Abstract
Transforming growth factor β (TGF-β) signaling facilitates metastasis in advanced malignancy. While a number of protein-encoding genes are known to be involved in this process, information on the role of microRNAs (miRNAs) in TGF-β-induced cell migration and invasion is still limited. By hybridizing a 515-miRNA oligonucleotide-based microarray library, a total of 28 miRNAs were found to be significantly deregulated in TGF-β-treated normal murine mammary gland (NMuMG) epithelial cells but not Smad4 knockdown NMuMG cells. Among upregulated miRNAs, miR-155 was the most significantly elevated miRNA. TGF-β induces miR-155 expression and promoter activity through Smad4. The knockdown of miR-155 suppressed TGF-β-induced epithelial-mesenchymal transition (EMT) and tight junction dissolution, as well as cell migration and invasion. Further, the ectopic expression of miR-155 reduced RhoA protein and disrupted tight junction formation. Reintroducing RhoA cDNA without the 3′ untranslated region largely reversed the phenotype induced by miR-155 and TGF-β. In addition, elevated levels of miR-155 were frequently detected in invasive breast cancer tissues. These data suggest that miR-155 may play an important role in TGF-β-induced EMT and cell migration and invasion by targeting RhoA and indicate that it is a potential therapeutic target for breast cancer intervention.
Metastasis accounts for the majority of deaths of cancer patients, and thus, it is crucial to understand the molecular and cellular mechanisms that cause primary tumors to metastasize. The most critical step in the conversion of primary tumors to metastases is attributed to the process known as epithelial-mesenchymal transition (EMT). EMT is a remarkable example of cellular plasticity that involves the dissolution of epithelial tight junctions, the intonation of adherens junctions, the remodeling of the cytoskeleton, and the loss of apical-basal polarity (49, 55). In cells undergoing EMT, the loss of epithelial cell adhesion and cytoskeletal components is coordinated with a gain of mesenchymal components and the initiation of a migratory phenotype.
Transforming growth factor β (TGF-β) has emerged as a key regulator of EMT in late-stage carcinomas, where it promotes invasion and metastasis (54). TGF-β binds to a heteromeric complex of transmembrane serine/threonine kinases, the type I and II TGF-β receptors (TβRI and TβRII). Following ligand binding to TβRII, the type I receptor is recruited to the ligand-receptor complex, where the constitutively active TβRII transactivates TβRI. Activated TβRI phosphorylates the receptor-specific Smad2 and Smad3. Phosphorylated Smad2/Smad3 associates with Smad4 as a heteromeric complex and translocates to the nucleus. This complex binds directly to Smad-binding elements and associates with a plethora of transcription factors, coactivators or corepressors, thus leading to the transcriptional induction or repression of a diverse array of genes (54). A number of genes that are associated with tumor growth and metastasis have been shown previously to be directly regulated by this pathway, the effects of which include the induction of COX2, Slug, Snail, and Twist and the repression of Id2 and Id3 (54). Recent reports have shown the importance of microRNA-200 (miR-200) family downregulation during EMT (2, 12, 21, 36); however, the functions of upregulated miRNAs during TGF-β-induced EMT remain uncharacterized.
miRNAs are a class of 22-nucleotide noncoding RNAs that are evolutionarily conserved and function as negative regulators of gene expression. Like conventional protein-encoding mRNA, miRNAs are transcribed by RNA polymerase II and controlled by transcription factors (1, 9, 16, 38). The primary transcript (pri-miRNA) is capped and polyadenylated. The pri-miRNA is processed by the nuclear RNase III Drosha and its cofactor DGCR8/Pasha to generate a precursor miRNA, a 60- to 70-nucleotide RNA that has a stem-loop structure (3, 13, 15). The precursor miRNA is rapidly exported to the cytoplasm by exportin-5 in a Ran-GTP-dependent manner, where it is further processed by a second RNase III, Dicer, to release a mature ∼22-nucleotide miRNA. Subsequently, the mature miRNA enters an RNA-induced silencing complex, guides this complex to regions of complementarity in the 3′ untranslated region (UTR) of target mRNAs, and triggers either their degradation or the inhibition of translation, depending on the degree of complementarity between the miRNA and its target mRNA (24, 44). Based on predictions by publicly available algorithms, each miRNA may have several hundreds to potentially thousands of target mRNAs (25, 32). miRNA profiling has shown the deregulation of miRNAs in different types of human malignancy, some of which are associated with late-stage and high-grade tumors as well as poor prognosis (29, 34, 35), implying that miRNA may play a pivotal role in tumorigenesis and in tumor progression to metastasis.
In the present study, we profiled the miRNA signature of EMT induced by the TGF-β/Smad pathway in normal murine mammary gland (NMuMG) epithelial cells. We further demonstrated that miR-155 is a direct transcriptional target of the TGF-β/Smad4 pathway and mediates TGF-β-induced EMT. The ectopic expression of miR-155 disrupted proper tight junction formation and promoted cell migration and invasion. The knockdown of miR-155 reduced the occurrence of TGF-β-induced EMT and cell migration and invasion. Moreover, RhoA is negatively regulated by miR-155. The restoration of RhoA by using an expression vector cloned without the 3′ UTR eliminated the effects of miR-155-induced phenotypes. Thus, we demonstrated for the first time that miR-155 is regulated by the TGF-β/Smad pathway and plays a role in mammary epithelial cell plasticity through the targeting of RhoA.
MATERIALS AND METHODS
Cell line, treatment, and tumor specimens.
NMuMG epithelial cells were purchased from the American Type Culture Collection (Manassas, VA). Stable Smad4 knockdown (pRetroSuper-Smad4-shRNA) and pRetroSuper (pRS) vector-transfected (parental) NMuMG cells were kindly provided by Peter ten Dijke (Leiden University Medical Center, The Netherlands) (5). The cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. Cells were treated with TGF-β at a concentration of 5 ng/ml for the times indicated in the figures and legends. Cell transfection experiments were performed with Lipofectamine 2000 (Invitrogen). Frozen human primary breast tumor tissues and normal breast tissues were procured anonymously from patients who underwent surgery at H. Lee Moffitt Cancer Center, and each tumor sample contained at least 70% tumor cells as confirmed by the microscopic examination of sections. The tissues were snap-frozen within 15 min of accrual to prevent RNA degradation and stored at −70°C.
miRNA microarray, Northern blot, and quantitative reverse transcription-PCR (qRT-PCR) analyses.
Total RNA from cell lines and breast tumor and normal tissues was isolated using Trizol reagent (Invitrogen). miRNA array profiling was performed as described previously (52). Briefly, oligonucleotide arrays were printed with trimer oligonucleotide probes carrying antisense sequences relative to 515 miRNAs specific to humans and mice on GeneScreen Plus (NEN) membranes. The miRNA expression profiling was performed by the hybridization of the array with [γ-32P]ATP-labeled small RNA probes prepared from TGF-β-treated and untreated NMuMG cells. To ensure the accuracy of the hybridizations, each experimental grouping was hybridized onto three separate membranes. In addition, eight oligonucleotides not matching any known miRNA were used as hybridization controls. Hybridization signals for each spot of the array and background values at 15 empty spots were measured. Raw data were further automatically processed in Microsoft Excel. Hybridization signals that failed to exceed the average background value by more than three standard deviations were excluded from analysis. The data were normalized, and an unsupervised hierarchical clustering analysis with average linkage algorithms was performed with GeneCluster. The results were visualized with TreeView. Differentially expressed miRNAs were identified by using the t test procedure within the significance analysis of the microarray.
