Abstract
The late endosome (MVB) plays a key role in coordinating vesicular transport of proteins between the Golgi complex, vacuole/lysosome, and plasma membrane. We found that deleting multiple genes involved in vesicle fusion at the MVB (class C/D vps mutations) impairs transcriptional activation by Gcn4, a global regulator of amino acid biosynthetic genes, by decreasing the ability of chromatin-bound Gcn4 to stimulate preinitiation complex assembly at the promoter. The functions of hybrid activators with Gal4 or VP16 activation domains are diminished in class D mutants as well, suggesting a broader defect in activation. Class E vps mutations, which impair protein sorting at the MVB, also decrease activation by Gcn4, provided they elicit rapid proteolysis of MVB cargo proteins in the aberrant late endosome. By contrast, specifically impairing endocytic trafficking from the plasma membrane, or vesicular transport to the vacuole, has a smaller effect on Gcn4 function. Thus, it appears that decreasing cargo proteins in the MVB through impaired delivery or enhanced degradation, and not merely the failure to transport cargo properly to the vacuole or downregulate plasma membrane proteins by endocytosis, is required to attenuate substantially transcriptional activation by Gcn4.
Regulation of amino acid biosynthesis in Saccharomyces cerevisiae involves the transcriptional activator Gcn4 in the regulatory response known as general amino acid control (GAAC). Gcn4 synthesis is induced by starvation for any amino acid through a translational control mechanism involving the protein kinase Gcn2 and its phosphorylation of eukaryotic translation initiation factor 2 (eIF2). The induced Gcn4 protein binds to the UASGCRE (enhancer) elements at amino acid biosynthetic genes, stimulating their transcription and elevating the protein biosynthetic capacity of the cell (29, 31). Increased binding of Gcn4 at the arginine biosynthetic gene ARG1 occurs within minutes of isoleucine/valine (Ile/Val) limitation imposed by the antimetabolite sulfometuron (SM), which inhibits the Ile/Val biosynthetic enzyme encoded by ILV2 (32). This is followed quickly by recruitment of multiple coactivators (SAGA, SWI/SNF, RSC, and Mediator) that stimulate the assembly of general transcription factors and RNA polymerase II (Pol II) at the promoter (21, 52, 53, 62). Gcn− (general control nonderepressible) mutants, impaired for derepression of all Gcn4 target genes, are sensitive to SM and other inhibitors of amino acid biosynthetic enzymes. We and others have previously identified numerous Gen− mutants defective in factors required for translational induction of GCN4 mRNA or lacking coactivators required for transcriptional activation by Gcn4 on the basis of their sensitivity to SM or other inhibitors of amino acid biosynthesis (28, 62).
To identify novel factors involved in the GAAC, we screened a library of haploid deletion mutants for SM sensitivity (SMs). Surprisingly, we identified numerous SMs/Gcn− strains with deletions of genes involved in vesicular protein trafficking at the late endosome/MVB. Many of these vps (vacuolar protein sorting) mutants were identified previously by their missorting of vacuolar hydrolase carboxypeptidase Y (CPY) or defective vacuolar protease activity (pep mutants). CPY is transported in vesicles from the Golgi apparatus to vacuoles via the MVB (Fig. 1A), as are other hydrolases of the vacuolar lumen, like carboxypeptidase S (Cps1) and proteinase A (PrA). Moreover, downregulation of plasma membrane receptors and transporters by endocytosis and degradation involves vesicular trafficking to the MVB before they reach the vacuole for destruction (8) (Fig. 1A). vps mutants are defective for an array of different molecules required for producing vesicles with the appropriate cargo proteins or for the tethering and fusion of vesicles at the correct target membranes.
FIG. 1.
