Abstract
A more complete assessment of ovine prion strain diversity will be achieved by complementing biological strain typing in conventional and ovine PrP transgenic mice with a biochemical analysis of the resultant PrPSc. This will provide a correlation between ovine prion strain phenotype and the molecular nature of different PrP conformers associated with particular prion strains. Here, we have compared the molecular and transmission characteristics of ovine ARQ/ARQ and VRQ/VRQ scrapie isolates following primary passage in tg338 (VRQ) and tg59 (ARQ) ovine PrP transgenic mice and the conventional mouse lines C57BL/6 (Prnpa), RIII (Prnpa), and VM (Prnpb). Our data show that these different genotypes of scrapie isolates display similar incubation periods of >350 days in conventional and tg59 mice. Facilitated transmission of sheep scrapie isolates occurred in tg338 mice, with incubation times reduced to 64 days for VRQ/VRQ inocula and to ≤210 days for ARQ/ARQ samples. Distinct genotype-specific lesion profiles were seen in the brains of conventional and tg59 mice with prion disease, which was accompanied by the accumulation of more conformationally stable PrPSc, following inoculation with ARQ/ARQ compared to VRQ/VRQ scrapie isolates. In contrast, the lesion profiles, quantities, and stability of PrPSc induced by the same inocula in tg338 mice were more similar than in the other mouse lines. Our data show that primary transmission of different genotypes of ovine prions is associated with the formation of different conformers of PrPSc with distinct molecular properties and provide the basis of a molecular approach to identify the true diversity of ovine prion strains.
Prion diseases, such as scrapie of sheep, bovine spongiform encephalopathy (BSE) of cattle, and variant Creutzfeldt-Jakob disease of humans are transmissible neurodegenerative disorders of the central nervous system. During the course of these diseases, host PrPC is converted into an abnormal isomer PrPSc. The protein-only hypothesis postulates that the transmissible agent comprises principally proteinaceous material (54). In this model, the infectious agent, or prion, is regarded as synonymous with PrPSc, which is also responsible for the conformational change in PrPC. However, different isolates of the prion agent obtained from individuals of the same species exhibit strain variation, reminiscent of strains of other conventional infectious agents, such as viruses. Different prion strains isolated upon serial passage through mice produce different disease phenotypes, including incubation periods and lesion distributions (11, 13, 27, 32). These disease phenotypes, a feature of both the strain of the infectious prion agent and genetically encoded factors in the host, are typically stable on repeated passage through individuals of the same species. The strain-specific information of prions is therefore independent of the host from which they were originally derived. The phenomenon of prion strain variation has been a challenge to the protein-only hypothesis for prion diseases. Consequently, an alternative, less favored, hypothesis is that the infectious prion agent carries a genome and is a virus (18) or virino (31, 70) and suggests that the information that dictates prion strain diversity encoded by a molecule is independent of PrPSc.
Scrapie disease of sheep has been reported to exhibit a significant diversity of prion strains. The identification of ovine prion strains has typically involved serial passage of scrapie isolates in a panel of conventional mice, including C57BL/6 and RIII (Prnpa) and VM (Prnpb) lines, and sometimes C57BL × VM F1 (10). This procedure has reportedly identified at least 14 scrapie prion strains and has enabled the BSE agent to be distinguished from the scrapie agent (10-12). Significant drawbacks of this conventional approach to ovine scrapie prion strain typing are the large numbers of mice required, long incubation periods, and the low efficiency of transmission in certain cases. Not all scrapie isolates cause the appearance of clinical signs of prion disease in conventional mice (13) (our unpublished data). To date, approximately 20% of classical scrapie and all atypical scrapie cases fail to transmit to the standard prion strain typing mouse lines. Even successful transmissions of classical scrapie samples may be characterized by low attack rates. The failure to see the onset of terminal disease may arise as a consequence of the low dose of infectivity in the original prion inoculum or represent “adaptation” or “selection” of prions crossing the species barrier into a new host (30, 37-39). The absence of terminal disease in mice inoculated with different ovine prion strains is problematic for conventional strain typing, as no incubation time can be recorded and neuropathology may not be evident. Efficient transmission of prions from different species has been achieved in mice that express a PrP transgene homologous to the host from which the prion isolates were obtained (9, 17, 24). Facilitated transmission of ovine scrapie strains to mice transgenic for either ovine ARQ (26) or VRQ PrP (68) has also been described. Mice transgenic for ovine VRQ PrP (tg338 mice) show enhanced susceptibility, in terms of reduced incubation times to terminal disease, compared to conventional mice after inoculation with prion inoculum from field cases of classical scrapie (42, 68). In addition, atypical strains of scrapie, which do not induce terminal prion disease in conventional mice, do induce terminal disease in tg338 mice. Significantly, different scrapie isolates induced terminal disease in tg338 mice with different incubation times and with distinct neuropathologies, which allows the potential for single-passage scrapie prion strain typing (6, 42). These features of ovine PrP transgenic mice highlight their potential utility in developing more rapid systems of ovine prion strain typing.
The qualitative features of different prion strains, which include clinical signs, lesion profile, distribution of PrPSc in the brain, and Western blot pattern of PrPSc, do not presently explain or correlate with their quantitative traits, such as incubation time and dose response. Safar et al. (58) have successfully used a conformation-dependent immunoassay (CDI) to correlate conformational characteristics of PrPSc and its rate of accumulation with incubation time in eight prion strains propagated in Syrian hamsters. Each prion strain was found to produce a substantial fraction of protease-sensitive PrPSc, determined by CDI measurement of PrPSc before and after limited proteinase K (PK) digestion. A significant correlation was found between the level of protease-sensitive PrPSc and the incubation time of the prion strain (58). This implies that different incubation times of various prion strains may arise as a consequence of distinct rates of PrPSc clearance rather than rate of PrPSc formation. This in turn suggests that different prion strains exhibit different conformational stability of PrPSc, which appears to be the case (52, 53). Determination of the conformational stability of an extensive panel of synthetic and naturally occurring prions passaged in mice showed that the concentration of guanidine hydrochloride (GdnHCl) required to induce half-maximal denaturation correlated positively with their incubation time (43). These studies have begun to address the correlation of the biological properties of different prion strains with the biochemical properties of PrPSc.
In order to obtain a more complete understanding of the diversity of ovine prion strains, it will be important to supplement the biological strain typing of scrapie isolates in mice with different PrP genotypes with a biochemical analysis of the resultant PrPSc. This will lead to a correlation between prion strain phenotype and the molecular nature of different PrP conformers associated with particular prion strains. Strain typing ovine scrapie isolates in wild-type mice may lead to the generation of new prion strains as the original prions undergo “adaptation” or “selection” as a consequence of the infectious agent crossing the species barrier into a new host (1, 22). This may mean that lesion profiles at first passage in conventional mice do not reveal properties of the infectious prion agent as they exist in the original host. This should be circumvented if ovine scrapie isolates are passaged in ovine PrP transgenic mice. In order to address this, we have for the first time compared the primary transmission characteristics of ovine ARQ/ARQ and VRQ/VRQ scrapie isolates in conventional and ovine PrP transgenic mice in order to investigate the biochemical and biophysical characteristics of PrPSc that originate from sheep prion isolates in hosts of different PrP genotypes. Our data show that although ARQ and VRQ homozygous scrapie isolates show similar incubation periods in conventional mice and ovine ARQ PrP transgenic mice, the quantity and nature of deposited PrPSc show significant variation. In contrast, these differences are not evident following transmission of the same inocula in ovine VRQ PrP transgenic mice. Our data show that primary transmission of different genotypes of ovine prions is associated with the formation of different conformers of PrPSc with distinct molecular properties and provide the basis of a molecular approach to identify the true diversity of ovine prion strains.
MATERIALS AND METHODS
Mice.
Breeding colonies of the conventional mouse lines C57BL/6, RIII, and VM and the ovine PrP transgenic mouse lines tg59 (25) and tg338 (68) were maintained at the Veterinary Laboratories Agency, Weybridge laboratory. All regulated procedures involving experimental animals were carried out under project and personal license authority issued in accordance with The Animals (Scientific Procedures) Act 1986 (2a).
Inoculation of mice with ovine scrapie prion inoculum.
Approximately 2 g of cerebral cortex brain material from confirmed positive cases of ARQ/ARQ (n = 2) (SE1848/0007 and SE1848/0008) and VRQ/VRQ (n = 2) (SE1848/0005 and SE1848/0006) sheep with scrapie were homogenized and subsequently diluted in normal saline to produce a 10% (wt/vol) homogenate. Each scrapie isolate was from a different farm. Following confirmation of sterility by aerobic culture, each scrapie isolate was injected into 20 C57BL/6, RIII, VM, tg59 and tg338 mice by combined intracranial (20-μl) and intraperitoneal (100-μl) routes. Mice were monitored for the development of clinical signs of mouse prion disease and euthanized at the time of appearance of terminal signs of disease.
Histopathology and immunohistochemistry.
The brains from prion-inoculated mice were isolated, a rostrolateral portion was removed and stored frozen, and the remainder was fixed in 10% formal saline. Hematoxylin-and-eosin-stained sections at five coronal levels were examined for vacuolar pathology to determine lesion severity and distribution as described by Fraser and Dickinson (33). The incubation time for terminal prion disease was calculated as the time from inoculation to euthanasia at terminal prion disease. The attack rate for each mouse line was calculated as the number of mice diagnosed positive divided by the number surviving after, and including, the first positive diagnosis for each inoculum in a particular mouse line. This avoids underestimation of the attack rate due to loss of mice as a result of intercurrent deaths early in the study. The incubation time for attack rate was calculated as the time from inoculation to death or euthanasia (for whatever reason) for prion disease-positive mice. Brain sections from prion-inoculated mice were immunostained for PrP using rabbit anti-bovine PrP antiserum Rb486 as previously described (34a). At least three mouse brains were examined from each line of mice. For each mouse, the patterns of disease-associated PrP deposition were recorded, and the presence of plaques was noted.
