next methionine in the sequence. When assessed by Western blotting, expression of UL84, IE2 86, and IE2 40 in the M170L mutant virus-infected cells remains comparable to that in the WT virus-infected cells (Fig. 7B). Furthermore, a revertant virus of this mutant fully restored the expression of IE2 60, indicating that the mutant virus created did not have any other defects or mutations (Fig. 7B, M170L R). These data indicate that IE2 60 might play a less important role than IE2 40 with regard to UL84 expression. However, we have not ruled out that IE2 60 has some ability to aid in the expression of UL84 when IE2 40 is not present.
FIG. 7.
Sequence and characterization of the M170L mutant. (A) A portion of the IE2 86 coding region is shown. The mutated initiator methionine (M170, originally ATG) for IE2 60 was changed to a leucine (CTC) to create this mutant virus (M170L). (B) Expression of IE2 86, IE2 60, and IE2 40 in the M170L mutant compared to the WT and the M170L revertant (M170L R) at 24 to 96 h p.i. in HFFs infected at an MOI of 3 PFU/cell. A mock-infected sample (M) is also shown. Western blot analyses were used to examine expression of UL84 as well. Cellular β-actin was used as a control for protein loading.
Since the above IP studies showed that UL84 was able to interact with IE2 60 and IE2 40 in the absence of full-length IE2 86, we next wanted to identify the individual contribution of IE2 60 and IE2 40 to the interaction with UL84. Sequential IPs were conducted as described previously. Cells were infected with either the WT, IE2 Δ40, or M170L mutant viruses at an MOI of 1 PFU/cell and then harvested at 96 h p.i., when IE2 60 and IE2 40 are most abundant. Cells were lysed, and IPs were carried out using the CH16.0 antibody. As described previously, the post-IP supernatant was then subjected to a subsequent IP with the C-terminal mIE2 86 antibody, and the presence of UL84, IE2 86, IE2 60, and IE2 40 was analyzed by Western blotting. As expected, UL84 is present in all of the IP lanes corresponding to the CH16.0 IP (Fig. 8, lanes 2, 5, and 8). Interestingly, UL84 is able to interact with both IE2 60 and IE2 40 alone, as judged by the IP lanes in the mIE2 86 IP corresponding to either IE2 Δ40 or the M170L viruses (Fig. 8, lanes 14 and 17). The majority of UL84 that is not in complex with full-length IE2 86 appears to be in complex with either IE2 60 or IE2 40, as indicated by the fact that very little UL84 is detected in the supernatant following the subsequent mIE2 86 IPs (Fig. 8, lanes 12, 15, and 18); UL84 is only observed upon long exposure of the Western blots (data not shown). This was also seen in the previous IPs (Fig. 6B). It was noteworthy that there was a direct relationship between the amount of UL84 and IE2 proteins in the IP lanes for all viruses. This is most apparent in comparing lane 14 with lanes 11 and 17. These results suggest that UL84 must be in a complex with the various IE2 proteins for proper UL84 function and that these interactions may play a key role in the regulatory functions of each of the IE2 proteins. Our finding that IE2 60 and IE2 40 were present in the IP lane corresponding to the CH16.0 IP with full-length IE2 86 and UL84 also confirms that IE2 60 and IE2 40 can interact with IE2 86 (Fig. 8, lanes 2, 5, and 8). Immunodepletion of the full-length IE2 86 protein was again confirmed by Western blotting with both the monoclonal IE2 86 antibody at 96 h p.i. (Fig. 8, lanes 10 to 18) and also with the CH16.0 antibody (data not shown).
FIG. 8.