For Northern blot analysis, 20 μg of RNA was separated on a 15% denaturing polyacrylamide gel and then electroblotted onto a Zeta-Probe GT blotting membrane (Bio-Rad). Following transfer, the membrane was dried and UV cross-linked. The probes were prepared using the Starfire oligonucleotide labeling system according to the protocol of the manufacturer (Integrated DNA Technologies). The blots were hybridized overnight at 42°C in a buffer containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 20 mM Na2HPO4 (pH 7.2), 7% sodium dodecyl sulfate, 1× Denhardt's solution, and 0.2 mg of salmon sperm DNA/ml and then washed with 1× SSC-1% sodium dodecyl sulfate buffer at 42°C (25). qRT-PCR was performed with the mirVana qRT-PCR detection kit according to the instructions of the manufacturer (Ambion).
Immunofluorescence and immunoblotting.
For immunofluorescence, cells grown to 60 to 80% confluence were washed with phosphate-buffered saline and fixed with 4% paraformaldehyde. Cells were permeabilized with 0.5% Triton X-100 in phosphate-buffered saline prior to the addition of primary and secondary antibodies. The visualization of E-cadherin was performed by staining with mouse anti-E-cadherin (BD Transduction Labs) and then with tetramethyl rhodamine isothiocyanate-conjugated goat secondary anti-mouse immunoglobulin G (IgG; Sigma). The visualization of ZO-1 was performed by staining with rabbit anti-ZO-1 (34) and then with fluorescein isothiocyanate-conjugated goat secondary anti-rabbit IgG (Sigma). Fluorescence imaging was performed by confocal microscopy (Leica), and phase-contrast imaging was performed with an inverted microscope (Nikon). Immunoblotting analysis was carried out as described previously (52).
Isolation and analysis of the miR-155 promoter.
miR-155 is found within the BIC gene on chromosome 21 in humans and chromosome 16 in mice. The genomic structure of human BIC consists of three exons, of which exon 3 encodes miR-155 (8, 43). Based on the transcription start site identified previously by other groups (33, 53), a 1.0-kb putative promoter was amplified by nested PCR using NMuMG cell genomic DNA as the template. The PCR products were cloned into the pGL3-basic vector (Promega) by utilizing the HindIII-MluI sites, and the sequence of the resulting construct was confirmed by DNA sequencing. All-in-one Seq-Analyzer software was used to identify putative Smad4-binding sites within the miR-155/BIC promoter.
ChIP assay.
NMuMG cells were cultured to 70 to 80% confluence and treated with TGF-β at 5 ng/ml for 24 h. Cells were harvested for chromatin immunoprecipitation (ChIP) analysis as described previously (35). Briefly, solubilized chromatin was prepared from a total of 2 × 107 cells. The chromatin solution was diluted 10-fold with ChIP dilution buffer and precleared with protein A beads and preimmune serum. The precleared chromatin solution was divided and utilized in immunoprecipitation assays with either an anti-Smad4 antibody or an anti-IgG antibody. Following washing, the antibody-protein-DNA complex was eluted from the beads. After cross-linking, protein and RNA were removed and the purified DNA was subjected to PCR with primers specific for two putative Smad4-binding sites within the BIC/pri-miR-155 promoter. The sequences of the PCR primers used were as follows: 5′-CCAAAGGAATCACTGGAGGA-3′ and 5′-CCCACAGGTCACTAGGCAAT-3′. Amplified PCR products were resolved by 1.5% agarose gel electrophoresis and visualized by BioImage.
Knockdown of miR-155.
The knockdown of miR-155 in NMuMG cells was achieved by transfection with antisense 2′-O-methyl oligoribonucleotides (ASO) against miR-155 by using Lipofectamine 2000 (Invitrogen). Transfection complexes were prepared according to the instructions of the reagent manufacturer and added directly to the cells at a final oligonucleotide concentration of 10 nmol/liter. Following 36 h of incubation, cells were treated with or without human TGF-β1 (2 ng/ml; RD Systems) for different times and evaluated for tight junctions and EMT phenotypes. ASO were composed entirely of 2′-O-methyl bases and were chemically synthesized by Integrated DNA Technologies (Coralville, IA) with the following sequences: 2′-O-Me-155, 5′-CCCCTATCACAATTAGCATTAA-3′, and 2′-O-Me-scrambled, 5′AAGGCAAGCUGACCCUGAAGU-3′.
Construction of expression plasmid and establishment of cell lines with stable miR-155 expression.
An miR-155 expression plasmid was created according to the protocol for the BLOCK-iT polymerase II miRNA RNA interference expression vector kit. Briefly, the following oligonucleotides were cloned into the pcDNA6.2-GW/miR vector (Invitrogen) and the resulting construct was designated pcDNA6.2-GW/miR-155: 5′-TGCTGTTAATGCTAATTGTGATAGGGGGTTTTGGCCACTGACTGACCCCCTATCAATTAGCATTA-3′and 5′CCTGTAATGCTAATTGATAGGGGGTCAGTCAGTGGCCAAAACCCCCTATCACAATTAGCATTAAC-3′. To generate cells stably expressing miR-155, NMuMG cells were transfected with pcDNA6.2-GW/miR-155 or pcDNA6.2-GW/miR-control vector by using Lipofectamine 2000 (Invitrogen). Following selection with blasticidin, stable clonal cell lines were established and examined for the expression of miR-155 by Northern analysis.
RhoA gene 3′ UTR luciferase reporter assay.
To create individual RhoA gene 3′ UTR luciferase reporter constructs, 60-bp sequences from putative miR-155 binding sites were synthesized and ligated into the pMIR-REPORT vector (Ambion) at SpeI and HindIII sites. To create a full-length RhoA gene 3′ UTR reporter, the following primers were used to amplify the 3′ UTR of the RhoA gene from a mouse cDNA library: 5′ACTAGTGCAGCCTCATGCGGTTAAT-3′ and 5′-AAGCTTTTTTTTTAGAAAACTGCCTTTATTCT-3′. The primers were digested and cloned into the pMIR-REPORT vector (Ambion) at SpeI and HindIII sites. To create a mutant 3′ UTR, point mutations were introduced at the first two miR-155-matching nucleotides within selected putative seeding sequence regions with the following rules: A was changed to T and vice versa, and G was changed to C and vice versa. NMuMG cells in 24-well plates were transfected with 0.10 μg of the pMIR-REPORT-3′UTR/RhoA luciferase reporter, 0.05 μg of the normalization plasmid pCMV-β-galactosidase, and 0.6 μg of miR-155 or non-green fluorescent protein-expressing control vector or 5 ng of TGF-β/ml. Luciferase assays were performed using a luciferase assay system (Promega), and activities were normalized to β-galactosidase activity (52).
RESULTS
Profiles of miRNA expression in TGF-β-induced EMT in NMuMG and Smad4 knockdown NMuMG cells.