Anti-PrP monoclonal antibodies.
The anti-PrP monoclonal antibodies used here have been described in detail elsewhere. Monoclonal antibody V24 recognizes an undefined epitope in the C-terminal region of PrP (66). Monoclonal antibody 683 recognizes the core sequence PVDQY (amino acid residues 168 to 172 [ovine PrP numbering]) (66). Monoclonal antibody 6H4 reacts with the epitope DYEDRYYRE (amino acid residues 144 to 152 [human PrP numbering]) (40) and was a generous gift from Prionics AG, Zürich, Switzerland. Monoclonal antibody P4 reacts with the core sequence WGQGGSH (amino acid residues 93 to 99 [ovine PrP numbering]) (67) and was purchased from R-Biopharm, Darmstadt, Germany. Monoclonal antibodies were biotinylated as described in detail previously (63).
CDI.
Murine sagittal-cut half-brain homogenates were prepared by two cycles of homogenization in phosphate-buffered saline (PBS) (pH 7.4) in a Bio-Rad TeSeE Precess 24 homogenizer, and individual samples were diluted to 10% in PBS supplemented with 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF) diluted in distilled water. Samples were centrifuged at 100 × g for 1 min at 20°C to remove gross debris, and the supernatants were retained. Sarkosyl in PBS (pH 7.4) was added to all samples to a final concentration of 2%, and the samples were incubated for 10 min at 37°C with shaking prior to treatment with and without sodium phosphotungstic acid (NaPTA) at a final concentration of 0.4% diluted in PBS (pH 7.4) for 1 h at 37°C with shaking. The samples were centrifuged at 21,000 × g for 30 min at 10°C, the supernatants were discarded, and the pellets were resuspended in 200 μl of 0.1% Sarkosyl in PBS (pH 7.4) and in 200 μl of 250 mM EDTA in water (pH 8.0). The samples were thoroughly mixed and then centrifuged at 21,000 × g for 15 min at 10°C. The supernatants were discarded, and the pellets were resuspended in 6 M GdnHCl or not resuspended in GdnHCl. All 6 M GdnHCl samples were then heated to 80°C for 5 min, cooled to 20°C, and then diluted as required. Capture antibody diluted in coating buffer (0.01 M PBS [pH 7.4] containing 0.1% sodium azide) was routinely coated at 0.5 μg/well in triplicate in Nunc Maxisorp 96-well flat-bottomed plates overnight at 4°C. The remainder of the CDI protocol was carried out as described in detail previously (63). Plates were finally incubated for 5 min at 20°C with shaking, and the fluorescence, measured as counts per second (cps), was determined in a Victor time-resolved fluorimeter (PerkinElmer).
Western blot detection of PrPSc.
Ovine brain homogenates (100%) were diluted to 10% in Prionics Check Western homogenization buffer prior to treatment with various concentrations of PK (Roche). Murine sagittal-cut half-brain homogenates were prepared by two cycles of homogenization in Prionics Check Western homogenization buffer in a Bio-Rad TeSeE Precess 24 homogenizer. Brain homogenates were diluted to 10% in the same buffer prior to treatment with PK at various concentrations for ovine scrapie samples (see Fig. 1) or at a final concentration of 32 μg/ml for C57BL/6, RIII, and VM homogenates and at a final concentration of 64 μg/ml for tg59 and tg338 homogenates. Digestion was terminated with the addition of 1 mM AEBSF, and samples were electrophoresed by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a minigel under reducing conditions and subsequently transferred onto a nitrocellulose membrane by semidry blotting. Membranes were blocked overnight at 4°C with TBS-T (10 mM Tris-HCl [pH 7.8], 100 mM NaCl, 0.05% Tween 20) containing 5% (wt/vol) nonfat milk and subsequently washed three times with TBS-T. Membranes were incubated with either purified anti-PrP monoclonal antibody 683 used at 5 μg/ml (for detection of murine PrP) or P4 (R-Biopharm, Darmstadt, Germany) used at 1 μg/ml (for the detection of ovine PrP) for 2 h at 20°C. The membranes were washed five times with TBS-T, incubated with horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (Sigma) at 1:2,000 for 2 h at 20°C, and washed five times with TBS-T. All of the antibody dilutions were prepared in 1% nonfat milk in TBS-T, and the duration of each wash step was 5 min. PrP bands were detected by enhanced chemiluminescence (Amersham Biosciences).
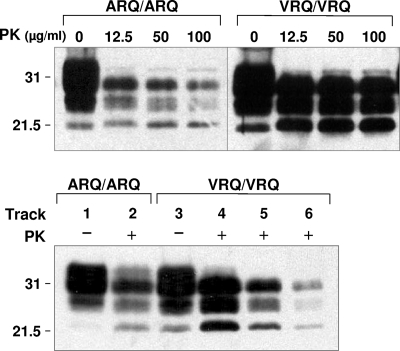
FIG. 1.
Western blot analysis of ARQ/ARQ and VRQ/VRQ ovine scrapie isolates. Homogenates of ARQ/ARQ and VRQ/VRQ ovine scrapie isolates were prepared as described in Materials and Methods and analyzed by SDS-PAGE and Western blotting using anti-PrP monoclonal antibody P4 at 1 μg/ml. (Top) Homogenates were treated with various concentrations of PK or not treated with PK. (Bottom) Tracks 1 to 4, 10% homogenate; track 5, 5% homogenate; track 6, 2.5% homogenate; tracks 1 and 3, no PK; tracks 2 and 4 to 6, PK at 32 μg/ml. The positions of molecular mass markers (in kilodaltons) are shown to the left of the blots.
Conformational stability Western blot.
Murine sagittal-cut half-brain homogenates were prepared by two cycles of homogenization in PBS (pH 7.4) in a Bio-Rad TeSeE Precess 24 homogenizer. Brain homogenates were diluted to 10% in PBS (pH 7.4) prior to treatment with GdnHCl (0 to 2 M final concentration) for 30 min at 20°C. GdnHCl was diluted to a final concentration of 0.1 M with PBS (pH 7.4), prior to incubation with PK at a final concentration of 32 μg/ml for C57BL/6, RIII and VM homogenates or at a final concentration of 64 μg/ml for tg59 or tg338 homogenates for 30 min at 37°C. The reaction was terminated by the addition of 1 mM AEBSF and centrifuged at 16,160 × g, and the pelleted material was subsequently analyzed by SDS-PAGE and Western blotting. PrP bands were detected by enhanced chemiluminescence (Amersham Biosciences).
Statistical analysis.
Statistical analyses of the data were performed using one-way analysis of variance with Tukey's honestly significant difference test for post hoc analysis or the two-tailed Student t test (unpaired samples) using the Prism 4 software package (GraphPad, San Diego, CA).
RESULTS
Transmission of ovine scrapie isolates to conventional and ovine PrP transgenic mice.
Over several years, numerous isolates of scrapie have been passaged as primary transmissions to a panel of conventional mice in order to determine strain diversity. Here we have compared primary transmission of sheep scrapie isolates in conventional and ovine PrP transgenic mice in order to determine whether the latter may contribute to the assessment of ovine prion strain diversity. The ovine ARQ/ARQ and VRQ/VRQ scrapie isolates used were from scrapie-positive sheep that showed typical vacuolar pathology in the medulla oblongata of the brain stem and that were positive for disease-associated PrP as judged by immunohistochemistry or Western blotting through routine statutory surveillance. Figure 1 shows a Western blot analysis of representative ARQ and VRQ homozygous scrapie isolates, with or without PK digestion, probed with monoclonal antibody P4. The top panel of Fig. 1 shows that the PK-resistant core of PrPSc, PrP27-30, was present in both ARQ and VRQ homozygous ovine scrapie isolates at all concentrations of proteolytic enzyme tested. The bottom panel of Fig. 1 shows that both genotypes of scrapie isolate were characterized by a predominance of diglycosylated PrP27-30. Although not formally quantified, it was apparent that the homozygous VRQ scrapie isolates exhibited more PrP27-30 than that seen in homozygous ARQ samples, which correlated with the greater level of total PrP in these samples.
The molecular and transmission characteristics of the various ARQ/ARQ and VRQ/VRQ scrapie isolates were determined following primary passage in C57BL/6 (Prnpa), RIII (Prnpa), VM (Prnpb), tg59 (ovine ARQ), and tg338 (ovine VRQ) mice. Following inoculation, animals from each of the various mouse lines developed prion disease, which was usually, but not always, accompanied by the appearance of typical clinical signs of this condition. Clinical signs of prion disease were least evident in prion-inoculated tg59 mice. Table 1 shows the mean incubation time for the onset of terminal prion disease and the mean incubation time for the attack rate. As shown in Table 1, most of the scrapie isolates produced similar incubation periods for the onset of terminal prion disease in the ovine ARQ PrP transgenic mouse line tg59 and the conventional mouse lines, although there were exceptions. Furthermore, all of the inocula produced significantly accelerated incubation periods following inoculation into tg338 mice, which are transgenic for ovine VRQ PrP. tg338 mice succumbed to terminal prion disease with very short incubation periods following inoculation with VRQ/VRQ scrapie isolates, approximately 64 days in both cases, which was significantly shorter than that seen for the same samples in C57BL/6 (P < 0.001), RIII (P < 0.001), or VM (P < 0.001) mice. In a similar manner, tg338 mice succumbed to terminal prion disease after relatively short incubation periods following inoculation with ARQ/ARQ scrapie isolates, 155 and 210 days, which was significantly shorter than that seen for the same samples in C57BL/6 (P < 0.001), RIII (P < 0.001), or VM (P < 0.001) mice. Furthermore, the incubation times for the VRQ/VRQ scrapie isolates in tg338 mice were significantly shorter than that seen for the ARQ/ARQ isolates in the same mouse line (P ≤ 0.01 in both cases). Despite similarities and differences in incubation periods for terminal prion disease, efficient attack rates were seen for all inocula in all of the mouse lines. The attack rate ranges were 93 to 100% in RIII mice and 67 to 100% in VM mice. The attack rate in C57BL/6 and tg59 mice was 100% in all four groups, and in tg338 mice, the range was 92 to 100%.