IE2 60 and IE2 40 interact individually with UL84 in the absence of IE2 86. As in Fig. 6, sequential IPs were performed, with the exception of the mutant viruses used, which were the IE2 Δ40 and M170L mutants, and the time points examined (only 96 h p.i. is shown here). At 96 h p.i., cells were harvested and the first IP (lanes 2, 5, and 8) was carried out using the CH16.0 antibody. Western blot analyses were performed to assess the interactions occurring and expression patterns of UL84, IE2 86, IE2 60, and IE2 40. The supernatants following IP (POST*, lanes 3, 6, and 9) were used for the next immunoprecipitation (mIE2 86 IPs, lanes 10 to 18). POST* (lanes 3, 6, and 9) and PRE* (lanes 10, 13, and 16) indicate that the post-binding supernatant for the first IP was used for the pre-binding sample for the second IP. The pre and post samples represent approximately 10% of the IP. The interaction of UL84 and IE2 86 were assessed in lanes 2, 5, and 8, while the interactions between UL84, IE2 60, and IE2 40 were assessed in lanes 11, 14, and 17. Lanes 14 and 17 represent the individual interactions with UL84 and IE2 60 or IE2 40, respectively. The arrow indicates the sequential nature of the IPs from the initial CH16.0 IPs to the mIE2 86 IPs.
A C-terminal portion of IE2 86 contributes to the interaction between IE2 86 and UL84.
The above results with the IE2 40 protein showed that a domain C-terminal to aa 242 of IE2 86 was sufficient to interact with UL84. The finding that IE2 86ΔSX, which lacks aa 136 to 290, was also able to form a complex with UL84 suggested that either the region sufficient for an interaction with UL84 resided C-terminal to aa 290 or that a second domain in exon 5 between aa 85 and 135 could contribute to the interaction with UL84 in the absence of aa 136 to 290.
To address these possibilities, two additional mutant viruses were tested for this interaction: one containing a deletion of aa 88 to 135 (Δ88-135), and the second with a deletion of aa 88 to 290 (Δ88-290). Cells were infected at an MOI of 3 PFU/cell and then harvested at 72 and 96 h p.i., when UL84 is most abundant. The lysates were incubated with beads conjugated to an antibody that recognizes the amino-terminal region of IE2 86 and IE1 72 (CH16.0). Following IP, the proportion of UL84 pulled down by the assay was analyzed by Western blotting with a monoclonal antibody specific for UL84 (Fig. 9A). Deletion of either of these regions from IE2 86 still did not result in a loss of interaction with UL84, indicating that the region between aa 88 and 290 was not required for this complex to form (Fig. 9A). It was notable, however, that significantly less UL84 accumulated in cells infected with the IE2 86Δ88-290 virus. Since less UL84 was present in the cells infected with this mutant, the amount of UL84 present in the pre-IP lane was not visible at this exposure of the blot. These results, coupled with the newly identified interaction between UL84 and IE2 60 and IE2 40, differ from those previously reported that identified the first 290 aa as the only region that binds UL84 (12). However, these experiments were conducted as cotransfection assays, which may behave differently than our experiments conducted in virus-infected cells. It is possible that both regions of IE2 86 have the ability to contribute to the interactions we observed.
FIG. 9.
Identification of a C-terminal domain involved in the interaction between IE2 86 and UL84. (A) HFFs were infected at an MOI of 3 PFU/cell with the WT, Δ88-135, or Δ88-290 viruses and harvested at either 72 or 96 h p.i. Lysates were prepared, a small aliquot was removed (PRE), and the remainder was incubated with beads conjugated with the CH16.0 antibody (which recognizes IE1 72 and IE2 86). Following incubation, an aliquot of the supernatant was removed (POST). The beads were then washed and IP products were eluted from the beads (IP). The PRE and POST lanes represent 10% of the protein loaded in the IP lanes. UL84 was analyzed by Western blotting as described in Materials and Methods. (B) Schematic of the IE2 86 coding region and all of the mutated forms assayed. Amino acid deletions: SX, Δ136-290; MN, Δ85-369; MX, Δ85-290; TM293, Δ290-579; XN, Δ290-369; MX364, Δ85-290 and Δ369-579; STOP, Δ85-579. (C) UL84 and IE2 86 binding was assessed as described in Materials and Methods. Eluted protein was analyzed by agarose gel electrophoresis, and the amount of UL84 bound was analyzed by autoradiography. The input was 10% of bound reaction mixtures. All mutants are described above. GST alone was used as a negative control for nonspecific binding. A representative of four experiments is shown.