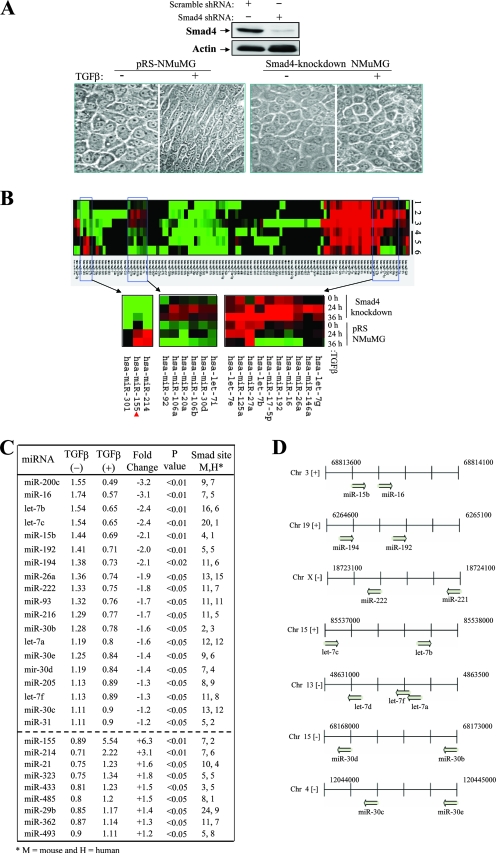
The TGF-β/Smad pathway plays a critical role in promoting cancer metastasis. Previous studies have identified a number of protein-encoding genes that are regulated by TGF-β/Smad and mediate TGF-β function (54). Since TGF-β-induced EMT in NMuMG cells is a frequently used cellular model to study the molecular mechanism of cancer metastasis (5, 34) and its miRNA expression signature is not completely understood, we proceeded to profile changes in mRNA expression by using an miRNA microarray in an attempt to identify possible miRNAs involved in TGF-β/Smad-induced EMT and cell migration and invasion. Results from previous studies have shown that different cell lines, including NMuMG cells, with the stable knockdown of Smad4 fail to undergo EMT in response to TGF-β treatment (5). As shown in Fig. 1A, the Smad4 level was decreased by 80% after the stable transfection of NMuMG cells with Smad4 short hairpin RNA. As expected, parental NMuMG cells but not Smad4 knockdown NMuMG cells underwent EMT after TGF-β treatment. Thus, we treated both cell lines with TGF-β for 0, 24, and 36 h to obtain an miRNA signature by using three separate hybridizations. Hybridization to a custom microarray, which contained 515 miRNAs, revealed 28 differentially regulated miRNAs in the parental cells but not in the Smad4 knockdown NMuMG cells between 0 and 24 or 36 h of TGF-β treatment, with a P value of ≤0.05 (Fig. 1B and C). Of the 28 listed miRNAs, 9 were upregulated and 19 were downregulated during TGF-β treatment as demonstrated at both the 24- and 36-h time points (Fig. 1C). Our array revealed that the members of the let-7 and miR-30 families of miRNAs were consistently downregulated and that clustered miRNAs were often regulated simultaneously (Fig. 1D). In agreement with previous findings, our array data also showed significant downregulation of miR-200c and miR-205 during the expression of a mesenchymal phenotype (12). miR-155, miR-214, miR-21, and miR-323 were all found to be significantly upregulated. We selected five significantly deregulated miRNAs for validation by Northern blot and qRT-PCR analyses to determine the accuracy of the array data (Fig. 2A and data not shown).
FIG. 1.
Profile of miRNA expression in TGF-β/Smad-induced EMT in NMuMG cells. (A) TGF-β induces the cellular morphological change of EMT in control (pRS vector-transfected) but not Smad4 knockdown NMuMG cells. (Upper panels) A Western blot analysis of control and Smad4-knockdown NMuMG cells was performed with anti-Smad4 and antiactin antibodies. shRNA, short hairpin RNA; +, present; −, absent. (Lower panels) The indicated cells were treated with (+) or without (−) TGF-β for 24 h and photographed. pRS-NMuMG, pRS vector-transfected NMuMG cells. (B) Heat map representation of miRNAs deregulated in control and Smad4 knockdown NMuMG cells during TGF-β treatment. The red arrowhead indicates hsa-miR-155 highest upregulated miRNA. (C) List of deregulated miRNAs induced by TGF-β in control but not Smad4 knockdown NMuMG cells. (D) Chromosomal representation of the locations of deregulated miRNAs within mouse genomic DNA. Clustered miRNAs were simultaneously downregulated or upregulated during TGF-β treatment. Chr, chromosome.
FIG. 2.
TGF-β/Smad transcriptionally regulates miR-155. (A) Verification of TGF-β-regulated miRNAs. Parental NMuMG cells were treated with TGF-β for the indicated times and subjected to Northern blot analysis with the indicated probes. The numbers between the gels represent the miR-155-to-U6 band density ratios. (B) TGF-β induces miR-155 promoter activity in parental but not Smad4 knockdown NMuMG cells. (Top panel) The diagram depicts the putative mouse miR-155 promoter construct containing two Smad4-binding sites and the individual Smad4-binding-site deletion mutant constructs cloned into the pGL3 plasmid. Parental and Smad4 knockdown cells were transfected with pGL3-miR-155-Luc and treated with or without TGF-β. Following 36 h of incubation, the cells were subjected to a luciferase reporter assay. The experiments were done three times with triplicate samples for each treatment. +, present; −, absent. (C) The first Smad4-binding site is required for TGF-β-induced miR-155 promoter activity. NMuMG cells were transfected with the indicated plasmids, treated (+) with TGF-β or left untreated (−), and assayed for luciferase activity. Values are expressed as relative luciferase units. (D) TGF-β induces Smad4 binding to the miR-155 promoter. NMuMG cells treated with or without TGF-β were evaluated by a ChIP assay. PCR was done with the eluted DNA fragments from anti-Smad4 immunoprecipitates by using a set of primers that detect the first Smad4-binding site, determined in the reporter assay to be important. IgG and antiactin antibody were used as negative controls. α-Smad4, anti-Smad4 antibody.
We next examined whether the promoters of TGF-β-regulated miRNAs may contain a Smad4-binding element. An analysis of the sequence of a 6-kb DNA region (5 kb upstream to 1 kb downstream from the position corresponding to the end of the pri-miRNA), designated the putative miRNA promoter, was conducted using MATCH and TRANSFAC (22, 29). As indicated in Fig. 1C, both mouse and human promoters of TGF-β-deregulated miRNAs contained one or more Smad4-binding sites, further suggesting that these miRNAs may be regulated by the TGF-β/Smad pathway and play important roles in TGF-β-induced EMT.
miR-155 is a direct target of the TGF-β/Smad pathway.
Since miR-155 is a highly upregulated miRNA in TGF-β-treated NMuMG cells (Fig. 1C and 2A) and its function in metastasis is currently unknown, we investigated if miR-155 is directly regulated by the TGF-β/Smad4 pathway. We cloned a mouse promoter 1.0 kb upstream from the transcriptional start site of the BIC (pri-miR-155) gene into the pGL3-basic vector (33, 53). Sequence analysis revealed two Smad response elements (CAGAC and CTGTCTGT) (23) located at bp −542 and −454 from the transcription start site, which are conserved in the human miR-155 promoter. A luciferase reporter assay revealed that miR-155 promoter activity is induced by TGF-β in parental but not Smad4 knockdown NMuMG cells (Fig. 2B). Deletion mapping showed that the first Smad4-binding site (i.e., bp −454) is responsible for TGF-β-induced miR-155 promoter activity (Fig. 2C). To determine whether Smad4 could directly bind to the Smad-binding site within the promoter in vivo, we carried out a ChIP assay. NMuMG cells were treated with or without TGF-β for 24 h and immunoprecipitated with anti-Smad4 antibody. Figure 2D shows that Smad4 specifically bound to the promoter following TGF-β treatment. These findings indicate that miR-155 is a transcriptional target of the TGF-β/Smad pathway.
miR-155 facilitates TGF-β-induced EMT and tight junction dissolution, as well as cell migration and invasion.
