Abstract
The H19/IGFf2 locus belongs to a large imprinted domain located on human chromosome 11p15.5 (homologue to mouse distal chromosome 7). The H19 gene is expressed from the maternal allele, while IGF2 is paternally expressed. Natural antisense transcripts and intergenic transcription have been involved in many aspects of eukaryotic gene expression, including genomic imprinting and RNA interference. However, apart from the identification of some IGF2 antisense transcripts, few data are available on that topic at the H19/IGF2 locus. We identify here a novel transcriptional activity at both the human and the mouse H19/IGF2 imprinted loci. This activity occurs antisense to the H19 gene and has the potential to produce a single 120-kb transcript that we called the 91H RNA. This nuclear and short-lived RNA is not imprinted in mouse but is expressed predominantly from the maternal allele in both mice and humans within the H19 gene region. Moreover, the transcript is stabilized in breast cancer cells and overexpressed in human breast tumors. Finally, knockdown experiments showed that, in humans, 91H, rather than affecting H19 expression, regulates IGF2 expression in trans.
H19 is located within a cluster of imprinted genes on the human chromosome 11 in p15.5 (homologue to the murine distal chromosome 7). The regulation of H19 and its closely linked and reciprocally imprinted neighbor, IGF2, has been studied intensively both because of its role in human diseases and as a model for understanding imprinting control mechanisms. Thereby, H19 is transcribed only from the maternal allele, whereas IGF2 expression is exclusively paternal (8). Over the years, H19 is considered as a regulatory RNA (5). It has been proposed to function in many different processes, ranging from transcriptional and posttranscriptional regulation (25, 26, 53) to tumor suppression (35, 45) and oncogenesis, more particularly in breast cancer (1, 27, 28).
Genomic imprinting plays a critical role in modulating gene expression during embryogenesis and normal development (36). It consists of an epigenetic modification that results in the silencing of a specific allele, depending on its parental origin. Most of the 80 mammalian imprinted genes identified thus far are organized into clusters (33). It has been suggested that the clustering of genes enables sharing of cis-acting elements located several kilobase pairs away (21). Differentially methylated regions associated with the imprinted clusters play a crucial role in the imprinted expression patterns, which are hence called imprinting control regions (ICRs) (22, 38). The H19/IGF2 locus is a well-characterized cluster. Both genes share a common set of enhancers located downstream from the H19 gene. The ICR, located 2 kb upstream of the H19 promoter, controls the monoallelic expression of the H19 and IGF2 genes by insulating communication between the 3′ enhancers and the IGF2 promoter. The chromatin insulator property of the H19/IGF2 ICR is regulated by a chromatin insulator protein, CTCF (for CCTC-binding factor), which binds only the nonmethylated maternal allele. On the paternal allele, the ICR methylation does not allow CTCF binding and leads to IGF2 expression (for a review, see reference 24). The IGF2 gene itself also contains differentially methylated regions (DMRs), with DMR1 being a methylation-sensitive silencer and DMR2 being a methylation-sensitive activator (7, 30). Elsewhere, the role of CTCF in chromatin loops is now well demonstrated. Indeed, CTCF binding in the maternal ICR is able to regulate its interaction with matrix attachment region 3 (MAR3) and DMR1 at IGF2, thus forming a tight loop around the maternal IGF2 locus which may contribute to its silencing. These interactions may contribute to restrict the physical access of distal enhancers to IGF2 promoter (19, 30, 50). Additional mechanisms exist for an imprint mark, such as chromatin composition, organization, and histone acetylation or methylation state (12, 14), even if DNA methylation is by far the best candidate (4).
A more recently identified characteristic of imprinted genes is their association, in some cases, with noncoding antisense transcripts (ncRNAs), which have been suggested to constitute a new epigenetic regulatory system. Indeed, for the maternally expressed Igf2r and UBE3A genes, overlapping antisense transcripts have been found and are oppositely imprinted with respect to the protein coding gene. It has been proposed that antisense transcripts serve to regulate overlapping genes by promoter or transcript occlusion or by competing with these loci for regulatory elements such as transcription factors or enhancers (39, 40, 44, 54). Elsewhere, in the imprinted cluster at the distal end of the mouse chromosome 7 (orthologous to the human chromosome 11), the ICR located in the KCNQ1 gene contains the promoter of a long paternally expressed antisense transcript called KCNQ1OT1. This transcript takes part in gene silencing of the KCNQ1 domain, and its interruption by the insertion of a polyadenylation sequence caused a loss of methylation spreading in cultured cells (31, 47).
These ncRNAs are not yet clearly classified, but categories with known gene regulatory functions are emerging. It includes intergenic transcripts that regulate local chromatin activity, cis-acting long ncRNAs such as Xist involved in chromosome inactivation, and the imprinted Air and KCNQ1OT1 ncRNAs involved in domain silencing (15, 32).
In the present study we have identified and characterized a new transcript within the human and mouse H19/IGF2 locus. This long intergenic RNA that we named 91H is antisense to H19. We assessed the effect of the 91H knockdown by RNA interference on genomic imprinting and on H19 and IGF2 gene expression. The results reported here add further complexity to the locus regulation and are attractive within the context of imprinting maintenance and/or cancer development.
MATERIALS AND METHODS
Cell culture and breast tissues.
The estrogen-sensitive MCF7 and T47D and the estrogen-insensitive BT20 human cancerous mammary epithelial cell lines were obtained from the American Type Culture Collection and maintained routinely in minimal essential medium containing 5% fetal calf serum (FCS). The prostate carcinoma PC3 cell line, provided by N. Prevarskaya, was grown in RPMI medium containing 5% FCS. All cell lines were incubated at 37°C in a humidified atmosphere with 5% CO2 and 95% air. Normal breast epithelial cells (NBEC) came from primary culture of normal breast tissue resections obtained from modeling surgery. The mouse skeletal muscle cell line C2C12 was grown in 50% Dulbecco modified Eagle medium, 50% Ham's F-12 containing 10% FCS. Myoblastic differentiation was induced by serum starvation. When cells reached 70% confluence, FCS was reduced to 1%, and the differentiated cells were recovered after 72 h. For RNA stability assays, actinomycin D at 5 μg/ml was added to the growth medium for the times indicated in the figure legends. Breast cancer tissues came from biopsies corresponding to intraductal cancers.
RNA extraction.
Total RNA was extracted with Tri-Reagent (Euromedex) and treated for 1 h at 25°C with amplification-grade RNase-free DNase I (Invitrogen) or isolated by using a Nucleospin RNAII isolation kit (Macherey-Nagel).
Reverse transcription.
Total reverse transcriptions were performed as follows: 1 μg of RNA, 4 μl of buffer containing random hexamers, and 1 μl of reverse transcriptase (RT; Bio-Rad) were incubated for 5 min at 70°C, for 30 min at 37°C, and for 5 min at 85°C in a final volume of 20 μl. For strand-specific reverse transcription, we used the Thermoscript RT-PCR System (Invitrogen). A total of 2 μg of total RNA was incubated 5 min at 65°C with a 1 μM concentration of the primer and a 10 μM concentration of deoxynucleoside triphosphate in a final volume of 12 μl. We then added 4 μl of buffer (5×), 1 μl of dithiothreitol (0.1 M), 1 μl of RNase Out (40 U/μl), and 2 μl of H2O. The sample was divided into two equal parts. Thermoscript (1 μl) was added in one of them (+RT), and 1 μl of H2O was added to the other to constitute the negative control (-RT). The reverse transcription was performed 20 min at 60°C to improve specificity. The reaction was stopped by 5 min at 85°C, and cDNA/RNA duplexes were removed by the addition of 1 μl of RNase H for 15 min at 37°C. The primer used for H19-specific RT is referred to as “H19-specific RT,” and the primer used for 91H specific RT is referred to as U'sense (Table S1 in the supplemental material).
Real-time RT-PCR.
Real-time PCR amplifications were performed by using a QuantiTect Sybr green PCR kit (Qiagen) with 2 μl of cDNA and 500 nM concentrations of the primers. The primers used for the H19 transcript, RPLP0 (for human acidic ribosomal phosphoprotein P0), GAPDH, U3snoRNA, MRPL23 mRNA, and 91H RNA (D and I) are described in Table S1 in the supplemental material. Other primer sequences are available upon request. The subsequent PCR conditions consisted of 40 cycles of 95°C for 15 s, 60°C for 20 s, and 72°C for 30 s. The data were analyzed by using an MX4000 PCR system software (Stratagene) with the Sybr green option (with dissociation curves). Standard curves were performed on serial dilutions of a PCR product for H19 and on serial dilutions of genomic human DNA for RPLP0. Values were obtained by the calculation methodology recommended by Pfaffl (34): ratio = (NC − b/a)target/(NC − b/a)reference, where a is the slope of the standard curve and b is the ordinate of origin.
