Abstract
SMAUG (SMG) is an RNA-binding protein that functions as a key component of a transcript degradation pathway that eliminates maternal mRNAs in the bulk cytoplasm of activated Drosophila melanogaster eggs. We previously showed that SMG destabilizes maternal Hsp83 mRNA by recruiting the CCR4-NOT deadenylase to trigger decay; however, the cis-acting elements through which this was accomplished were unknown. Here we show that Hsp83 transcript degradation is regulated by a major element, the Hsp83 mRNA instability element (HIE), which maps to a 615-nucleotide region of the open reading frame (ORF). The HIE is sufficient for association of a transgenic mRNA with SMG protein as well as for SMG-dependent destabilization. Although the Hsp83 mRNA is translated in the early embryo, we show that translation of the mRNA is not necessary for destabilization; indeed, the HIE functions even when located in an mRNA's 3′ untranslated region. The Hsp83 mRNA contains eight predicted SMG recognition elements (SREs); all map to the ORF, and six reside within the HIE. Mutation of a single amino acid residue that is essential for SMG's interaction with SREs stabilizes endogenous Hsp83 transcripts. Furthermore, simultaneous mutation of all eight predicted SREs also results in transcript stabilization. A plausible model is that the multiple, widely distributed SREs in the ORF enable some SMG molecules to remain bound to the mRNA despite ribosome transit through any individual SRE. Thus, SMG can recruit the CCR4-NOT deadenylase to trigger Hsp83 mRNA degradation despite the fact that it is being translated.
During animal oogenesis and early embryogenesis, posttranscriptional gene regulation represents an essential mechanism for the control of gene expression. Mature oocytes and early embryos are transcriptionally quiescent and rely upon maternally supplied mRNAs deposited during oogenesis (5, 37). Often, multiple mechanisms of posttranscriptional control act coordinately to precisely regulate the spatial and temporal expression of proteins derived from these maternal mRNAs (23, 25).
In Drosophila melanogaster, the first 2 hours of embryogenesis are programmed by maternal mRNAs (38). A surprisingly large fraction of the protein coding genome is present in this maternal transcript pool, with between one-half and two-thirds of all mRNAs being represented (16, 21, 35). Between a quarter and a third of these maternal mRNAs are eliminated during the maternal-to-zygotic transition that occurs prior to cellularization 3 hours after fertilization (16, 21, 35, 37). Two degradation pathways direct elimination of these transcripts (3, 4). The first is termed the “maternal degradation pathway” because it is comprised of an exclusively maternally encoded machinery that is triggered upon egg activation, prior to fertilization (4, 36). The second, termed the “zygotic degradation pathway” since it requires both fertilization and zygotic transcription, initiates approximately 2 hours into embryonic development (4). The identification of these two decay activities allows maternal mRNAs to be categorized according to whether they are targeted exclusively by the maternal degradation pathway (e.g., nanos and polar granule component), exclusively by the zygotic degradation pathway (e.g., hunchback), by both activities (e.g., Hsp83 and string), or by neither (e.g., rpA1, rp49, and αTub84B) (4, 16, 30, 31, 35, 36).
A major regulator of transcript destabilization is the RNA-binding protein SMAUG (SMG), whose function is required for elimination of two-thirds of the mRNAs attacked by the maternal pathway (35). Analyses of Hsp83 mRNA have shown that SMG acts as a specificity factor, recruiting the CCR4-NOT deadenylase, thus initiating decay through deadenylation (30). SMG's sterile-alpha motif (SAM) domain binds to specific stem-loops termed SMG recognition elements (SREs) (2, 11-13, 32, 33). However, mutation of four computationally predicted SREs in the open reading frame (ORF) of Hsp83 had no effect on the kinetics of transcript decay (30), raising several possibilities: that additional SREs might reside in the Hsp83 mRNA, that SMG recognizes a novel cis element in the Hsp83 mRNA, or that SMG's interaction with Hsp83 mRNA is indirect. Subsequent to those analyses it was shown that VTS1, the Saccharomyces cerevisiae homolog of SMG, can bind to SREs with a loop up to 7 nucleotides (nt) long, CNGGN0-3 (1). In addition, the crystal structure indicated that SRE binding involves limited sequence specificity, through recognition of the G residue at position 3 of the loop, in combination with recognition of the tertiary structure of the SRE (1). Thus, sequences that adopt a similar shape and contain a G residue in the correct context—but do not match the current SRE consensus—could, in principle, also be bound by VTS1/SMG (1).
Given the complexity of VTS1/SMG transcript recognition, here we took an unbiased approach to mapping the cis elements in the Hsp83 mRNA that are required for destabilization, using hybrid transgenic mRNAs that fused parts of the Hsp83 mRNA to parts of a stable mRNA, rpA1. These hybrid mRNAs enabled us to test different parts of the Hsp83 transcript for a role in mRNA destabilization via the maternal degradation machinery (i.e., in unfertilized eggs) and via SMG (i.e., in smg mutants). Using this strategy, we mapped a major instability element—which we have termed the Hsp83 mRNA instability element (HIE)—within the ORF. The HIE is sufficient to strongly destabilize rpA1 mRNA when inserted into its ORF. The presence of the HIE within the ORF suggested that translation of the Hsp83 mRNA might be required for transcript destabilization. However, the HIE retained its activity as an instability element even when inserted into a 3′ untranslated region (UTR), indicating that translation is not needed. We show that the HIE functions together with auxiliary elements in the more 5′ part of the Hsp83 ORF as well as in its 3′ UTR to direct transcript destabilization. One of these auxiliary elements is the previously identified Hsp83 degradation element (HDE), a 97-nt region within the Hsp83 3′ UTR (4).
Eight computationally predicted SREs are present in the Hsp83 mRNA; all reside in its ORF, and six map to the HIE. We show that the HIE is sufficient to direct association with SMG protein as well as to confer SMG-dependent transcript instability. A single-amino-acid substitution that abrogates SMG's ability to bind SREs prevents Hsp83 transcript destabilization. Furthermore, simultaneous mutation of all eight predicted SREs (without substantially altering the coding capacity of the ORF) stabilizes the transcript. We conclude that SMG binds directly to the Hsp83 mRNA's ORF, thus recruiting the CCR4-NOT deadenylase and triggering transcript elimination despite the fact that Hsp83 transcripts are actively translated.
MATERIALS AND METHODS
Fly stocks.
Drosophila melanogaster wild-type stocks included y w1118 and w1118. Both of these genetic backgrounds were used for the generation of transgenic lines. Expression of pUASP constructs was induced using P[GAL4::VP16-nos.UTR] (41). Due to the presence of a recessive lethal mutation(s) on the smg1 chromosome, smg mutants were smg1/Df(3L)ScfR6 (12). In some cases, a transgenic P element insertion and the smg1 allele were recombined onto the same chromosome, which resulted in the loss of the recessive lethal mutation, and thus, smg mutants were smg1/smg1. These genetic backgrounds are noted in the text.
Construction of transgenes. (i) H-H-H.
The construct H-H-H was designed by Arash Bashirullah and contains the full-length Hsp83 transcript with a p53 epitope tag inserted 3′ to the start codon. This transgene was constructed by Vent polymerase PCR of three fragments. The first Hsp83 fragment, H**, contained the 5′ genomic region (876 nt), the 5′ UTR (149 nt), the intron (1,129 nt), and start codon, flanked with a 5′ BamHI site and a 3′ ApaI site. The second Hsp83 fragment, *H*, was designed to contain the entire ORF with no start codon but with the stop codon (2,151 nt) and the first 6 nt of the 3′ UTR. This fragment was flanked with a 5′ ApaI site while two nucleotides, GC, were introduced between the +2 and +3 positions of the 3′ UTR to generate a 3′ AatII site. The third Hsp83 fragment, **H, was designed to contain the 3′ UTR (407 nt) and downstream genomic DNA (1,281 nt total) flanked by 5′ AatII and 3′ KpnI sites. These three PCR fragments were subcloned into pBAAK, which is a modified pBluescript-SK vector that contained ApaI and AatII sites between the BamHI and KpnI sites, by digestion with HindIII/KpnI and insertion of annealed oligonucleotides Apa-Aat-F and Aat-Apa-R (all oligonucleotide sequences are listed in Table S1 in the supplemented material). The 5′ fragment was first cloned into pBAAK at the BamHI/ApaI sites, generating pBAAK-H**, and the 3′ fragment was subsequently cloned at AatII/KpnI sites to generate pBAAK-H*H. The final fragment, the ORF, was then cloned into pBAAK-H*H at ApaI/AatII sites to generate pBAAK-H-H-H. An N-terminal p53 tag was introduced at the ApaI site using annealed oligonucleotides p53-F and p53-R, thus generating an 8-amino-acid insertion immediately downstream of the start codon (the p53 epitope contains five residues). The insertion of this oligonucleotide duplex destroyed the first ApaI site and maintained the second site.
(ii) R-R-R.
The construct R-R-R was designed with a strategy similar to that for H-H-H and used a full-length 2.7-kb genomic BamHI-BamHI rpA1 fragment (gift of Marcello Jacobs-Lorena) which was subcloned into pBluescript-SK (collapsed at SalI/XhoI sites) and subsequently used as a PCR template for the following fragments. The first rpA1 fragment, R**, was generated to contain the 5′ BamHI site, 5′ flanking genomic DNA (∼1 kb), the 5′ UTR (89 nt), the start codon, the p53 tag, and an ApaI site. This fragment was PCR generated by Accuprime Pfx polymerase (Invitrogen) using primers R5-Bam-5gen-F and R5-Apa-p53-R and subsequently cloned into pBAAK at BamHI/ApaI sites to generate pBAAK-R**. The second rpA1 fragment, *R*, contained the entire ORF, minus the start codon but including the stop codon (339 nt), and was flanked by a 5′ ApaI site and a 3′ AatII site. This second fragment was PCR generated via Elongase (Invitrogen) utilizing the primers RO-Apa-F and RO-Aat-R and was cloned into pBAAK at ApaI/AatII sites to generate pBAAK-*R*. The third rpA1 fragment,**R, contained the entire 3′ UTR (168 nt) and 3′ flanking genomic DNA (∼1 kb) flanked by 5′ AatII and 3′ KpnI sites and was PCR generated using Accuprime Pfx polymerase (Invitrogen) and primers R3-Aat-F and R3-Bam-R. This third fragment contained a BamHI site at the 3′ end just upstream of the flanking KpnI site. The third fragment was cloned into pBAAK at ApaI/KpnI sites to generate pBAAK-**R. Once all three fragments were individually cloned into pBAAK, the vector pBAAK-R-R-R was constructed via sequential digestion and ligation of ApaI-AatII-digested pBAAK-*R* and AatII-KpnI digested pBAAK-**R fragments into the ApaI-KpnI-digested pBAAK-R** backbone.
(iii) H/R hybrid constructs.