We next assessed whether miR-155 plays a role during TGF-β-induced EMT. First, NMuMG cells were transfected with ASO against miR-155 and control ASO and then treated with or without TGF-β. Figure 3A shows that miR-155 ASO effectively knocked down miR-155 induced by TGF-β. Accordingly, TGF-β-driven morphological changes were reduced by the knockdown of miR-155 for up to 24 h (Fig. 3B). Immunofluorescent staining for E-cadherin and ZO-1 at the 24-h time point revealed that tight junction assembly was disrupted by TGF-β in control ASO-transfected NMuMG cells but still intact in miR-155 knockdown cells (Fig. 3C). In addition, the TGF-β downregulation of E-cadherin was abolished by the knockdown of miR-155 (Fig. 3D). To further demonstrate the effect of miR-155 on TGF-β function, we established a cell line stably transfected with miR-155 by the transfection of NMuMG cells with pcDNA6.2-GW/miR-155 followed by selection with blasticidin (Fig. 3A). The ectopic expression of miR-155 alone was not sufficient to induce EMT but did cause disruption in cell polarity and tight junction formations (Fig. 3E and F). Moreover, cells overexpressing miR-155 underwent a complete TGF-β-induced EMT by 12 h compared to the 24 to 36 h it normally takes for the parental control cells in response to TGF-β (Fig. 3E and F). In addition, the E-cadherin level in miR-155-transfected cells was much lower than that in vector-treated cells by 12 h of TGF-β treatment (Fig. 3G).
FIG. 3.
miR-155 mediates the effect of TGF-β on EMT. (A) Ectopic expression and knockdown of miR-155. NMuMG cells were transfected with the indicated plasmids and oligonucleotides. Following treatment with (+) or without (−) TGF-β, cells were subjected to Northern blot analysis with [α-32P]dATP-labeled miR-155 and U6 probes. Pre miR-155, pcDNA6.2-GW/miR-155; Pre miR-ctrl, pcDNA6.2-GW/miR-control. (B and C) The knockdown of miR-155 inhibited TGF-β-induced EMT and tight junction dissolution. NMuMG cells were transfected with miR-155 ASO and control ASO and then treated with or without TGFβ for 24 h. Cell morphologies were documented using a phase-contrast microscope (B), and cells were stained with anti-ZO-1 and anti-E-cadherin antibodies conjugated to fluorescein isothiocyanate and tetramethyl rhodamine isothiocyanate, respectively (C). Arrows in panel C indicate the restoration of TGF-β-disrupted tight junctions by the knockdown of miR-155. (D) The knockdown of miR-155 inhibits the TGF-β downregulation of E-cadherin (E-Cad). NMuMG cells were transfected with control and miR-155 ASO, treated with TGF-β for 24 h, and then immunoblotted with the indicated antibodies. (E and F) The overexpression of miR-155 disrupted proper tight junction formations and accelerated TGF-β-induced EMT. Stably miR-155-transfected and control NMuMG cells were treated with or without TGF-β for 12 h and photographed (E) and immunofluorescence-stained with the indicated antibodies (F). The tight junction dissolution promoted by the ectopic expression of miR-155 is indicated by arrows. (G) Western blot analysis of E-cadherin in NMuMG cells that were transfected with miR-155 and then treated with TGF-β for 12 h.
Furthermore, we assessed the role of miR-155 in cell mobility and invasion. Cell migration was examined using Boyden chambers and “wound healing” assays. Cell invasion was measured using Matrigel-coated Boyden chambers as previously described (28). Triplicate experiments showed that the stable expression of miR-155 nearly doubled cell migration, as illustrated by the results of Boyden chamber and wound healing assays (Fig. 4A, B, and D). Further, cell invasion was also significantly enhanced by the stable overexpression of miR-155 (Fig. 4A and D). In contrast, the knockdown of miR-155 considerably reduced cell migration and invasion (Fig. 4C and D). Taken collectively, these results indicate that miR-155 plays an important role in TGF-β-induced EMT, as well as in cell migration and invasion.
FIG. 4.
miR-155 plays a significant role in cell migration and invasion. (A and B) The ectopic expression of miR-155 induces cell migration and invasion. NMuMG cells stably transfected with pcDNA6.2-GW/miR-control (Pre miR-ctrl) and pcDNA6.2-GW/miR-155 (Pre miR-155) were examined for cell migration and invasion by using Boyden chamber and wound healing assays as previously described (28). (C) The knockdown of miR-155 reduces cell migration and invasion. NMuMG cells were transfected with miR-155 ASO and control ASO. Following 36 h of incubation, cells were subjected to chamber cell migration and invasion assays using Boyden chambers with or without the inclusion of coating Matrigel. (D) Statistical analysis. The experiments depicted in panels A to C were repeated three times. P values for comparisons are indicated. Values are arbitrary units.
RhoA is negatively regulated by miR-155.
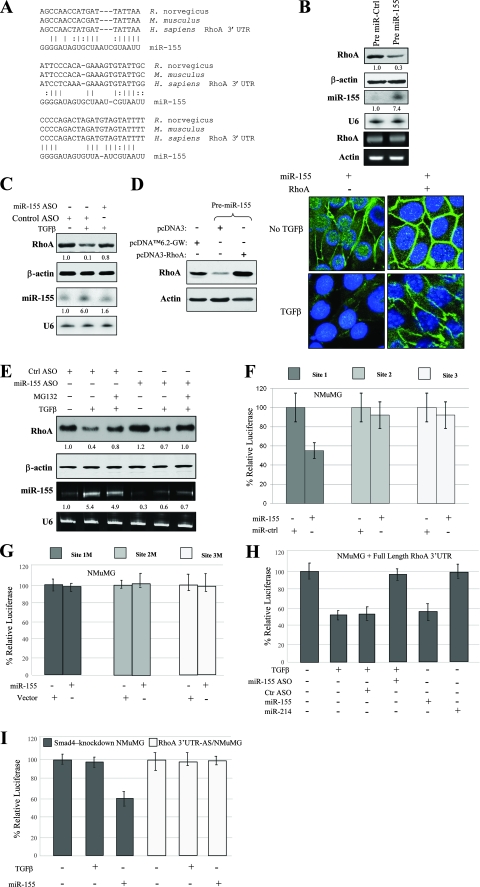
Since miR-155 was significantly upregulated in NMuMG cells after treatment with TGF-β (Fig. 1C and 2A) and mediates TGF-β function in EMT (Fig. 3) and cell mobility and invasion (Fig. 4), we proceeded to identify potential targets known to play a role in EMT by using RNA22 miRNA target detection and miRBase databases. Among the candidates surveyed, we found that the 3′ UTR of the RhoA gene, which plays an important role in cell junction formation and stabilization (34, 37, 46, 49), contains three highly conserved regions that may serve as a binding site for miR-155 as determined by the RNA22 algorithm (Fig. 5A).
FIG. 5.