5′RACE- and 3′RACE-PCR.
Transcription start site and 3′ end of the 91H transcript were determined by using RACE (rapid amplification of cDNA ends)-PCR experiments. A Smart RACE cDNA amplification kit (Clontech) was used according to the manufacturer's instructions with cDNA from T47D cells. Primers used for PCR and nested PCR were available upon request. PCR products were amplified with Titanium Taq DNA polymerase. RACE-PCR products were characterized by cloning and sequencing. For the 5′RACE, since the transcript is 120 kb in length, we performed the reverse transcription with a specific primer and not with the conventional oligo(dT). The genomic sequence upstream of the transcription start site was tested for promoter elements by using the TFsearch (http://molsun1.cbrc.aist.go.jp/research/db/TFSEARCH.html) (16) and AliBaba (http://www.gene-regulation.com/pub/programs.html) (13) programs.
Analysis of H19 and Igf2 restriction fragment length polymorphisms.
PCRs encompassing AluI and ApaI sites for H19 and IGF2, respectively, were performed on genomic DNA (gDNA), cDNA of nontransfected cells, or cDNA of small interfering RNA (siRNA)-transfected cells. The primers used were HP1 and HP2 for H19 and Igf2ApaI sense and Igf2ApaI antisense for IGF2 (see Table S1 in the supplemental material). PCR conditions were carried out in the following manner: 38 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s for IGF2 and 38 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 40 s for H19. Then, 15 μl of the PCR product was used for digestions with 2 μl of enzyme buffer 10×, 0.5 μg of bovine serum albumin/μl, and 10 U of the appropriate enzyme for 1 h at 37°C. For AluI polymorphism, digestion of the gDNA-derived PCR product yielded 228-, 128-, and 100-bp bands indicating heterozygosity. The cDNA restriction pattern corresponds to a 148-bp fragment in the allele that lacks the AluI site or two bands of 100 and 48 bp for the allele with the recognition site. For IGF2, ApaI digestion generates a 292-bp fragment in the allele without the recognition site or 218- and 74-bp fragments in the allele with the site.
Isolation of nuclei.
Nuclei were isolated from human T47D cells and from proliferating and confluent murine C2C12 cells in growth medium and after 1 and 3 days in differentiation medium as follows. First, 106 cells were washed twice with phosphate-buffered saline (PBS). The cell layer was scraped into 500 μl of PBS and centrifuged 5 min at 1,200 × g. The supernatant was cleared, and this wash step was performed twice. The pellet was estimated to a volume (V) and resuspended in 5V of buffer A (TEA, 10 mM; KCl, 10 mM, MgCl2, 2.5 mM; dithiothreitol, 0.5 mM). After centrifugation under previously described conditions, the pellet was resuspended in 2V of buffer A and disrupted by using a Dounce homogenizer. The separation of nuclei from cytoplasmic compartments was done by centrifugation for 5 min at 3,000 × g. The supernatant containing cytoplasmic extracts was resuspended in 5V of Tri-Reagent. The pellet containing the nuclear extracts was washed in 5V buffer A and centrifuged as previously. The pellet was finally resuspended in 5V of Tri-Reagent. Both extracts were maintained at 4°C before RNA extractions. The efficiency of the fractionation was assessed by quantifying two cytoplasmic mRNAs (RPLP0 and GAPDH) and a nuclear RNA (snoRNA U3). We analyzed the RNA levels of two cytoplasmic control genes. About 80% of these RNA are detected in the cytoplasm. These data demonstrate the efficient separation of cellular compartments and the absence of contamination from one compartment to the other.
gDNA isolation.
A total of 107 cells were washed twice with PBS. The cell layer was scraped into 500 μl of physiologic water and then centrifuged for 5 min at 1,000 × g. The supernatant was resuspended in 3 ml of STE buffer (Tris-HCl, 10 mM [pH 8.0]; NaCl, 100 mM; EDTA, 1 mM [pH 8.0]). Proteinase K was added at 200 μg/ml. The sample was incubated for 10 min at 37°C. Sodium dodecyl sulfate at a final concentration of 1% was added, and the sample was incubated again for 20 min. gDNA was extracted by addition of an equal volume of saturated phenol and centrifugation for 10 min at 10,000 × g. The aqueous phase was collected, and 0.2 M sodium acetate at and 2 volumes of ethanol were added. DNA was rolled up with a Pasteur pipette by mixing progressively the two phases. DNA was finally resuspended in 300 μl of H2O and stoked at 4°C.
RNA interference.
RNA interference was carried out by using synthetic siRNA duplexes, as described by Elbashir et al. (10). Two synthetic siRNA duplexes (si91H 1 and si91H 2) corresponding to the 91H RNA sequences 5′-GGCGUCAUUCUGAUGGGACTT-3′ and 5′-UUCAGGAGCUUAAGAUGCUTT-3′, respectively were used to inhibit 91H RNA expression. A synthetic siRNA duplex (siGFP) corresponding to the green fluorescent protein mRNA sequence (5′-GCUGACCCUGAAGUUCAUCTT-3′) and a sequence corresponding to an intergenic region located at the H19/IGF2 locus (+108603 bp of the H19 transcription start site; 5′-CGUGGGUGGAUGCAUGGAUTT-3′) were used as a negative control. The siRNA duplexes were purchased from Eurogentec. Cells were grown on coverslips in six-well plates and transfected with 400 pmol of siRNAs using Jetsi (Eurogentec), as recommended by the manufacturer. To monitor the transfection efficacy, a tagged-siRNA duplex was transfected in parallel, and the transfection rate was evaluated by fluorescence-activated cell sorting and corresponds to 80 to 90% of transfected cells. After transfection, cells were lysed for total RNA isolation.
RESULTS
Intergenic transcriptional activity at the human H19 locus.
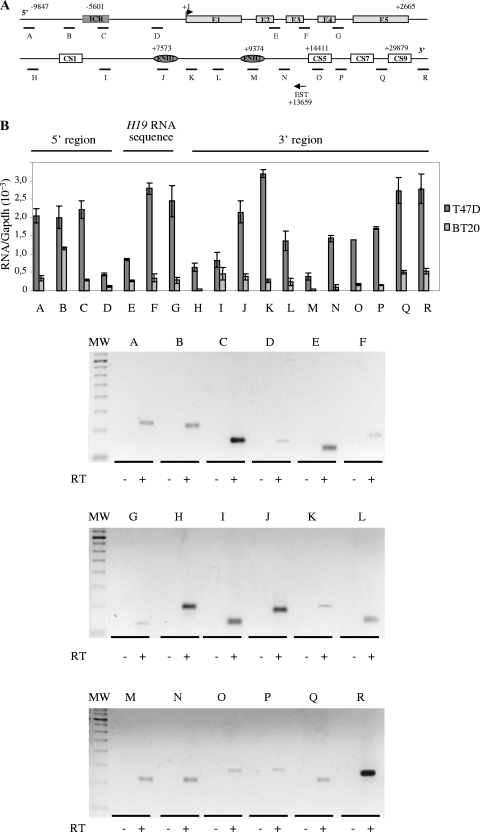
Natural antisense transcripts already identified often overlap the 5′ and 3′ intergenic regions of coding sequences. A quick database search allowed us to identify an expressed sequence tag (EST) that corresponds to intergenic sequences that mapped at the human H19 locus and possessed an antisense orientation (Fig. 1A) (EST, BX 377296, 976 bp, cDNA source placenta). We thus decided to analyze intergenic transcriptional activity at the human H19 locus. Using a quantitative RT-PCR analysis, we scanned the H19 locus from sequences upstream of the ICR to sequences downstream of the endodermic enhancers (Fig. 1A). A transcriptional activity was detected throughout the entire region in both the estrogen-sensitive T47D and the estrogen-insensitive BT20 breast cancer cell lines. The produced transcripts have various expression levels according to the cell line (compared T47D and BT20, Fig. 1B). The variations observed throughout the locus in a given cell line were most likely due to differences in the local reverse transcription efficiency. To verify that there were no transcription breaks, we performed near these regions additional specific reverse transcriptions followed by PCR detection. The results obtained indicated that there is no interruption of the transcription and that the variations were due to RT skews (Fig. 1C).
FIG. 1.