The following constructs were generated from the appropriate digestions and ligations of the BamHI-ApaI 5′ fragment, the BamHI-AatII 5′+ ORF fragment, the ApaI-AatII ORF fragment, the ApaI-KpnI ORF +3′ fragment, or the AatII-KpnI 3′ fragment from the vectors pBAAK-H-H-H and pBAAK-R-R-R: pBAAK-H-H-R, pBAAK-H-R-R, pBAAK-H-R-H, pBAAK-R-R-H, pBAAK-R-H-R, and pBAAK-R-H-H.
(iv) H-H-HΔHDE and H-HΔHORF4-HΔHDE.
The AatII-KpnI fragment of pBAAK-H-H-H and that of pBAAK-H-HΔHORF4-H (see below), respectively, each containing the full-length Hsp83 3′ UTR, were replaced by a corresponding AatII-KpnI fragment from pBluescript-ΔHDE vector. This fragment contained the 97-nt deletion of the HDE, and its construction has been described previously (4). For R-RlacZ-R, a 621-nt fragment of the lacZ coding sequence was PCR generated with flanking SalI sites and inserted in frame into the unique SalI site of the rpA1 ORF within the full-length 2.7-kb genomic BamHI-BamHI rpA1 fragment in pBluescript-SK (collapsed at SalI/XhoI sites) as described above. For R-RlacZ-RHDE, the BamHI-BamHI fragment of R-RlacZ-R was subcloned into pUC18. In this vector backbone, a polylinker was inserted into the rpA1 3′ UTR at the NgoMI site, which introduced AvrII and BglII sites using the annealed oligonucleotides AvrII-BglII-F and BglII-AvrII-R. The HDE was PCR generated with flanking 5′ AvrII and 3′ BglII sites and subcloned into the same sites in pUC18-R-RlacZ-R to generate pUC18-rpA1-lacZ+HDE.
(v) R-RHORF-R.
Four overlapping fragments of the Hsp83 ORF (named HORF1 through HORF4 shifting 5′ to 3′) were PCR generated using Elongase (Invitrogen) and were flanked with XhoI sites. The vector pCSR4-H-H-H was used as the template (see below for construction of pCSR4 vectors). Each fragment contained 612 nt of Hsp83 ORF sequence with the exception of HORF4, which contained 615 nt of the 3′-most region of coding sequence. The primers Xho-HORF1-F and Xho-HORF1-R were used for generating HORF1, the primers Xho-HORF2-F and Xho-HORF2-R were used for generating HORF2, the primers Xho-HORF3-F and Xho-HORF3-R were used for generating HORF3, and the primers Xho-HORF4-F and Xho-HORF4-R were used for generating HORF4. Each XhoI-digested PCR fragment was cloned into the unique SalI site contained within the rpA1 ORF of pBAAK-R-R-R to generate the following constructs: pBAAK-R-RHORF1-R, pBAAK-R-RHORF2-R, pBAAK-R-RHORF3-R, and pBAAK-R-RHORF4-R. The use of flanking XhoI sites in the primer sequences introduced amino acids VE at the 5′ junction between rpA1 and Hsp83 coding sequences (i.e., at the 5′ collapsed SalI site) and amino acids LD at the 3′ junction between Hsp83 and rpA1 coding sequences (i.e., at the preserved SalI site). These insertions maintained both the rpA1 and Hsp83 reading frames.
(vi) R-RHORF4-X-R.
Six overlapping subfragments (∼120 nt each) of the HORF4 coding region were PCR generated using Taq polymerase (Amersham) and flanked with XhoI sites (fragment 4-7 was synthesized using Elongase [Invitrogen]). The vector pCSR4-H-H-H was used as the template (see “P-element transformation” below for construction of pCSR4 vectors). These six fragments overlap by ∼20 nt with one another and were named HORF4-1 through HORF4-6 shifting 5′ to 3′ along the HORF4 region. The primers Xho-HORF4-F and Xho-HORF4-1-R were used for generating HORF4-1. The primers Xho-HORF4-n-F and Xho-HORF4-n-R were used to generate HORF4-n, where 2 ≤ n ≤ 5. The primers Xho-HORF4-6-F and Xho-HORF4-R were used for generating HORF4-6. The HORF4 coding region was also subdivided into two larger overlapping fragments that were ∼320 nt each and contained a ∼20-nt overlap. These subfragments were named HORF4-7 and HORF4-8 (with the former representing the 5′-most fragment of the HORF4 region), were also PCR generated via Taq polymerase (Amersham), and were flanked with XhoI sites. The primers Xho-HORF4-F and Xho-HORF4-3-R were used for generating HORF4-7, and the primers Xho-HORF4-4-F and Xho-HORF4-R were used for generating HORF4-8. Each XhoI-digested PCR fragment was cloned into the unique SalI site contained within the rpA1 ORF of pBAAK-R-R-R to generate the following constructs: pBAAK-R-RHORF4-1-R, pBAAK-R-RHORF4-2-R, pBAAK-R-RHORF4-3-R, pBAAK-R-RHORF4-4-R, pBAAK-R-RHORF4-5-R, pBAAK-R-RHORF4-6-R, pBAAK-R-RHORF4-7-R, and pBAAK-R-RHORF4-8-R. The use of flanking XhoI sites in the primer sequences introduced amino acids VE at the 5′ junction between rpA1 and Hsp83 coding sequences (i.e., at the 5′ collapsed SalI site) and amino acids LD at the 3′ junction between Hsp83 and rpA1 coding sequences (i.e., at the preserved SalI site). These insertions maintained both the rpA1 and Hsp83 reading frames.
(vii) R-R-RHORF4.
The HORF4 fragment was inserted in both sense and antisense orientations at two different unique restriction sites within the rpA1 3′ UTR in the vector pBAAK-R-R-R. The HORF4 fragment was generated by Elongase PCR (Invitrogen) and was flanked either with AatII sites using primers Aat-HORF4-F and Aat-HORF4-R or with FseI sites using primers Fse-HORF4-F and Fse-HORF4-R. The vector pCSR4-H-H-H was used as the PCR template (see below for construction of pCSR4 vectors). These two fragments were digested with their appropriate restriction enzymes, i.e., either digested with AatII and subsequently ligated into the AatII site in pBAAK-R-R-R (which is immediately following the stop codon) or digested with FseI and subsequently ligated into the FseI site just upstream of the poly(A) signal in the rpA1 3′ UTR in pBAAK-R-R-R. This resulted in the construction of pBAAK-R-R-RsHORF4(Aat), pBAAK-R-R-RasHORF4(Aat), pBAAK-R-R-RsHORF4(Fse), and pBAAK-R-R-RasHORF4(Fse).
(viii) H-HΔHORF4-H.
The construct pBAAK-H-H-H was digested at unique Bsu36I and AatII sites, which deleted a 528-nt fragment of the Hsp83 coding region containing the majority of HORF4 (615 nt). The oligonucleotides H4-Bsu-Aat-F and H4-Aat-Bsu-R were annealed and ligated into the digested pBAAK-H-H-H vector to generate pBAAK-H-HΔHORF4-H. A stop codon in the oligonucleotide duplex truncated the Hsp83 ORF after codon 541 of the 717-amino-acid HSP83 protein. For R-HΔHORF4-R, the ApaI-AatII fragment of pBAAK-H-HΔHORF4-H was ligated into ApaI/AatII-digested pBAAK-R-R-R to generate pBAAK-R-HΔHORF4-R. For H-HΔHORF4+lacZ-H, a 528-nt lacZ ORF fragment that contained flanking 5′ Bsu36I and 3′ AatII sites was PCR generated using Taq polymerase (Amersham) and primers LacZ-Bsu-F and LacZ-Aat-R. This PCR lacZ fragment and the vector pBAAK-H-H-H were both digested with Bsu36I and AatII and ligated together to generate pBAAK-H-HΔHORF4+lacZ-H. The lacZ PCR fragment was partially digested with Bsu36I, as it also contains a Bsu36I site internal to the primer sequences, and thus, the full-length lacZ fragment was maintained. The construct Hsp83-lacZ (H-HΔ1.8kb+lacZ-H) has been previously described (4, 10). Briefly, the Hsp83 5′ genomic region, 5′ UTR, intron, and first 333 nt were fused to a 603-nt truncated fragment of the lacZ ORF. The 3′ end of the lacZ ORF was flanked with AatII and KpnI sites in which the same AatII-KpnI fragment described for the generation of H-H-H was used to insert the full-length Hsp83 3′ UTR and downstream genomic region. Thus, this construct created a ∼1.8-kb deletion of the Hsp83 ORF which was replaced with a lacZ fragment. The lacZ fragment did not, however, contain a stop codon and thus allowed translation to proceed into the Hsp83 3′ UTR.
Translation constructs.
Oligonucleotide pairs were annealed and inserted at the unique BsiWI site within the Hsp83 5′ UTR of pCSR4-H-H-H. All oligonucleotide duplexes maintained the 5′ BsiWI site but eliminated the 3′ BsiWI site when inserted into BsiWI-digested pCSR4-H-H-H. For the generation of the Hhairpin-H-H construct, the 75-nt oligonucleotides Hsp83-5′HP-F and Hsp83-5′HP-R were designed to include the strong hp7 hairpin sequence (20, 29) and annealed according to the Promega protocol for cloning a hairpin insert into pGeneClip vectors (Promega). These oligonucleotides contained NotI sites immediately internal to the BsiWI sites and also introduced a unique SacII site into the vector. Hrandom-H-H was produced by inserting the BsiWI-digested product of an Elongase PCR-amplified fragment of pCSR4 using the oligonucleotides Bsi-CSR-F and Bsi-CSR-R into the BsiWI site of pCSR-H-H-H. The 75-nt random sequence from the pCSR4 vector introduced an additional HindIII site which aided detection of positive transformants.
pUASP-SMG rescue constructs.
The generation of the SMGRBD point mutation A642H has been previously described (2). Both wild-type and point-mutated (A642H) smg cDNA fragments containing the ORF and 3′ UTR were cloned into the pUASP vector (27).
H-H8xSRE(-)-H.
Four overlapping fragments of the Hsp83 ORF were PCR generated using Accuprime Pfx (Invitrogen). The vector 4xSRE− pCSR4-H-H-H (30) was used as the template. The primers Apa-HORF1-F and H-SRE 3&4-R were used to generate 1.1614, H-SRE 3&4-F and H-SRE 6-R were used for 1567.1953, H-SRE 6-F and H-SRE 8-R were used for 1929.2100, and H-SRE 8-F and Hsp83 3U100-3H were used for 2095.3U-100. Accuprime Pfx, primer Apa-HORF1-F, Hsp83 3U100-3H, and 1 μl of each gel-purified fragment were used to PCR generate 8xSRE(-) Hsp83 ORF+3U-100. Gel-purified PCR fragment was cloned into the pCR21.TOPO vector. ApaI- and AatII-digested 8xSRE(-) HORF fragment was subcloned into the same site in pBAAK-H-H-H to replace the wild-type HORF and generate the 8xSRE(-) pBAAK-H-H-H, the mRNA from which is referred to in the text as H-H8xSRE(-)-H.