To further define the region within IE2 86 necessary for interaction with UL84, we conducted in vitro GST binding assays. Here, IE2 86 and several mutant forms of the protein were expressed as GST fusion proteins in bacteria. Following induction and lysis of bacteria, the GST protein was bound to glutathione-agarose beads. Figure 9B shows the mutants tested.
To test UL84 binding to IE2 86, 35S-labeled UL84 was synthesized in an in vitro transcription/translation reaction and then analyzed for binding following incubation with bound GST-IE2 86. The UL84/IE2 86 complex was then analyzed by gel electrophoresis and autoradiography. The amount of GST protein bound to the beads was normalized based on visualization of the GST protein in the gel following staining and to bovine serum albumin as a standard (data not shown). UL84 bound efficiently to WT GST-IE2 86, as well as to a number of the mutants (Fig. 9C). The minimal domain involved in the interaction with UL84 was localized to aa 290 to 369, based on the finding that the binding of UL84 to a GST-IE2 86 mutant containing only this region plus the N-terminal 85 aa (MX364) was comparable to that observed for the WT GST-IE2 86 construct. As expected, a mutant containing only the N-terminal 85 aa (IE2 86-Stop) was unable to bind to UL84, since IE1 72, which shares the amino-terminal 85 aa with IE2 86, does not interact with UL84 (17). Interestingly, loss of the region between aa 290 and 369 (XN) did not abolish binding, indicating that there is some contribution from another region that is able to aid in the binding of UL84 and IE2 86 in vitro (Fig. 9C). However, deletion of aa 290 to 579 (TM 293) resulted in a loss of binding comparable to that of the mutant that is missing the region between aa 85 to 369 (MN), indicating that the C-terminal domain (290 to 369) is the primary region that is important for binding to UL84. Loss of aa 136 to 290 (SX) or 85 to 290 (MX) also did not significantly disrupt binding, confirming the interaction study results with the mutant virus infections (Fig. 9C). It should be noted that the domain defined here (aa 290 to 369) is also contained within IE2 60 and IE2 40, further implicating these proteins and this region of IE2 86 as being important for this interaction.
DISCUSSION
Multiple functions have been ascribed to IE2 86, the majority of which converge on its effects on both viral and cellular transcription (9, 17, 22, 24, 25, 28-30, 38, 39, 48-50, 53, 58, 64). However, the involvement of IE2 86 in regulating the steady-state levels of many of the proteins expressed at early and late times postinfection has not been investigated. Our previous characterization of a mutant virus that is missing aa 136 to 290 of IE2 86, termed IE2 86ΔSX, showed that this region was important for potential functions of IE2 86 during the late stages of the infection. Since this mutant is also missing the initiator methionines for IE2 60 and IE2 40, these proteins are not expressed in the infected cells. The IE2 86ΔSX virus displays delayed expression of IE2 86, shows severe dysregulation of proteins important for late stages of the infection, and grows to reduced titers (45, 60). To further identify the relative contribution due to the loss of the IE2 60 and IE2 40 proteins versus the loss of the region between aa 136 and 290 of full-length IE2 86, we previously constructed a virus that does not express any IE2 60 or IE2 40 but expresses full-length IE2 86. This virus displayed very similar characteristics to the IE2 86ΔSX virus at late times postinfection with respect to expression of UL84 (60). This observation pointed to a role for both full-length IE2 86 and the late IE2 60 and IE2 40 in UL84 expression.