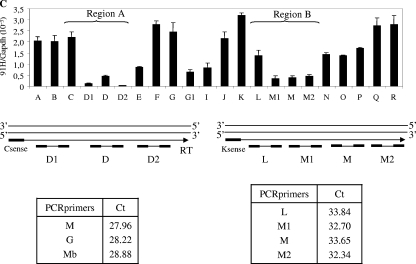
Detection of an intergenic transcriptional activity at the H19 locus in cancer cell lines. (A) Map of the H19 gene locus. The relative positions of the real-time PCR amplification fragments (A to R) are depicted in the map. CS represents conserved regions between mice and humans that possess enhancer activities (17). Black arrow represents a known EST that was identified after alignment of the human chromosome 11 sequence with EST databases. EST, BX 377296 cDNA source placenta. (B) Quantification of RNA levels were determined by real-time RT-PCR on total RNA samples in T47D and BT20 cell lines. The results were normalized to GAPDH expression levels. Agarose gel shows the amplification products obtained after the real-time PCR on T47D cells. Note that amplifications E to G may also potentially quantify H19 precursors RNAs. (C) Strand-specific RT in two regions showing a decrease of 91H transcription (region 1 and region 2). Antisense-specific RT analyses were performed with the primer C sense in region 1 and K sense in region 2. Transcription was detected by PCR using several contiguous primer pairs (D1, D, and D2 for region 1 and L, M1, M, and M2 for region 2). Results, expressed in terms of the cycle threshold (Ct) are indicated in the lower part of panel C. Cycle threshold values that are equivalent for all primer pairs indicate that there is no transcriptional arrest in both tested regions. Primer sequences are available in Table S1 in the supplemental material.
Therefore, in both cell lines, this transcriptional activity overlaps the H19 RNA sequence and spreads through the 5′ and 3′ intergenic regions, including the ICR and the enhancer regions.
Transcriptional orientation of the intergenic transcriptional activity.
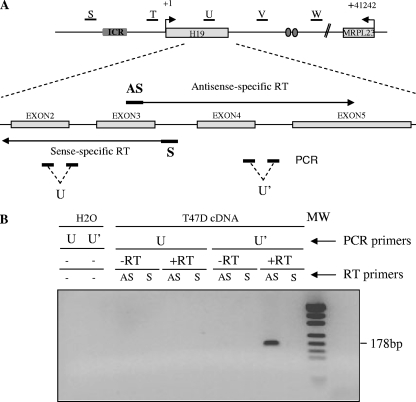
We analyzed the transcriptional orientation at four positions flanking the H19 gene, as well as within the H19 gene. The principle of the experiments that we performed is described in Fig. 2A (see also Fig. S1A in the supplemental material). Briefly, primer AS allows the detection of transcripts in an antisense orientation to H19, while primer S is sense specific. These strand-specific primers were used for reverse transcription on total RNA samples from T47D cells. Real-time PCR amplifications were then performed on each side of the RT primers. At all positions analyzed around (see Fig. S2A in the supplemental material) or within (Fig. 2B and see Fig. S1B in the supplemental material) the H19 gene, only antisense-specific RT primers (AS primer) gave PCR amplifications. We conclude that, in humans, the detected transcripts are antisense to the H19 gene. This result is consistent with the data available from the EST described above. It should be noted that, within the H19 gene, no precursor of H19 (S primer) could be detected in the same experimental conditions where the antisense transcription is found (Fig. 2B).
FIG. 2.
Transcriptional orientation of the intergenic transcriptional activity at the H19 locus. (A) Map of the H19 locus. Black lines indicate the five regions (named S to W) where antisense transcription has been established using strand-specific reverse transcription and real-time PCR. Below the map of the H19 locus, the strategy used to analyze transcriptional orientation at the U region is depicted. The same strategy was used to analyze the S, T, V, and W regions. The locations of strand-specific primers used for reverse transcription (primer AS is sense to H19, and primer S is antisense to H19) and PCR amplifications (U and U' amplifications) are indicated. Strand-specific primers were used for reverse transcription on total RNA samples from T47D cells. Real-time PCRs were then performed on each side of the RT priming region to identify the orientation of 91H (U and U' PCR amplifications). (B) Agarose gel of PCR products obtained for the U region with the different RT-PCR combinations. Only RT analyses performed to detect antisense RNA (primer AS) gave significant PCR amplification and only when PCR amplified by the U' primer pair. To exclude any gDNA contaminations, we analyzed RT reactions performed without RT (-RT), and we ascertained the absence of self-priming analyzing reverse transcription performed without any strand-specific primers (H2O). The results obtained for the S, T, V, and W regions can be seen in Fig. S2A in the supplemental material. The primer sequences are available in Table S1 in the supplemental material.
Finally, an in situ hybridization signal can be detected with an H19 antisense-specific probe in breast cancer cell lines and tumor biopsies but not in normal breast cells or normal biopsies (data not shown).
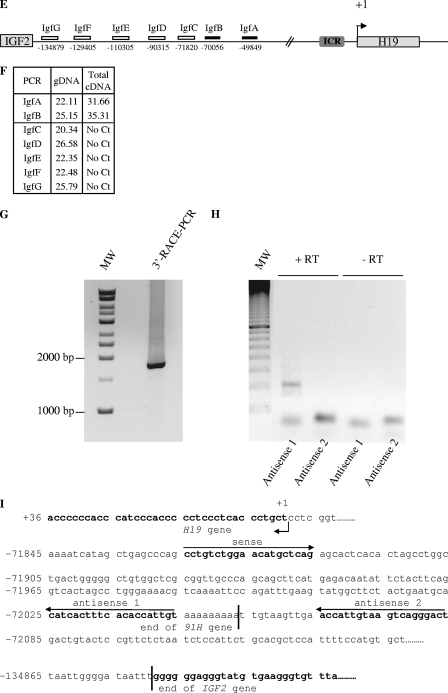
The antisense H19 transcription defines a new transcript: the 91H RNA.
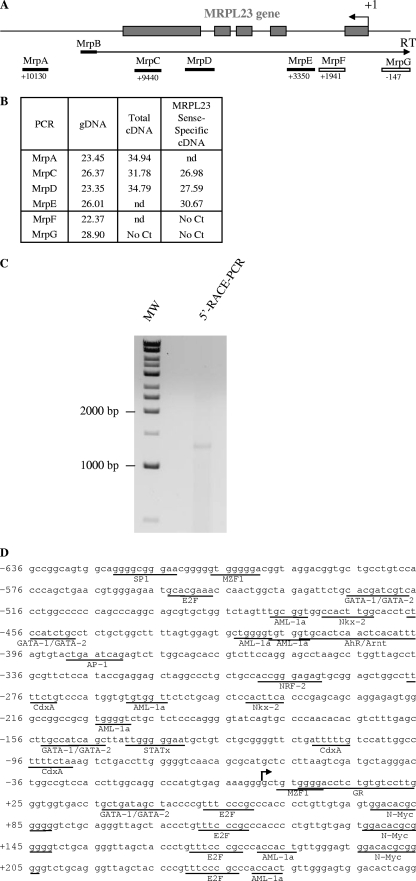
To further characterize the antisense H19 transcription, we then mapped its 5′ and 3′ limits. In humans, the 5′ limit is located within intron 1 of the MRPL23 gene (Fig. 3A and B), a gene located ∼40 kb downstream of H19, which is transcribed in an opposite transcriptional orientation (Fig. 2A). Indeed, on total cDNA, we detected transcripts in the 3′ flanking region of MRPL23 gene (MrpA amplification), as well as within the MRPL23 region (MrpC and MrpD amplifications), but not in the promoter region of MRPL23 gene (MrpG amplification) (Fig. 3B, total cDNA column). PCR amplifications performed on strand-specific RT with a MrpB primer (Fig. 3A and B) formally exclude any interference with MRPL23 pre-messengers and showed that the 5′ limit of the antisense H19 transcription is located within MRPL23 intron 1, between MrpE and MrpF amplifications, about 3,000 to 1,900 bp downstream of the MRPL23 transcription start site. To define the precise beginning of the 91H transcript, we performed 5′RACE-PCR experiments. We performed PCR with specific primers, and we verified the specificity of the amplification by nested PCR. Cloning and sequencing of the PCR fragments obtained (Fig. 3C) confirmed that the transcription start site was located in intron 1 of the MRPL23 gene at bp 47348 of the transcription start site of H19. To identify transcriptional regulatory sequences, we analyzed the 5′-flanking region from 500 bp upstream of the transcription start site with a predicting computer program (TFsearch and AliBaba). The sequence analysis failed to identify a TATA-box (Fig. 3D), but we identified several putative E2F binding sites that are also known to regulate H19 expression (3). AML-1a, GATA-1/2, N-Myc, and other putative binding sites were also detected (Fig. 3D). Elsewhere, we found that the 3′ limit was located about 70 kb upstream of H19 (Fig. 3E and F). Indeed, transcriptional activity has been detected on total cDNA with amplifications IgfA and IgfB but not with amplifications IgfC to IgfG. 3′RACE-PCR experiments mapped the 3′ end at bp -72044 relative to the H19 transcription start site (Fig. 3G and I). We confirmed these data by PCR using primers located on each side of the 3′ end, and we obtained amplification only with the sense and antisense 1 primer pair, but no fragment could be detected with the sense and antisense 2 primer pair (see Fig. 3H). Thus, antisense H19 transcription was detected throughout a large portion of the H19/IGF2 locus and has the potential to produce a single 119.392-kb transcript that we named the 91H RNA.