P-element transformation.
All NotI-KpnI inserts from the pBAAK vectors described above (i.e., containing the 5′, p53 tag, ORF, and 3′ fragments) were then subcloned into pCSR4 (39) at NotI/KpnI sites. All inserts were sequenced in pBAAK and/or pCSR4 vectors (except for the full-length p53-tagged Hsp83 construct, H-H-H, which was tested for its ability to rescue an Hsp83 RNA null mutation and for wild-type mRNA decay kinetics [30]). The BamHI fragment from rpA1-lacZ in pBluescript-SK was subcloned into pCSR2 (39) at the BamHI site, while the BamHI fragment of rpA1-lacZ+HDE (R-RlacZ-RHDE) in pUC18 was subcloned into pCSR4 at its BamHI site. Germ line transformants were recovered using standard methods (28).
Northern, RNA dot blot, and real-time RT-PCR analysis.
Fertilized embryo and unfertilized egg collections were performed at 25°C as previously described (4). Unfertilized eggs were derived from adult virgin females mated to T(Y,2) sterile males. RNase H cleavage assays, Northern blotting, and RNA dot blotting techniques were performed as previously described (4, 30). Briefly, total RNA was extracted from eggs/embryos collected at various time periods after egg lay (AEL) by homogenizing it in Trizol (Invitrogen) containing 250 μg/ml glycogen and following standard reagent protocols. For Northern blot analyses, samples containing either an equivalent number of eggs/embryos or an equivalent amount of total RNA were electrophoresed using 1% agarose-formaldehyde gels in 1× morpholinepropanesulfonic acid (MOPS) buffer and transferred overnight onto Hybond-N nylon membranes. For RNase H cleavage assays, samples containing 10 μg of total RNA were incubated with the oligonucleotide Hsp83ORF.3 (for detection of H-H-HΔHDE mRNA) or the oligonucleotide p53-Apa24.3 (for detection of R-R-R mRNA) in the presence of RNase H according to standard methods (15). Cleaved RNA samples containing H-H-HΔHDE mRNA were run on a 4% denaturing urea-polyacrylamide gel and electrotransferred overnight using the Trans-blot cell (Bio-Rad Laboratories) at 30 V while R-R-R mRNA samples were resolved using a 1.2% agarose-formaldehyde gel and transferred as described above. For RNA dot blots, total RNA was extracted from single embryos and blotted directly onto Hybond-N membrane in SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) buffer using the Schleicher & Schuell Minifold-I dot blot system according to the manufacturer's protocol (Schleicher & Schuell). All blots were UV cross-linked, hybridized overnight with random-primed radiolabeled probes, and exposed to a storage phosphor screen, which was then scanned using the STORM860 (Molecular Dynamics) phosphorimager. Quantification was performed using Imagequant software (Molecular Dynamics) and Excel (Microsoft Office). Real-time reverse transcription-PCR (RT-PCR) was performed as described previously (30).
To detect H-H8xSRE(-)-H transcripts, a 19-base locked nucleic acid (LNA) probe specific to the sequence encoding the p53 tag was synthesized (5′-CCA CCA CGG AGT GGC GGC C-3′, where underlines indicate locked bases; the 3′ end was modified with digoxigenin). Gel running and transfer steps for RNA were performed as described above. Hybridization followed the Exiqon microRNA LNA probe Northern protocol. Briefly, the membrane was prehybridized in RNA hybridization buffer (50% formamide, 0.5% sodium dodecyl sulfate [SDS], 5× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA (pH 7.7)], 5× Denhardt's solution, and 20 μg/ml sheared, denatured, salmon sperm DNA) for 30 to 60 min. Forty-eight picomoles of p53-LNA probe was heated for 1 minute at 95°C before being added to 7 ml hybridization buffer. The membrane was hybridized overnight with the LNA probe at 45°C in the same solution. After hybridization, membranes were washed with 2× SSC-0.1% SDS at 45°C twice for 5 minutes, then with 0.1× SSC-0.1% SDS at 59°C twice for 10 minutes. The digoxigenin luminescent detection kit (Roche 1363514) was used following Roche's protocol. Briefly, the membranes were blocked and incubated with antidigoxigenin-alkaline phosphatase antibody and then developed with CSPD. Membranes were exposed to an imaging device or to X-ray film. Quantification was performed using Imagequant software and Excel (Microsoft Office).
Real-time RT-PCR was performed as described previously (30). Embryos were disrupted in a minimal volume of buffer A (150 mM KCl, 20 mM HEPES, pH 7.4, 1 mM MgCl2, 1 mM dithiothreitol [DTT], 1 mM aminoethylbenzenesulfonyl fluoride, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 2 mM benzamidine). After centrifugation the supernatant was supplemented with glycerol to a final concentration of 10%, and 9 M urea (supplemented with 1 mM DTT, 1 mM aminoethylbenzenesulfonyl fluoride, 2 μg/ml leupeptin, 2 μg/ml pepstatin, 2 μM benzamidine) was added to 75 μl of extract to a 2 M final concentration. The extract was recentrifuged and added to 5 μl of protein G beads carrying either rat anti-SMG antibody or normal rat serum. Beads were incubated for 2 h at 4°C and washed several times in buffer A supplemented with urea. RNA was then purified from the beads using Trizol reagent (Invitrogen) and reverse transcribed using random primers and Superscript II (Invitrogen). Samples were subjected to real-time PCR using Sybr green master mix and the ABI Prism 7900 sequence detection system (Applied Biosystems).
Western blot analysis.
Western blot analyses were carried out as previously described (30). Briefly, eggs/embryos were dechorionated in 50% bleach, rinsed and homogenized in lysis buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 1% NP-40, 1 mM DTT, 1× EDTA-free protease inhibitor cocktail [Roche]), incubated on ice for 5 min, and then centrifuged for 5 min at 12,000 × g at 4°C. The supernatant was combined with 2× SDS loading dye, loaded, and subjected to SDS-polyacrylamide gel electrophoresis (7.5% acrylamide). The Bio-Rad Trans-blot SD semidry transfer cell was used to transfer proteins to a polyvinylidene difluoride membrane (Invitrogen). Blots were blocked in 1% skim milk powder before incubations with primary and secondary antibodies. Primary antibodies used were mouse monoclonal anti-p53 at 1:1,000 (Santa Cruz Biotechnology; SC-99), guinea pig polyclonal anti-SMG at 1:10,000 (35), and guinea pig polyclonal anti-DDP1 at 1:10,000 (24, 35). Secondary horseradish peroxidase-conjugated antibodies were used at 1:5,000 (Jackson ImmunoResearch Laboratories, Inc.). Chemiluminescence was detected using Pierce enhanced chemiluminescence Western blotting reagent and standard X-ray film.
RESULTS
The Hsp83 ORF functions as a transcript instability element.
To map cis-acting instability elements in the Hsp83 mRNA, chimeric transgenes were constructed, each containing combinations of the 5′ UTR, ORF, and 3′ UTR of Hsp83 and rpA1, a highly stable mRNA. The constructs are listed in Table 1; these, and additional ones used in subsequent experiments, encoded a five-residue p53 epitope tag immediately after the amino-terminal methionine unless otherwise noted. The stability profiles of the hybrid mRNAs were examined using Northern blot assays of total RNA extracted from staged unfertilized eggs laid by transgenic females at 0 to 2, 2 to 4, and 4 to 6 h AEL, thus enabling us to assess the role of the cis elements in the maternally encoded transcript degradation pathway.
TABLE 1.
The Hsp83 ORF acts as a strong instability element
| Transcript | % Degraded by 4-6 h AEL | Destabilizationa |
|---|---|---|
| Endogenous and control transgenic transcripts | ||
| Hsp83 | 92 ± 6 | Strong |
| HΔintron-H-H | 87 ± 16 | Strong |
| R-R-R | 26 ± 5 | None |
| rpA1 | 5 ± 21 | None |
| Necessity hybrid transcripts | ||
| R-H-H | 78 ± 7 | Strong |
| H-H-R | 88 ± 4 | Strong |
| HΔintron-H-R | 97 ± 1 | Strong |
| H-R-H | NAb | |
| Sufficiency hybrid transcripts | ||
| R-H-R | 72 ± 7 | Strong |
| R-R-H | 36 ± 15 | Weak |
| H-R-R | NA |
Destabilization categories: strong, >50% degraded by 4 to 6 h AEL; weak, 30 to 50% degraded by 4 to 6 h AEL; none, <30% degraded by 4 to 6 h AEL.
NA, not assayed. H-R-R and H-R-H were not testable because artifactual destabilization was found to occur when an exogenous 5′ UTR (either from Hsp83 or from a stable mRNA, αTub84B) was fused to the rpA1 ORF (data not shown). Constructs with the rpA1 5′ UTR are intronless. To confirm that the lack of an intron did not alter transcript stability, intronless constructs HΔintron-H-H and HΔintron-H-R were made and shown to be strongly unstable.
The control rpA1 transgenic transcript, R-R-R (where the first letter denotes the source of the 5′ UTR, the second the source of the ORF, and the third the source of the 3′ UTR), was highly stable, although less so than endogenous rpA1 mRNA (Table 1) (for endogenous rpA1, 95% ± 21% of the transcripts remained at 4 to 6 h AEL, while for R-R-R 74% ± 5% remained). The R-H-R and R-R-H constructs were used to assess whether the Hsp83 ORF or Hsp83 3′ UTR, respectively, was sufficient to destabilize rpA1 mRNA while R-H-H and H-H-R were used to ask whether the corresponding regions were necessary for Hsp83 mRNA decay. For technical reasons, the sufficiency of the Hsp83 5′ UTR could not be assayed in these experiments (see footnotes to Table 1); however, our subsequent experiments showed definitively that the 5′ UTR is not required for destabilization. Since 26% ± 5% of R-R-R mRNA was degraded by 4 to 6 h AEL, throughout this study we have used the following terminology to describe the results: instability was defined as “strong” if >50% of the mRNA was eliminated by 4 to 6 h AEL, “weak” if 30 to 50% was eliminated, and “none” if <30% was eliminated.
The extent of mRNA destabilization produced by the necessity constructs, R-H-H (78% ± 7% degraded by 4 to 6 h AEL) and H-H-R (88% ± 4% degraded), was strong and comparable to that observed for endogenous Hsp83 mRNA (92% ± 6%) (Table 1). With respect to sufficiency, the Hsp83 ORF inserted into R-H-R was sufficient to strongly destabilize the hybrid transcripts, eliminating 72% ± 7% of the transcripts by 4 to 6 h AEL. In contrast, the effects of the Hsp83 3′ UTR on R-R-H were weak, eliminating 36% ± 15% of transcripts by the same stage. The region common to all of the hybrid transcripts that exhibited strong destabilization is the Hsp83 ORF, consistent with the hypothesis that the ORF contains one or more transcript instability elements. The data are also consistent with the possibility that the 3′ UTR contains a weak instability element.