Previous studies showed that the levels of UL84 protein were markedly decreased throughout the infection in IE2 86ΔSX virus-infected cells, but there appeared to be no decrease in the UL84 RNA as measured by real-time RT-PCR (60). Here, we have confirmed by Northern blot analysis that the amount of UL84 mRNA is not affected at either early or late times in IE2 86ΔSX infection. Importantly, these data also documented that there is no disruption of the other transcripts (UL85 and UL86) that are 3′-coterminal with the UL84. The UL85 and UL86 transcripts encode two capsid proteins (minor and major, respectively) and do not seem to share any of the same functions as UL84. We showed that, similar to other late viral proteins in the mutant virus-infected cells, there is a delay in the accumulation of the UL85 and UL86 proteins at 72 h p.i. (45). However, in contrast to the UL84 protein levels, which remain low throughout the IE2 86ΔSX infection, the levels of the UL85 and UL86 proteins in the WT and mutant virus-infected cells were comparable by 96 h p.i.
Addition of a proteasome or calpain inhibitor did not increase the levels of the UL84 protein; thus, this loss in IE2 86ΔSX infection appears to be mediated through a proteasome- and calpain-independent pathway. Moreover, studies with cycloheximide that addressed the stability of UL84 indicated that the protein is not degraded more rapidly in the IE2 86ΔSX infection than in the WT virus infection. However, we cannot exclude the possibility that the IE2 86ΔSX and UL84 proteins measured in these assays were representative of only those proteins in complex with one another. It is possible that the interaction with any of the IE2 proteins aids in the stability of UL84 and that without this interaction, the protein is very rapidly degraded via some proteasome- and calpain-independent mechanism.
To further elucidate the mechanisms of regulation between this family of IE2 proteins and UL84, we also assessed the nuclear-to-cytoplasmic export and distribution and loading of the UL84 mRNA onto polysomes. Surprisingly, these analyses indicated that both processes were functioning in a comparable fashion to that in the WT virus-infected cells. Similarly, the IE2 86 mRNA did not exhibit any defects in loading and distribution on polysomes in IE2 86ΔSX-infected cells. These data further indicate that these proteins are regulated in a posttranscriptional fashion. Interestingly, the distribution of the UL84 RNA was different than other transcripts of the same size that were assessed, which could indicate a difference in the translation of this transcript. It is also possible that IE2 86, IE2 40, and/or IE2 60 functions cotranslationally to ensure the synthesis and release of full-length UL84 and IE2 86 proteins. Alternatively, the IE2 proteins may be important for some type of posttranslational regulation. Although the simplest explanation is that a rapid interaction of UL84 and IE2 proteins during or immediately after protein synthesis is necessary for the accumulation of both proteins, there is no a priori reason that the IE2 proteins do not affect UL84 expression indirectly through some other viral or cellular protein. Studies are under way to assess these possibilities.
To further understand which regions of the IE2 proteins might be involved in the regulation of UL84 protein levels throughout the infection, we conducted IP and GST-binding studies to determine the domains that contributed to the IE2-UL84 complex formation. Interestingly, IE2 86, IE2 60, and IE2 40 are each able to interact individually with UL84, as shown by the IP studies. These data also revealed that IE2 60 and IE2 40 are able to form heterodimers with IE2 86, which is not surprising given that the dimerization domain of IE2 86 is present in these two smaller proteins (1, 10, 15).
Further evidence for the above interactions observed in vivo is provided by the in vitro experiments that indicate that the region between aa 290 and 369, which is also contained in the IE2 60 and IE2 40 proteins, contributes to the interaction with UL84. Although, at least in vitro, this region is not essential for binding and there appears to be a contribution from other regions, a construct containing this domain alone was able to interact with UL84. The data from the IE2-UL84 interaction studies were surprising, since it had previously been proposed that the N terminus (aa 85 to 290) was important for UL84 binding (12). Using our viral mutants and GST constructs that removed either aa 136 to 290 (IE2 86ΔSX), 88 to 135, or 88 to 290, a strong interaction still occurred between UL84 and IE2 86 in the context of the actual infection and in the in vitro assays. It is possible that these differences in experimental procedure—transfection versus infection—may explain some of the discrepancies between these results. Although the region between aa 85 and 290 of IE2 86 may have the ability to interact with UL84 in some in vitro studies, the data clearly show that loss of this region does not prevent formation of the IE2 86-UL84 complex during the infection. Further confirming the in vivo data presented here, we show that a GST mutant containing only the first 290 aa of IE2 86 displays greatly reduced binding to UL84, which also indicates that a site outside of this region may be important for the IE2 86-UL84 interaction. Finally, IE2 60 and IE2 40, which do not contain the amino-terminal region of IE2 86, are able to interact with UL84.