FIG. 3.
Determination of the 5′ and 3′ limits of the antisense H19 transcription. (A) Map of the MRPL23 gene region. The relative positions of the PCR amplifications and strand-specific primer used for reverse transcription (MrpB) are indicated. (B) Table indicates cycle thresholds obtained in real-time PCR for each PCR amplifications on total cDNA (random priming of the RT) or after strand-specific reverse transcription with MrpB primer. gDNA is used as a positive amplification control to assess for the amplification efficiency. (C) Determination of the transcription start site of the 91H gene by 5′RACE-PCR. RACE-PCR using T47D cell cDNA and 91H specific primers resulted in the amplification of an ∼1,300-bp DNA fragment. This fragment was cloned into TOPO plasmid for sequencing. (D) Nucleotide sequence of the 5′ region of the 91H transcript. The transcription start site is indicated by a black arrow. Putative transcription factor binding sites identified by computer analysis are underlined. (E) Map of the H19/IGF2 locus. Boxes indicate PCR amplifications used to determine the 3′ limits of the antisense transcription. Successful amplifications are depicted by full boxes, whereas abortive amplifications are shown as open boxes. (F) A table indicates the cycle thresholds (Ct) obtained for each PCR amplification. (G) Determination of the 3′ end of the 91H RNA by 3′RACE-PCR. RACE-PCR using T47D cell cDNA resulted in the amplification of an ∼1,750 bp-DNA fragment. This fragment was cloned into the TOPO plasmid for sequencing. (H) Confirmation of the 3′ end of 91H. RT-PCRs were performed with the primer pairs depicted in panel I: sense plus antisense 1 or sense plus antisense 2. (I) Nucleotide sequence of the 3′ region of the 91H transcript. The H19 and IGF2 genes are indicated in boldface, and the 3′ limit of 91H is pointed out by a black line. Positions relative to the H19 transcription start site (+1) are indicated. Primer sequences can be seen in Table S1 in the supplemental material.
91H RNA is a nuclear transcript.
RNA levels from nuclear and cytoplasmic extracts of T47D cells were assessed by quantitative RT-PCR to test the cellular localization of 91H. The efficiency of the fractionation was checked by quantifying two cytoplasmic mRNAs (RPLP0 and GAPDH) and a nuclear RNA (snoRNA U3). Using G and H primer pairs (Fig. 1), we found that 91H RNA is almost exclusively located in the nucleus (>80%). As a control, MRPL23 mRNA localization was analyzed and found to be mainly cytoplasmic. However, the H19 RNA had an equal repartition in both compartments (Table 1).
TABLE 1.
Cellular localization of human 91Ha
| Gene | Nuclear cell extract | Cytoplasmic extract |
|---|---|---|
| Rplp0 | 20.44 | 79.56 |
| Gapdh | 21.4 | 78.6 |
| U3 snoRNA | 73.6 | 26.4 |
| H19 | 44.4 | 55.6 |
| 91H (G primers) | 81.82 | 18.18 |
| 91H (H primers) | 80.12 | 19.88 |
| MrpL23 | 20.83 | 79.17 |
Relative RNA levels were determined by quantitative RT-PCR after separation of the nuclear and cytoplasmic compartments. MrpL23, Rplp0, Gapdh, and H19 RNAs were used as cytoplasmic control transcripts, and U3 snoRNA was used as a nuclear control RNA. 91H was quantified independently at the G and H regions (see Fig. 1). H19 and MrpL23 were detected using primers designed over exon/exon junctions. Values in boldface indicate the preferential localization of the transcript.
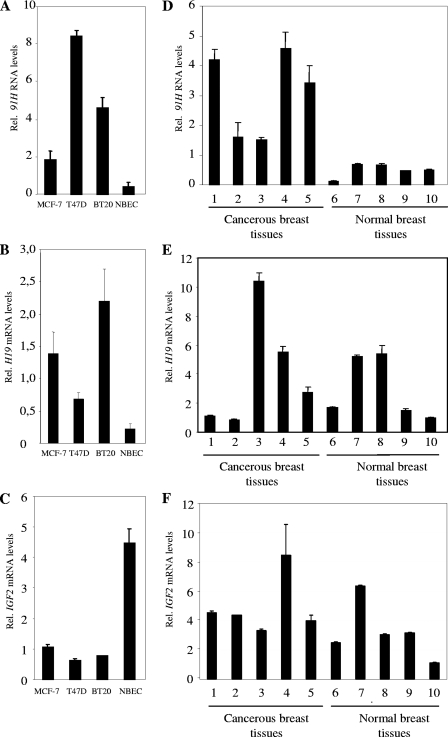
91H overexpression in breast cancer.
91H, H19, and IGF2 expression levels were determined by real-time RT-PCR on total RNA samples from three cancer cell lines (MCF-7, T47D, and BT20) and NBEC coming from three patients (Fig. 4A to C). 91H and H19 RNAs were preferentially expressed in cancer cell lines relative to noncancerous cells (Fig. 4A and B), whereas IGF2 was strongly expressed in normal epithelial cells and displayed lower expression levels in cancerous cell lines (Fig. 4C).
FIG. 4.
91H expression is upregulated in breast cancer. (A to C) 91H RNA levels (A), H19 mRNA levels (B), and IGF2 mRNA levels (C) in human breast cells. RNA levels were determined by real-time RT-PCR on total RNA samples from three cancer cell lines (MCF-7, T47D, and BT20) and NBEC. (D to E) 91H RNA levels (D), H19 mRNA levels (E), and IGF2 mRNA levels (F) in human normal and cancer breast tissues. Expressions were normalized on RPLP0 gene expression. H19 RNA sense levels were quantified by using primers designed over exon/exon junctions, and 91H was detected with the H primer pair previously described.
We carried on our investigations by an in vivo study in normal and cancerous human breast tissues (Fig. 4D to F). H19 and IGF2 expression was found to be heterogeneous (Fig. 4E and F) but, more interestingly, 91H was strongly and systematically overexpressed in all cancerous tissues examined compared to normal breast tissues (Fig. 4D). Altogether, these results indicate that 91H RNA is consistently overexpressed in breast cancer.
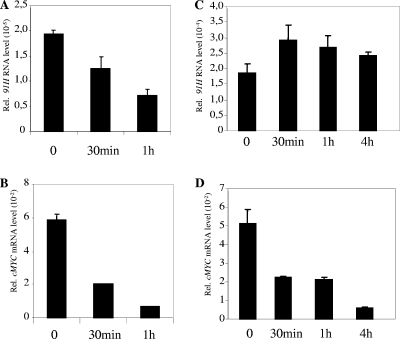
91H stabilization in human cancer cells.
One possibility to explain 91H RNA accumulation in cancer cells may be that it results from a stabilization of the 91H transcripts in these cells. To determine the stability of 91H RNA, inhibition of total cellular transcription by actinomycin D was used. MYC was chosen as a control for short half-life mRNA (Fig. 5B and D). Figure 5A and C shows the results from a quantitative RT-PCR analysis in normal and cancer cells treated from 30 min to 4 h with actinomycin D demonstrating that 91H RNA half-lives were dramatically increased in T47D cancer cells (>4 h) compared to normal cells (about 30 min). The same results were obtained in BT20 cells (data not shown). Therefore, 91H RNA accumulation in cancer cells likely results from transcript stabilization.
FIG. 5.
91H RNA stability in cancer and normal breast cells. Cells were treated with 5 μg of actinomycin D/ml, and RNA levels were determined by real-time RT-PCR at the indicated times. cMYC was used as a positive control for actinomycin D treatment since this mRNA is known to be very unstable. 91H (A) and cMYC (B) RNA levels were determined in NBEC. 91H (C) and cMYC (D) RNA levels were determined in T47D cancer cells.
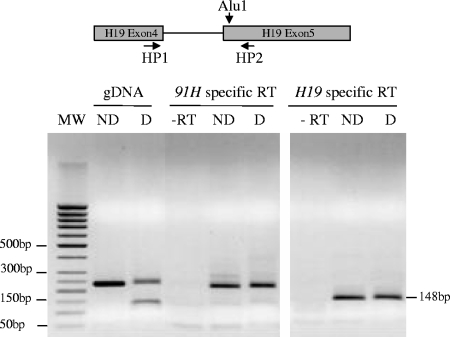
91H RNA is monoallelically expressed.