We note that an alternative to the “instability element” interpretation presented here is that the rpA1 mRNA houses stabilization elements. For example, the R-H-R transcript could be unstable due to loss of such elements. rpA1 has been used previously as a backbone with which to map instability elements within the bicoid and fushi tarazu mRNAs (17, 19, 26, 34). Since the bicoid and fushi tarazu instability elements function in the presence of the rpA1 5′ UTR and/or ORF, we favor—and subsequent experiments presented below support—the interpretation that the Hsp83 mRNA contains instability elements.
A 615-nt HIE resides within the ORF.
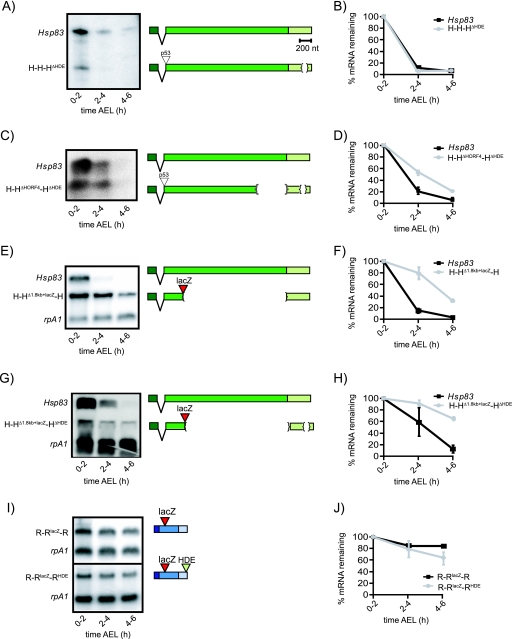
To refine the mapping of the strong instability element(s) within the Hsp83 ORF, four overlapping fragments that together spanned the entire 2,154 nt were inserted in frame into the ORF of R-R-R (Fig. 1A). These fragments were each ∼615 nt long and overlapped by ∼100 nt. The stability of the resulting hybrid mRNAs was assayed in unfertilized eggs as described above. The fourth fragment, HORF4, was sufficient to strongly destabilize R-R-R mRNA, eliminating 68% ± 1% of transcripts by 4 to 6 h AEL, while the other fragments—HORF1 (12% ± 14%), HORF2 (25% ± 6%), and HORF3 (15% ± 9%)—had no significant destabilizing activity (Fig. 1B and C). That all four fragments of the Hsp83 ORF were inserted at the same site within the R-R-R ORF and yet only HORF4 destabilized the hybrid mRNA supports the hypothesis that the instability of R-RHORF4-R transgenic mRNA is due to the presence of the HORF4 sequence and not to an effect of the insertion site. Consistent with this conclusion, insertion of a piece of the lacZ ORF into this same site did not significantly destabilize the mRNA (see R-RlacZ-R below and Fig. 4I and J).
FIG. 1.
A strong instability element—the HIE—maps within the Hsp83 ORF. (A) Schematic representation of the Hsp83 ORF sufficiency constructs. Four overlapping fragments (∼615 nt each with 100-nt overlap) of the Hsp83 coding sequence (HORF1 to HORF4) were independently inserted in frame into the transgenic R-R-R construct at a unique SalI restriction site within the rpA1 coding sequence. Constructs are drawn to scale. (B) Northern blot analysis of the Hsp83 ORF sufficiency constructs in unfertilized eggs. Blots were probed with the rpA1 3′ UTR to detect mRNA from both the transgene and the loading control, rpA1. (C) Quantification of Northern blot data in panel B. Average normalized transgenic mRNA levels between two or more independently derived transgenic fly lines are shown. Transgenic mRNA levels were normalized to loading controls. Error bars represent standard deviations calculated from at least two independent experiments. All experiments were performed using unfertilized eggs from transgenic mothers, collected at 0 to 2 h, 2 to 4 h, and 4 to 6 h AEL. Transcript levels at 2 to 4 h and 4 to 6 h AEL are represented as a percentage of the initial amount detected at 0 to 2 h (i.e., % mRNA remaining).
FIG. 4.
Mapping of auxiliary ORF elements and the role of the HDE. (A and B) Test of the necessity of the HDE to mediate Hsp83 mRNA decay via the maternal degradation pathway. (A) RNase H cleavage and Northern blot analysis of H-H-HΔHDE transgenic mRNA. Hsp83 mRNA and H-H-HΔHDE mRNA were cleaved with RNase H in the presence of an oligonucleotide that anneals to the 3′ end of the ORF and oligo(dT), thus generating 407-nt and 307-nt 3′ UTR fragments without their poly(A) tails, respectively. The blot was probed with full-length Hsp83 3′ UTR. (B) Quantification of Northern blot data in panel B revealed similar degradation rates for Hsp83 and H-H-HΔHDE transcripts. H-H-HΔHDE females were heterozygous for the H-H-H transgene, thus accounting for the differences in initial levels relative to Hsp83 mRNA. Raw mRNA levels are shown. Experiments were performed in duplicate. (C and D) Deletion of the both the HIE and HDE does not stabilize Hsp83 transcripts. Northern blots were probed with Hsp83 probe, thus detecting both endogenous Hsp83 transcripts and the transgenic RNA (C). Quantification (D) included normalization relative to rpA1 transcripts. Experiments were performed in duplicate, and the averages ± standard deviations are shown. For H-HΔHORF4-H, 22% ± 13% of transcripts remained at 4 to 6 h AEL while, for H-HΔHORF4-HΔHDE, 21% ± 0% remained at 4 to 6 h AEL compared with <10% of endogenous mRNA. (E and F) Deletion of a ∼1.8-kb 3′ region of the Hsp83 ORF weakly stabilizes the transcript. The H-HΔ1.8kb+lacZ-H transgene contains a ∼1.8-kb 3′ deletion of the Hsp83 coding sequence (including deletion of the entire HIE) along with a 603-nt lacZ ORF substitution. No p53 tag was present. For the Northern blot analysis of H-HΔ1.8kb+lacZ-H mRNA in unfertilized eggs, both endogenous Hsp83 and H-HΔ1.8kb+lacZ-H mRNAs were detected by probing for Hsp83. rpA1 mRNA was used as a loading control. Quantification values are the averages of three independent replicates for H-HΔ1.8kb+lacZ-H. (G and H) Role of the HDE is revealed in a sensitized background. Northern blots were probed with Hsp83 probe, thus detecting both endogenous Hsp83 transcripts and the transgenic RNA. Quantification (H) included normalization relative to rpA1 transcripts. Experiments were performed in duplicate, and the averages ± standard deviations are shown. For H-HΔ1.8kb+lacZ-H, 31% ± 3% of transcripts remained at 4 to 6 h AEL (F) while, for H-HΔ1.8kb+lacZ-HΔHDE, 66% ± 3.7% remained at 4 to 6 h AEL (H). (I and J) Sufficiency of the HDE. R-RlacZ-R and R-RlacZ-RHDE were derived from the same genomic rpA1 DNA as R-R-R but lacked the p53 epitope tag and restriction sites at the 5′-UTR/ORF and ORF/3′-UTR junctions. Instead, these constructs contained a lacZ tag within their coding sequence. For Northern blot analysis of transgenic R-RlacZ-R and R-RlacZ-RHDE mRNA, blots were probed with full-length rpA1 to detect both endogenous and transgenic mRNAs. For quantification of Northern blot data of R-RlacZ-R and R-RlacZ-RHDE, experiments were replicated two and five times, respectively, and the averages of those experiments ± standard deviations are shown. Constructs are drawn to scale.
These results demonstrate that the Hsp83 ORF houses a strong instability element within its 3′-most region (viz., nucleotide positions 1537 to 2154 in reference to the published Drosophila Hsp83 cDNA sequence [6]). Since there was a ∼100-nt overlap between HORF3 and HORF4, and HORF4 but not HORF3 could trigger strong destabilization, the major instability element is likely to reside within the last 516 nt of the Hsp83 ORF. However, since we cannot rule out the possibility that the 3′ limit of HORF3 fragment disrupts an instability element that spans this boundary, we have called the entire 615-nt HORF4 fragment the HIE. The identification of the HIE is consistent with a conclusion reached above: that a major instability element resides within the Hsp83 mRNA coding region. Furthermore, if there are stabilization elements present within the rpA1 transcript, the HIE is able to override their stabilizing functions.
To dissect the HIE further, six subfragments spanning the entire HIE were generated and named HORF4-1 through HORF4-6 (Fig. 2A). These subfragments were each ∼120 nt long with an ∼20-nt overlap and were inserted into R-R-R as described above. In addition, two larger subfragments of the HIE, which were each ∼320 nt long with ∼20 nt of overlap, were inserted in frame into the R-R-R ORF (HORF4-7 and HORF4-8) (Fig. 2A). None of the subfragments of the HIE was capable of recapitulating the strong destabilization accomplished by insertion of the entire HIE (Fig. 2B and C). A weak destabilizing effect was observed by 4 to 6 h upon insertion of HORF4-1 (50% ± 5% mRNA eliminated), HORF4-6 (39% ± 20% eliminated), HORF4-7 (34% ± 5% eliminated), and HORF4-8 (33% ± 9% eliminated). Transcripts carrying HORF4-2, HORF4-3, HORF4-4, and HORF4-5 were stable since only 16% ± 7%, 13% ± 5%, 10% ± 4%, and 21% ± 11% of the respective transcripts were eliminated by 4 to 6 h AEL.
FIG. 2.
Several subfragments of the HIE confer moderate instability. (A) Schematic representation of the HORF4 subfragments utilized to study the HIE. Six overlapping fragments (∼100 nt each with 20-nt overlap) as well as two larger overlapping fragments (∼320 nt each with 20-nt overlap) of the HORF4 region (HORF4-1 to HORF4-8) were independently inserted into the transgenic R-R-R construct at a unique SalI restriction site within the rpA1 coding sequence. All HORF fragments were inserted in frame into R-R-R. Constructs are drawn to scale. (B) Northern blot analysis of the HORF4 subfragment sufficiency constructs in unfertilized eggs. Blots were probed with the HORF4 fragment of the Hsp83 ORF to detect transgenic mRNAs and with the loading control, rp49. (C) Quantification of Northern blot data in panel B. Average normalized transgenic mRNA levels between two independently derived transgenic fly lines are shown. Egg collection time points were the same as those for Fig. 1, and normalization was performed as described for Fig. 1. Light grey, 0 to 2 h AEL; dark grey, 2 to 4 h AEL; black, 4 to 6 h AEL. The red lines indicate the cutoffs defined in the text for lack of destabilizing ability (above the upper red line), weak destabilizing ability (between the red lines), and strong destabilizing ability (below the lower red line) at the 4- to 6-h time point.
The inability to uncover a single subfragment of the HIE that produced a strong decay profile resembling that of the entire HIE, along with the weak destabilizing effect of several subfragments, suggests that the HIE contains two or more subelements that function in combination, possibly additively, to mediate strong transcript degradation. The weak destabilizing activity of HORF4-1 and HORF4-6 supports the notion that most of these subelements reside toward the ends, rather than the central region, of the HIE. Consistent with this hypothesis, weak degradation was also induced by each of the larger subfragments, HORF4-7 and HORF4-8, which include HORF4-1 and HORF4-6, respectively.