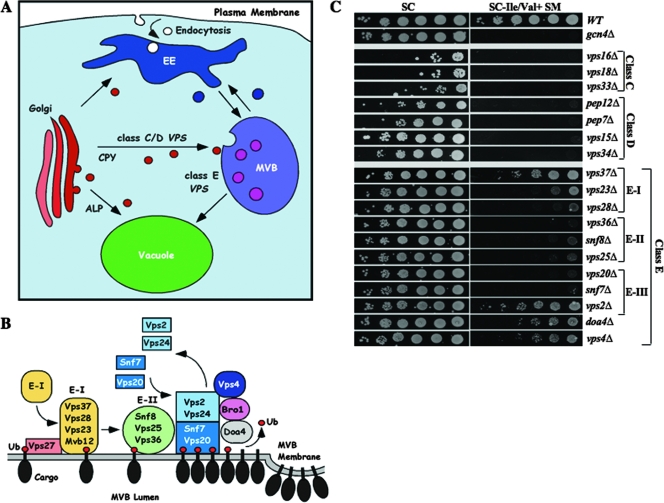
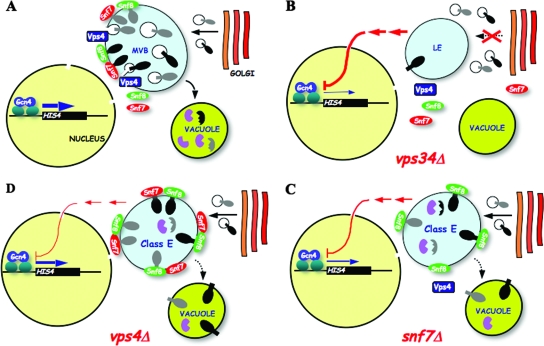
Multiple vps mutants impaired for vesicular trafficking at the late endosome exhibit Gcn− phenotypes. (A) Multiple vesicular trafficking pathways in yeast connect the late Golgi complex to vacuole and plasma membrane via early endosome (EE) and MVB. Adapted from reference 8. (B) Functions of ESCRT complexes (E-I, E-II, and E-III) in sorting ubiquitinated transmembrane proteins at the MVB outer membrane, adapted from reference 2. Ub, ubiquitin. (C) Serial 10-fold dilutions of vps mutants and the gcn4Δ mutant, derived from WT strain BY4741, were spotted to SC medium or SC lacking Ile and Val and containing SM at 1.0 μg/ml and incubated for 2 to 3 days at 30°C. The mutant strains examined were 249, 2783, 4105, 5305, 1812, 3682, 3236, 5149, 2730, 3416, 2763, 5325, 2826, 2580, 6211, 1580, 4850, 4004, and 5588 (described in Table 3).
We found that Gcn4 function is impaired to the greatest extent in class C and D vps mutants defective for various aspects of vesicle fusion at the endosome (Fig. 1A) (8). Our results indicate that mutations in these factors impair activation of Gcn4 target promoters and reduce preinitiation complex (PIC) assembly at ARG1, without reducing the amount of Gcn4 bound to the UASGCRE in vivo. SMs phenotypes also were observed for class E vps mutants, which lack factors needed to sort cargo proteins into intralumenal vesicles (ILVs) at the MVB for subsequent transport to the vacuole lumen (Fig. 1A). This sorting function is carried out by the heteromeric protein complexes ESCRT-I, -II, and -III (abbreviated below as E-I, E-II, and E-III), which bind to ubiquitinated cargo proteins on the MVB outer membrane. The AAA-ATPase Vps4 then recycles the ESCRT factors and segregates the cargo into ILVs (Fig. 1B). Class E vps mutants accumulate MVB cargo proteins in aberrant structures lacking ILVs, called class E compartments, and also mislocalize a proportion of the cargo destined for the vacuolar lumen to the vacuolar outer membrane (reviewed in references 2 and 8). The missorted proteins include vacuolar hydrolyases, which are improperly matured and capable of proteolyzing other cargoes that accumulate in the class E compartment (3, 54).
Our detailed analysis of two class E mutants lacking a key component of ESCRT complex E-II (snf8Δ) or E-III (snf7Δ) revealed significant reductions in activation by nucleus-localized Gcn4. Subsequent genetic analysis of class E mutants suggested that transcriptional attenuation in snf7Δ cells likely results from proteolysis of cargo proteins in the class E compartment, rather than the inability to transport cargo via ILVs per se. This and other findings described below led us to propose that impaired delivery of MVB cargo originating in the Golgi complex (class C/D mutants) or having this cargo rapidly proteolyzed in the aberrant class E compartment (snf7Δ and snf8Δ mutants) are key conditions of MVB dysfunction that lead to a strong reduction in transcriptional activation by Gcn4.