The possibility exists that any of the three versions of IE2 can contribute to efficient expression of the UL84 protein and that it is the levels of the IE2 proteins present in the infection that are important for proper UL84 regulation. Although we previously showed that the loss of IE2 40 resulted in a partial loss of UL84 protein expression (60), the data presented here with the M170L virus revealed that total loss of the IE2 60 protein did not have an effect on UL84 expression, indicating that IE2 40 and IE2 86 likely play a more important role in UL84 expression. It is possible, however, that IE2 60 can partially compensate for the loss of IE2 40, or vice versa.
There are many potential sites within both the region of aa 136 to 290 and of aa 290 to 369 of IE2 86 that may be important for proper expression of and interaction with UL84. For example, there are serine-rich regions and sites of sumoylation and phosphorylation within these domains that are important for efficient viral replication (3-5, 8, 13, 17, 19, 21, 34). Furthermore, mutations of the serines within IE2 86 aa 258 to 275 have been previously shown to influence the expression of some late proteins in HCMV-infected cells (5). Understanding whether any of these sites are important for the regulatory mechanisms described here will be a focus of future studies.
In summary, these data have provided a strong basis for understanding the regulation of UL84 expression in HCMV infection. Although the mechanism of regulation remains to be fully characterized, we have identified many of the important players responsible for governing proper expression of UL84. Clearly, the IE2 proteins are governing the expression of UL84 protein in a novel fashion that has yet to be described for this family of proteins. We have eliminated the possibilities that this regulation is at the level of transcription of the UL84 mRNA, transport of the mRNA to the cytoplasm, or loading of the mRNA onto polysomes. The lack of accumulation of UL84 protein also does not appear to be due to proteasome- or calpain-dependent degradation. It is possible that translation of the mRNA to the termination codon is impaired or that proteolytic degradation involves one of the many other cellular proteases, and studies are in progress to address these questions.
It is interesting that UL84 is the only viral protein that IE2 86 with which has been shown to interact, and yet very little UL84 is actually needed to facilitate the production of functional virus. Elucidating the mechanisms governing the interactions between IE2 86, IE2 60, IE2 40, and UL84 should provide insight into how these proteins are regulated throughout infection and the contribution of each of the protein complexes to efficient viral replication.
Acknowledgments
We are thankful to E. S. Huang for providing antibodies to UL84 and to Greg Pari for providing the PTARGET-HA vector containing the UL84 cDNA as well as the antibodies to UL84. We also thank Mark Lawson and Minh-Ha Do (University of California, San Diego) for assistance with the polysome experiments, as well as members of the Spector laboratory for their helpful input throughout the course of this work.
This work was supported by NIH grants CA073490 and CA034729.
Footnotes
Published ahead of print on 10 September 2008.