The HIE is not necessary for the destabilization of full-length Hsp83 mRNA.
To assess the role of the HIE in the context of the Hsp83 mRNA, we constructed a transgene, H-HΔHORF4-H, in which the majority of the HIE was removed (i.e., the 3′-most 528 nt of the 615-nt HIE) (Fig. 3A). To ensure that any observed instability was not a consequence of shortening the length of the ORF, a “substitution” construct was also made, in which the deleted 528 nt were replaced with 528 nt of the lacZ coding sequence fused in frame to the Hsp83 ORF (H-HΔHORF4+lacZ-H) (Fig. 3A). Both the H-HΔHORF4-H and H-HΔHORF4+lacZ-H transgenic mRNAs were strongly unstable: 78% ± 13% of the H-HΔHORF4-H transcripts were eliminated by 4 to 6 h AEL while 78% ± 0% of H-HΔHORF4+lacZ-H transcripts were eliminated (Fig. 3B and C). Thus, the 528-nt fragment of the HIE is not necessary for H-H-H mRNA degradation, likely because of additional instability elements elsewhere in the Hsp83 mRNA.
FIG. 3.
The HIE is not necessary for destabilization of full-length Hsp83 transgenic mRNA but is required for the destabilization of R-H-R transgenic mRNA. (A) Schematic representation of necessity constructs. These constructs all contained a deletion within the Hsp83 coding sequence that removes the majority of the HIE (the 5′-most 87 nt of HIE are still present). This 3′-most 528-nt region of the Hsp83 coding sequence was deleted within full-length H-H-H (top), was replaced with 528-nt lacZ coding sequence within full-length H-H-H (middle), and was deleted within the transgenic construct R-H-R (bottom). (B) Northern blot analysis of H-HΔHORF4-H, H-HΔHORF4+lacZ-H, and R-HΔHORF4-R mRNAs in unfertilized eggs. H-HΔHORF4-H and H-HΔHORF4+lacZ-H mRNAs were detected by probing for the p53 sequence, and rpA1 mRNA was used as a loading control. For R-HΔHORF4-R, probing was with the rpA1 3′ UTR, which detected both the transgenic mRNA and the loading control, rpA1. (C) Quantification of Northern blot data in panel B. Average normalized transgenic mRNA levels between two independently derived transgenic fly lines are shown.
To assess the necessity of the HIE in the absence of additional Hsp83 mRNA instability elements, we next examined the 528-nt deletion of the HIE within the context of the R-H-R transgene, which lacks the Hsp83 5′ UTR and 3′ UTR (Fig. 3A). Transgenic R-HΔHORF4-R transcripts were substantially stabilized relative to R-H-R but did remain weakly unstable (39% ± 3% eliminated versus 72% ± 7%, respectively) (Fig. 3B and C).
Together, our results support the hypothesis that the HIE is a major instability element that can function to strongly destabilize an otherwise stable mRNA (R-R-R). The weak instability of R-HΔHORF4-R mRNA is consistent with additional, minor instability elements residing in the undeleted part of the Hsp83 ORF. Furthermore, the fact that the HIE is necessary to mediate strong degradation in the R-H-R context but not in the full-length H-H-H context is consistent with additional instability elements mapping outside the ORF, possibly in the 3′ UTR, which exerts weak destabilizing activity (Table 1).
A large deletion in the Hsp83 ORF substantially stabilizes the mRNA.
Given the inference that weak instability elements might reside in the Hsp83 ORF upstream of the HIE, we next assessed whether a large deletion within the ORF would stabilize the mRNA to a greater extent than deletion of the HIE alone. We previously reported that an Hsp83-lacZ hybrid mRNA containing the Hsp83 5′ UTR, the first 333 nt of the ORF (fused in frame to a fragment of the lacZ ORF), and the full-length Hsp83 3′ UTR is unstable in early embryos (4). To assess whether this 1,821-nt deletion, which removes more than 80% of the ORF, affects transcript stability via the maternal decay pathway, we examined the Hsp83-lacZ reporter mRNA—here renamed H-HΔ1.8kb+lacZ-H to be consistent with the nomenclature used in the present study—in unfertilized eggs and found that it was partially stabilized (Fig. 4 E and F): at 2 to 4 h AEL, 79% ± 10% of H-HΔ1.8kb+lacZ-H mRNA remained compared to 14% ± 3% of endogenous Hsp83 transcripts while, by 4 to 6 h, 31% ± 3% of H-HΔ1.8kb+lacZ-H transcripts remained compared with only 2% ± 1% of endogenous transcripts. Thus, deletion of most of the Hsp83 ORF, including the HIE, substantially (but incompletely) abrogates mRNA degradation via the maternal decay pathway.
The fact that H-HΔ1.8kb+lacZ-H transcripts were partially stabilized relative to H-HΔHORF4-H and H-HΔHORF4+lacZ-H transcripts is consistent with the hypothesis that instability elements additional to the HIE reside in the more-5′ region of the Hsp83 ORF. The fact that the H-HΔ1.8kb+lacZ-H transcript was only weakly stabilized is consistent with additional instability elements mapping in the 3′ UTR.
An auxiliary instability element—the previously identified HDE—resides in the Hsp83 3′ UTR.
We previously reported that deletion of a 97-nt region within the 407-nt 3′ UTR of Hsp83—the HDE—within the context of the H-HΔ1.8kb+lacZ-H reporter mRNA (i.e., H-HΔ1.8kb+lacZ-HΔHDE) resulted in strong stabilization of this mRNA in unfertilized eggs (4). The HDE is, however, not necessary for degradation of Hsp83 transcripts: deletion of the HDE in the context of a full-length Hsp83 transgene (H-H-HΔHDE) had no effect on transcript destabilization (Fig. 4A and B). That the HDE is not required for the decay of the Hsp83 transcripts is consistent with our finding that H-H-R transcripts, in which the entire Hsp83 3′ UTR was replaced, remained highly unstable (Table 1).
We next assayed whether simultaneous deletion of the HIE and HDE (H-HΔHORF4-HΔHDE) had an effect on stability. Our results clearly indicated that there is no difference between the decay kinetics of H-HΔHORF4-H and those of H-HΔHORF4-HΔHDE transcripts: in the case of H-HΔHORF4-H, 22% ± 13% of transcripts remained while, for H-HΔHORF4-HΔHDE, 21% ± 0% remained at 4 to 6 h AEL compared with <10% of endogenous mRNA (Fig. 4C and D).
As described in the previous section, transcripts carrying the large, 1.8-kb ORF deletion, H-HΔ1.8kb+lacZ-H, are weakly stabilized, with 31% ± 3% remaining 4 to 6 h AEL (Fig. 4E and F). However, together with the HDE deletion (H-HΔ1.8kb+lacZ-HΔHDE), these were strongly stabilized with 66% ± 4% remaining at 4 to 6 h AEL (Fig. 4G and H).
Together, these data support the hypothesis that several instability elements for Hsp83 mRNA—including a major element, the HIE—reside in its ORF and that the 3′-UTR-located HDE functions as an auxiliary instability element whose role is revealed only in the context of a sensitized transcript in which most, but not all, of the ORF-located elements are deleted.
The sufficiency experiments documented above are consistent with the hypothesis that the HDE is an auxiliary instability element, since weak destabilization was observed for R-R-H transgenic mRNA (Table 1). To specifically address the role of the HDE in mediating this destabilization, we inserted the HDE into the 3′ UTR of a transgene, R-RlacZ-R, which comprises full-length genomic rpA1 with a portion of the lacZ ORF inserted in frame into the rpA1 coding sequence (to distinguish the transgenic transcripts from endogenous ones). R-RlacZ-R was stable, with only 14% ± 20% eliminated by 4 to 6 h AEL (Fig. 4I and J). Insertion of the HDE (R-RlacZ-RHDE) had a weak destabilizing effect: 37% ± 14% of transgenic mRNA was eliminated by 4 to 6 h AEL (Fig. 4I and J), essentially identical to that seen for R-R-H (36% ± 15% eliminated) (Table 1).
We conclude that the HDE is an auxiliary instability element that functions together with the major instability elements, which are located in the Hsp83 ORF.
Translation of the Hsp83 mRNA and/or of the HIE is not required for transcript destabilization.
We showed previously that Hsp83 mRNA is actively translated in early embryos (30). To investigate whether translation of an Hsp83 transcript is required to trigger its degradation, we inserted a strong, stable hairpin within the 5′ UTR of the full-length H-H-H transgene (position 18 of the 149-nt-long 5′ UTR) (Fig. 5 A). The identical hairpin inserted anywhere between a 5′ cap and an initiation codon has been shown previously to block translation by preventing 40S ribosome subunit scanning in vitro and, thus, to efficiently block translation of c-fos mRNA in mouse NIH 3T3 cells (20, 29). To control for any potential stabilizing effect of disruption of the Hsp83 5′ UTR per se, an additional construct was made in which a random sequence of identical length was inserted at the same position (Fig. 5A). Both of these transgenic transcripts, Hhairpin-H-H and Hrandom-H-H, were examined for translational status and stability. Western blot analysis showed that, as expected, Hrandom-H-H mRNA was translated while Hhairpin-H-H was not (Fig. 5B). Thus, the insertion of the hairpin efficiently and specifically blocked translation. We next assessed the decay kinetics of Hrandom-H-H and Hhairpin-H-H mRNA (Fig. 5C and D). Both mRNAs were rapidly degraded over a 6-hour time course and exhibited decay kinetics similar to those of endogenous Hsp83 mRNA. These results demonstrate that Hsp83 mRNA translation in cis is not required for transcript degradation.
FIG. 5.
Hsp83 mRNA destabilization does not require translation in cis. (A to D) Insertion of a strong stable hairpin into full-length Hsp83 transgenic mRNA blocks translation in cis but does not prevent mRNA degradation. (A) Schematic representation of the insertion of either a strong stable hairpin or random nonhairpin sequence (inverted triangle) into the 5′ UTR of H-H-H. Constructs are drawn to scale. (B) Western blot analysis using anti-p53 antibody demonstrates that protein was not detectable from 0- to 2-h unfertilized egg collections from Hhairpin-H-H transgenic mothers in contrast to Hrandom-H-H transgenic mothers. Specificity of the anti-p53 antibody and the proper sizes of the p53-tagged transgenic protein were demonstrated by looking at additional p53-tagged constructs such as H-HΔHORF4-H and H-HΔHORF4+lacZ-H. (C) Northern blot analysis of Hhairpin-H-H and Hrandom-H-H mRNA. Transgenic mRNAs were detected using a p53-specific probe, and rpA1 mRNA was used as a loading control. Lanes marked “−” denote unfertilized eggs lacking any transgene. (D) Quantification of Northern blot data in panel C. Average normalized transgenic mRNA levels from two independently derived transgenic fly lines are shown. The endogenous Hsp83 mRNA decay profile from Fig. 1 is included as a reference. Egg collection time points and normalization were as described for Fig. 1. (E and F) The HIE mediates the decay of stable rpA1 transgenic mRNA in a position-independent but orientation-dependent manner. (E) The HIE was inserted in sense and antisense orientations at two different locations within the rpA1 3′ UTR (and thus downstream of the stop codon). Northern blot analysis of transgenic mRNAs in unfertilized eggs is shown below the schematic representations (drawn to scale). Blots were probed with the full-length rpA1 3′ UTR, which detected both transgenic (experimental) and rpA1 (loading control) transcripts. (F) Quantification of Northern blot data in panel E. Average normalized transgenic mRNA levels from two independently derived transgenic fly lines are shown. Egg collection time points were the same, and normalizations were performed, as described in Materials and Methods. The decay profile of R-RHORF4-R from Fig. 1C is included as a reference.