Using a polymorphic AluI restriction site available in T47D cells, we performed allelic expression analyses of both the H19 and 91H RNAs. Strand-specific reverse transcriptions were performed on total RNA from T47D cells and PCR amplified with primers flanking the AluI polymorphic restriction site as indicated in Fig. 6 (and Fig. S3 in supplemental material). Amplification fragment on 91H-specific RT is resistant to AluI digestion showing that this RNA is exclusively expressed from the parental allele that does not have the AluI site (Fig. 6, left panel). Amplifications performed on H19-specific RT are not digested (Fig. 6, right panel) showing that H19 RNA expression occurs on the same allele. Therefore, since H19 RNA is known to be maternally expressed, we conclude that 91H RNA is also produced from the maternal allele.
FIG. 6.
Allelic expression analysis of the human 91H RNA. Strand-specific reverse transcription was performed on total RNA from T47D cells with a reverse primer for H19-specific RT and a forward primer for 91H-specific RT (see the Materials and Methods). PCR were performed with primers located on each side of the AluI polymorphic restriction site as indicated in the figure (HP1 and HP2). The PCR products were then digested with AluI and separated into agarose gel. Note that, as expected, the size of the H19 amplification fragment is smaller than those obtained for 91H RNA amplification because of the removal of the 81-bp H19 intron 5 in the H19 RNA. ND, not digested; D, AluI digestion; -RT, amplifications on control reactions made without reverse transcription. gDNA, control amplification on T47D cell gDNA.
91H RNA is conserved in mice.
Using RT-PCRs on total RNAs extracted from perinatal mouse liver and heart tissue, we showed that seven intergenic regions of the H19 locus (mA to mG, Fig. 7A) are transcribed (data not shown). In the livers of neonates, the regions located between the H19 gene and the endodermic enhancers display expression patterns similar to those of the H19 RNA itself (see Fig. S4A in the supplemental material). In the heart, relative to GAPDH, the 91H RNA levels are similar to those observed in liver, but they appear to be almost constant from embryonic day 18.5 to 12.5 days after birth (see Fig. S4B in the supplemental material).
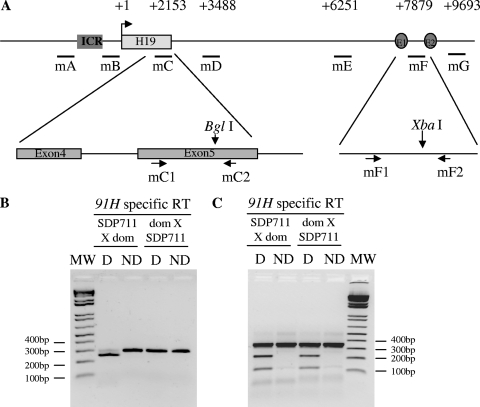
FIG. 7.
Allelic expression of the murine 91H RNA. (A) Map of H19 region indicating positions of PCR primer pairs used in real-time PCR (mC1 and mC2; mF1 and mF2). (B) RT using a H19-sense-intron 4 primer was performed on total RNA extracted from 5-day-old mouse liver issued from SDP711XM.m.dom. (left panel) or M.m.dom.XSDP711 hybrid mice (right panel). A 301-bp PCR fragment was then obtained by using the mC primer pair and digested (D) or undigested (ND) with BglI at an M. spretus-specific restriction site. (C) Reverse transcription using an mF1 sense primer was performed on the samples described above. A 344-bp PCR fragment was then obtained using the mF primer pair and cut by XbaI at a M. musculus domesticus-specific restriction site.
We also examined the transcriptional orientation of the intergenic transcription in both postnatal mouse livers and hearts. The region upstream of the endodermic enhancers (mE) was tested for transcriptional orientation (see Fig. S2B in the supplemental material). Similar to the case in humans, the transcript is antisense to H19 in both tissues. We conclude that the 91H RNA is conserved in the mouse.
It was then of interest to determine whether, as observed in humans, the mouse 91H RNA was transcribed preferentially from one parental allele. We used a hybrid mouse (Mus musculus domesticus female × SDP711 or the reverse cross) to provide allele-specific restriction sites. SDP711 is a congenic mouse strain that has the distal part of chromosome 7 from Mus spretus origin). We first assessed 91H RNA allelic expression in H19 exon 5. In agreement with data obtained in humans, an M. spretus-specific BglI restriction site (Fig. 7A) allowed us to show that, in 5-day-old mouse liver, 91H RNA is expressed almost exclusively from the maternal allele (Fig. 7B). However, using an XbaI M. domesticus-specific restriction site located between the two endodermic enhancers, we show that the 91H RNA is expressed on both alleles in postnatal liver (Fig. 7C) and heart (data not shown). Elsewhere, as the regions located near the ICR (mA and mB) were recently found to be paternally transcribed (9, 42), we also determined the allelic expression in this region. While, within the ICR, some expression can be detected on the paternal allele, 91H RNA expression is only detected on the maternal allele in the intergenic region located between the ICR and the H19 gene (see Fig. S5 in the supplemental material).
In C2C12 myoblasts, 91H RNA is upregulated upon induction of myoblastic cell differentiation (see Fig. S4C in the supplemental material). 91H RNA levels are always much lower than those of the H19 RNAs. They are 4 to 5 orders of magnitude less abundant than H19 RNA levels and 10 to 100 times less abundant than H19 precursor RNAs. In mouse myoblastic C2C12 cells, the half-life of 91H was found to be very short since 91H RNA is largely depleted after 15 min of actinomycin D treatment, whereas H19 RNA shows no significant changes after 4 h of treatment (data not shown). Therefore, because of its short half-life and even if the 91H RNA levels are rather weak, 91H transcriptional activity may be important. Finally, in mouse proliferating and differentiated myoblastic C2C12 cells, we confirmed the nuclear localization of 91H, whereas the H19 RNA was mainly cytoplasmic (Table 2).
TABLE 2.
Cellular localization of mouse H19 in proliferative and differentiated C2C12 myoblastic cellsa
| Gene | C2C12 cells (relative % total expression)
|
|||
|---|---|---|---|---|
| Proliferating
|
Differentiated
|
|||
| Nuclear cell extract | Cytoplasmic cell extract | Nuclear cell extract | Cytoplasmic cell extract | |
| U3 snoRNA | 68 | 32 | 94.2 | 5.8 |
| H19 | 23.6 | 76.4 | 16.6 | 83.4 |
| 91H (mE primers) | 100 | <0.05 | 99.5 | <0.5 |
| 91H (mG primers) | 100 | <0.05 | 100 | <0.05 |
RNA levels were determined as described for Table 1. The results are expressed in relative percentages of total expression. Values in boldface indicate the preferential localization of the transcript.
91H silencing decreases IGF2 expression.
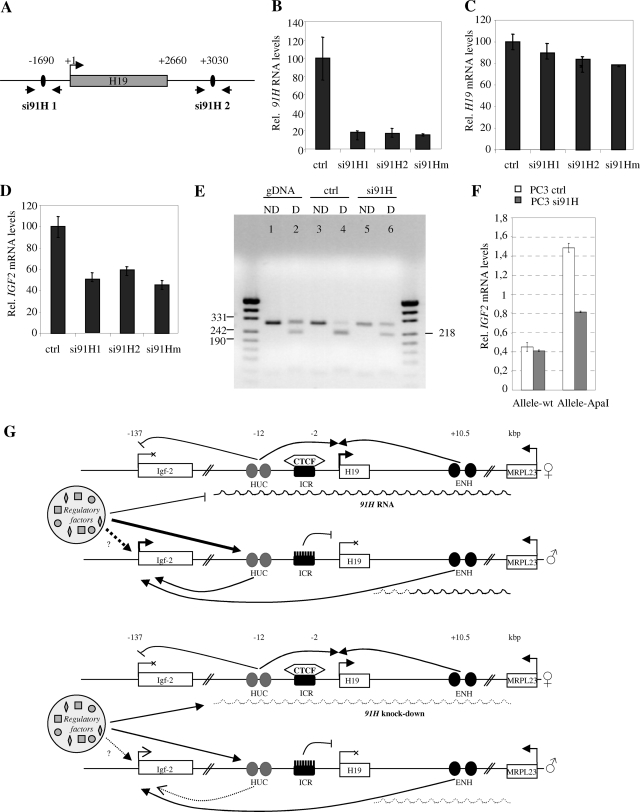
To check whether the antisense RNA had a role in the imprinting or gene expression at the H19/IGF2 locus, 91H RNA levels were downregulated by using two siRNAs (si91H 1 and 2 [Fig. 8A]). Cotransfection of the siRNA alone or in combination in T47D cells induces a strong decrease of 91H RNA levels (Fig. 8B) and a slight decrease in H19 RNA levels (Fig. 8C). Similar results were obtained in BT20 and PC3 cell lines (data not shown). Using a polymorphic AluI site for H19 in T47D cells, we assessed that H19 imprinting was not altered (see Fig. S6 in the supplemental material). Interestingly, IGF2 mRNA levels were twofold lower after si91H treatment (Fig. 8D). These results suggest thus that the reduced expression of IGF2 in siRNA-treated cells depends on interference directed against 91H nuclear transcripts. Since no informative polymorphic restriction site was available to investigate IGF2 allelic expression in T47D cells, we used PC3 cells in which we found a polymorphic ApaI site located in exon 9 of the IGF2 gene. In these cells, we also observed an ∼50% reduction of IGF2 expression after si91H treatment (data not shown). Combining RT-PCR performed on cDNA from siRNA-treated cells and ApaI restriction, we showed that both IGF2 parental alleles are expressed (Fig. 8E). This result may be explained by a loss of IGF2 imprinting on the maternal allele. However, since IGF2 mRNA levels are weak in these cells and because they are decreased in siRNA-treated cells, we suspected that it may rather reflect a reduction of IGF2 expression on the nonimprinted paternal allele, thus uncovering a transcriptional leakage observed on the maternal allele. Using a previously described allele-specific quantitative RT-PCR technique (51), we demonstrated that downregulation of the 91H RNA by siRNA leads to a decrease of IGF2 expression exclusively on the paternal ApaI allele (Fig. 8F). Since this allele is the one that is preferentially expressed in PC3 cells, we can thus reasonably assume that it corresponds to the paternal allele.