REFERENCES
- 1.Ahn, J. H., C. J. Chiou, and G. S. Hayward. 1998. Evaluation and mapping of the DNA binding and oligomerization domains of the IE2 regulatory protein of human cytomegalovirus using yeast one and two hybrid interaction assays. Gene 21025-36. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J. H., Y. Xu, W. J. Jang, M. J. Matunis, and G. S. Hayward. 2001. Evaluation of interactions of human cytomegalovirus immediate-early IE2 regulatory protein with small ubiquitin-like modifiers and their conjugation enzyme Ubc9. J. Virol. 753859-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreoni, K. A., X. Wang, S. M. Huang, and E. S. Huang. 2002. Human cytomegalovirus hyperimmune globulin not only neutralizes HCMV infectivity, but also inhibits HCMV-induced intracellular NF-κB, Sp1, and PI3-K signaling pathways. J. Med. Virol. 6733-40. [DOI] [PubMed] [Google Scholar]
- 4.Barrasa, M. I., N. Harel, Y. Yu, and J. C. Alwine. 2003. Strain variations in single amino acids of the 86-kilodalton human cytomegalovirus major immediate-early protein (IE2) affect its functional and biochemical properties: implications of dynamic protein conformation. J. Virol. 774760-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrasa, M. I., N. Y. Harel, and J. C. Alwine. 2005. The phosphorylation status of the serine-rich region of the human cytomegalovirus 86-kilodalton major immediate-early protein IE2/IEP86 affects temporal viral gene expression. J. Virol. 791428-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett, V. D., and S. L. Adams. 1987. Characterization of the translational control mechanism preventing synthesis of alpha 2(I) collagen in chicken vertebral chondroblasts. J. Biol. Chem. 26214806-14814. [PubMed] [Google Scholar]
- 7.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 738320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castillo, J. P., F. M. Frame, H. A. Rogoff, M. T. Pickering, A. D. Yurochko, and T. F. Kowalik. 2005. Human cytomegalovirus IE1-72 activates ataxia telangiectasia mutated kinase and a p53/p21-mediated growth arrest response. J. Virol. 7911467-11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherrington, J. M., E. L. Khoury, and E. S. Mocarski. 1991. Human cytomegalovirus IE2 negatively regulates α gene expression via a short target sequence near the transcription start site. J. Virol. 65887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiou, C.-J., J. Zong, I. Waheed, and G. S. Hayward. 1993. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J. Virol. 676201-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colletti, K. S., K. E. Smallenburg, Y. Xu, and G. S. Pari. 2007. Human cytomegalovirus UL84 interacts with an RNA stem-loop sequence found within the RNA/DNA hybrid region of oriLyt. J. Virol. 817077-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colletti, K. S., Y. Xu, S. A. Cei, M. Tarrant, and G. S. Pari. 2004. Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication. J. Virol. 789203-9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colletti, K. S., Y. Xu, I. Yamboliev, and G. S. Pari. 2005. Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J. Biol. Chem. 28011955-11960. [DOI] [PubMed] [Google Scholar]
- 14.Fortunato, E. A., and D. H. Spector. 1999. Regulation of human cytomegalovirus gene expression. Adv. Virus Res. 5461-128. [DOI] [PubMed] [Google Scholar]
- 15.Furnari, B. A., E. Poma, T. F. Kowalik, S.-M. Huong, and E.-S. Huang. 1993. Human cytomegalovirus immediate-early gene 2 protein interacts with itself and with several novel cellular proteins. J. Virol. 674981-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gawn, J. M., and R. F. Greaves. 2002. Absence of IE1 p72 protein function during low-multiplicity infection by human cytomegalovirus results in a broad block to viral delayed-early gene expression. J. Virol. 764441-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gebert, S., S. Schmolke, G. Sorg, S. Floss, B. Plachter, and T. Stamminger. 1997. The UL84 protein of human cytomegalovirus acts as a transdominant inhibitor of immediate-early-mediated transactivation that is able to prevent viral replication. J. Virol. 717048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harel, N. Y., and J. C. Alwine. 1998. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2. J. Virol. 725481-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, Y. S., L. Xu, and E. S. Huang. 1992. Characterization of human cytomegalovirus UL84 early gene and identification of its putative protein product. J. Virol. 661098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofmann, H., S. Floss, and T. Stamminger. 2000. Covalent modification of the transactivator protein IE2-p86 of human cytomegalovirus by conjugation to the ubiquitin-homologous proteins SUMO-1 and hSMT3b. J. Virol. 742510-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, L., and M. F. Stinski. 1995. Binding of cellular repressor protein or the IE2 protein to a cis-acting negative regulatory element upstream of a human cytomegalovirus early promoter. J. Virol. 697612-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins, D. E., C. L. Martens, and E. S. Mocarski. 1994. Human cytomegalovirus late protein encoded by ie2: a transactivator as well as a repressor of gene expression. J. Gen. Virol. 752337-2348. [DOI] [PubMed] [Google Scholar]