Our results using Hhairpin-H-H mRNA suggested that the HIE might not need to reside within the ORF for it to function as an instability element. Constructs were therefore made to assess the position dependence of the HIE by inserting it in both the sense and antisense orientations at two different sites within the 3′ UTR of R-R-R (Fig. 5E). In each of these, since the HIE was downstream of the translation stop codon, it was “protected” from ribosome transit. Insertion of the HIE at either site in its sense orientation conferred strong transcript instability: 77% ± 5% of R-R-RsHORF4(Aat) and 77% ± 9% of R-R-RsHORF4(Fse) transcripts were degraded by 4 to 6 h AEL (Fig. 5E and F). In contrast, control antisense HIE insertions caused only 2% ± 30% and 19% ± 27% of the mRNA to be eliminated, respectively, for R-R-RasHORF4(Aat) and R-R-RasHORF4(Fse) mRNAs. Thus, the HIE can mediate transcript degradation even when located in the 3′ UTR, and ribosome transit through the HIE is not necessary for its function.
We conclude that translation in cis of the Hsp83 mRNA and, specifically, of the HIE is not necessary for transcript degradation and that the HIE mediates transcript decay in a position-independent manner.
The HIE mediates SMG-dependent mRNA degradation.
We have shown previously that SMG function is required for destabilization of endogenous Hsp83 mRNA (30). We therefore next assessed whether HIE-dependent destabilization depends on SMG function. To do so, we determined the degradation profiles of R-RHORF4-R, R-R-RsHORF4(Fse), and R-R-RasHORF4(Fse) transgenic mRNA in unfertilized eggs produced by smg mutant females.
The degradation of R-RHORF4-R and R-R-RsHORF4(Fse) mRNA was severely compromised—in a dose-dependent manner—by loss of SMG (Fig. 6 shows R-RHORF4-R). In an smg1 heterozygous background (i.e., in the presence of one dose of the smg+ gene), R-RHORF4-R mRNA was partially stabilized relative to the wild type (55% ± 13% versus 32% ± 1%, respectively, remaining by 4 to 6 h AEL in unfertilized eggs). In an smg1 homozygous mutant background (i.e., zero doses of the smg+ gene), transgenic R-RHORF4-R mRNA was almost completely stabilized with 89% ± 2% of the mRNA remaining by 4 to 6 h AEL. This is almost identical to the stabilization of endogenous Hsp83 mRNA in 2- to 3-h embryos from smg mutant females, in which 92% ± 4% of endogenous Hsp83 transcripts remain (30). Likewise, R-R-RsHORF4(Fse) transcripts were stabilized in smg1 homozygotes (data not shown). We conclude that the HIE functions in an SMG-dependent manner, thus reflecting a true endogenous major instability element.
FIG. 6.
SMG is required for the destabilization of R-RHORF4-R transgenic mRNA. (A) Northern blot analysis of R-RHORF4-R mRNA as assayed in smg1 heterozygous and smg mutant backgrounds. smg1 heterozygous and smg1 mutant eggs were both derived from mothers containing a single copy of the R-RHORF4-R transgene. smg mutant eggs were derived from females containing the smg1 allele and the deficiency Df(3L)ScfR6, which uncovers the smg region. Blots were probed as previously described for Fig. 3. (B) Quantification of Northern blot data in panel A. Average normalized transgenic mRNA levels between two independently derived transgenic fly lines are shown. The decay profile of R-RHORF4-R in a +/+ background was taken from Fig. 1C and is included as a reference. Egg collection time points were the same, and normalizations were performed, as described for Fig. 1 and in Materials and Methods.
The HIE directs association with SMG protein.
We previously used real-time RT-PCR to show that endogenous Hsp83 transcripts are present in an SMG-containing messenger ribonucleoprotein complex (mRNP): after immunoprecipitation with anti-SMG antibody, Hsp83 transcripts were fivefold enriched relative to normal rat serum; a positive control, nanos, was also fivefold enriched; and a negative control, rp49, was only 1.7-fold enriched (30). To assess whether the HIE is sufficient to target a hybrid mRNA to the SMG-containing mRNP, here we again used real-time RT-PCR to study the R-RHORF4-R and R-R-RsHORF4(Fse) mRNAs. We found that the R-RHORF4-R and R-R-RsHORF4(Fse) transcripts were enriched 3.3- ± 0.5-fold and 5.6- ± 1.2-fold, respectively, while endogenous Hsp83 transcripts were enriched 5.6- ± 0.5-fold and 4.5- ± 0.3-fold and rp49 transcripts were enriched 1.3- ± 0.0-fold and 1.6- ± 0.4-fold in their respective experiments.
We conclude that the HIE can direct transcripts into an SMG-containing mRNP.
SMG's RNA-binding ability is necessary for Hsp83 mRNA destabilization.
Alanine498 of the budding yeast homolog of SMG, VTS1, is found in its RNA-binding domain (RBD) and makes direct contact with the third base in the loop of the SRE (1). A single amino acid mutation (to histidine) of either alanine498 within the VTS1 SAM domain or the analogous residue within the SMG SAM domain, alanine642, completely abolishes the ability of these SAM domains to bind consensus SREs in vitro but does not disrupt protein folding (2).
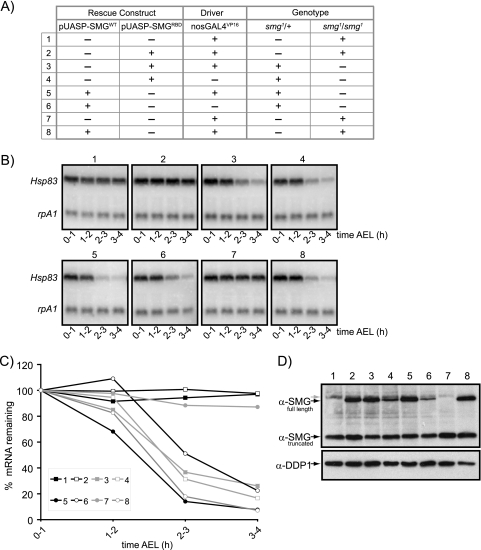
To assess whether direct binding of SMG to Hsp83 mRNA is required for transcript destabilization, we therefore made full-length SMG “rescue” constructs expressing either wild-type protein (SMGWT) or an A642H mutant protein (SMGRBD, for RBD mutant). Using the modified GAL4/upstream activation sequence (UAS) system to drive expression of the transgenic mRNAs during oogenesis (27, 41), we assessed the ability of SMGRBD and SMGWT to rescue the severe Hsp83 mRNA degradation defects observed in smg mutants. In these experiments, because we had shown that Hsp83 mRNA degradation is SMG dependent in both unfertilized and fertilized eggs (30) and the amount of material was limiting, embryos rather than unfertilized eggs were used.
Expression of SMGWT in embryos from smg mutant females fully rescued endogenous Hsp83 mRNA destabilization (Fig. 7A to C, genotype 8). In sibling controls, Hsp83 mRNA decay was observed in smg1 heterozygous backgrounds (Fig. 7A to C, genotypes 5 and 6), while Hsp83 mRNA was completely stabilized in embryos from smg1 mutant mothers lacking the SMGWT transgene (Fig. 7A to C, genotype 7). In striking contrast, embryos expressing SMGRBD failed to degrade endogenous Hsp83 mRNA (Fig. 7A to C, genotype 2); this decay profile was indistinguishable from that observed in the smg1 mutant background alone (Fig. 7A to C, genotype 1). Embryos from their smg1 heterozygous siblings, in contrast, displayed normal Hsp83 mRNA decay (Fig. 7A to C, genotypes 3 and 4).
FIG. 7.
RNA-binding ability of SMG is necessary for degradation of endogenous Hsp83 mRNA. Full-length SMG transgenes driven by the GAL4/UAS system were tested for their ability to rescue the Hsp83 mRNA degradation phenotype in smg1 mutant embryos. The function of the wild-type rescue construct (SMGWT) was compared to that of a mutated SMG construct (SMGRBD) which contains a single point mutation within the sequence encoding the SMG RBD, which is predicted to abolish SMG's ability to bind its cognate RNA sequence, the SRE. Rescue constructs were expressed using pUASP vectors and were driven via the maternal driver, nosGAL4VP16. (A) Genetic background of transgenic mothers with sibling control lines included. (B) Northern blot analysis of total RNA extracted from fertilized embryos over a 4-hour time course. Embryos were examined at 0 to 1, 1 to 2, 2 to 3, and 3 to 4 h AEL. Blots were probed for Hsp83 (experimental) and rpA1 (loading control) mRNAs. Genotypes are described in the legend to panel A. (C) Quantification of Northern blot data in panel B. Only a single experiment was performed. Normalization was as described for Fig. 1. (D) Western blot analysis of 0- to 3-h embryos demonstrating the production of SMG protein from the rescue constructs SMGWT and SMGRBD. The anti-SMG antibody recognizes both the endogenous and the transgenic SMG proteins, as well as the truncated form of the protein derived from the smg1 allele. Anti-DDP1 antibody was used as a loading control. The gray arrow represents a cross-reacting band that migrates just above the SMG protein.
Western blot analysis confirmed that pUASP-SMGWT and pUASP-SMGRBD produced equivalent levels of SMG protein (Fig. 7D); therefore, the lack of rescue by SMGRBD was not attributable to lower protein levels. The results shown in Fig. 7 derive from single transgenic lines for SMGWT and SMGRBD; almost-identical results were obtained for two additional transgenic lines for each construct (data not shown). Thus, a point mutation that abrogates the SRE binding ability of SMG also blocks the ability of SMG to mediate Hsp83 mRNA destabilization, consistent with direct binding of SMG to the Hsp83 mRNA being a prerequisite for transcript destabilization.
Simultaneous mutation of all eight computationally predicted SREs in the Hsp83 ORF stabilizes the transcript.