RhoA is a target of miR-155. (A) Sequence alignment of miR-155 with the 3′ UTR of the RhoA gene. The seed sequence of miR-155 matches three regions of the RhoA gene 3′ UTR, which are highly conserved among humans (Homo sapiens), mice (Mus musculus), and rats (Rattus norvegicus). Vertical lines and colons indicate Watson-Crick and wobble base pairing, respectively. (B) miR-155 reduces RhoA protein but not mRNA expression. NMuMG cells were transfected with pcDNA6.2-GW/miR-155 (Pre miR-155) and control vector (Pre miR-Ctrl). After selection with blasticidin, the expression level of RhoA was determined using Western blot analysis (first panel). The same blot was reprobed with antiactin antibody (second panel). The expression of miR-155 from the same set of cells was examined by Northern blot analysis (third panel). RT-PCR was performed to determine RhoA mRNA levels (fifth panel). U6 and actin were used for loading controls (fourth and sixth panels). The numbers between the gels represent the miR-155-to-U6 band density ratios. (C) The knockdown of miR-155 inhibits the TGF-β downregulation of RhoA. NMuMG cells were transfected with miR-155 ASO or control ASO. After 36 h of incubation, cells were treated with (+) or without (−) TGF-β for 24 h and immunoblotted with the indicated antibodies (first and second panels). A Northern blot was hybridized with the indicated probes (third and fourth panels). (D) The ectopic expression of RhoA cDNA lacking the 3′ UTR overrides the effects of miR-155 on cell tight junction dissolution. Clonal cells stably expressing miR-155 were transfected with pcDNA3-RhoA or control vector. Following treatment with or without TGF-β, cells were immunostained with anti-ZO-1 antibody. (E) The inhibition of the ubiquitin-proteasome pathway and miR-155 hinders TGF-β downregulation of RhoA. NMuMG cells were transfected with control (ctrl) or miR-155 ASO. Following 48 h of incubation, cells were treated with or without MG132 and/or TGF-β for 12 h and then subjected to immunoblotting (first and second panels) and qRT-PCR (third and fourth panels). (F and G) miR-155 inhibits RhoA gene 3′ UTR luciferase activity. Cells were transfected with individual site constructs in pGL3 (F) and the corresponding mutant constructs (G), together with pCMV-β-galactosidase, pcDNA6.2-GW/miR-155 (miR-155), or pcDNA6.2-GW/miR-control vector (miR-ctrl). Luciferase activities were normalized to β-galactosidase activity. (H and I) TGF-β and miR-155 repress the full-length RhoA gene 3′ UTR but not its antisense strand in parental NMuMG cells. Cells were transfected with the indicated plasmids and treated with or without TGF-β for 24 h. Luciferase activities were normalized to β-galactosidase activity. The full-length RhoA gene 3′ UTR responds to TGF-β and miR-155 in parental (H) but not Smad4 knockdown (I) NMuMG cells. TGF-β-inhibited reporter activity was inhibited by the knockdown of miR-155. miR-214 was used as a control. Experiments were done in triplicate for standard deviation calculations. RhoA 3′ UTR-AS/NMuMG, NMuMG cells transfected with the antisense strand of the RhoA gene 3′ UTR.
To examine whether the RhoA gene is indeed a target of miR-155, NMuMG cells were transfected with pcDNA6.2-GW/miR-155 and pcDNA6.2-GW/miR-control and selected with blasticidin. Immunoblotting and RT-PCR analyses revealed that RhoA protein but not RhoA mRNA was considerably decreased in miR-155-transfected cells (Fig. 5B). As expected, TGF-β treatment reduced the expression level of RhoA in NMuMG cells (Fig. 5C). However, the knockdown of miR-155 largely abolished the inhibitory effects of TGF-β on the RhoA protein level (Fig. 5C). Since miR-155 downregulates RhoA to drive EMT progression, it is reasoned that the ectopic expression of RhoA can reverse this phenomenon. Indeed, progression toward EMT was hindered when pcDNA3-RhoA, in which the RhoA gene lacked a 3′ UTR, was introduced into stably miR-155-transfected NMuMG cells that were subsequently treated with TGF-β for 24 h. As shown in Fig. 5D, the ectopic expression of RhoA decreased tight junction dissolution induced by miR-155. These results further indicate that RhoA is a target of miR-155 and mediates the contribution of miR-155 to the control of epithelial cell plasticity.
Previous studies have shown that TGF-β induces RhoA protein degradation through the ubiquitin-proteasome pathway (34, 49). Thus, we further examined the significance of protein degradation and miR-155 in the regulation of RhoA by TGF-β. NMuMG cells were transfected with control or miR-155 ASO and then treated with or without TGF-β and proteasome inhibitor MG132. Figure 5E shows that neither MG132 nor the knockdown of miR-155 alone fully inhibited the TGF-β downregulation of RhoA. However, the combined treatment with MG132 and miR-155 ASO abolished the TGF-β repression of RhoA expression. These results indicate that both the ubiquitin-proteasome pathway and miR-155 mediate the TGF-β regulation of RhoA.
To further demonstrate that RhoA is negatively regulated by miR-155, we generated luciferase reporters that contained each of the three highly conserved seeding sites, along with corresponding mutant forms, and a construct that contained the full length of the RhoA gene 3′ UTR. To determine if any of the three seeding sites respond to miR-155, the reporter plasmids were introduced into NMuMG cells together with the pcDNA6.2-GW/miR-155 or pcDNA6.2-GW/miR-control vector. Following 36 h of incubation, cells were subjected to a luciferase assay. Results from triplicate experiments showed that reporter activity for each site was reduced by the ectopic expression of miR-155, with site 1 exhibiting a more significant response than the two other sites. When these sites were mutated, the luciferase reporter was no longer inhibited by miR-155 (Fig. 5F and G). We then examined if the full-length RhoA gene 3′ UTR is regulated by TGF-β through miR-155. Results from triplicate experiments showed that the full-length reporter responded to TGF-β and that this response was largely abolished when miR-155 ASO was introduced simultaneously into the cells (Fig. 5H). When the same experiments were carried out with the Smad4 knockdown NMuMG cell line, the full-length reporter did not respond to TGF-β (Fig. 5I). Furthermore, the antisense full-length 3′ UTR of the RhoA gene failed to respond to miR-155 and TGF-β (Fig. 5I).
miR-155 expression in invasive breast cancer.
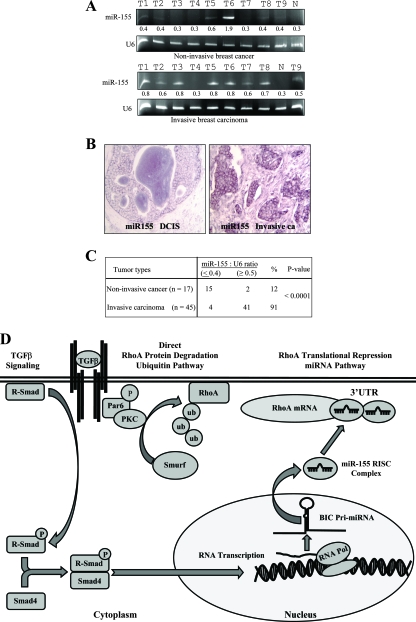
Having observed that miR-155 mediates TGF-β-induced EMT and cell migration and invasion, we asked if the expression of miR-155 is associated with cancer invasiveness in human primary breast carcinoma. A total of 62 breast cancer specimens (17 noninvasive and 45 invasive breast carcinomas) and 5 normal breast tissue samples were examined for the expression of miR-155. qRT-PCR (Fig. 6A) and miRNA locked nucleic acid in situ hybridization (LNA-ISH) (Fig. 6B) analyses revealed high levels of miR-155 in 41 of 45 invasive tumors but in only 2 of 17 noninvasive cancer tissues (Fig. 6C). The level of expression of miR-155 in normal breast tissue was very low (Fig. 6A and B). These data further support the findings demonstrating the involvement of miR-155 in EMT and invasion as observed in NMuMG cells and suggest that miR-155 may play a pivotal role in breast cancer metastasis.