- 24.Klucher, K. M., M. Sommer, J. T. Kadonaga, and D. H. Spector. 1993. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol. Cell. Biol. 131238-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang, D., and T. Stamminger. 1994. Minor groove contacts are essential for an interaction of the human cytomegalovirus IE2 protein with its DNA target. Nucleic Acids Res. 223331-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, H.-R., and J.-H. Ahn. 2004. Sumoylation of the major immediate-early IE2 protein of human cytomegalovirus Towne strain is not required for virus growth in cultured human fibroblasts. J. Gen. Virol. 852149-2154. [DOI] [PubMed] [Google Scholar]
- 27.Lee, J. M., H. J. Kang, H. R. Lee, C. Y. Choi, W. J. Jang, and J. H. Ahn. 2003. PIAS1 enhances SUMO-1 modification and the transactivation activity of the major immediate-early IE2 protein of human cytomegalovirus. FEBS Lett. 555322-328. [DOI] [PubMed] [Google Scholar]
- 28.Liu, B., T. W. Hermiston, and M. F. Stinski. 1991. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J. Virol. 65897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macias, M. P., and M. F. Stinski. 1993. An in vitro system for human cytomegalovirus immediate early 2 protein (IE-2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate-early promoter. Proc. Natl. Acad. Sci. USA 90707-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 641498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 751870-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding ie1 (491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 9311321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mocarski, E. S., T. Shenk, and R. F. Pass. 2007. Cytomegaloviruses, p. 2701-2772. In D. M. Knipe and P. M. Howley (ed.), Fields' virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 34.Nevels, M., W. Brune, and T. Shenk. 2004. SUMOylation of human cytomegalovirus 72-kilodalton IE1 protein facilitates expression of the 86-kilodalton IE2 protein and promotes viral replication. J. Virol. 787803-7812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 676979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petrik, D. T., K. P. Schmitt, and M. F. Stinski. 2007. The autoregulatory and transactivating functions of the human cytomegalovirus IE86 protein use independent mechanisms for promoter binding. J. Virol. 815807-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrik, D. T., K. P. Schmitt, and M. F. Stinski. 2006. Inhibition of cellular DNA synthesis by the human cytomegalovirus IE86 protein is necessary for efficient virus replication. J. Virol. 803872-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 646154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pizzorno, M. C., M.-A. Mullen, Y.-N. Chang, and G. S. Hayward. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 653839-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plachter, B., W. Britt, R. Vornhagen, T. Stamminger, and G. Jahn. 1993. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology 193642-652. [DOI] [PubMed] [Google Scholar]
- 41.Puchtler, E., and T. Stamminger. 1991. An inducible promoter mediates abundant expression from the immediate-early 2 gene region of human cytomegalovirus at late times after infection. J. Virol. 656301-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinhardt, J., G. B. Smith, C. T. Himmelheber, J. Azizkhan-Clifford, and E. S. Mocarski. 2005. The carboxyl-terminal region of human cytomegalovirus IE1491aa contains an acidic domain that plays a regulatory role and a chromatin-tethering domain that is dispensable during viral replication. J. Virol. 79225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudolph, S. A., T. Stamminger, and G. Jahn. 1990. Transcriptional analysis of the eight-kilobase mRNA encoding the major capsid protein of human cytomegalovirus. J. Virol. 645167-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samaniego, L. A., M. J. Tevethia, and D. J. Spector. 