SMG recognizes its target mRNAs via stem-loop structures called SREs (12, 32, 33). Initially, the consensus SRE sequence was defined as either a 4- or a 5-nt loop with the sequence CNGG or CNGGN, respectively, on a nonspecific stem (2, 11). Four copies of this consensus SRE, present within the ORF of endogenous Hsp83 mRNA, were shown by us to be dispensable since simultaneous mutation of all four elements did not affect transcript instability (30). Subsequent to those analyses, it was shown that the length of an SRE loop can be increased to 7 nt (CNGGN0-3) and the SRE can remain fully functional (1). In addition, structural analyses revealed that the VTS1 RBD makes contact with the phosphate groups between the third and fourth bases of the stem, suggesting that the length of the stem may also be important for RNA recognition (1). A computational search of the entire Hsp83 transcript for the revised SRE consensus (CNGGN0-3 on a stem of at least 4 bp) revealed an additional four predicted SREs within the Hsp83 mRNA. Thus, there are a total of eight predicted SREs in the Hsp83 mRNA: all map within the ORF, six of the eight reside within the HIE, and seven of the eight are removed by the 1.8-kb deletion used in our analyses reported above (in Fig. 9 the four potential SREs that were mutated in our 2005 study are indicated with asterisks; see also Fig. S1 in the supplemental material for the complete sequence of the ORF and location of the SREs).
FIG. 9.
Potential SREs in the Hsp83 mRNA and their relationships to fragments of the ORF that are sufficient to destabilize the rpA1 transcript. The diagrams and color coding are as in Fig. 1 to 4. Predicted SREs (defined as in the text) are shown at their approximate locations in the Hsp83 mRNA but are drawn larger than scale. Asterisks indicate the predicted SREs that were mutated in our previous study (30). The braces indicate the 1,821-nt deletion that incompletely abrogates instability of the Hsp83 transcript. The ability of the full-length ORF and its subfragments to confer instability as defined in Table 1 is indicated to the right: ++, strong instability element; +, moderate instability element; −, no destabilizing activity.
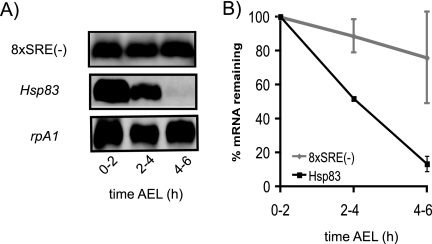
To assess whether destabilization of Hsp83 mRNA requires binding to these predicted SREs, we constructed a transgene encoding an mRNA—H-H8xSRE(-)-H—in which all eight sites were mutated simultaneously (Fig. 8; see also Fig. S1 in the supplemental material). In seven out of eight cases, third-base mutations ensured that the transgene retained identical amino acid coding capacity; in one case it was necessary to introduce a conservative substitution (see Materials and Methods and Fig. S1 in the supplemental material).
FIG. 8.
Mutation of eight predicted SREs in the Hsp83 ORF results in transcript stabilization. (A) Northern blot analysis of H-H8xSRE(-)-H transcripts [8xSRE(-)] and endogenous Hsp83 mRNAs in unfertilized eggs. H-H8xSRE(-)-H transcripts were detected by probing with the p53-LNA-3′-digoxigenin probe described in Materials and Methods. Endogenous Hsp83 mRNA from non-8xSRE(-) eggs served as a positive control for instability, and endogenous rpA1 mRNA was used as a loading control for normalization. H-H8xSRE(-)-H transcripts were strongly stabilized: by 4 to 6 h AEL in unfertilized eggs, 76% ± 27% of transgenic transcripts were present compared with only 13% ± 4% of endogenous Hsp83 transcripts. (B) Quantification of Northern blot data in panel A. Error bars represent standard deviations calculated from at least two independent experiments.
Northern blot analysis of H-H8xSRE(-)-H mRNA showed that it was strongly stabilized: by 4 to 6 h AEL in unfertilized eggs 76% ± 27% of transgenic transcripts remained, compared with only 13% ± 4% of endogenous Hsp83 transcripts in controls (Fig. 8). Strong stabilization also occurred in embryos, with 68% ± 12% of the H-H8xSRE(-)-H transcripts remaining at 3 to 4 h AEL (data not shown).
Together with our results obtained by point mutation of SMG's RBD, we conclude that SMG interacts directly with the SREs in the Hsp83 ORF to destabilize the mRNA. Since mutation of four out of the eight predicted SREs did not abrogate transcript destabilization (30) while mutation of all eight did, we conclude that SMG is likely to bind to more than four SREs to destabilize the Hsp83 transcript.
DISCUSSION
We have shown here that the Drosophila Hsp83 mRNA contains one major and several minor instability elements that direct transcript degradation upon egg activation via what we previously termed the “maternal degradation pathway” (4). The major instability element (the HIE) resides in the 3′-most 615 nt of the ORF but does not require ribosome transit for its destabilizing function. The HIE contains six of the eight predicted SREs in the Hsp83 transcript and is able to direct an mRNA to associate with an SMG-containing mRNP, thus conferring SMG-dependent destabilization on that transcript. Two additional predicted SREs are present outside the HIE, in the more-5′ part of the Hsp83 ORF. Mutation of the SMG RBD or of all eight SREs results in stabilization of the Hsp83 transcript. Thus, SMG directly interacts with multiple SREs in the Hsp83 ORF to recruit the CCR4-NOT deadenylase and trigger decay. The only previously identified Drosophila transcript shown to be directly bound by SMG is nanos, which has two SREs in its 3′ UTR (11, 12, 32, 33). However, precedent for an ORF-located SRE comes from budding yeast, where it has been shown that the ORF of the NNF1 mRNA contains a single SRE, which confers VTS1-dependent instability (1).
Our analyses have also shown that an auxiliary instability element (the HDE [4]) resides in the Hsp83 3′ UTR. The HDE is capable of weakly destabilizing an otherwise stable mRNA (rpA1); however, its role in the context of the Hsp83 mRNA is revealed only in a situation in which more than 80% of the ORF is deleted—including seven of the eight predicted SREs—thus partially stabilizing the transcript and sensitizing it to deletion of the HDE.
Mechanism of HIE function.
Studies of c-fos and c-myc mRNA decay in mammalian cells and MATα1 mRNA decay in S. cerevisiae have provided a detailed mechanistic understanding of how coding region determinants (CRDs) act to destabilize their mRNAs (7-9, 18, 22, 29). The rapid decay of c-fos mRNA involves the formation of a pre-translation-initiation mRNP that acts to bridge the coding region instability element with the poly(A) tail via the RNA-binding proteins UNR, PABP, PAIP, hnRNP D, and NSAP. Upon ribosome transit, mRNP reorganization permits access of the CCR4-NOT deadenylase to the poly(A) tail, leading to mRNA degradation. In contrast, degradation of c-myc and MATα1 mRNAs is due to ribosome pausing at rare codons found in the vicinity of the CRDs. It is believed that ribosome stalling creates a downstream region devoid of ribosomes, which can be accessed by the degradation machinery unless the regions downstream of the rare codons are bound by protective RNA-binding factors.
Translation-independent CRDs, which function either to stabilize or to destabilize mRNAs, have been identified in several transcripts. Stabilizing elements reside in the ORF of the yeast OLE1 mRNA (42), and destabilizing elements map to the ORFs of mammalian PAI-2 and MnSOD mRNAs (14, 40). The CRDs in PAI-2 and MnSOD mRNAs do not require ribosome transit and function when inserted into a 3′ UTR. trans-acting RNA-binding proteins required for the function of the CRDs of PAI-2 and MnSOD have yet to be identified, and their decay mechanisms remain unknown.
Formally, the HIE resembles the PAI-2 and MnSOD CRDs. Maternally loaded Hsp83 mRNA is actively translated in the early Drosophila embryo (30), and we have shown here that a 5′ UTR hairpin that inhibits translation has no effect on Hsp83 transcript instability. Despite the fact that the HIE maps within the coding region, it continues to function when inserted into a 3′ UTR. Thus, ribosome transit is not required for HIE function. Below we present a model for the role of the HIE—and its resident SREs—in transcript destabilization.
The role of the HDE.
Our previous work identified the 97-nt HDE in the Hsp83 3′ UTR as an instability element that functions in the maternal degradation pathway (3, 4). We have shown here that these earlier results were a consequence of the use of a reporter mRNA (H-HΔ1.8kb+lacZ-H) lacking most of the ORF-located instability elements (seven of eight SREs) and sensitized to deletion of the auxiliary element. Deletion of the HDE in this context (H-HΔ1.8kb+lacZ-HΔHDE) thus resulted in strong transcript stabilization while the same reporter without the HDE deletion was only weakly unstable.
Our previous analyses also suggested that the HDE might function both as an instability element and as a translational enhancer (4). We have recently identified a multiprotein complex that binds the HDE and mediates translational enhancement (24). Three proteins—DDP1, HRP48, and PABP—function together to stimulate translation. To date, none of these proteins has been implicated in the transcript destabilization function of the HDE.
SMG, SREs, and Hsp83 mRNA destabilization.
In a previous study we identified and simultaneously mutated four potential SREs in the Hsp83 ORF without affecting transcript destabilization (30). Identification of those SREs was based on the loop consensus CNGGN0-1 (2). More recently, a revised loop consensus sequence (CNGGN0-3) (1) enabled us to identify four additional predicted SREs within the Hsp83 transcript. Thus, there are a total of eight predicted SREs in the Hsp83 mRNA, all in the ORF (Fig. 9; see also Fig. S1 in the supplemental material). Of the large fragments of the ORF that were tested for sufficiency, only HORF4 (the HIE) was able to confer strong instability, and it contains six of the predicted SREs. The other three large fragments (HORF1, HORF2, and HORF3) each contain a single predicted SRE and did not, by themselves, confer instability. Of the subfragments of HORF4 that were sufficient to confer weak instability, two contain three predicted SREs (HORF4-7 and HORF4-8), one contains two predicted SREs (HORF4-1), and one (HORF4-6) carries a single predicted SRE. In contrast, of the four subfragments of ORF4 that were not sufficient to confer instability, three contain a single predicted SRE (HORF4-3, HORF4-4, and HORF4-5) and one (HORF4-2) contains no predicted SRE. Thus, there is a good correlation between the number of predicted SREs in a fragment and its ability to destabilize a hybrid mRNA. Based on this correlation, a plausible hypothesis is that multiple SREs in the Hsp83 mRNA's ORF function together to bind strongly to SMG and recruit the transcript to an SMG-containing mRNP, following which deadenylation by the CCR4-NOT deadenylase triggers decay.
Two complementary approaches were used to prove that SMG binding to multiple SREs in the Hsp83 ORF is required to trigger destabilization. First, a single-amino-acid substitution, A462H, in the RBD of SMG completely stabilized the endogenous Hsp83 mRNA. Second, simultaneous mutation of all eight predicted SREs (without significantly affecting the coding capacity of the ORF) stabilized a transgenic Hsp83 transcript.