MATERIALS AND METHODS
Plasmid constructions.
All plasmids used in this study are listed in Table 1. Primers used in plasmid or yeast strain constructions are listed in Table 2. To construct the high-copy-number plasmid with GCN4-EGFP3, a SalI-EcoRI fragment with GCN4-EGFP3 from pKN85/p3233 was cloned into YEplac195 to produce pHQ1483. pKN85 was constructed by inserting a PCR-amplified BglII fragment containing the EGFP3 open reading frame (ORF) at the BglII site located just before the GCN4 stop codon in p1203 (pCD48-2). The resulting GCN4-EGFP fusion contains a silent T-to-G change at Ala codon 72 in EGFP3. pHQ1377 was constructed by inserting the EcoRI-SalI GCN4 fragment from p1208 into the corresponding sites of YEplac181.
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| YCplac 111 | sc LEU2 | 20 |
| pRS316-VPS15 | lcaURA3 VPS15 | 61 |
| pRS316-VPS15E200R | lc URA3 VPS15E200R | 61 |
| pRS316-VPS34 | lc URA3 VPS34 | 61 |
| pRS316-VPS15N736K | lc URA3 VPS34N736K | 61 |
| pNG9 | sc LEU2 VPS15 | This study |
| pNG10 | sc LEU2 VPS15E200R | This study |
| pNG11 | sc LEU2 VPS34 | This study |
| pNG12 | sc LEU2 VPS34N736K | This study |
| pRS316 | lc URA3 | 59 |
| pJT4 | lc URA3 SNF7 | 63 |
| pHQ1240 | GCN4 deletion plasmid | 71 |
| pHYC2 | hc URA3 with UASGCRE::CYC1-lacZ reporter | 30 |
| pKN7 | lc URA3 with HIS3-GUS reporter | 44 |
| p367 | lc URA3 with HIS4-lacZ reporter | 15 |
| p1208/pCD35-1 | sc URA3 with SalI-EcoRI fragment containing wild-type GCN4 | 16 |
| pHQ1377 | hc LEU2 with GCN4 | This study |
| pNKY1009 | TRP1 disruption plasmid | 1 |
| pRS424 | hc TRP1 | 59 |
| pRS424-HOG1 | hc TRP1 with HOG1 | 42 |
| pTXZA-GCN4-LexA | lc TRP1 with PGCN4-GCN41-281-lexA1-202 | G. Santangelo |
| pSH17-4 | hc HIS3 with PADH1-lexA1-202-GAL474-881 | 25 |
| pDB198 | sc TRP1 with PADH1-lexA1-202-VP16 | 6 |
| YEplac181 | hc LEU2 | 20 |
| pNG13 | hc LEU2 with PGCN4-GAL4768-881-GCN4210-281 | This study |
| pNG14 | hc LEU2 with PGCN4-VP16413-490-GCN4210-281 | This study |
| pSH18-34 | hc URA3 with lexAOP-GAL1-lacZ | 23 |
| YEplac195 | hc URA3 | 20 |
| p1203/pCD48-2 | sc URA3 with BglII site just before GCN4 stop codon | 16 |
| pKN85/p3233 | sc URA3 with SalI-EcoRI fragment containing GCN4-EGFP3 | This study |
| pHQ1483 | YEplac195 (hc URA3) with SalI-EcoRI fragment containing GCN4-EGFP3 from pKN85 | This study |
| pCGS286 | hc URA3 with GAL1-lacZ | Gerald Fink |
| pFA6a-HIS3MX6 | HIS3MX6 cassette | 40 |
lc, low copy number.
TABLE 2.