FIG. 6.
Elevated levels of miR-155 are associated with invasive breast cancer. (A) qRT-PCR analysis of miR-155 expression in normal human breast tissue (lanes N), noninvasive breast cancer tissue, and invasive breast carcinoma. U6 was used as a control. The numbers between the gels represent the miR-155-to-U6 band density ratios. T1 to T9 indicate the tumor samples. (B) LNA-ISH analyses. miR-155 was labeled with digoxigenin-ddUTP by using the Dig 3′-end labeling kit (Roche) and hybridized to paraffin sections of breast cancer tissue. Representative photomicrographs of sections of ductal carcinoma in situ (DCIS) breast tissue (left) and invasive breast tumor tissue (right) are shown. (C) Summary of qRT-PCR and LNA-ISH analyses. miR-155 was detected more frequently in invasive breast carcinoma tissue than in noninvasive tumor tissue. (D) Schematic illustration of the transcriptional induction of miR-155 by the TGF-β/Smad pathway and the TGF-β downregulation of RhoA through the ubiquitin-proteasome and miR-155 cascades. PKC, protein kinase C; RISC, RNA-induced silencing complex; RNA Pol, RNA polymerase.
DISCUSSION
Many studies have demonstrated previously that the TGF-β pathway plays a critical role in breast cancer metastasis (18, 31, 39, 50). Several TGF-β/Smad-regulated genes have been shown to mediate TGF-β signaling in the control of cellular processes associated with breast cancer metastasis (18, 31, 39, 50). In this study, we report an miRNA expression signature of TGF-β-induced EMT in NMuMG epithelial cells. Twenty-eight miRNAs were found to be significantly deregulated by TGF-β in parental but not Smad4 knockdown NMuMG cells. Further, we showed that miR-155, the most significantly upregulated miRNA, plays an important role in TGF-β-induced EMT and cell migration and invasion. In addition, RhoA was negatively regulated by miR-155 and the restoration of RhoA in miR-155-overexpressing cells decreased TGF-β/miR-155-induced tight junction dissolution. Moreover, high expression levels of miR-155 correlate with invasive breast carcinomas. These findings are important for several reasons. First, they provide an miRNA expression signature of the TGF-β/Smad pathway in mammary gland epithelial cells. Second, the findings of this study establish a direct link between TGF-β/Smad4 and miR-155 during EMT. Finally, this is the first study to describe RhoA as a direct target of miR-155.
miR-155, a product of the BIC gene, is overexpressed in a number of human malignancies, which include B-cell lymphoma and carcinomas of the breast, colon, lung, and ovary (8, 17, 48, 51, 52). Eμ-mmu-miR-155 transgenic mice develop B-cell malignancies (4), whereas miR-155 knockout mice exhibit impaired immune function (40, 45). The transcription factor Pu.1 has been validated previously as a direct target of the miR-155-mediated immunoresponse (47). Moreover, miR-155 represses tumor protein 53-induced nuclear protein 1 (TP53NP1), leading to pancreatic tumor development (10). In addition, the NF-κB and AP-1 transcription factors have been shown previously to regulate miR-155 expression (20, 33, 53). However, the functions of miR-155 in cell migration and invasion have not been investigated. Thus, our study provides the first evidence that miR-155 is upregulated by the TGF-β/Smad4 pathway and mediates TGF-β-induced EMT and cell invasion.
Previous computational and experimental studies have focused on the quality of sequence matching between miRNA and the target (6, 7, 14, 26, 42). miRNAs negatively regulate their target mRNAs through base-pairing interactions, which lead to either mRNA degradation or translational inhibition, depending on the degree of matching between the seed sequence (positions 2 to 7 on the 5′ side) of miRNA and the 3′ UTR of mRNA; e.g., miRNA induces mRNA degradation when the seed sequence perfectly matches the target 3′ UTR or inhibits translation when the sequences are partially identical (6, 7, 14, 26, 42). In addition, recent reports indicate that mRNA secondary structures may contribute to target recognition due to the fact that there is an energy cost associated with the un-base pairing of the messenger required to make the target site accessible for miRNA binding (27, 30). Kertesz et al. showed that site accessibility is as important as base pairing within the seeding region. Effective miRNA function requires nucleic acids flanking the target site, as well as the target itself, to be unpaired in a thermodynamically stable fashion (19). Through the RNA22 algorithm, we found that the seed sequence of miR-155 has the potential to bind in multiple regions within the 3′ UTR of the RhoA gene. Of these sites, we selected three sites that fit the above-mentioned criteria and are highly conserved among species. Further, RhoA gene 3′ UTR reporter assays showed that miR-155 at all three sites significantly diminished luciferase activity. To mimic endogenous conditions more closely, we cloned the full-length RhoA gene 3′ UTR and performed experiments using TGF-β instead of the ectopic expression of miR-155. As expected, the full-length 3′ UTR responded to TGF-β, which was inhibited by the knockdown of miR-155 in the parental NMuMG line but not in the Smad4 knockdown line.
RhoA is the prototypical member of the Rho GTPase family, which regulates many cellular processes, including cellular adhesion, motility, and polarity, and is an important modulator of cell junction formation and stability (34, 37, 46, 49). Previous studies showed that TGF-β induces the disruption of tight junctions, cell polarity, and EMT through the ubiquitination and degradation of RhoA by Smurf1 E3 ligase that is activated by Par6 (34). Our study demonstrated that TGF-β downregulated RhoA protein expression through the upregulation of miR-155 and thus provided an additional molecular mechanism of TGF-β regulation of RhoA (Fig. 6D). Regulation by miRNAs provides a means for cells to prevent protein translation, a mechanism to quickly prevent the accumulation of proteins by translational inhibition; our findings go hand in hand with earlier findings that TGF-β ubiquitinates RhoA for degradation. In this scenario, the induction of miR-155 halts the translation of RhoA while ubiquitination degrades translated RhoA proteins. Based on computational program predictions, each miRNA may negatively regulate hundreds of protein-encoding mRNAs (6, 7, 14, 26, 42). Two recent studies using gene expression microarray analyses showed that miR-155 and its viral orthologue Kaposi's sarcoma-associated herpesvirus miR-K12-11 negatively regulate more than 180 mRNAs, some of which encode proteins involved in cell migration and invasion, including GSK3, PCSK5, and Rho GTPase-activating protein 21 (11, 41). Thus, RhoA is a major but not the only target that mediates miR-155 function in the control of cell polarity, EMT, and cell invasion contributing to cancer metastasis.