The fact that mutation of four of the predicted SREs (two in the HIE) did not stabilize the mRNA (30) whereas mutation of all eight predicted SREs (six in the HIE) did suggests a requirement for multiple SREs in Hsp83 transcript destabilization. Using metabolic labeling with [35S]methionine, we showed previously that, in early embryos, Hsp83 transcripts are in fact translated as well as destabilized (30). In light of this fact, the location of all of the predicted SREs in the Hsp83 ORF rather than its UTRs has mechanistic implications for transcript destabilization since SMG cannot remain bound while a ribosome transits through an SRE. First, it is possible that only a fraction of Hsp83 mRNA is translated in early embryos; thus, most of the maternal Hsp83 transcripts may exist in an untranslated state with SMG bound and, therefore, able to recruit the deadenylase. Alternatively, most Hsp83 transcripts may be translated and their destabilization may result from the distribution of multiple SREs over a large domain rather than clustered as a pair as in the 3′ UTR of the nanos mRNA (11, 12, 32, 33). A plausible model is that multiple, widely spaced SREs ensure that some SMG molecules remain bound to an Hsp83 mRNA molecule even as ribosomes transit its ORF. The Hsp83 mRNA could, then, be targeted for deadenylation and decay even though it was simultaneously being translated. Thus, the SREs in the ORF permit transcript destabilization in spite of—but not because of—translation.
Supplementary Material
Acknowledgments
We thank Arash Bashirullah for designing and Tyler Davies for making the H-H-HΔHDE construct. Najeeb Siddiqui and Wael Tadros kindly provided critical comments on a draft of the manuscript.
Trainee support came from the following sources: a Natural Sciences and Engineering Research Council of Canada (NSERC) Graduate Scholarship (J.L.S.), a Canada Graduate Scholarship from the Canadian Institutes for Health Research (CIHR) (J.L.S.), studentships from the Ontario Student Opportunity Trust-Hospital for Sick Children Foundation Student Scholarship Program (J.L.S. and R.L.C.), and a scholarship from the CIHR (R.L.C.). H.D.L. is Canada Research Chair (Tier 1) in Developmental Biology at the University of Toronto. This research was supported by an operating grant to C.A.S. from the National Cancer Institute of Canada with funds from the Terry Fox Run and an operating grant (MOP-14409) to H.D.L. from the CIHR.
Footnotes
Published ahead of print on 15 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aviv, T., Z. Lin, G. Ben-Ari, C. A. Smibert, and F. Sicheri. 2006. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat. Struct. Mol. Biol. 13168-176. [DOI] [PubMed] [Google Scholar]
- 2.Aviv, T., Z. Lin, S. Lau, L. M. Rendl, F. Sicheri, and C. A. Smibert. 2003. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat. Struct. Biol. 10614-621. [DOI] [PubMed] [Google Scholar]
- 3.Bashirullah, A., R. L. Cooperstock, and H. D. Lipshitz. 2001. Spatial and temporal control of RNA stability. Proc. Natl. Acad. Sci. USA 987025-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bashirullah, A., S. R. Halsell, R. L. Cooperstock, M. Kloc, A. Karaiskakis, W. W. Fisher, W. Fu, J. K. Hamilton, L. D. Etkin, and H. D. Lipshitz. 1999. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster. EMBO J. 182610-2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettegowda, A., and G. W. Smith. 2007. Mechanisms of maternal mRNA regulation: implications for mammalian early embryonic development. Front. Biosci. 123713-3726. [DOI] [PubMed] [Google Scholar]
- 6.Blackman, R. K., and M. Meselson. 1986. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J. Mol. Biol. 188499-515. [DOI] [PubMed] [Google Scholar]
- 7.Caponigro, G., D. Muhlrad, and R. Parker. 1993. A small segment of the MATα1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 135141-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caponigro, G., and R. Parker. 1996. mRNA turnover in yeast promoted by the MATalpha1 instability element. Nucleic Acids Res. 244304-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, T. C., A. Yamashita, C. Y. Chen, Y. Yamashita, W. Zhu, S. Durdan, A. Kahvejian, N. Sonenberg, and A. B. Shyu. 2004. UNR, a new partner of poly(A)-binding protein, plays a key role in translationally coupled mRNA turnover mediated by the c-fos major coding-region determinant. Genes Dev. 182010-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooperstock, R. L. 2002. Mechanisms of transcript regulation in the early Drosophila embryo: degradation, localization and translational regulation. Ph.D. thesis. University of Toronto, Toronto, Ontario, Canada.
- 11.Crucs, S., S. Chatterjee, and E. R. Gavis. 2000. Overlapping but distinct RNA elements control repression and activation of nanos translation. Mol. Cell 5457-467. [DOI] [PubMed] [Google Scholar]
- 12.Dahanukar, A., J. A. Walker, and R. P. Wharton. 1999. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol. Cell 4209-218. [DOI] [PubMed] [Google Scholar]
- 13.Dahanukar, A., and R. P. Wharton. 1996. The Nanos gradient in Drosophila embryos is generated by translational regulation. Genes Dev. 102610-2620. [DOI] [PubMed] [Google Scholar]
- 14.Davis, C. A., J. M. Monnier, and H. S. Nick. 2001. A coding region determinant of instability regulates levels of manganese superoxide dismutase mRNA. J. Biol. Chem. 27637317-37326. [DOI] [PubMed] [Google Scholar]
- 15.Decker, C. J., and R. Parker. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 71632-1643. [DOI] [PubMed] [Google Scholar]
- 16.De Renzis, S., O. Elemento, S. Tavazoie, and E. F. Wieschaus. 2007. Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS Biol. 5e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fontes, A. M., A. Riedl, and M. Jacobs-Lorena. 2001. Developmental regulation of an instability element from the Drosophila fushi tarazu mRNA. Genesis 3059-64. [DOI] [PubMed] [Google Scholar]
- 18.Grosset, C., C. Y. Chen, N. Xu, N. Sonenberg, H. Jacquemin-Sablon, and A. B. Shyu. 2000. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 10329-40. [DOI] [PubMed] [Google Scholar]
- 19.Ito, J., and M. Jacobs-Lorena. 2001. Functional mapping of destabilizing elements in the protein-coding region of the Drosophila fushi tarazu mRNA. J. Biol. Chem. 27623525-23530. [DOI] [PubMed] [Google Scholar]
- 20.Kozak, M. 1989. Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol. Cell. Biol. 95134-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lecuyer, E., H. Yoshida, N. Parthasarathy, C. Alm, T. Babak, T. Cerovina, T. R. Hughes, P. Tomancak, and H. M. Krause. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131174-187. [DOI] [PubMed] [Google Scholar]
- 22.Lemm, I., and J. Ross. 2002. Regulation of c-myc mRNA decay by translational pausing in a coding region instability determinant. Mol. Cell. Biol. 223959-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipshitz, H. D., and C. A. Smibert. 2000. Mechanisms of RNA localization and translational regulation. Curr. Opin. Genet. Dev. 10476-488. [DOI] [PubMed] [Google Scholar]
- 24.Nelson, M. R., H. Luo, H. K. Vari, B. J. Cox, A. J. Simmonds, H. M. Krause, H. D. Lipshitz, and C. A. Smibert. 2007. A multiprotein complex that mediates translational enhancement in Drosophila. J. Biol. Chem. 28234031-34038. [DOI] [PubMed] [Google Scholar]
- 25.Richter, J. D. 2007. CPEB: a life in translation. Trends Biochem. Sci. 32279-285. [DOI] [PubMed] [Google Scholar]
- 26.Riedl, A., and M. Jacobs-Lorena. 1996. Determinants of Drosophila fushi tarazu mRNA instability. Mol. Cell. Biol. 163047-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rorth, P. 1998. Gal4 in the Drosophila female germline. Mech. Dev. 78113-118. [DOI] [PubMed] [Google Scholar]
- 28.Rubin, G. M., and A. C. Spradling. 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218348-353. [DOI] [PubMed] [Google Scholar]
- 29.Schiavi, S. C., C. L. Wellington, A. B. Shyu, C. Y. Chen, M. E. Greenberg, and J. G. Belasco. 1994. Multiple elements in the c-fos protein-coding region facilitate mRNA deadenylation and decay by a mechanism coupled to translation. J. Biol. Chem. 2693441-3448. [PubMed] [Google Scholar]
- 30.Semotok, J. L., R. L. Cooperstock, B. D. Pinder, H. K. Vari, H. D. Lipshitz, and C. A. Smibert. 2005. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr. Biol. 15284-294. [DOI] [PubMed] [Google Scholar]
- 31.Semotok, J. L., and H. D. Lipshitz. 2007. Regulation and function of maternal mRNA destabilization during early Drosophila development. Differentiation 75482-506. [DOI] [PubMed] [Google Scholar]
- 32.Smibert, C. A., Y. S. Lie, W. Shillinglaw, W. J. Henzel, and P. M. Macdonald. 1999. Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. RNA 51535-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smibert, C. A., J. E. Wilson, K. Kerr, and P. M. Macdonald. 1996. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev. 102600-2609. [DOI] [PubMed] [Google Scholar]
- 34.Surdej, P., and M. Jacobs-Lorena. 1998. Developmental regulation of bicoid mRNA stability is mediated by the first 43 nucleotides of the 3′ untranslated region. Mol. Cell. Biol. 182892-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tadros, W., A. L. Goldman, T. Babak, F. Menzies, L. Vardy, T. Orr-Weaver, T. R. Hughes, J. T. Westwood, C. A. Smibert, and H. D. Lipshitz. 2007. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev. Cell 12143-155. [DOI] [PubMed] [Google Scholar]
- 36.Tadros, W., S. A. Houston, A. Bashirullah, R. L. Cooperstock, J. L. Semotok, B. H. Reed, and H. D. Lipshitz. 2003. Regulation of maternal transcript destabilization during egg activation in Drosophila. Genetics 164989-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tadros, W., and H. D. Lipshitz. 2005. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev. Dyn. 232593-608. [DOI] [PubMed] [Google Scholar]
- 38.Tadros, W., J. T. Westwood, and H. D. Lipshitz. 2007. The mother-to-child transition. Dev. Cell 12847-849. [DOI] [PubMed] [Google Scholar]
- 39.Thummel, C. S., and V. Pirrotta. 1991. New pCaSpeR P element vectors. Drosophila Info. Newsl. 219. [Google Scholar]
- 40.Tierney, M. J., and R. L. Medcalf. 2001. Plasminogen activator inhibitor type 2 contains mRNA instability elements within exon 4 of the coding region. Sequence homology to coding region instability determinants in other mRNAs. J. Biol. Chem. 27613675-13684. [DOI] [PubMed] [Google Scholar]
- 41.Van Doren, M., A. L. Williamson, and R. Lehmann. 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8243-246. [DOI] [PubMed] [Google Scholar]
- 42.Vemula, M., P. Kandasamy, C. S. Oh, R. Chellappa, C. I. Gonzalez, and C. E. Martin. 2003. Maintenance and regulation of mRNA stability of the Saccharomyces cerevisiae OLE1 gene requires multiple elements within the transcript that act through translation-independent mechanisms. J. Biol. Chem. 27845269-45279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.