Primers used in this study
| Primer group and name | Purpose | Sequence | Reference or source |
|---|---|---|---|
| Primers for strain verification and construction | |||
| SNF7 A | Verification and construction of snf7Δ::kanMX4 | 5′-GTAAAGTAGTTATTGAGGTGGGGGT-3′ | 66 |
| SNF7 B | Verification of snf7Δ::kanMX4 | 5′-TATTTCATCTCCTAATTCGACTTGC-3′ | 66 |
| SNF7 C | Verification of snf7Δ::kanMX4 | 5′-GCAAGTCGAATTAGGAGATGAAATA-3′ | 66 |
| SNF7 D | Verification and construction of snf7Δ::kanMX4 | 5′-TTTTTCGTTATTTGGGTTTTAGTCA-3′ | 66 |
| HQ1431 | Forward primer for verification of snf7Δ::kanMX4 | 5′-CTAGTGATTTCGCCTCTATAAA-3′ | This study |
| HQ1432 | Reverse primer for verification of snf7Δ::kanMX4 | 5′-GAAGTGGGAAATCTCTTCCAA-3′ | This study |
| VPS4 A | Verification of vps4Δ::kanMX4 and vsp4Δ::URA3 | 5′-TAAGAGCAGTAAACCCGTTAGTGAC-3′ | 66 |
| VPS4 B | Verification of vps4Δ::kanMX4 | 5′-TGTAAACTTCGCTCTTATCAAATCC-3′ | 66 |
| VPS4 C | Verification of vps4Δ::kanMX4 | 5′-AAACAAGAAAACTAACACCATGCTC-3′ | 66 |
| VPS4 D | Verification of vps4Δ::kanMX4 and vsp4Δ::URA3 | 5′-TTTGTTACAGGAGTTAAATCAAGCC-3′ | 66 |
| VPS34 A | Verification and construction of vps34Δ::kanMX4 | 5′-TGAGGGTTTTATAGGATGTGTCATT-3′ | 66 |
| VPS34 B | Verification of vps34Δ::kanMX4 | 5′-AGAAGGGAATACATTTGACCCTATC-3′ | 66 |
| VPS34 C | Verification of vps34Δ::kanMX4 | 5′-TATTCGATTTTAAGAAGGAATGCAG-3′ | 66 |
| VPS34 D | Verification and construction of vps34Δ::kanMX4 | 5′-TGCTAATCTTGTTTGTTTAGCCTTC-3′ | 66 |
| VPS34 C1 | Forward primer for Verification of vps34Δ::kanMX4 in NGY11 | 5′-CTAATGTCTCCTGTATTTCCAATC-3′ | This study |
| VPS34 C2 | Reverse priemr for Verification of vps34Δ::kanMX4 in NGY11 | 5′-GATACGATGCTGCCGACTTGAG-3′ | This study |
| Kan B | Verification of kanMX4 associated deletions | 5′-CTGCAGCGAGGAGCCGTAAT-3′ | 66 |
| Kan C | Verification of kanMX4 associated deletions | 5′-TGATTTTGATGACGAGCGTAAT-3′ | 66 |
| FZP190 | Forward primer for verification of pep4::HIS3 | 5′-AGTAAAGAAGTTTGGGTAATTCGCT-3′ | This study |
| FZP191 | Reverse primer for verification of pep4::HIS3 | 5′-AGTGTTCTATGTTTGCCTTGATTTC-3′ | This study |
| FZP192 | Forward primer for amplification of pep4::HIS3 | 5′-CCTTTCTTCACTGAAGGTGGTCAC-3′ | This study |
| FZP193 | Reverse primer for amplification of pep4::HIS3 | 5′-AATGGTATCGTAACCCAAACCCAA-3′ | This study |
| FZP203 | Verification of HIS3MX associated deletions | 5′-GACGTTCCCTCAACCAAAGGTG-3′ | This study |
| FZP59 | Verification of HIS3MX associated deletions | 5′-AAATTCGCTTATTTAGAAGTGGCGC-3′ | This study |
| HQ1172 | Verification of pep4 deletion | 5′-CGCGAGCTCGCCCAATGGTACCAAC-3′ | This study |
| HQ521 | Verification of pep4 deletion | 5′-CGGAGATCTTTGCCTTCGTTTATCTTGCC-3′ | This study |
| N1 | Forward primer for PCR amplification of N-terminal portion of GCN4 ORF | 5′-GATCGGTCGACCCCGTCCTGTGGAT-3′ | This study |
| N3 | Reverse primer for PCR amplification of N-terminal portion of GCN4 ORF | 5′-GATCGAAGCTTAGCAAATAAACTTGG CTGATATTCG-3′ | This study |