1994. The human cytomegalovirus 86-kilodalton immediate-early 2 protein: synthesis as a precursor polypeptide and interaction with a 75-kilodalton protein of probably viral origin. J. Virol. 68720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez, V., C. L. Clark, J. Y. Yen, R. Dwarakanath, and D. H. Spector. 2002. Viable human cytomegalovirus recombinant virus with an internal deletion of the IE2 86 gene affects late stages of viral replication. J. Virol. 762973-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanders, R. L., C. L. Clark, C. S. Morello, and D. H. Spector. 7 May 2008. Development of cell lines that provide tightly controlled temporal translation of the human cytomegalovirus IE2 proteins for complementation and functional analyses of growth-impaired and nonviable IE2 mutant viruses. J. Virol. doi: 10.1128/JVI.00675-08. [DOI] [PMC free article] [PubMed]
- 47.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 707398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz, R., M. H. Sommer, A. Scully, and D. H. Spector. 1994. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J. Virol. 685613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scully, A. L., M. H. Sommer, R. Schwartz, and D. H. Spector. 1995. The human cytomegalovirus IE2 86 kDa protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 696533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sommer, M. H., A. L. Scully, and D. H. Spector. 1994. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J. Virol. 686223-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spector, D. J., and M. J. Tevethia. 1994. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J. Virol. 687549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stenberg, R. M., A. S. Depto, J. Fortney, and J. A. Nelson. 1989. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J. Virol. 632699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 641556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stenberg, R. M., D. R. Thomsen, and M. F. Stinski. 1984. Structural analysis of the major immediate early gene of human cytomegalovirus. J. Virol. 49190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stenberg, R. M., P. R. Witte, and M. F. Stinski. 1985. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J. Virol. 56665-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stinski, M. F., D. R. Thomsen, R. M. Stenberg, and L. C. Goldstein. 1983. Organization and expression of the immediate-early genes of human cytomegalovirus. J. Virol. 461-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsujinaka, T., Y. Kajiwara, J. Kambayashi, M. Sakon, N. Higuchi, T. Tanaka, and T. Mori. 1988. Synthesis of a new cell penetrating calpain inhibitor (calpeptin). Biochem. Biophys. Res. Commun. 1531201-1208. [DOI] [PubMed] [Google Scholar]
- 58.Waheed, I., C. J. Chiou, J. H. Ahn, and G. S. Hayward. 1998. Binding of the human cytomegalovirus 80-kDa immediate-early protein (IE2) to minor groove A/T-rich sequences bounded by CG dinucleotides is regulated by protein oligomerization and phosphorylation. Virology 252235-257. [DOI] [PubMed] [Google Scholar]
- 59.White, E. A., C. L. Clark, V. Sanchez, and D. H. Spector. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 781817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.White, E. A., C. J. Del Rosario, R. L. Sanders, and D. H. Spector. 2007. The IE2 60-kilodalton and 40-kilodalton proteins are dispensable for human cytomegalovirus replication but are required for efficient delayed early and late gene expression and production of infectious virus. J. Virol. 812573-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White, E. A., and D. H. Spector. 2005. Exon 3 of the human cytomegalovirus major immediate-early region is required for efficient viral gene expression and for cellular cyclin modulation. J. Virol. 797438-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu, Y., S. A. Cei, A. R. Huete, K. S. Colletti, and G. S. Pari. 2004. Human cytomegalovirus DNA replication requires transcriptional activation via an IE2- and UL84-responsive bidirectional promoter element within oriLyt. J. Virol. 7811664-11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu, Y., S. A. Cei, A. R. Huete, and G. S. Pari. 2004. Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth. J. Virol. 7810360-10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeung, K. C., C. M. Stoltzfus, and M. F. Stinski. 1993. Mutations of the human cytomegalovirus immediate-early 2 protein defines regions and amino acid motifs important in transactivation of transcription from the HIV-1 LTR promoter. Virology 195786-792. [DOI] [PubMed] [Google Scholar]
- 65.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 10012396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]