TABLE 1.
Primary passage incubation times for sheep scrapie inocula in conventional and ovine PrP transgenic micea
| Sheep scrapie inoculum | Incubation time for the onset of terminal disease in miceb:
|
Incubation time for the attack rate in micec:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| C57BL/6 | RIII | VM | tg59 | tg338 | C57BL/6 | RIII | VM | tg59 | tg338 | |
| ARQ | ||||||||||
| SE1848/0007 | 468 ± 16 (16/16) | 458 ± 6d (3/4) | 617 ± 8e (3/12) | NA (0/16) | 210 ± 197g,h (5/14) | 468 ± 16 (16/16) | 450 ± 17 (4/4) | 586 ± 54e (12/14) | 470 ± 56d (16/16) | 318 ± 235h,i,j (14/14) |
| SE1848/0008 | 395 ± 50 (14/15) | 422 ± 11 (12/14) | 565 ± 30f (12/17) | 392 ± 50 (2/14) | 155 ± 4g,k (12/12) | 387 ± 56 (15/15) | 398 ± 52 (14/15) | 565 ± 34f (17/17) | 331 ± 61d (14/14) | 155 ± 4g,j (12/13) |
| VRQ | ||||||||||
| SE1848/0005 | 549 ± 43 (15/16) | 487 ± 28e (13/15) | 551 ± 61 (14/17) | 434 ± 0 (1/14) | 64 ± 2g (16/16) | 548 ± 42 (16/16) | 486 ± 28 (15/16) | 557 ± 76 (17/19) | 440 ± 60d,e (14/14) | 64 ± 2g,j (16/16) |
| SE1848/0006 | 527 ± 49 (13/13) | 492 ± 23 (8/10) | 524 ± 59 (9/12) | 403 ± 0 (1/12) | 63 ± 1g (13/15) | 527 ± 49 (13/13) | 489 ± 24 (10/10) | 526 ± 66 (12/18) | 460 ± 57 (12/12) | 63 ± 1g,j (15/15) |
Mice were inoculated with ARQ/ARQ and VRQ/VRQ sheep scrapie isolates by a combined intracranial and intraperitoneal route. Inoculated mice were monitored for clinical signs of mouse prion disease. Mice scored positive for terminal prion disease were those that displayed clinical signs for this condition as described by Dickinson et al. in 1968 (28). The majority of prion-inoculated tg59 mice showed a much more subtle form of terminal prion disease. Mice were euthanized at the point of neurological disease and dysfunction, and prion disease was confirmed by histopathology, immunohistochemistry for disease-associated PrP, or Western blotting for PK-resistant PrP27-30. The data shown are incubation times for the onset of terminal prion disease and attack rate. Statistical analysis of the data was performed by one-way analysis of variance together with Tukey's honestly significant difference test for post hoc analysis.
Incubation time for terminal prion disease was calculated as the time from inoculation to euthanasia at terminal prion disease (mean ± standard deviation). The number of mice with terminal prion disease/total number of mice positive for prion disease for each group is shown in parentheses. NA, not available.
Incubation time for attack rate was calculated as the time from inoculation to death or euthanasia (for whatever reason) for prion disease-positive mice (mean ± standard deviation). The number of prion disease-positive mice/number of mice surviving and including the first positive diagnosis for each group is shown in parentheses.
Significantly different from the value for VM mice (P ≤ 0.01).
Significantly different from the value for C57BL/6 mice (P ≤ 0.05).
Significantly different from the values for C57BL/6 and RIII mice (P < 0.001).
Significantly different from the values for C57BL/6, RIII, and VM mice (P < 0.001).
Significantly different from the value for tg338 mice inoculated with the VRQ scrapie isolates (P < 0.001).
Significantly different from the values for C57BL/6 and VM mice (P < 0.001).
Significantly different from the value for tg59 mice (P < 0.001).
Significantly different from the value for tg338 mice inoculated with the VRQ scrapie isolates (P < 0.01).
All prion-inoculated mice were evaluated according to predetermined protocols established from the study of prion disease in wild-type mouse lines (28). Clinical signs of terminal prion disease were clearly evident in C57BL/6, RIII, VM, and tg338 mice, which all showed a clear demarcation between early clinical disease and signs of terminal prion disease. With respect to prion-inoculated tg59 mice, these animals showed signs of prion disease that persisted over a long period of time and did not show the same signs of terminal prion disease as all of the other mouse lines did. tg59 prion-inoculated mice did show a much more subtle form of terminal prion disease. However, based on PrPSc accumulation assessed by Western blotting or immunohistochemistry and spongiform changes assessed by histopathology, the majority of prion-inoculated tg59 mice were sacrificed or died around the recorded incubation time for terminal prion disease for this line of mice. This is reflected by the fact that the incubation period for the attack rate was similar to that for the incubation period to terminal disease in tg59 mice. A similar correlation was seen between the incubation period for the attack rate and the incubation period for the onset of terminal disease in all of the other mouse lines as shown by the data in Table 1. As a consequence, we consider that prion-inoculated tg59 mice were culled or died at the terminal stage of prion disease, although they were not initially scored as such according to the criteria of clinical signs described by Dickinson et al. in 1968 (28).
Lesion profiles of primary-passaged scrapie isolates.
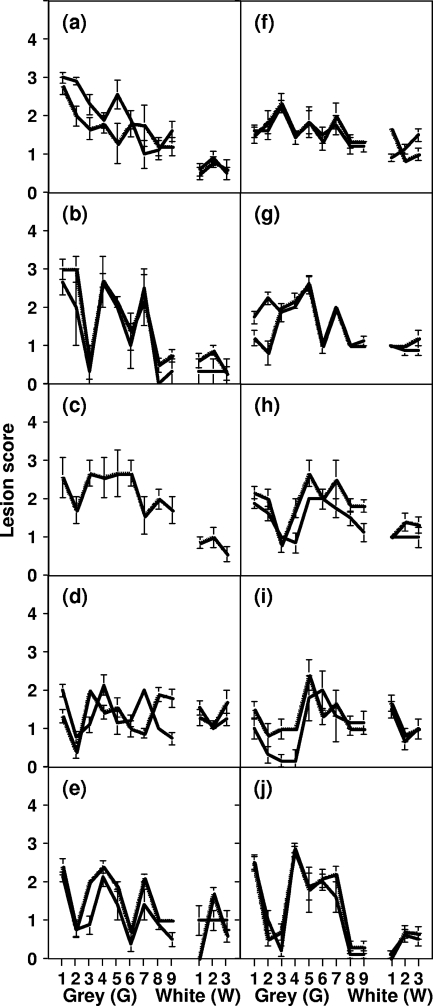
The brains of mice inoculated with the various scrapie isolates were subjected to neuropathological analysis at terminal prion disease to determine the resultant lesion profile. The data in Fig. 2 show the average lesion profile for each scrapie inoculum passaged in conventional or ovine PrP transgenic mice. While the different genotypes of scrapie inocula induced different lesion profiles in each mouse line, the vacuolation patterns for replicate scrapie samples were similar, although this was less true for ARQ/ARQ inocula, especially in tg59 mice. The greatest degree of similarity between grey matter lesion profiles induced by ARQ/ARQ and VRQ/VRQ inocula was seen in tg338 mice (Fig. 2e and j, respectively), where the most apparent difference in pathology score was seen in the thalamic region. In RIII mice, ARQ/ARQ scrapie inocula induced peak pathology scores in the medulla, hypothalamus, and septal brain regions with one profile corresponding closely and another with an alternative 1, 4, 7 grey matter pathology score seen for other ARQ inocula upon primary passage in Prnpa mice (5). The 1, 4, 7 grey matter lesion profile for ARQ/ARQ inocula passaged in RIII mice was also seen following inoculation of the same samples into tg338 mice.
FIG. 2.
Lesion profiles induced by primary transmissions of ovine scrapie isolates. Prion-infected brains were harvested from mice that had developed terminal prion disease and were subjected to neuropathological examination for the presence and severity of spongiform neurodegeneration. The lesion profiles for C57BL/6 (a and f), RIII (b and g), VM (c and h), tg59 (d and i), and tg338 (e and j) mice inoculated with ARQ/ARQ (a to e) or VRQ/VRQ (f to j) sheep scrapie isolates are shown. The lesion profiles were induced by SE1848/0007 (ARQ/ARQ) (thick line in panels a to e), SE1848/0008 (ARQ/ARQ) (dashed line in panels a to e), and SE1848/0005 (VRQ/VRQ) (thick line in panels f to j), and SE1848/0006 (VRQ/VRQ) (dashed line in panels f to j). The data shown are mean lesion profile scores (three or more brains examined) for the following areas of the brain: for grey (G) matter, G1, dorsal medulla nuclei; G2, cerebellar cortex of the folia, including the granular layer, adjacent to the fourth ventricle; G3, cortex of the superior colliculus; G4, hypothalamus; G5, thalamus; G6, hippocampus; G7, septal nuclei of the paraterminal body; G8, cerebral cortex (at the level of G4 and G5); G9, cerebral cortex (at the level of G7); for white (W) matter, W1, cerebellar peduncles; W2, white matter in lateral tegmentum; W3, cerebellar peduncle/internal capsule. No data were obtained for SE1848/0007 in VM mice (c).