FIG. 8.
91H silencing by RNA interference reduces IGF2 expression on the paternal allele. (A) Map of H19 region indicating the positions of siRNA sequences and the PCR primer pairs used in real-time PCR. T47D cells were transfected with siRNA targeting either the green fluorescent protein (ctrl) or the 91H RNA (si91H 1 and 2 alone or in combination). 91H (B), H19 (C), and IGF2 (D) RNA levels were determined by real-time RT-PCR 24 h after transfection. The results were normalized to RPLP0 expression levels. (E) PC3 cells were transfected with si91H, and the imprinting status of the IGF2 alleles was examined using a RT-PCR amplification digested (D) or undigested (ND) with an ApaI polymorphic restriction site available in this cell line. (F) The IGF2 expression levels were determined on each parental allele using an allele-specific RT-PCR amplification method (51) from PC3 cells transfected with control siRNA or with si91H. (G) Model for 91H trans effect on IGF2 expression. Two sets of enhancers (HUC and ENH sequences) regulate IGF2 expression. Both would be required for full expression of the gene. In addition, the two IGF2 alleles would be competing for a common limited stock of regulatory elements (methylation/acetylation/transcription?). On the maternal allele, 91H would block the locus and prevent the HUC sequences from interaction with any regulatory factors. These would be then directed on the paternal allele, in the HUC region, and/or in the IGF2 promoter region and would cooperate with the cis endodermic enhancers, resulting in IGF2 enhanced expression. Upon siRNA treatment and 91H knockdown, the competition would be lost and, as a consequence, both IGF2 alleles would be now able to interact with the regulatory factors with similar efficiencies. Because these factors are in a limiting amount, a part would be depleted from the paternal IGF2 allele to interact with the maternal allele. This would lead to a paternal IGF2 expression decrease but unchanged maternal IGF2 expression because of the absence of functional ENH sequences. Arrows indicates positive regulations, whereas lines with bars correspond to inhibitions.
DISCUSSION
Antisense transcription is a widespread phenomenon in eukaryote genome. In recent years, natural noncoding antisense transcripts have been implicated in many aspects of eukaryotic gene expression, including genomic imprinting, RNA interference, translational regulation, alternative splicing, X-inactivation, and RNA editing (20). We have identified here a new antisense transcript that we named 91H, at the human and mouse H19/IGF2 locus. It shares similar characteristics with well-known ncRNAs such as the Air transcript (43). Indeed, 91H RNA corresponds to a short-lived and 120-kb-long transcript, overlapping the H19 imprinted gene, the nonimprinted flanking gene MRPL23 (55), and intergenic regions including the ICR. Its transcription start site is located at bp 47348 relative to the transcription start site of the H19 gene, and the 3′ end is found at bp −72044 relative to the transcription start site of the H19 gene. It is localized to the nucleus, and its promoter lies in an intron of an active host mRNA gene, such as the Air promoter that is located in the second intron of the Igf2r promoter, showing that transcription of the ncRNA promoter does not interfere with expression of the host mRNA gene.
Interestingly, 91H RNA shows an increased stability in cancer cells, leading to its accumulation in cancer cell lines and breast cancer tissues. This transcript stabilization is reminiscent of the previously described stabilization of the H19 ncRNA, which is responsible for its accumulation during in vitro muscle cell differentiation (29).
We noticed a preferential 91H expression on the maternal allele in both mice and humans within the H19 gene region (Fig. 6 and 7) and between the ICR and the H19 cap site (Fig. S5B in the supplemental material). However, we detected a biallelic transcription in the 3′ endodermic enhancers region (Fig. 7C) and in the ICR in mouse (Fig. S5A in the supplemental material) according to the results obtained by Drewell et al. (9) and Schoenfelder et al. (42), who demonstrated biallelic sense and antisense transcription in the mouse H19 ICR. These data show that, in the mouse, some regions, and particularly regulatory regions, probably generate transcription independently of 91H. However, in humans, using a HhaI polymorphic site, we only detected monoallelic antisense transcription in the ICR (Fig. S7 in the supplemental material).
The developmental expression patterns of 91H are very similar to those of the H19 RNA in murine liver, and both RNAs are upregulated upon induction of myoblastic cell differentiation. This suggests that both transcripts are subject to a common regulation of the locus which allows gene transcription in a time-restricted manner.
To address the functional role of the antisense transcript, we knocked down its expression by RNA interference assays in human cells. The results indicated that, whereas 91H silencing does not modify H19 imprinted status and slightly affects H19 expression, it strongly reduces the overall IGF2 mRNA levels. It was therefore somewhat surprising that semiquantitative RT-PCR detected transcriptional activity on both IGF2 alleles in siRNA-treated cells. To reconcile these data, we assumed that this IGF2 reexpression from the maternal allele may rather reflect a transcriptional leakage that was underestimated in untreated cells because of a high expression level on the paternal allele. Indeed, allele-specific quantitative RT-PCR demonstrated that downregulation of the 91H RNA leads to a decrease of IGF2 expression exclusively on the paternal allele. Therefore, we conclude that, in humans, 91H RNA produced from the maternal chromosome does not affect genomic imprinting but is involved in the maintenance of IGF2 gene expression in trans on the paternal allele. Importantly, two distinct siRNA probes, targeting different regions of the 91H transcript and used in separate experiments have identical effects on Igf2 mRNA levels. Therefore, the possibility that the siRNA probes interfere with known cis-acting regulatory DNA elements is very unlikely. IGF2 upregulation through a trans effect by ectopic expression of H19 antisense transcripts has been suggested (53). The fact that 91H knockdown only slightly affects H19 gene expression from the maternal allele seems to indicate that the 91H effect is not mediated through the ICR/CTCF complex. However, other candidates such as enhancer DNA regions may be considered for interacting with the antisense RNA. Indeed, 91H encompasses a region including a previously described nuclease hypersensitive site (approximately 30 kb upstream of the H19 gene) that can be found on both parental alleles (2) and that have enhancer functions in both imprinted and nonimprinted tissues (6). In addition, the biallelically transcribed H19 upstream conserved (HUC) sequences are regulatory elements located just upstream of the ICR that act as strong mesodermal enhancers and are supposed to activate IGF2 expression on the paternal allele (9).
The 91H trans effect may reflect a competition model that explains a communication between both parental chromosomes that we evoked in a previous work (11). Indeed, deletions of the active maternal allele of the H19 gene reduce the paternal IGF2 DMR2 methylation level and therefore probably have both cis and trans effects on IGF2 gene expression. These results are consistent with the present study and may be due to 91H transcript disruption. Elsewhere, it has been demonstrated that ncRNAs are able to guide the epigenetic regulation of genes. For the human HOX loci, their expression demarcates broad chromosomal domains of differential histone methylation and RNA polymerase accessibility (37). In particular, the HOTAIR ncRNA located in a regulatory boundary in the HOXC cluster represses transcription in trans across 40 kb of the HOXD locus. This ncRNA binds to an epigenetic regulatory complex and changes the methylation patterns of the target locus. We then proposed a model to explain the 91H trans effect on IGF2 expression in which cooperation between two sets of enhancer sequences would be required for full expression of the gene. In addition, both parental alleles would be competing for a common limiting pool of regulatory factors. In this model, 91H would be essentially involved in modifying the accessibility of HUC sequences to regulating proteins. Indeed, maternal expression of 91H would preferentially direct regulatory elements on the paternal allele within the HUC region. This region would cooperate with ENH sequences, resulting in paternal IGF2 enhanced expression (Fig. 8G). We cannot exclude the intervention of other enhancer sequences that may be responsible for the trans effect of 91H RNA on IGF2 expression (18).