| N7 | Forward primer for PCR amplification of GAL4AD coding sequences | 5′-GATCGAAGCTTGCCAATTTTAATCAAAGT GGGAATA-3′ | This study |
| N8 | Reverse primer for PCR amplification of GAL4AD coding sequences | 5′-GATCGAGATCTCTCTTTTTTTGGGTTTGG TGGG-3′ | This study |
| N4 | Forward primer for PCR amplification of C-terminal portion of GCN4 ORF | 5′-GATCGAGATCTAAACAGCGTTCGATTCCA CTTTCT-3′ | This study |
| N2 | Reverse primer for PCR amplification of C-terminal portion of GCN4 ORF | 5′-GATCGGAATTCTCTAGCTTAAAATGAATA G-3′ | This study |
| N9 | Forward primer for PCR amplification of VP16AD coding sequences | 5′-GATCGAAGCTTACCGCCCCCATTACCGAC GTC-3′ | This study |
| N10 | Reverse primer for PCR amplification of VP16AD coding sequences | 5′-GATCGAGATCTCCCCCCAAAGTCGTCAAT GCC-3′ | This study |
| PEP7A | Verification and construction of pep7Δ::kanMX4 | 5′-GCTAATGTAAAATAGCCAAGCACAT-3′ | 66 |
| PEP7B | Verification of pep7Δ::kanMX4 | 5′-GTCATACTGTGCGGAAAGATTAAGT-3′ | 66 |
| PEP7C | Verification of pep7Δ::kanMX4 | 5′-TTGCTAAATATGATAGCATGCAAAA-3′ | 66 |
| PEP7D | Verification and construction of pep7Δ::kanMX4 | 5′-CTGCTCCTCTTTCTTCTTAGCATTA-3′ | 66 |
| PEP7C1 | Forward primer for Verification of pep7Δ::kanMX4 in NGY13 and NGY14 | 5′-AAGGCTATCATACTCTAAGGGC-3′ | This study |
| PEP7C2 | Reverse primer for Verification of vps34Δ::kanMX4 in NGY13 and NGY14 | 5′-GTTACTACAGGGTGTAGCTTGG-3′ | This study |
| VPS15A | Verification and construction of vps15Δ::kanMX4 | 5′-AGACAGTACCATTGGAAAACTTGAG-3′ | 66 |
| VPS15B | Verification of vps15Δ::kanMX4 | 5′-CTTATACGTTGGAGAAAAGGTCGTA-3′ | 66 |
| VPS15C | Verification of vps15Δ::kanMX4 | 5′-TCAGTGACGTTTTTATTCCTACACA-3′ | 66 |
| VPS15D | Verification and construction of vps15Δ::kanMX4 | 5′-AATCTTTGTCCTCAACAAAATCAAC-3′ | 66 |
| VPS15C1 | Forward primer for verification of vps15Δ::kanMX4 in NGY12 | 5′-TGACCAAATACGTCCTTAAGGAC-3′ | This study |
| VPS15C2 | Reverse primer for verification of vps15Δ::kanMX4 in NGY12 | 5′-CAGATTTTCTTTGCGGTGATGATG-3′ | This study |
| FZP255 | Forward primer for verification of vps2Δ::kanMX4 in FZY720 | 5′-CGTATTCAGTTGAAGCGTATTTGTG-3′ | This study |
| FZP256 | Reverse primer for verification of vps2Δ::kanMX4 in FZY720 | 5′-CGACCAGAAGACGGTTGAAGAA-3′ | This study |
| FZP201 | Forward primer for verification of vps4Δ::kanMX4 in FZY718 | 5′-TTGAGCGAGACAACCTCAAACC-3′ | This study |
| FZP202 | Reverse primer for verification of vps4Δ::kanMX4 in FZY718 | 5′-CTCCGACGCCGACTTCTATTCC-3′ | This study |
| FZP251 | Forward primer for construction of snf7Δ::HIS3 | 5′-GGAAGTACGAGCTTCTAAAGGGTAAGATATTGTATTTCGGACGGAAGCAGCAGAAACATAACAGTATTGATAAATAAGGCCGGATCCCCGGGTTAATTAA-3′ | This study |
| FZP137 | Reverse primer for construction of snf7Δ::HIS3 | 5′-GTATATAAAAGAGCGTATACAGAACATGGAAAGTAAGAACACCTTTTTTTTTTCTTTCATCTAAACCGCATAGAACACGTGAATTCGAGCTCGTTTAAAC-3′ | This study |
| FZP253 | Forward primer for construction of vps20Δ::HIS3 | 5′-AAAAGAACAAAACATAGATAGTTTGGAAAAATAGTACAGACTGCTGAATTAACTCCACTTGGTGCTTTTGTATATATCGACGGATCCCCGGGTTAATTAA-3′ | This study |