Genotypic differences in PrPSc distribution in prion-diseased mouse brains.
Distinct morphological patterns of PrPSc deposition were evident in the brains of mice that had succumbed to terminal prion disease, and differences were evident between PrPSc distribution induced by ARQ/ARQ and VRQ/VRQ scrapie inocula in some lines of mice. A summary of the ovine scrapie-induced PrPSc profile in the various mouse lines is shown in Table 2, and representative photographs are shown in Fig. 3. In C57BL/6 mice inoculated with VRQ homozygous scrapie material, widespread particulate PrPSc staining was observed in all coronal sections of the brain examined (frontal cortex, thalamic section, midbrain, medulla, and cerebellum). A similar distribution of particulate PrPSc staining was seen in C57BL/6 mice inoculated with ARQ homozygous scrapie material, but in addition, amyloid plaques and PrPSc aggregates were also present, most commonly in the thalamus. Similar patterns of PrPSc deposition were also seen in the brains of RIII, VM, and tg59 mice inoculated with either ARQ or VRQ homozygous scrapie isolates, although in tg59 mice inoculated with VRQ homozygous inocula, the particulate deposition of PrPSc was notably absent in the cerebellar cortex. In contrast to the results seen with all of the other mouse lines, inoculation of tg338 mice with VRQ/VRQ or ARQ/ARQ scrapie inocula resulted in similar depositions of PrPSc, evident as fine particulate deposits in the hippocampus, thalamus, hypothalamus, midbrain, and medulla but not the cortical grey matter of the cerebral and cerebellar cortex or the cerebellar white matter. Significantly, the brains of tg338 mice inoculated with either genotype of sheep scrapie inocula were characterized by a general absence of amyloid plaques and PrPSc aggregates, commonly seen in all of the other lines of mice that had received the same inocula. There was a low level of intraneuronal PrPSc deposition in all five mouse lines inoculated with either genotype of scrapie inocula.
TABLE 2.
Disease-associated PrP distribution in the brains of mice with terminal prion disease
| Coronal section | Distribution of PrPSc in the brains of micea
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C57BL/6
|
VM
|
tg59
|
tg338
|
|||||||||||||
| ARQ
|
VRQ
|
ARQ
|
VRQ
|
ARQ
|
VRQ
|
ARQ
|
VRQ
|
|||||||||
| Part | Agg | Part | Agg | Part | Agg | Part | Agg | Part | Agg | Part | Agg | Part | Agg | Part | Agg | |
| Frontal | + | + | + | − | + | + | + | − | + | + | + | − | +/− | −/+ | − | −/+ |
| Thalamic | + | + | + | − | + | + | + | −/+ | + | + | + | −/+ | + | −/+ | + | +/− |
| Midbrain | + | + | + | − | + | + | + | − | + | + | + | − | + | −/+ | + | − |
| Medulla | + | −/+ | + | − | + | −/+ | + | − | + | −/+ | + | − | + | −/+ | + | − |
Immunohistochemistry summary of particulate (Part) and aggregated (Agg) PrPSc in the brains of prion-infected mice. C57BL/6, VM, tg59, and tg338 mice were infected with ARQ and VRQ scrapie inocula. Symbols: +, present in all mice; −, absent in all mice; +/−, present in the majority of mice; −/+, present in a minority of mice.
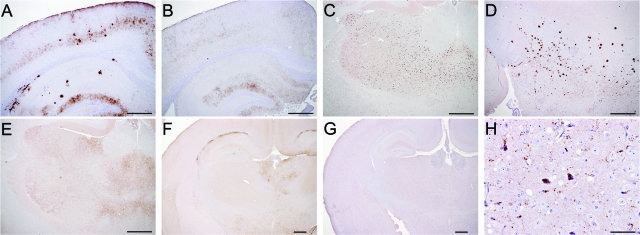
FIG. 3.
Immunohistochemistry of prion-infected mouse brains. (A and B) PrPSc patterns in the cortex and hippocampus of C57BL/6 mice inoculated with ovine ARQ/ARQ and VRQ/VRQ “good” transmitter isolates, respectively. Granular deposits were evident in both cases, but aggregates and plaques were observed only in the mice inoculated with ARQ/ARQ isolates. (C) PrPSc patterns in the thalamus of tg59 mice inoculated with ARQ/ARQ scrapie isolates showing granular PrPSc deposits and larger aggregates distributed throughout the thalamic region. (D) Granular PrPSc patterns in the hypothalamus of tg59 mice inoculated with ARQ/ARQ scrapie isolates. Large aggregates and plaques were also evident in the ventral thalamus. Plaques were the predominant PrPSc deposits in the ventral thalamus, while only rarely were small granular PrPSc deposits observed in the rest of the thalamic region. (E) PrPSc patterns in the thalamus of tg59 mice inoculated with VRQ/VRQ isolates showing granular PrPSc. Aggregates and plaques were observed only in the mice inoculated with ARQ/ARQ isolates. (F and G) PrPSc patterns in the cortex, hippocampus, thalamus, and dorsal hypothalamus of tg338 mice inoculated with ARQ/ARQ and VRQ/VRQ scrapie isolates, respectively. Fine granular deposits, not obvious at this magnification, were evident in both cases, but aggregates of PrPSc along the corpus callosum were observed only in the mice inoculated with ARQ/ARQ isolates. (H) High magnification of the hypothalamus from the same tg338 animal as in panel G to demonstrate PrPSc deposits located in the neuron, glial cells, and neurophil. Bars, 500 μm (A to G) and 50 μm (H).
Quantitation of PrPSc in terminal prion-diseased mouse brains.
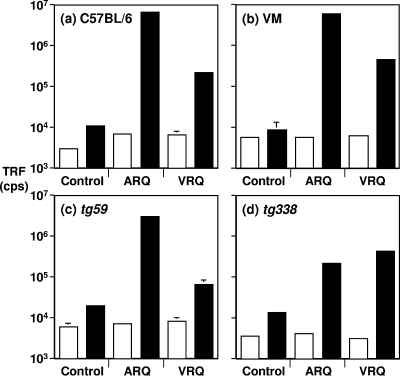
The distinct patterns of PrPSc deposition in mice inoculated with ARQ/ARQ or VRQ/VRQ scrapie isolates were suggestive of quantitative differences in total PrPSc and its subtypes in the brains of these animals. This was investigated by measuring the level of PrPSc in the brains of mice infected with terminal prion disease using a CDI capable of recognition of PK-sensitive and -resistant disease-associated PrP (62). The capture detector CDI used anti-PrP monoclonal antibody V24 for capture and monoclonal antibody 6H4 for detection of PrP. In order to quantify disease-associated PrP in the brains of mice with terminal prion disease, PrPSc was extracted by Sarkosyl in the absence or presence of NaPTA precipitation. Native PrPSc (not treated with GdnHCl) and denatured PrPSc (treated with 6 M GdnHCl) was subsequently quantified by the CDI. Figure 4 shows that significant levels of PrPSc were present in terminal prion-diseased brains of mice inoculated with ARQ or VRQ homozygous scrapie isolates. This was evidenced by the increase in fluorescence counts for Sarkosyl-extracted PrPSc treated with 6 M GdnHCl compared to that obtained in the absence of denaturant. In contrast, the fluorescence counts for both denatured and native samples obtained from control mice were similar. This reflects the exposure of previously buried epitopes within PrPSc that were no longer accessible in denatured PrPC and confirmed the presence of disease-associated PrP in scrapie isolate-inoculated mice. The level of PrPSc that accumulated in C57BL/6, RIII, VM, and tg59 mice following inoculation with ARQ/ARQ scrapie inocula was significantly greater than that which accumulated after inoculation with VRQ/VRQ inocula (P < 0.001). In contrast, this trend was reversed in tg338 mice where the level of PrPSc induced by VRQ homozygous scrapie isolates was higher than that induced by ARQ samples, although the differences were not always statistically significant. Similar trends with elevated levels were seen in all mice when PrPSc was quantified following extraction with NaPTA precipitation (data not shown). These qualitative differences in PrPSc levels measured by CDI correlated with the level of PrPSc described by immunohistochemistry of brain sections from scrapie-inoculated mice.
FIG. 4.
Relative levels of total PrPSc in prion-infected C57BL/6, VM, tg59, and tg338 mouse brains measured by CDI. Prion-infected brains were harvested from mice that had developed terminal prion disease, and homogenates were prepared in Sarkosyl as described in Materials and Methods. Native PrPSc (not treated with GdnHCl) and denatured PrPSc (treated with 6 M GdnHCl) was captured by anti-PrP monoclonal antibody V24 and detected by biotinylated anti-PrP monoclonal antibody 6H4 followed by europium-labeled streptavidin. The data shown are the mean time-resolved fluorescence (TRF) counts per second (cps) plus standard deviation (error bar) for each treatment group. The brain homogenates were treated with 6 M GdnHCl (black bars) or not treated with GdnHCl (white bars). Statistical analysis of the data, with P values shown in the text, was performed using the two-tailed Student t test (unpaired samples).
Molecular profiles of ARQ- and VRQ-induced PrPSc and PrP27-30.