In addition, a previous study described a profile of miRNA expression in human keratinocytes treated with TGF-β. Four miRNAs were upregulated and another four were downregulated by TGF-β (55). Of the eight deregulated miRNAs, only one miRNA (e.g., miR-21) showed a change consistent with our results. This discrepancy may be due to the use of different cell types and the duration of TGF-β treatment. Recent reports have indicated the importance of downregulation for miR-200 family miRNAs during TGF-β-induced EMT (2, 12, 22, 36). In congruence to previous findings, our array also showed the downregulation of miR-200c and miR-205 during a mesenchymal transition (Fig. 1C). However, the remaining members of the miR-200 family were not detected in our array. This result may be due to the use of different cell lines for the miRNA array analyses in our study and those described in previous reports (2, 12, 36). In addition, we showed frequent upregulation of miR-155 in primary invasive breast cancer tissues. Consistent with this finding, a previous study reported that miR-155 is elevated in a metastatic breast cancer cell line, MDA-MB-231, but not in a nonmetastatic line, MCF-7 (28). We also observed that the knockdown of miR-155 inhibited the TGF-β downregulation of E-cadherin but that the ectopic expression of miR-155 enhanced the TGF-β effects on E-cadherin expression (Fig. 3D and G). Sequence analysis showed no match between the seed sequence of miR-155 and the 3′ UTRs of the E-cadherin, ZEB1, and SIP1 genes (12). Further investigation is required to determine the mechanism of miR-155 downregulation of E-cadherin, although it is likely that the effects are indirect but serve as a useful EMT indicator.
In summary, we demonstrated the miRNA expression signature of TGF-β/Smad-induced EMT in mammary epithelial cells. All 28 deregulated miRNAs contained at least one Smad4-binding site within their putative promoters. miR-155 mediated TGF-β/Smad pathway-induced EMT and cell migration and invasion through the targeting of RhoA. Further, the expression of miR-155 was associated with the invasive phenotype of breast cancer. Thus, miR-155 may be a potential metastatic/prognostic marker and therapeutic target for breast cancer metastasis intervention.
Acknowledgments
We thank Peter ten Dijke for the Smad4 knockdown NMuMG cell lines. We also thank the tissue procurement and DNA sequencing facilities at H. Lee Moffitt Cancer Center for providing cancer specimens and sequencing.
This work was supported by grants from the National Institutes of Health (CA77935 and CA107078) and the Department of Defense (DAMD17-02-1-0671) and Bankhead-Coley grant 07BB-01.
Footnotes
Published ahead of print on 15 September 2008.
REFERENCES
- 1.Ambros, V. 2004. The functions of animal microRNAs. Nature 431350-355. [DOI] [PubMed] [Google Scholar]
- 2.Burk, U., J. Schubert, U. Wellner, O. Schmalhofer, E. Vincan, S. Spaderna, and T. Brabletz. 2008. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 9582-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai, X., C. H. Hagedorn, and B. R. Cullen. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 101957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costinean, S., N. Zanesi, Y. Pekarsky, E. Tili, S. Volinia, N. Heerema, and C. M. Croce. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in Eμ-miR155 transgenic mice. Proc. Natl. Acad. Sci. USA 1037024-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deckers, M., M. van Dinther, J. Buijs, I. Que, C. Lowik, G. van der Pluijm, and P. ten Dijke. 2006. The tumor suppressor Smad4 is required for transforming growth factor beta-induced epithelial to mesenchymal transition and bone metastasis of breast cancer cells. Cancer Res. 662202-2209. [DOI] [PubMed] [Google Scholar]
- 6.Didiano, D., and O. Hobert. 2006. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 13849-851. [DOI] [PubMed] [Google Scholar]
- 7.Doench, J. G., and P. A. Sharp. 2004. Specificity of microRNA target selection in translational repression. Genes Dev. 18504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eis, P. S., W. Tam, L. Sun, A. Chadburn, Z. Li, M. F. Gomez, E. Lund, and J. E. Dahlberg. 2005. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. USA 1023627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farh, K. K., A. Grimson, C. Jan, B. P. Lewis, W. K. Johnston, L. P. Lim, C. B. Burge, and D. P. Bartel. 2005. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 3101817-1821. [DOI] [PubMed] [Google Scholar]
- 10.Gironella, M., M. Seux, M. J. Xie, C. Cano, R. Tomasini, J. Gommeaux, S. Garcia, J. Nowak, M. L. Yeung, K. T. Jeang, A. Chaix, L. Fazli, Y. Motoo, Q. Wang, P. Rocchi, A. Russo, M. Gleave, J. C. Dagorn, J. L. Iovanna, A. Carrier, M. J. Pebusque, and N. J. Dusetti. 2007. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. USA 10416170-16175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottwein, E., N. Mukherjee, C. Sachse, C. Frenzel, W. H. Majoros, J. T. Chi, R. Braich, M. Manoharan, J. Soutschek, U. Ohler, and B. R. Cullen. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 4501096-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory, P. A., A. G. Bert, E. L. Paterson, S. C. Barry, A. Tsykin, G. Farshid, M. A. Vadas, Y. Khew-Goodall, and G. J. Goodall. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10593-601. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman, and A. J. Enright. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34D140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grun, D., Y. L. Wang, D. Langenberger, K. C. Gunsalus, and N. Rajewsky. 2005. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 1e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, J., Y. Lee, K. H. Yeom, Y. K. Kim, H. Jin, and V. N. Kim. 2004. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 183016-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Houbaviy, H. B., M. F. Murray, and P. A. Sharp. 2003. Embryonic stem cell-specific microRNAs. Dev. Cell 5351-358. [DOI] [PubMed] [Google Scholar]
- 17.Iorio, M. V., M. Ferracin, C. G. Liu, A. Veronese, R. Spizzo, S. Sabbioni, E. Magri, M. Pedriali, M. Fabbri, M. Campiglio, S. Menard, J. P. Palazzo, A. Rosenberg, P. Musiani, S. Volinia, I. Nenci, G. A. Calin, P. Querzoli, M. Negrini, and C. M. Croce. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 657065-7070. [DOI] [PubMed] [Google Scholar]
- 18.Kang, Y., W. He, S. Tulley, G. P. Gupta, I. Serganova, C. R. Chen, K. Manova-Todorova, R. Blasberg, W. L. Gerald, and J. Massague. 2005. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc. Natl. Acad. Sci. USA 10213909-13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kertesz, M., N. Iovino, U. Unnerstall, U. Gaul, and E. Segal. 2007. The role of site accessibility in microRNA target recognition. Nat. Genet. 391278-1284. [DOI] [PubMed] [Google Scholar]
- 20.Kluiver, J., A. van den Berg, D. de Jong, T. Blokzijl, G. Harms, E. Bouwman, S. Jacobs, S. Poppema, and B. J. Kroesen. 2007. Regulation of pri-microRNA BIC transcription and processing in Burkitt lymphoma. Oncogene 263769-3776. [DOI] [PubMed] [Google Scholar]