Elsewhere, 91H may attract repressive chromatin modifications to the maternal allele by trapping factors responsible for DNA or histone modifications that become available for the paternal allele when 91H is disrupted, resulting in IGF2 expression decrease. Yu et al. (52) recently showed that many tumor suppressor genes have nearby antisense RNAs. These authors focused on the p15 antisense RNA, which is able to induce p15 gene silencing through heterochromatin formation (increase in dimethylation of H3K9 and decrease in dimethylation of H3K4). Thus, it will be useful to analyze histone methylation and/or acetylation patterns of the paternal IGF2 region in the presence or in the absence of the 91H RNA.
Other authors have already evoked this type of trans regulation within an imprinted locus. Takeda et al. (46) examined the callipyge phenotype (postnatal muscular hypertrophy of shape) caused by a mutation in the DLK1-GTL2 locus. The mutation and its “polar overdominance” mode of inheritance imply that only the heterozygous animals that have received the paternal mutation display the phenotype. The mutation, when paternally inherited, increases DLK1 and PEG11 protein levels, which results in the callipyge phenotype. However, in the case of a double maternal and paternal inherited mutation, it was suggested that the overexpression of ncRNAs from the maternal allele could inhibit in trans the translation of DLK1 and PEG11 genes, resulting in a normal phenotype. This trans inhibition have been formally demonstrated for PEG11 through a maternal miRNA. For DLK1, these authors propose that the posttranscriptional inhibition may be mediated by a long ncRNA which could, for instance, compete toward a trans factor responsible for DLK1 RNA localization or translation. Our model shares interesting similarities, although the trans regulation affecting DLK1 protein levels is different from those affecting IGF2 transcripts accumulation after 91H knockdown.
Interestingly, and in agreement with the role of 91H displayed in PC3 cells, we noticed an inverse correlation between 91H and IGF2 transcription in normal and cancer mammary cell lines. However, in breast cancer tissues, whereas 91H is always overexpressed, IGF2 and H19 expression profiles are more heterogeneous; this is probably due to various co- or posttranscriptional regulations. These data provide a link between antisense transcription and cancer.
Finally, 91H RNA discovery leads us to reconsider some previous results. Indeed, a set of studies in the mouse described deletions of sequences within the H19/IGF2 locus that did not affect H19 and IGF2 expression: deletion of the H19 promoter and structural gene (41), deletion of a G-rich repetitive element located immediately 3′ to the ICR (48), deletion of a conserved direct tandem repeat 1 kb upstream of the ICR (23), and deletion of a 4.2-kb domain between the H19 transcription unit and the enhancers (49). We presume that these deletions did not impede 91H transcription and generated a transcript that could still assume its regulatory function.
In conclusion, the antisense transcript reported here adds further complexity to the cluster of imprinted genes in the human imprinted 11p15.5 region. 91H RNA is involved in the control of IGF2 expression in trans, suggesting that, beyond imprinting, other sophisticated mechanisms involved in the control of gene expression arise at this locus. Cooperation between individual regulatory elements (i.e., enhancers, silencers, antisense RNA, DNA, and histone modifications) seems to be required for fine-tuning H19 and IGF2 gene expression.
Supplementary Material
Acknowledgments
We thank A. Béthouart, N. Nguyen, and C. Cathelineau for their collaboration in the collect of breast cancer tissues. We thank P. Pellerin (Lille Hospital, Lille, France) for its collaboration to collect breast normal tissues. We thank P. Delannoy (USTL, Lille, France) for the use of MX4000 apparatus and L. Brunet and G. Courtand (CCMIC and USTL) for their help with the text illustrations.
J.-J.C., E.A., and T.D. hold grants from the Fédération des Groupements des Entreprises Françaises dans la Lutte contre le Cancer, the Comités du Nord et de l'Aisne de la Ligue Nationale contre le Cancer, and the Association pour la Recherche sur le Cancer (ARC). N.B. is the recipient of fellowships from the Ministère de l'Education Nationale et de la Recherche, the ARC, and the Institut National du Cancer. This study was also supported by grants from the ARC (grant 4868), the GIS Longévité (contract GISLO401), the Fond National de la Science (ACI Jeune Chercheur), and the Agence Nationale de La Recherche (ANR-07-BLAN-0052-02 -T.F.-) and by funds from the Centre National de la Recherche Scientifique.
We dedicate this report to the memory of our friend Jean Coll.
Footnotes
Published ahead of print on 15 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adriaenssens, E., L. Dumont, S. Lottin, D. Bolle, A. Leprêtre, A. Delobelle, F. Bouali, T. Dugimont, J. Coll, and J. J. Curgy. 1998. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am. J. Pathol. 1531597-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ainscough, J. F., T. Koide, M. Tada, S. Barton, and M. A. Surani. 1997. Imprinting of IGF2 and H19 from a 130-kb YAC transgene. Development 1243621-3632. [DOI] [PubMed] [Google Scholar]
- 3.Berteaux, N., S. Lottin, D. Monté, S. Pinte, B. Quatannens, J. Coll, H. Hondermarck, J. J. Curgy, T. Dugimont, and E. Adriaenssens. 2005. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J. Biol. Chem. 28029625-29636. [DOI] [PubMed] [Google Scholar]
- 4.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 92395-2402. [DOI] [PubMed] [Google Scholar]
- 5.Brannan, C. I., E. C. Dees, R. S. Ingram, and S. M. Tilghman. 1990. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1028-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charalambous, M., T. R. Menheniott, W. R. Bennett, S. M. Kelly, G. Dell, L. Dandolo, and A. Ward. 2004. An enhancer element at the IGF2/H19 locus drives gene expression in both imprinted and non-imprinted tissues. Dev. Biol. 271488-497. [DOI] [PubMed] [Google Scholar]
- 7.Constancia, M., W. Dean, S. Lopes, T. Moore, G. Kelsey, and W. Reik. 2001. Deletion of a silencer element in IGF2 results in loss of imprinting independent of H19. Nat. Genet. 26203-206. [DOI] [PubMed] [Google Scholar]
- 8.DeChiara, T. M., E. J. Robertson, and A. Efstratiadis. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64849-859. [DOI] [PubMed] [Google Scholar]
- 9.Drewell, R. A., K. L. Arney, T. Arima, S. C. Barton, J. D. Brenton, and M. A. Surani. 2002. Novel conserved elements upstream of the H19 gene are transcribed and act as mesodermal enhancers. Development 1291205-1213. [DOI] [PubMed] [Google Scholar]
- 10.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411494-498. [DOI] [PubMed] [Google Scholar]
- 11.Forne, T., J. Oswald, W. Dean, J. R. Saam, B. Bailleul, L. Dandolo, S. M. Tilghman, J. Walter, and W. Reik. 1997. Loss of the maternal H19 gene induces changes in IGF2 methylation in both cis and trans. Proc. Natl. Acad. Sci. USA 9410243-10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuks, F. 2003. DNA methyltransferases: from chromatin remodeling to cancer. Med. Sci. 19477-480. [DOI] [PubMed] [Google Scholar]
- 13.Grabe, N. 2002. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. 21-15. [PubMed] [Google Scholar]
- 14.Grandjean, V., L. O'Neill, T. Sado, B. Turner, and A. Ferguson-Smith. 2001. Relationship between DNA methylation, histone H4 acetylation and gene expression in the mouse imprinted IGF2-H19 domain. FEBS Lett. 488165-169. [DOI] [PubMed] [Google Scholar]
- 15.Heard, E. 2004. Recent advances in X-chromosome inactivation. Curr. Opin. Cell Biol. 16247-255. [DOI] [PubMed] [Google Scholar]