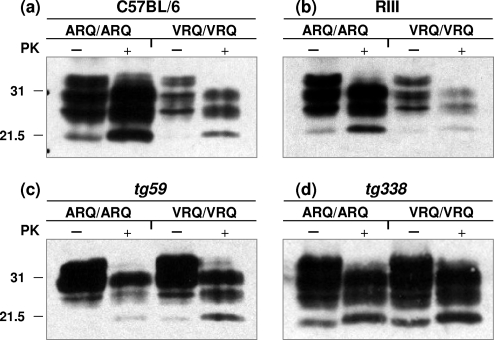
Different subtypes of PrPSc, distinguished by properties such as PK-resistant fragment length and glycoform ratio, have been associated with different prion strains or with different phenotypes of prion disease in the same species (7, 8, 24, 42, 50, 51, 53, 61). In order to investigate the molecular profile of PrP that arose as a consequence of the primary passage of either ARQ or VRQ homozygous sheep scrapie isolates, the brains of mice with terminal prion disease were homogenized and treated with PK or not treated with PK prior to Western blot analysis with anti-PrP monoclonal antibodies. Figure 5 shows that brain homogenates from scrapie-inoculated mice at terminal prion disease contained significant levels of PK-resistant PrPSc in the form of PrP27-30. Brain tissue from scrapie agent-infected C57BL/6 mice (Fig. 5a), RIII mice (Fig. 5b) and VM mice (data not shown) inoculated with ARQ/ARQ scrapie isolates contained significantly higher levels of total PrP and displayed more PrP27-30 than in brain tissue following inoculation with VRQ/VRQ inocula, although the different scrapie isolates were associated with similar incubation times to terminal prion disease in these mice. In contrast, brain tissue from terminal prion-diseased tg59 (Fig. 5c) and tg338 (Fig. 5d) mice inoculated with either ARQ or VRQ sheep scrapie isolates contained similar levels of total PrP and displayed similar levels of PrP27-30. PrPSc induced by either ARQ/ARQ or VRQ/VRQ sheep scrapie isolates was subjected to a titration of PK enzyme in order to determine the susceptibility of PrP27-30 to proteolytic digestion. Despite the significantly different levels of PrP27-30 generated by PK cleavage of PrPSc in C57BL/6 mouse brains following inoculation with ARQ and VRQ homozygous scrapie isolates, this material showed equal resistance to PK digestion over a wide range of enzyme concentrations (1 to 1,000 μg/ml) (data not shown). Similar results were seen when the same titration of PK was used to treat tg59 and tg338 prion-infected mouse brain homogenates (data not shown).
FIG. 5.
Western blot detection of PrP. Prion-infected brains were harvested from mice that had developed terminal prion disease, and homogenates were prepared as described in Materials and Methods. Brain homogenates were treated with PK (+) or not treated with PK (−) and analyzed by SDS-PAGE and Western blotting with anti-PrP monoclonal antibody 683 for C57BL/6 and RIII mouse brains and monoclonal antibody P4 for tg59 and tg338 mouse brains. Each track was loaded with 25 μg of total protein. The positions of molecular mass markers (in kilodaltons) are shown to the left of the blots.
Difference in conformational stability of ARQ- and VRQ-induced PrPSc.
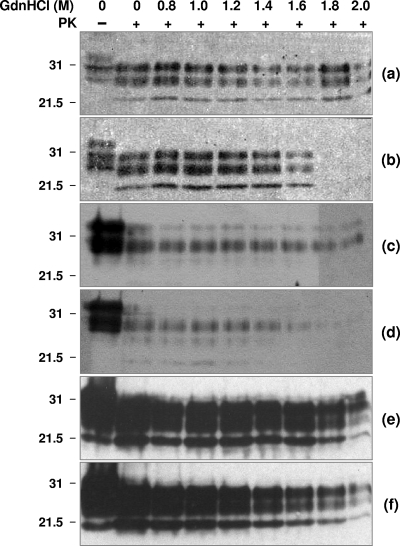
Prion strains have been characterized by the relative stability of their associated PrPSc, measured by resistance to proteolysis following exposure to GdnHCl (43, 52, 53, 63). Exposure of PrPSc to increasing concentrations of GdnHCl leads to a transition from the native to denatured state, measured as loss of resistance to protease digestion. We have used this approach here to examine the relative stability of PrPSc induced by ARQ and VRQ homozygous scrapie inocula following primary passage in conventional and ovine PrP transgenic mice. Accordingly, aliquots of terminal prion-diseased mouse brain homogenate were incubated with increasing amounts of GdnHCl for 30 min followed by limited proteolysis with PK. The samples were subsequently analyzed by Western blotting with anti-PrP monoclonal antibody to detect PrP27-30, the protease-resistant core of PrPSc. Figure 6 shows that the amount of PrP27-30 detected in PK-treated ARQ- and VRQ-inoculated C57BL/6 mouse brains remained constant following treatment with up to 1.6 M GdnHCl. Exposure to increasing amounts of GdnHCl had little effect on the level of PrP27-30 detected in ARQ-inoculated mice (Fig. 6a) but caused an increase in the proteolytic sensitivity of VRQ-induced PrPSc as evidenced by the reduction in PrP27-30 levels on the Western blot (Fig. 6b). VRQ-induced PrPSc was completely denatured by treatment with >1.6 M GdnHCl (Fig. 6b), as evidenced by its complete proteolysis at these concentrations of denaturant. In contrast, significant amounts of PrP27-30 were still evident following treatment of ARQ-induced PrPSc with concentrations of GdnHCl of >1.6 M (Fig. 6a). These data indicate that VRQ-induced PrPSc in the brains of C57BL/6 mice was more unstable than its ARQ-induced counterpart. Similar trends were seen in tg59 mice (Fig. 6c and d) and in RIII and VM mice (data not shown). In contrast, the amount of PrP27-30 detected in PK-treated ARQ- and VRQ-inoculated tg338 mouse brains remained constant following treatment with up to 2.0 M GdnHCl, which indicated a similar stability for PrPSc induced by different genotypes of scrapie inocula in this particular mouse line (Fig. 6e and f, respectively). Replicate ARQ and VRQ homozygous sheep scrapie inocula showed similar trends in the stability of denaturant-induced PrPSc following transmission in each of the mouse lines tested (three mouse brains analyzed from each mouse line in which the inoculum was passaged). These data show that the similar levels of PrPSc induced by ARQ/ARQ and VRQ/VRQ scrapie inocula in tg338 mice possessed similar denaturant-induced stability, whereas the converse was seen for PrPSc induced by the same inocula in C57BL/6, RIII, VM, and tg59 mice.
FIG. 6.
Conformational stability of ARQ- and VRQ-induced PrPSc. Prion-infected mouse brain homogenates were prepared as described in Materials and Methods. Brain homogenates were treated with GdnHCl at the final concentrations shown, incubated in the presence (+) or absence (−) of PK, and subsequently analyzed by SDS-PAGE and Western blotting with anti-PrP monoclonal antibody 683 (a and b) or monoclonal antibody P4 (c to f). The Western blots show PrP from C57BL/6 mice (a and b), tg59 mice (c and d), or tg338 mice (e and f). The mice were inoculated with ARQ/ARQ (a, c, and e) or VRQ/VRQ (b, d, and f) sheep scrapie isolates. The positions of molecular mass markers (in kilodaltons) are shown to the left of the blots.
DISCUSSION
Different isolates of the transmissible scrapie prion agent can exhibit differences in the length of incubation period, lesion profile, and PrPSc deposition in the brains of affected hosts in which the isolates have been passaged (11, 13, 27, 32). The concept of prion strains has emerged in order to account for this variation in transmission properties exhibited by different scrapie isolates, during serial passage, when variance due to dose, inoculation route, and most importantly, host and donor PrP genotype are taken into account. Historically, ovine prion strain typing has been carried out by serial passage of scrapie isolates in a panel of conventional mice (11). This strategy has allowed identification of a number of different ovine prion strains and discrimination between scrapie and BSE (10, 12). However, the failure of some scrapie isolates to transmit to conventional mice is likely to underestimate the true diversity of ovine scrapie strains. Furthermore, passage of ovine prions across a species or transmission barrier may not reveal properties of the infectious prion agent as they exist in the original host. Both of these issues can potentially be circumvented through the use of mice transgenic for ovine PrP. Although facilitated transmission of ovine prions is seen in ovine PrP transgenic mice (4, 26, 42, 68), the utility of these hosts for ovine prion strain typing has not been fully established. Here we have compared the molecular and transmission characteristics of scrapie isolates of different genotypes in ovine PrP transgenic mice and conventional mice in order to begin to address this issue.
The ARQ and VRQ homozygous scrapie isolates used here showed similar glycoform profiles but distinctly different levels of PrP27-30, and presumably therefore different levels of prion infectivity. Despite this, both genotypes of scrapie isolate showed high primary transmission attack rates in C57BL/6 (Prnpa), RIII (Prnpa), and VM (Prnpb) mouse lines and in tg59 and tg338 mice, those transgenic for ovine ARQ or VRQ PrP, respectively. The attack rates for the scrapie isolates in the different mouse lines ranged from 67 to 100%. Other studies have reported a failure of transmission by some ARQ homozygous isolates in C57BL/6 mice (4, 12). Despite the efficient attack rates seen here for both genotypes of scrapie inocula, similarities and differences in prion disease incubation time were seen between the different prion-inoculated mouse lines. Both genotypes of scrapie isolate showed similar primary transmission incubation times in C57BL/6, RIII, and VM mice. However, significantly shorter incubation times were seen for all of the scrapie isolates in tg338 mice. The accelerated rate of prion disease in tg338 mice may reflect a lower transmission barrier for ovine prion inocula passaged in mice that express ovine PrP compared to that for similar inocula in conventional mouse lines. The fact that a similar level of facilitated transmission was not seen in tg59 mice may indicate differences in the expression level or location of ovine PrP between the two different ovine PrP transgenic mice. tg338 mice express ovine VRQ PrP under the control of the ovine PrP promoter, and tg59 mice express ovine ARQ under the control of the neuron-specific enolase promoter (25, 68). Furthermore, tg338 mice express approximately fivefold-more ovine PrP protein than do tg59 mice (our unpublished observations). However, while tg338 mice allowed facilitated transmission for both genotypes of scrapie isolates, VRQ/VRQ inocula showed significantly shorter incubation times in this transgenic mouse line than did ARQ/ARQ inocula. Although the relative susceptibility of tg338 mice for the ARQ or VRQ homozygous inocula used here could not be assessed because of their potential difference in prion infectivity, as indicated by their different levels of PrP27-30, it was clear that rapid transmission of different genotypes of scrapie inocula occurred in tg338 mice.