| FZP254 | Reverse primer for construction of vps20Δ::HIS3 | 5′-ACAAATTCTATCAACGAAAAACCTGGAAGGAACCTATTTACATTCCCTTTATTTTTAATTTTGAAGCTACATACAGACATGAATTCGAGCTCGTTTAAAC-3′ | This study |
| Primers for ChIP assays | |||
| ON273 | POL1 forward PCR primer | 5′-GACAAAATGAAGAAAATGCTGATGCACC | 62 |
| ON274 | POL1 reverse PCR primer | 5′-TAATAACCTTGGTAAAACACCCTG | 62 |
| HQ605 | ARG1 UAS forward PCR primer | 5′-ACGGCTCTCCAGTCATTTAT | 62 |
| HQ607 | ARG1 UAS reverse PCR primer | 5′-GCAGTCATCAATCTGATCCA | 62 |
| ON265 | ARG1 TATA forward PCR primer | 5′-TAATCTGAGCAGTTGCGAGA | 52 |
| ON266 | ARG1 TATA reverse PCR primer | 5′-ATGTTCCTTATCGCTGCACA | 52 |
| HQ614 | ARG1 3′ ORF forward PCR primer | 5′-CAGATCTATGATCCAACCATC | 52 |
| HQ615 | ARG1 3′ ORF reverse PCR primer | 5′-CTCATCCATAGAGGATTCTGT | 52 |
To construct pNG9, pNG10, pNG11, and pNG12, SacI/SmaI-digested VPS15, VPS15E200R, VPS34, and VPS34N736K fragments from plasmids pRS316-VPS15, pRS316-VPS15E200R, pRS316-VPS34, and pRS316-VPS34N736K (61) were cloned into SacI/SmaI-digested single-copy (sc) plasmid vector YCplac111 to produce pNG9, pNG10, pNG11, and pNG12, respectively.
Construction of pNG13, encoding the Gal4AD-Gcn4DB fusion encoded by PGCN4-GAL4AD-GCN4DB involved PCR amplification of (i) an 890-nucleotide (nt) SalI-HindIII fragment containing the promoter, translational control element (TCE), and first 10 codons of the GCN4 coding sequence, from pHQ1377 using primers N1 and N3 (harboring the SalI and HindIII restriction sites, respectively), (ii) a 339-nt HindIII-BglII fragment encoding the Gal4 activation domain (AD; amino acids [aa] 768 to 881) from genomic DNA using primers N7 and N8 (harboring HindIII and BglII restriction sites, respectively), and (iii) a 1,348-nt BglII-EcoRI fragment encoding the Gcn4 DNA binding domain (DBD) from aa 210 to the stop codon from plasmid pHQ1377, using primers N4 and N2 (containing BglII and EcoRI restriction sites, respectively). The PCR-amplified fragments, digested with the appropriate enzymes, were gel purified and cloned into the SalI/EcoRI-digested vector YEplac181 to produce pNG13.
Construction of pNG14, encoding the VP16AD-Gcn4DB fusion encoded by PGCN4-VP16AD-GCN4DB, involved PCR amplification of (i) an 890-nt SalI-HindIII fragment containing the promoter, TCE, and first 10 codons of the GCN4 coding sequence from pHQ1377 using primers N1 and N3 (harboring the SalI and HindIII restriction sites, respectively), (ii) a 231-nt HindIII-BglII fragment encoding the VP16 AD (aa 413 to 490) from pDB198 using primers N9 and N10 (harboring HindIII and BglII restriction sites, respectively), and (iii) a 1,348-nt BglII-EcoRI fragment encoding the Gcn4 DBD from aa 210 to the stop codon from plasmid pHQ1377 using primers N4 and N2 (containing BglII and EcoRI restriction sites, respectively). The PCR-amplified fragments, digested with the appropriate enzymes, were gel purified and cloned into SalI/EcoRI-digested vector YEplac181 to produce pNG14. All the plasmid constructions were confirmed by restriction digestion and DNA sequencing of the complete PCR-amplified fragments.