- 16.Heinemeyer, T., E. Wingender, I. Reuter, H. Hermjakob, A. E. Kel, O. V. Kel, E. V. Ignatieva, E. A. Ananko, O. A. Podkolodnaya, F. A. Kolpakov, N. L. Podkolodny, and N. A. Kolchanov. 1998. Database on transcriptional regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 26362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishihara, K., N. Hatano, H. Furuumi, R. Kato, T. Iwaki, K. Miura, Y. Jinno, and H. Sasaki. 2000. Comparative genomic sequencing identifies novel tissue-specific enhancers and sequence elements for methylation-sensitive factors implicated in IGF2/H19 imprinting. Genome Res. 10664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaffer, C. R., A. Grinberg, and K. Pfeifer. 2001. Regulatory mechanisms at the Igf2/H19 locus. Mol. Cell. Biol. 218189-8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurukuti, S., V. K. Tiwari, G. Tavoosidana, E. Pugacheva, A. Murrell, Z. Zhao, V. Lobanenkov, W. Reik, and R. Ohlsson. 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to IGF2. Proc. Natl. Acad. Sci. USA 10310684-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavorgna, G., D. Dahary, B. Lehner, R. Sorek, C. M. Sanderson, and G. Casari. 2004. In search of antisense. Trends Biochem. Sci. 2988-94. [DOI] [PubMed] [Google Scholar]
- 21.Leighton, P. A., J. R. Saam, R. S. Ingram, C. L. Stewart, and S. M. Tilghman. 1995. An enhancer deletion affects both H19 and IGF2 expression. Genes Dev. 92079-2089. [DOI] [PubMed] [Google Scholar]
- 22.Leighton, P. A., R. S. Ingram, J. Eggenschwiler, A. Efstratiadis, and S. M. Tilghman. 1995. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 37534-39. [DOI] [PubMed] [Google Scholar]
- 23.Lewis, A., K. Mitsuya, M. Constancia, and W. Reik. 2004. Tandem repeat hypothesis in imprinting: deletion of a conserved direct repeat element upstream of H19 has no effect on imprinting in the Igf2-H19 region. Mol. Cell. Biol. 245650-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis, A., and A. Murrel. 2004. Genomic imprinting: CTCF protects the boundaries. Curr. Biol. 14284-286. [DOI] [PubMed] [Google Scholar]
- 25.Li, Y. M., G. Franklin, H. M. Cui, K. Svensson, X. B. He, G. Adam, R. Ohlsson, and S. Pfeifer. 1998. The H19 transcript is associated with polysomes and may regulate IGF2 expression in trans. J. Biol. Chem. 27328247-28252. [DOI] [PubMed] [Google Scholar]
- 26.Lottin, S., A. S. Vercoutter-Edouart, E. Adriaenssens, X. Czeszak, J. Lemoine, M. Roudbaraki, J. Coll, H. Hondermarck, T. Dugimont, and J. J. Curgy. 2002. Thioredoxin posttranscriptional regulation by H19 provides a new function to mRNA-like non-coding RNA. Oncogene 211625-1631. [DOI] [PubMed] [Google Scholar]
- 27.Lottin, S., E. Adriaenssens, T. Dupressoir, N. Berteaux, C. Montpellier, J. Coll, T. Dugimont, and J. J. Curgy. 2002. Overexpression of an ectopic H19 gene enhances the tumorigenic properties of breast cancer cells. Carcinogenesis 231885-1895. [DOI] [PubMed] [Google Scholar]
- 28.Lottin, S., E. Adriaenssens, N. Berteaux, A. Leprêtre, M. O. Vilain, E. Denhez, J. Coll, T. Dugimont, and J. J. Curgy. 2005. The human H19 gene is frequently overexpressed in myometrium and stroma during pathological endometrial proliferative events. Eur. J. Cancer 41168-177. [DOI] [PubMed] [Google Scholar]
- 29.Milligan, L., E. Antoine, C. Bisbal, M. Weber, C. Brunel, T. Forne, and G. Cathala. 2000. H19 gene expression is up-regulated exclusively by stabilization of the RNA during muscle cell differentiation. Oncogene 195810-5816. [DOI] [PubMed] [Google Scholar]
- 30.Murrell, A., S. Heeson, and W. Reik. 2004. Interaction between differentially methylated regions partitions the imprinted genes IGF2 and H19 into parent-specific chromatin loops. Nat. Genet. 36889-893. [DOI] [PubMed] [Google Scholar]
- 31.Pandey, R. R., M. Ceribelli, P. B. Singh, J. Ericsson, R. Mantovani, and C. Kanduri. 2004. NF-Y regulates the antisense promoter, bidirectional silencing, and differential epigenetic marks of the Kcnq1 imprinting control region. J. Biol. Chem. 27952685-52693. [DOI] [PubMed] [Google Scholar]
- 32.Pauler, F. M., and D. P. Barlow. 2006. Imprinting mechanisms-it only takes two. Genes Dev. 201203-1206. [DOI] [PubMed] [Google Scholar]
- 33.Peters, J., and C. Beechey. 2004. Identification and characterisation of imprinted genes in the mouse. Brief Funct. Genomic Proteomic 2320-333. [DOI] [PubMed] [Google Scholar]
- 34.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reik, W., K. W. Brown, H. Schneid, Y. Le Bouc, W. Bickmore, and E. R. Maher. 1995. Imprinting mutations in the Beckwith-Wiedemann syndrome suggested by altered imprinting pattern in the IGF2-H19 domain. Hum. Mol. Genet. 42379-2385. [DOI] [PubMed] [Google Scholar]
- 36.Reik, W. 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447425-432. [DOI] [PubMed] [Google Scholar]
- 37.Rinn, J. L., M. Kertesz, J. K. Wang, S. L. Squazzo, X. Xu, S. A. Brugmann, L. H. Goodnough, J. A. Helms, P. J. Farnham, E. Segal, and H. Y. Chang. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 1291311-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ripoche, M. A., C. Kress, F. Poirier, and L. Dandolo. 1997. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 111596-1604. [DOI] [PubMed] [Google Scholar]
- 39.Rougeulle, C., and E. Heard. 2002. Antisense RNA in imprinting: spreading silence through Air. Trends Genet. 18434-437. [DOI] [PubMed] [Google Scholar]
- 40.Rougeulle, C., C. Cardoso, M. Fontes, L. Colleaux, and M. Lalande. 1998. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat. Genet. 1915-16. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt, J. V., J. M. Levorse, and S. M. Tilghman. 1999. Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc. Natl. Acad. Sci. USA 969733-9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoenfelder, S., G. Smits, P. Fraser, W. Reik, and R. Paro. 2007. Non coding transcripts in the H19 imprinting control region mediate gene silencing in transgenic Drosophila. EMBO Rep. 81068-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seidl, C., S. H. Stricker, and D. P. Barlow. 2006. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J. 253565-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sleutels, F., R. Zwart, and D. P. Barlow. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415810-813. [DOI] [PubMed] [Google Scholar]
- 45.Steenman, M. J., S. Rainier, C. J. Dobry, P. Grundy, I. L. Horon, and A. P. Feinberg. 1994. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms' tumour. Nat. Genet. 7433-439. [DOI] [PubMed] [Google Scholar]
- 46.Takeda, H., F. Caiment, M. Smit, S. Hiard, X. Tordoir, N. Cockett, M. Georges, and C. Charlier. 2006. The callipyge mutation enhances bidirectional long-range DLK1-GTL2 intergenic transcription in cis. Proc. Natl. Acad. Sci. USA 108119-8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thakur, N., V. K. Tiwari, H. Thomassin, R. R. Pandey, M. Kanduri, A. Gondor, T. Grange, R. Ohlsson, and C. Kanduri. 2004. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol. Cell. Biol. 247855-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thorvaldsen, J. L., M. R. Mann, O. Nwoko, K. L. Duran, and M. S. Bartolomei. 2002. Analysis of sequence upstream of the endogenous H19 gene reveals elements both essential and dispensable for imprinting. Mol. Cell. Biol. 222450-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verona, R. I., and M. S. Bartolomei. 2004. Role of H19 3′ sequences in controlling H19 and Igf2 imprinting and expression. Genomics 8459-68. [DOI] [PubMed] [Google Scholar]
- 50.Weber, M., H. Hagege, A. Murrell, C. Brunel, W. Reik, G. Cathala, and T. Forne. 2003. Genomic imprinting controls matrix attachment regions in the IGF2 gene. Mol. Cell. Biol. 238953-8959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber, M., H. Hagege, G. Lutfalla, L. Dandolo, C. Brunel, G. Cathala, and T. Forne. 2003. A real-time polymerase chain reaction assay for quantification of allele ratios and correction of amplification bias. Anal. Biochem. 320252-258. [DOI] [PubMed] [Google Scholar]
- 52.Yu, W., D. Gius, P. Onyango, K. Muldoon-Jacobs, J. Karp, A. P. Feinberg, and H. Cui. 2008. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451202-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilkin, F., J. Paquette, E. Ledru, C. Hamelin, M. Pollak, and C. L. Deal. 2000. H19 sense and antisense transgenes modify insulin-like growth factor-II mRNA levels. Eur. J. Biochem. 2674020-4027. [DOI] [PubMed] [Google Scholar]
- 54.Wutz, A., O. W. Smrzka, N. Schweifer, K. Schellander, E. F. Wagner, and D. P. Barlow. 1997. Imprinted expression of the IGF2r gene depends on an intronic CpG island. Nature 389745-749. [DOI] [PubMed] [Google Scholar]
- 55.Zubair, M., K. Hilton, J. R. Saam, M. A. Surani, S. M. Tilghman, and H. Sasaki. 1997. Structure and expression of the mouse L23mrp gene downstream of the imprinted H19 gene: biallelic expression and lack of interaction with the H19 enhancers. Genomics 45290-296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.