Lesion profiles within terminal prion-diseased ovine PrP transgenic and conventional mouse brains were determined in order to assess the spongiform neurodegeneration induced by transmission of the ARQ/ARQ and VRQ/VRQ inocula. Lesion profiles provide a semiquantitative assessment of neuropathology in different brain regions and have been used to discriminate between different prion strains (13, 32, 33). The different lesion profiles reported here for ARQ/ARQ and VRQ/VRQ scrapie inocula are indicative of different prion strains, although the generally distinct neuropathologies resulted from primary transmission of different genotypes of ovine prion inocula. Different strains of the infectious prion agent are revealed, or produced, by passage of prions through hosts of different PrP genotypes (57). To account for this, it is proposed that prion strains comprise an ensemble of PrPSc conformations characterized by a dominant conformer for a given PrP amino acid sequence (22). Accordingly, the ability of prions to replicate within a new host of a different PrP primary amino acid sequence will be influenced by the degree of overlap between the allowed repertoires of PrPSc conformations for both donor and host PrP genotype. In this scheme, passage of prions between hosts occurs more readily when the range of PrPSc conformers overlap but less readily when there is little overlap, with the latter situation leading to the emergence of a new dominant PrPSc conformer or strain. The propagation of prions between species, or more precisely between different PrP genotypes, may therefore be viewed as a function of the thermodynamic stability of the PrPSc conformation and the kinetics of its formation and clearance (21, 22). Generally, there was good correlation between lesion profiles induced by replicate ARQ/ARQ or VRQ/VRQ inocula in most of the mouse lines. In addition, lesion profiles for ARQ/ARQ inocula in tg338 mice were similar to that seen in RIII mice. However, in tg59 mice, somewhat different profiles were obtained during transmission of ARQ/ARQ inocula, which is consistent with selection of different prion strains from the different replicates or the effect of mouse background differences on strain selection. The tg59 mouse line is produced on a C57BL/6J, 129Sv, and OF1 mixed genetic background (26), and individual mouse variation to particular prion strain susceptibility may occur. Variation in individual genetic backgrounds is considered a possible contributing factor in the apparent selection of different human prion strains following transmission of bovine BSE to mice overexpressing human PrP (3). In contrast to the other mouse lines analyzed here, lesion profiles in tg338 mice induced by ARQ/ARQ and VRQ/VRQ inocula were very similar, differing most qualitatively in the thalamic grey matter area. This was despite the fact that total PrPSc levels in tg338 mice measured by CDI following inoculation with ARQ/ARQ or VRQ/VRQ scrapie isolates were fairly similar, whereas in all other mouse lines, ARQ/ARQ inocula induced more PrPSc than VRQ/VRQ samples did.
We investigated the deposition of PrPSc in the brains of terminal prion-diseased mice by immunohistochemistry. The relatively high levels of total PrPSc in prion-diseased C57BL/6, RIII, VM, and tg59 mouse brains induced by ARQ/ARQ inocula correlated with the presence of aggregated PrP deposition, which was rarely seen in mice that had received VRQ/VRQ inocula. In cases of natural sheep scrapie, there is a genotype-specific differentiation in the patterns and types of disease-associated PrP deposition in the central nervous system (44, 59). In addition to the PrP genotype of the host, it has been shown that the nature of the inoculum may also significantly affect the pattern of disease-associated PrP deposition (34). Here we have also observed genotype-related differences in the deposition of disease-associated PrP during primary transmissions of ovine scrapie inocula in mice. The increased deposition of PrPSc induced by ARQ/ARQ scrapie isolates seen here in conventional and tg59 mice may be the result of an increased efficiency in PrPSc formation in these animals. It is worth noting that ovine and murine PrP show significant amino acid homology. Ovine wild-type ARQ and VRQ PrP allelic variants differ only in the A→V polymorphism at amino acid position 136 (ovine PrP numbering), and the equivalent amino acid in both Prnpa and Prnpb genotypes of murine PrP (amino acid residue 133 [murine PrP numbering]) is an alanine residue (71). Identity at amino acid residue 136 of ovine PrP and at the equivalent amino acid residue in murine PrP may predispose to a more efficient conversion and aggregation of PrPC into PrPSc. Alternatively, the increased deposition of PrPSc aggregates in the brains of mice inoculated with ARQ/ARQ scrapie isolates may suggest that the clearance of PrPSc occurred at a slower rate, or with lower efficiency, than in mice inoculated with VRQ/VRQ isolates. One reason to account for this could be that PrPSc induced by ARQ/ARQ scrapie isolates in Prnpa, Prnpb, and tg59 mice was more resistant to metabolism than PrPSc that accumulated in similar mice inoculated with VRQ/VRQ scrapie material. This appeared to be the case, since ARQ/ARQ-induced PrPSc from these mice was found to be more stable than PrPSc induced by VRQ/VRQ inocula. Studies of prion proteins in other biological systems, such as Saccharomyces cerevisiae, have suggested that the strain-specific properties correlate with the physical properties of prion protein aggregates, such as frangibility, which may contribute to their metabolic fate (60).
It has been a long-standing assumption that PrPSc is primarily responsible for spongiform neurodegeneration in prion diseases because of the correlation between its anatomical and temporal accumulation and the ensuing neuropathology. The fact that different levels of PrPSc accumulate in the brains of mice that succumbed to terminal prion disease with similar incubation times suggests that the level of PrPSc measured here does not directly correlate with the extent of pathology or neurotoxicity. Conversion of PrPC to PrPSc is a fundamental feature of prion disease pathogenesis, since PrP−/− mice are resistant to prion infection (15, 47). However, the mechanism of neurotoxicity remains to be elucidated (19). While it is established that neurons must express PrPC to be susceptible to toxicity, there are several examples of prion disease or experimental transmissions where the levels of PrPSc are low or apparently absent (23, 36, 41, 48, 49). In contrast, high levels of PrPSc may accumulate in the absence of full-blown clinical disease (35, 55, 64, 65). In addition, ablation of neuronal PrPC expression during ongoing prion disease in mice can prevent the onset of clinical signs and reverse early spongiform neuropathology (45, 46). An emerging view is that large aggregated forms of PrPSc are relatively inert and innocuous, and smaller, potentially labile, oligomeric species of PrP generated as an intermediate during prion replication constitute the actual neurotoxic agent (2, 14, 20, 35, 69). According to this scheme, a critical level of neurotoxic PrP must accumulate in order for the prion-infected individual to develop spongiform neurodegeneration. In the context of the protein-only hypothesis, the relatively long incubation times seen for sheep scrapie inocula in conventional mice can be interpreted as the result of an inefficient conversion of PrPC to PrPSc across the sheep-mouse species barrier, with a corresponding slow accumulation of the neurotoxic form of PrP. In contrast, the short incubation times for sheep scrapie inocula in tg338 mice can be seen to occur as a result of their high level of ovine PrPC expression, leading to an efficient conversion of PrP coupled with fast accumulation of the neurotoxic PrP intermediate.
Although tg338 mice showed short incubation periods for the onset of terminal prion disease following inoculation with sheep scrapie isolates, these mice displayed a more uniform phenotype with regard to those parameters typically used to distinguish different ovine prion strains. Scrapie-inoculated tg338 mice showed similar lesion profiles that were accompanied by similar PrPSc depositions following inoculation with either ARQ or VRQ homozygous scrapie isolates, whereas different patterns were seen in similarly inoculated tg59 and conventional mouse lines. These observations imply that tg338 mice may reveal a different range of ovine prion strains than do tg59 mice and that tg59 mice may more closely resemble conventional mice in the range of ovine prion strains that they can replicate. This difference between tg338 and tg59 mice in their potential to distinguish or generate ovine prion strains also correlates with the conformational variation of PrPSc that accumulates in these VRQ and ARQ ovine PrP transgenic mice. PrPSc that accumulated in ARQ tg59 mice showed different denaturant-induced stability following inoculation with either ARQ or VRQ homozygous inocula, whereas PrPSc that accumulated in similarly inoculated tg338 mice showed similar stability. We and others have previously shown that ovine VRQ PrP is more stable than ovine ARQ PrP (16, 29, 56), and this may reflect that ovine PrP with alanine at amino acid residue 136 is more flexible in its ability to adopt different conformations than is ovine PrP with valine at amino acid residue 136. This would in turn suggest that host PrP genotype plays a dominant role in distinguishing or generating ovine prion strains. Differences in prion disease phenotype were observed in the ovine PrP transgenic mouse lines after inoculation with scrapie isolates from sheep of different PrP genotypes. Whether these differences represent stable variations, and hence strain variation, can be confirmed only by further serial passage. If strain differences are confirmed, this would suggest that these transgenic lines may allow for a stable strain phenotype to be observed on first passage. This would be a significant advantage for the use of ovine PrP transgenic animals in ovine prion strain typing. Subsequent serial passage in tg338, tg59, and conventional mice of the different scrapie isolates used here will be required in order to determine the validity of this approach, and these studies are currently under way.
Acknowledgments
This work was supported by funds from Department for Environment, Food & Rural Affairs (Defra). L.H. is in receipt of a Defra Ph.D. studentship.