Yeast strains.
All yeast strains used in this study are listed in Table 3. Haploid wild-type (WT) strains BY4741 and BY4742, diploid WT BY4743, and deletion derivatives thereof and isogenic homozygous deletion mutants (19) were purchased from Research Genetics. For all mutations summarized in Fig. 2, except for class E mutations, the deletions were confirmed in the haploid mutants by PCR amplification of genomic DNA (with the few exceptions indicated below in Table 4), and the SMs phenotypes were shown to be nearly identical in the haploid and homozygous diploid deletion mutants, providing strong evidence that the SMs phenotypes are conferred by the vps deletions. If not listed in Table 2, the primers used to verify all deletions were described previously (66). The SMs phenotypes of the haploid class E mutants were shown to be indistinguishable from those of the corresponding mutants constructed independently in isogenic strain JBY46 (9). We also verifed the snf7Δ and snf8Δ alleles by PCR analysis and showed that the SMs phenotypes of vps15Δ, vps34Δ, snf7Δ, and snf8Δ were complemented by episomal wild-type alleles.
TABLE 3.
Yeast strains used in this study
| Strain | Parent | Relevant genotypeb | Reference or source |
|---|---|---|---|
| BY4743a | NAd | MATα/MATahis3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0 | Research Genetics |
| BY4742a | NA | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Research Genetics |
| BY4741a | NA | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| 249a | BY4741a | gcn4Δ::kanMX4 | Research Genetics |
| 2783a | BY4741a | vps16Δ::kanMX4 | Research Genetics |
| 4105a | BY4741a | vps18Δ::kanMX4 | Research Genetics |
| 5305a | BY4741a | vps33Δ::kanMX4 | Research Genetics |
| 1812a | BY4741a | vps6Δ/pep12::kanMX4 | Research Genetics |
| 3682a | BY4741a | vps19Δ/pep7::kanMX4 | Research Genetics |
| 3236a | BY4741a | vps15Δ::kanMX4 | Research Genetics |
| 5149a | BY4741a | vps34Δ::kanMX4 | Research Genetics |
| 1580a | BY4741a | snf7Δ::kanMX4 | Research Genetics |
| 2826a | BY4741a | snf8Δ::kanMX4 | Research Genetics |
| 5381a | BY4741a | vps27Δ::kanMX4 | Research Genetics |
| 2730a | BY4741a | vps37Δ::kanMX4 | Research Genetics |
| 3416a | BY4741a | vps23Δ::kanMX4 | Research Genetics |
| 2763a | BY4741a | vps28Δ::kanMX4 | Research Genetics |
| 5325a | BY4741a | vps36Δ::kanMX4 | Research Genetics |
| 2580a | BY4741a | vps25Δ::kanMX4 | Research Genetics |
| 6211a | BY4741a | vps20Δ::kanMX4 | Research Genetics |
| 4850a | BY4741a | vps2Δ::kanMX4 | Research Genetics |
| 2744a | BY4741a | bro1Δ::kanMX4 | Research Genetics |
| 4004a | BY4741a | doa4Δ::kanMX4 | Research Genetics |
| 5588a | BY4741a | vps4Δ::kanMX4 | Research Genetics |
| 3043 | BY4741a | vps10Δ::kanMX4 | Research Genetics |
| 975a | BY4741a | vps29Δ::kanMX4 | Research Genetics |
| 2132a | BY4741a | vps30Δ::kanMX4 | Research Genetics |