- 21.Korpal, M., E. S. Lee, G. Hu, and Y. Kang. 2008. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J. Biol. Chem. 28314910-14914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulshreshtha, R., M. Ferracin, S. E. Wojcik, R. Garzon, H. Alder, F. J. Agosto-Perez, R. Davuluri, C. G. Liu, C. M. Croce, M. Negrini, G. A. Calin, and M. Ivan. 2007. A microRNA signature of hypoxia. Mol. Cell. Biol. 271859-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusanagi, K., H. Inoue, Y. Ishidou, H. K. Mishima, M. Kawabata, and K. Miyazono. 2000. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol. Biol. Cell 11555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425415-419. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 12015-20. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, B. P., I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel, and C. B. Burge. 2003. Prediction of mammalian microRNA targets. Cell 115787-798. [DOI] [PubMed] [Google Scholar]
- 27.Long, D., R. Lee, P. Williams, C. Y. Chan, V. Ambros, and Y. Ding. 2007. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 14287-294. [DOI] [PubMed] [Google Scholar]
- 28.Ma, L., J. Teruya-Feldstein, and R. A. Weinberg. 2007. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449682-688. [DOI] [PubMed] [Google Scholar]
- 29.Matys, V., E. Fricke, R. Geffers, E. Gossling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel, O. V. Kel-Margoulis, D. U. Kloos, S. Land, B. Lewicki-Potapov, H. Michael, R. Munch, I. Reuter, S. Rotert, H. Saxel, M. Scheer, S. Thiele, and E. Wingender. 2003. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 31374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muckstein, U., H. Tafer, J. Hackermuller, S. H. Bernhart, P. F. Stadler, and I. L. Hofacker. 2006. Thermodynamics of RNA-RNA binding. Bioinformatics 221177-1182. [DOI] [PubMed] [Google Scholar]
- 31.Nam, J. S., A. M. Suchar, M. J. Kang, C. H. Stuelten, B. Tang, A. M. Michalowska, L. W. Fisher, N. S. Fedarko, A. Jain, J. Pinkas, S. Lonning, and L. M. Wakefield. 2006. Bone sialoprotein mediates the tumor cell-targeted prometastatic activity of transforming growth factor beta in a mouse model of breast cancer. Cancer Res. 666327-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Negrini, M., M. Ferracin, S. Sabbioni, and C. M. Croce. 2007. MicroRNAs in human cancer: from research to therapy. J. Cell Sci. 1201833-1840. [DOI] [PubMed] [Google Scholar]
- 33.O'Connell, R. M., K. D. Taganov, M. P. Boldin, G. Cheng, and D. Baltimore. 2007. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 1041604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozdamar, B., R. Bose, M. Barrios-Rodiles, H. R. Wang, Y. Zhang, and J. L. Wrana. 2005. Regulation of the polarity protein Par6 by TGFβ receptors controls epithelial cell plasticity. Science 3071603-1609. [DOI] [PubMed] [Google Scholar]
- 35.Park, S., D. Kim, S. Kaneko, K. M. Szewczyk, S. V. Nicosia, H. Yu, R. Jove, and J. Q. Cheng. 2005. Molecular cloning and characterization of the human AKT1 promoter uncovers its up-regulation by the Src/Stat3 pathway. J. Biol. Chem. 28038932-38941. [DOI] [PubMed] [Google Scholar]
- 36.Park, S. M., A. B. Gaur, E. Lengyel, and M. E. Peter. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perez-Moreno, M., C. Jamora, and E. Fuchs. 2003. Sticky business: orchestrating cellular signals at adherens junctions. Cell 112535-548. [DOI] [PubMed] [Google Scholar]
- 38.Reinhart, B. J., F. J. Slack, M. Basson, A. E. Pasquinelli, J. C. Bettinger, A. E. Rougvie, H. R. Horvitz, and G. Ruvkun. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403901-906. [DOI] [PubMed] [Google Scholar]
- 39.Roberts, A. B., and L. M. Wakefield. 2003. The two faces of transforming growth factor beta in carcinogenesis. Proc. Natl. Acad. Sci. USA 1008621-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez, A., E. Vigorito, S. Clare, M. V. Warren, P. Couttet, D. R. Soond, S. van Dongen, R. J. Grocock, P. P. Das, E. A. Miska, D. Vetrie, K. Okkenhaug, A. J. Enright, G. Dougan, M. Turner, and A. Bradley. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316608-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skalsky, R. L., M. A. Samols, K. B. Plaisance, I. W. Boss, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 8112836-12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stark, A., J. Brennecke, N. Bushati, R. B. Russell, and S. M. Cohen. 2005. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 1231133-1146. [DOI] [PubMed] [Google Scholar]
- 43.Tam, W. 2001. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene 274157-167. [DOI] [PubMed] [Google Scholar]
- 44.Tang, G. 2005. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 30106-114. [DOI] [PubMed] [Google Scholar]
- 45.Thai, T. H., D. P. Calado, S. Casola, K. M. Ansel, C. Xiao, Y. Xue, A. Murphy, D. Frendewey, D. Valenzuela, J. L. Kutok, M. Schmidt-Supprian, N. Rajewsky, G. Yancopoulos, A. Rao, and K. Rajewsky. 2007. Regulation of the germinal center response by microRNA-155. Science 316604-608. [DOI] [PubMed] [Google Scholar]
- 46.Vaezi, A., C. Bauer, V. Vasioukhin, and E. Fuchs. 2002. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell 3367-381. [DOI] [PubMed] [Google Scholar]
- 47.Vigorito, E., K. L. Perks, C. Abreu-Goodger, S. Bunting, Z. Xiang, S. Kohlhaas, P. P. Das, E. A. Miska, A. Rodriguez, A. Bradley, K. G. Smith, C. Rada, A. J. Enright, K. M. Toellner, I. C. Maclennan, and M. Turner. 2007. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 27847-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Volinia, S., G. A. Calin, C. G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris, and C. M. Croce. 2006. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 1032257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, H. R., Y. Zhang, B. Ozdamar, A. A. Ogunjimi, E. Alexandrova, G. H. Thomsen, and J. L. Wrana. 2003. Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 3021775-1779. [DOI] [PubMed] [Google Scholar]
- 50.Xie, W., J. C. Mertens, D. J. Reiss, D. L. Rimm, R. L. Camp, B. G. Haffty, and M. Reiss. 2002. Alterations of Smad signaling in human breast carcinoma are associated with poor outcome: a tissue microarray study. Cancer Res. 62497-505. [PubMed] [Google Scholar]
- 51.Yanaihara, N., N. Caplen, E. Bowman, M. Seike, K. Kumamoto, M. Yi, R. M. Stephens, A. Okamoto, J. Yokota, T. Tanaka, G. A. Calin, C. G. Liu, C. M. Croce, and C. C. Harris. 2006. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9189-198. [DOI] [PubMed] [Google Scholar]
- 52.Yang, H., W. Kong, L. He, J. J. Zhao, J. D. O'Donnell, J. Wang, R. M. Wenham, D. Coppola, P. A. Kruk, S. V. Nicosia, and J. Q. Cheng. 2008. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 68425-433. [DOI] [PubMed] [Google Scholar]
- 53.Yin, Q., X. Wang, J. McBride, C. Fewell, and E. K. Flemington. 2008. B-cell receptor activation induces BIC/MIR-155 expression through a conserved AP-1 element. J. Biol. Chem. 2832654-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zavadil, J., and E. P. Bottinger. 2005. TGFβ and epithelial-to-mesenchymal transitions. Oncogene 245764-5774. [DOI] [PubMed] [Google Scholar]
- 55.Zavadil, J., M. Narasimhan, M. Blumenberg, and R. J. Schneider. 2007. Transforming growth factor-β and microRNA:mRNA regulatory networks in epithelial plasticity. Cells Tissues Organs 185157-161. [DOI] [PubMed] [Google Scholar]