We thank INRA Jouy-en-Josas for supplying the tg338 mouse line. We thank Thierry Baron for supplying the tg59 mouse line. We thank support staff in the Departments of Histology and Neuropathology and the Animal Services Unit of the Veterinary Laboratories Agency, Weybridge laboratory, for their skilled sample preparation and handling.
Footnotes
Published ahead of print on 3 September 2008.
REFERENCES
- 1.Aguzzi, A., M. Heikenwalder, and M. Polymenidou. 2007. Insights into prion strains and neurotoxicity. Nat. Rev. Mol. Cell Biol. 8552-561. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, M., O. V. Bocharova, N. Makarava, L. Breydo, V. V. Salnikov, and I. V. Baskakov. 2006. Polymorphism and ultrastructural organization of prion protein amyloid fibrils: an insight from high resolution atomic force microscopy. J. Mol. Biol. 358580-596. [DOI] [PubMed] [Google Scholar]
- 2a.The Animals (Scientific Procedures) Act. 1986. Eliz. 2, chapter 14. Her Majesty's Stationery Office, London, United Kingdom.
- 3.Asante, E. A., J. M. Linehan, M. Desbruslais, S. Joiner, I. Gowland, A. L. Wood, J. Welch, A. F. Hill, S. E. Lloyd, J. D. Wadsworth, and J. Collinge. 2002. BSE prions propagate as either variant CJD-like or sporadic CJD-like prion strains in transgenic mice expressing human prion protein. EMBO J. 216358-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, T., C. Crozet, A. G. Biacabe, S. Philippe, J. Verchere, A. Bencsik, J. Y. Madec, D. Calavas, and J. Samarut. 2004. Molecular analysis of the protease-resistant prion protein in scrapie and bovine spongiform encephalopathy transmitted to ovine transgenic and wild-type mice. J. Virol. 786243-6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck, K., R. Salis, S. Simonini, and J. Spiropoulos. 2007. Primary isolation of the scrapie agent in RIII mice: identification of sheep PrP genotype-associated profiles, abstr. P02.20. Prion 2007. NeuroPrion European Network of Excellence.
- 6.Beringue, V., A. Bencsik, A. Le Dur, F. Reine, T. L. Lai, N. Chenais, G. Tilly, A. G. Biacabe, T. Baron, J. L. Vilotte, and H. Laude. 2006. Isolation from cattle of a prion strain distinct from that causing bovine spongiform encephalopathy. PLoS Pathog. 2e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bessen, R. A., and R. F. Marsh. 1992. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol. 662096-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessen, R. A., and R. F. Marsh. 1994. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 687859-7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browning, S. R., G. L. Mason, T. Seward, M. Green, G. A. Eliason, C. Mathiason, M. W. Miller, E. S. Williams, E. Hoover, and G. C. Telling. 2004. Transmission of prions from mule deer and elk with chronic wasting disease to transgenic mice expressing cervid PrP. J. Virol. 7813345-13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruce, M. E. 1993. Scrapie strain variation and mutation. Br. Med. Bull. 49822-838. [DOI] [PubMed] [Google Scholar]
- 11.Bruce, M. E. 2003. TSE strain variation. Br. Med. Bull. 6699-108. [DOI] [PubMed] [Google Scholar]
- 12.Bruce, M. E., A. Boyle, S. Cousens, I. McConnell, J. Foster, W. Goldmann, and H. Fraser. 2002. Strain characterization of natural sheep scrapie and comparison with BSE. J. Gen. Virol. 83695-704. [DOI] [PubMed] [Google Scholar]
- 13.Bruce, M. E., and H. Fraser. 1991. Scrapie strain variation and its implications. Curr. Top. Microbiol. Immunol. 172125-138. [DOI] [PubMed] [Google Scholar]
- 14.Bucciantini, M., E. Giannoni, F. Chiti, F. Baroni, L. Formigli, J. Zurdo, N. Taddei, G. Ramponi, C. M. Dobson, and M. Stefani. 2002. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 416507-511. [DOI] [PubMed] [Google Scholar]
- 15.Bueler, H., A. Aguzzi, A. Sailer, R. A. Greiner, P. Autenried, M. Aguet, and C. Weissmann. 1993. Mice devoid of PrP are resistant to scrapie. Cell 731339-1347. [DOI] [PubMed] [Google Scholar]
- 16.Bujdoso, R., D. F. Burke, and A. M. Thackray. 2005. Structural differences between allelic variants of the ovine prion protein revealed by molecular dynamics simulations. Proteins 61840-849. [DOI] [PubMed] [Google Scholar]
- 17.Buschmann, A., E. Pfaff, K. Reifenberg, H. M. Muller, and M. H. Groschup. 2000. Detection of cattle-derived BSE prions using transgenic mice overexpressing bovine PrP(C). Arch. Virol. Suppl. 200075-86. [DOI] [PubMed] [Google Scholar]
- 18.Chesebro, B. 1998. BSE and prions: uncertainties about the agent. Science 27942-43. [DOI] [PubMed] [Google Scholar]
- 19.Chiesa, R., and D. A. Harris. 2001. Prion diseases: what is the neurotoxic molecule? Neurobiol. Dis. 8743-763. [DOI] [PubMed] [Google Scholar]
- 20.Chiesa, R., P. Piccardo, E. Quaglio, B. Drisaldi, S. L. Si-Hoe, M. Takao, B. Ghetti, and D. A. Harris. 2003. Molecular distinction between pathogenic and infectious properties of the prion protein. J. Virol. 777611-7622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collinge, J. 1999. Variant Creutzfeldt-Jakob disease. Lancet 354317-323. [DOI] [PubMed] [Google Scholar]
- 22.Collinge, J., and A. R. Clarke. 2007. A general model of prion strains and their pathogenicity. Science 318930-936. [DOI] [PubMed] [Google Scholar]
- 23.Collinge, J., M. S. Palmer, K. C. Sidle, I. Gowland, R. Medori, J. Ironside, and P. Lantos. 1995. Transmission of fatal familial insomnia to laboratory animals. Lancet 346569-570. [DOI] [PubMed] [Google Scholar]
- 24.Collinge, J., K. C. Sidle, J. Meads, J. Ironside, and A. F. Hill. 1996. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature 383685-690. [DOI] [PubMed] [Google Scholar]
- 25.Cordier, C., A. Bencsik, S. Philippe, D. Betemps, F. Ronzon, D. Calavas, C. Crozet, and T. Baron. 2006. Transmission and characterization of bovine spongiform encephalopathy sources in two ovine transgenic mouse lines (TgOvPrP4 and TgOvPrP59). J. Gen. Virol. 873763-3771. [DOI] [PubMed] [Google Scholar]
- 26.Crozet, C., F. Flamant, A. Bencsik, D. Aubert, J. Samarut, and T. Baron. 2001. Efficient transmission of two different sheep scrapie isolates in transgenic mice expressing the ovine PrP gene. J. Virol. 755328-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickinson, A. G., and V. M. Meikle. 1971. Host-genotype and agent effects in scrapie incubation: change in allelic interaction with different strains of agent. Mol. Gen. Genet. 11273-79. [DOI] [PubMed] [Google Scholar]
- 28.Dickinson, A. G., V. M. Meikle, and H. Fraser. 1968. Identification of a gene which controls the incubation period of some strains of scrapie agent in mice. J. Comp. Pathol. 78293-299. [DOI] [PubMed] [Google Scholar]
- 29.Eghiaian, F., J. Grosclaude, S. Lesceu, P. Debey, B. Doublet, E. Treguer, H. Rezaei, and M. Knossow. 2004. Insight into the PrPC→PrPSc conversion from the structures of antibody-bound ovine prion scrapie-susceptibility variants. Proc. Natl. Acad. Sci. USA 10110254-10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Espinosa, J. C., O. Andreoletti, J. Castilla, M. E. Herva, M. Morales, E. Alamillo, F. D. San-Segundo, C. Lacroux, S. Lugan, F. J. Salguero, J. Langeveld, and J. M. Torres. 2007. Sheep-passaged bovine spongiform encephalopathy agent exhibits altered pathobiological properties in bovine-PrP transgenic mice. J. Virol. 81835-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farquhar, C. F., R. A. Somerville, and M. E. Bruce. 1998. Straining the prion hypothesis. Nature 391345-346. [DOI] [PubMed] [Google Scholar]
- 32.Fraser, H., and A. G. Dickinson. 1973. Scrapie in mice. Agent-strain differences in the distribution and intensity of grey matter vacuolation. J. Comp. Pathol. 8329-40. [DOI] [PubMed] [Google Scholar]
- 33.Fraser, H., and A. G. Dickinson. 1968. The sequential development of the brain lesion of scrapie in three strains of mice. J. Comp. Pathol. 78301-311. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez, L., S. Martin, I. Begara-McGorum, N. Hunter, F. Houston, M. Simmons, and M. Jeffrey. 2002. Effects of agent strain and host genotype on PrP accumulation in the brain of sheep naturally and experimentally affected with scrapie. J. Comp. Pathol. 12617-29. [DOI] [PubMed] [Google Scholar]
- 34a.Green, R. B., C. Horrocks, A. Wilkinson, S. A. C. Hawkins, and S. J. Ryder. 2005. Primary isolation of the bovine spongiform encephalopathy agent in mice: agent definition based on a review of 150 transmissions. J. Comp. Pathol. 132117-131. [DOI] [PubMed] [Google Scholar]
- 35.Hill, A. F., S. Joiner, J. Linehan, M. Desbruslais, P. L. Lantos, and J. Collinge. 2000. Species-barrier-independent prion replication in apparently resistant species. Proc. Natl. Acad. Sci. USA 9710248-10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsiao, K. K., M. Scott, D. Foster, D. F. Groth, S. J. DeArmond, and S. B. Prusiner. 1990. Spontaneous neurodegeneration in transgenic mice with mutant prion protein. Science 2501587-1590. [DOI] [PubMed] [Google Scholar]
- 37.Kimberlin, R. H., S. Cole, and C. A. Walker. 1987. Temporary and permanent modifications to a single strain of mouse scrapie on transmission to rats and hamsters. J. Gen. Virol. 681875-1881. [DOI] [PubMed] [Google Scholar]
- 38.Kimberlin, R. H., and C. A. Walker. 1978. Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J. Gen. Virol. 39487-496. [DOI] [PubMed] [Google Scholar]
- 39.Kimberlin, R. H., C. A. Walker, and H. Fraser. 1989. The genomic identity of different strains of mouse scrapie is expressed in hamsters and preserved on reisolation in mice. J. Gen. Virol. 702017-2025. [DOI] [PubMed] [Google Scholar]
- 40.Korth, C., B. Stierli, P. Streit, M. Moser, O. Schaller, R. Fischer, W. Schulz-Schaeffer, H. Kretzschmar, A. Raeber, U. Braun, F. Ehrensperger, S. Hornemann, R. Glockshuber, R. Riek, M. Billeter, K. Wuthrich, and B. Oesch. 1997. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature 39074-77. [DOI] [PubMed] [Google Scholar]
- 41.Lasmezas, C. I., J. P. Deslys, O. Robain, A. Jaegly, V. Beringue, J. M. Peyrin, J. G. Fournier, J. J. Hauw, J. Rossier, and D. Dormont. 1997. Transmission of the BSE agent to mice in the absence of detectable abnormal prion protein. Science 275402-405. [DOI] [PubMed] [Google Scholar]
- 42.Le Dur, A., V. Beringue, O. Andreoletti, F. Reine, T. L. Lai, T. Baron, B. Bratberg, J. L. Vilotte, P. Sarradin, S. L. Benestad, and H. Laude. 2005. A newly identified type of scrapie agent can naturally infect sheep with resistant PrP genotypes. Proc. Natl. Acad. Sci. USA 10216031-16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Legname, G., H. O. Nguyen, D. Peretz, F. E. Cohen, S. J. DeArmond, and S. B. Prusiner. 2006. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc. Natl. Acad. Sci. USA 10319105-19110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ligios, C., G. Dexter, J. Spiropoulos, C. Maestrale, A. Carta, and M. M. Simmons. 2004. Distribution of vascular amyloid in scrapie-affected sheep with different genotypes. J. Comp. Pathol. 131271-276. [DOI] [PubMed] [Google Scholar]
- 45.Mallucci, G., A. Dickinson, J. Linehan, P. C. Klohn, S. Brandner, and J. Collinge. 2003. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 302871-874. [DOI] [PubMed] [Google Scholar]
- 46.Mallucci, G. R., M. D. White, M. Farmer, A. Dickinson, H. Khatun, A. D. Powell, S. Brandner, J. G. Jefferys, and J. Collinge. 2007. Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron 53325-335. [DOI] [PubMed] [Google Scholar]
- 47.Manson, J. C., A. R. Clarke, P. A. McBride, I. McConnell, and J. Hope. 1994. PrP gene dosage determines the timing but not the final intensity or distribution of lesions in scrapie pathology. Neurodegeneration 3331-340. [PubMed] [Google Scholar]
- 48.Manson, J. C., E. Jamieson, H. Baybutt, N. L. Tuzi, R. Barron, I. McConnell, R. Somerville, J. Ironside, R. Will, M. S. Sy, D. W. Melton, J. Hope, and C. Bostock. 1999. A single amino acid alteration (101L) introduced into murine PrP dramatically alters incubation time of transmissible spongiform encephalopathy. EMBO J. 186855-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medori, R., P. Montagna, H. J. Tritschler, A. LeBlanc, P. Cortelli, P. Tinuper, E. Lugaresi, and P. Gambetti. 1992. Fatal familial insomnia: a second kindred with mutation of prion protein gene at codon 178. Neurology 42669-670. [DOI] [PubMed] [Google Scholar]
- 50.Monari, L., S. G. Chen, P. Brown, P. Parchi, R. B. Petersen, J. Mikol, F. Gray, P. Cortelli, P. Montagna, and B. Ghetti. 1994. Fatal familial insomnia and familial Creutzfeldt-Jakob disease: different prion proteins determined by a DNA polymorphism. Proc. Natl. Acad. Sci. USA 912839-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parchi, P., R. Castellani, S. Capellari, B. Ghetti, K. Young, S. G. Chen, M. Farlow, D. W. Dickson, A. A. Sima, J. Q. Trojanowski, R. B. Petersen, and P. Gambetti. 1996. Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann. Neurol. 39767-778. [DOI] [PubMed] [Google Scholar]
- 52.Peretz, D., M. R. Scott, D. Groth, R. A. Williamson, D. R. Burton, F. E. Cohen, and S. B. Prusiner. 2001. Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci. 10854-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peretz, D., R. A. Williamson, G. Legname, Y. Matsunaga, J. Vergara, D. R. Burton, S. J. DeArmond, S. B. Prusiner, and M. R. Scott. 2002. A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron 34921-932. [DOI] [PubMed] [Google Scholar]
- 54.Prusiner, S. B. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216136-144. [DOI] [PubMed] [Google Scholar]
- 55.Race, R., A. Raines, G. J. Raymond, B. Caughey, and B. Chesebro. 2001. Long-term subclinical carrier state precedes scrapie replication and adaptation in a resistant species: analogies to bovine spongiform encephalopathy and variant Creutzfeldt-Jakob disease in humans. J. Virol. 7510106-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rezaei, H., Y. Choiset, F. Eghiaian, E. Treguer, P. Mentre, P. Debey, J. Grosclaude, and T. Haertle. 2002. Amyloidogenic unfolding intermediates differentiate sheep prion protein variants. J. Mol. Biol. 322799-814. [DOI] [PubMed] [Google Scholar]
- 57.Ridley, R. M., and H. F. Baker. 1996. To what extent is strain variation evidence for an independent genome in the agent of the transmissible spongiform encephalopathies? Neurodegeneration 5219-231. [DOI] [PubMed] [Google Scholar]
- 58.Safar, J., H. Wille, V. Itri, D. Groth, H. Serban, M. Torchia, F. E. Cohen, and S. B. Prusiner. 1998. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 41157-1165. [DOI] [PubMed] [Google Scholar]
- 59.Spiropoulos, J., C. Casalone, M. Caramelli, and M. M. Simmons. 2007. Immunohistochemistry for PrPSc in natural scrapie reveals patterns which are associated with the PrP genotype. Neuropathol. Appl. Neurobiol. 33398-409. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka, M., S. R. Collins, B. H. Toyama, and J. S. Weissman. 2006. The physical basis of how prion conformations determine strain phenotypes. Nature 442585-589. [DOI] [PubMed] [Google Scholar]
- 61.Telling, G. C., P. Parchi, S. J. DeArmond, P. Cortelli, P. Montagna, R. Gabizon, J. Mastrianni, E. Lugaresi, P. Gambetti, and S. B. Prusiner. 1996. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 2742079-2082. [DOI] [PubMed] [Google Scholar]
- 62.Thackray, A. M., L. Hopkins, and R. Bujdoso. 2007. Proteinase K-sensitive disease-associated ovine prion protein revealed by conformation-dependent immunoassay. Biochem. J. 401475-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thackray, A. M., L. Hopkins, M. A. Klein, and R. Bujdoso. 2007. Mouse-adapted ovine scrapie prion strains are characterized by different conformers of PrPSc. J. Virol. 8112119-12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thackray, A. M., M. A. Klein, A. Aguzzi, and R. Bujdoso. 2002. Chronic subclinical prion disease induced by low-dose inoculum. J. Virol. 762510-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thackray, A. M., M. A. Klein, and R. Bujdoso. 2003. Subclinical prion disease induced by oral inoculation. J. Virol. 777991-7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thackray, A. M., S. Yang, E. Wong, T. J. Fitzmaurice, R. J. Morgan-Warren, and R. Bujdoso. 2004. Conformational variation between allelic variants of cell-surface ovine prion protein. Biochem. J. 381221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thuring, C. M., J. H. Erkens, J. G. Jacobs, A. Bossers, L. J. Van Keulen, G. J. Garssen, F. G. Van Zijderveld, S. J. Ryder, M. H. Groschup, T. Sweeney, and J. P. Langeveld. 2004. Discrimination between scrapie and bovine spongiform encephalopathy in sheep by molecular size, immunoreactivity, and glycoprofile of prion protein. J. Clin. Microbiol. 42972-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vilotte, J. L., S. Soulier, R. Essalmani, M. G. Stinnakre, D. Vaiman, L. Lepourry, J. C. Da Silva, N. Besnard, M. Dawson, A. Buschmann, M. Groschup, S. Petit, M. F. Madelaine, S. Rakatobe, A. Le Dur, D. Vilette, and H. Laude. 2001. Markedly increased susceptibility to natural sheep scrapie of transgenic mice expressing ovine PrP. J. Virol. 755977-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walsh, D. M., I. Klyubin, J. V. Fadeeva, W. K. Cullen, R. Anwyl, M. S. Wolfe, M. J. Rowan, and D. J. Selkoe. 2002. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature 416535-539. [DOI] [PubMed] [Google Scholar]
- 70.Weissmann, C. 1991. A ‘unified theory’ of prion propagation. Nature 352679-683. [DOI] [PubMed] [Google Scholar]
- 71.Wopfner, F., G. Weidenhofer, R. Schneider, A. von Brunn, S. Gilch, T. F. Schwarz, T. Werner, and H. M. Schatzl. 1999. Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J. Mol. Biol. 2891163-1178. [DOI] [PubMed] [Google Scholar]