| 5269a | BY4741a | vps38Δ::kanMX4 | Research Genetics |
| 1845a | BY4741a | vps5Δ::kanMX4 | Research Genetics |
| 2388a | BY4741a | vps17Δ::kanMX4 | Research Genetics |
| 4015a | BY4741a | vps41Δ::kanMX4 | Research Genetics |
| 6797a | BY4741a | vps53Δ::kanMX4 | Research Genetics |
| 3966a | BY4741a | vps54Δ::kanMX4 | Research Genetics |
| 4329a | BY4741a | vps3Δ::kanMX4 | Research Genetics |
| 405a | BY4741a | vps8Δ::kanMX4 | Research Genetics |
| 6495a | BY4741a | vps9Δ::kanMX4 | Research Genetics |
| 1865a | BY4741a | vps21Δ::kanMX4 | Research Genetics |
| 4462a | BY4741a | vps45Δ::kanMX4 | Research Genetics |
| 5072a | BY4741a | vps1Δ::kanMX4 | Research Genetics |
| 34063a | BY4743a | sac6Δ::kanMX4 | Research Genetics |
| 4572a | BY4741a | chc1Δ::kanMX4 | Research Genetics |
| 4797a | BY4741a | clc1Δ::kanMX4 | Research Genetics |
| 33266a | BY4743a | vma2Δ::kanMX4 | Research Genetics |
| 4929a | BY4741a | vma5Δ::kanMX4 | Research Genetics |
| 3267a | BY4741a | apg14Δ::kanMX4 | Research Genetics |
| 575a | BY4741a | vam4Δ::kanMX4 | Research Genetics |
| 33774a | BY4743a | vam6Δ::kanMX4 | Research Genetics |
| 4578a | BY4741a | vam7Δ::kanMX4 | Research Genetics |
| 2362a | BY4741a | vam3Δ::kanMX4 | Research Genetics |
| 1323a | BY4741a | lsb6Δ::kanMX4 | Research Genetics |
| 4320a | BY4741a | vps60Δ::kanMX4 | Research Genetics |
| 5981a | BY4741a | vps46Δ::kanMX4 | Research Genetics |
| 4130a | BY4741a | vta1Δ::kanMX4 | Research Genetics |
| 4836a | BY4741a | mvb12Δ::kanMX4 | Research Genetics |
| 1974a | BY4741a | kex2Δ::kanMX4 | Research Genetics |
| 1097a | BY4741a | apl5Δ::kanMX4 | Research Genetics |
| 5913a | BY4741a | apl6Δ::kanMX4 | Research Genetics |
| 7174a | BY4741a | apm3Δ::kanMX4 | Research Genetics |
| 3214a | BY4741a | gse1Δ::kanMX4 | Research Genetics |
| 5078a | BY4741a | gse2Δ::kanMX4 | Research Genetics |
| 6522a | BY4741a | gtr1Δ::kanMX4 | Research Genetics |
| 4793a | BY4741a | gtr2Δ::kanMX4 | Research Genetics |
| 4993a | BY4741a | ltv1Δ::kanMX4 | Research Genetics |
| H1486 | NA | MATα his1-29 leu2-3,112 ura3-52 ino1 <HIS4::lacZ ura3-52> | 65 |
| H1485 | NA | MATα his1-29 leu2-3,112 ura3-52 ino1 <HIS4::lacZ ura3-52> gcn2-508 | 65 |
| FZY512 | H1486 | MATα his1-29 leu2-3, −112 ura3-52 ino1 <HIS4::lacZ, ura3-52> snf7Δ::kanMX4 | This study |
| FZY718 | H1486 | MATα his1-29 leu2-3,112 ura3-52 ino1 <HIS4::lacZ ura3-52> vps4Δ::kanMX4 | This study |
| FZY720 | H1486 | MATα his1-29 leu2-3,112 ura3-52 ino1 <HIS4::lacZ ura3-52> vps2Δ::kanMX4 | This study |
| NGY11 | H1486 | MATα his1-29 leu2-3,112 ura3-52 ino1 <HIS4::lacZ ura3-52> vps34Δ::kanMX4 | This study |
| NGY12 | H1486 | MATα his1-29 leu2-3,112 ura3-52 ino1 <HIS4::lacZ ura3-52> vps15Δ::kanMX4 | This study |
| NGY1 | BY4741a | trp1Δ::hisGc | This study |
| NGY3 | 1580a | snf7Δ::kanMX4 trp1Δ::hisGc | This study |
| NGY4 | 3682a | vps19Δ/pep7::kanMX4 trp1Δ::hisGc | This study |
| NGY5 | 1812a | vps6Δ/pep12::kanMX4 trp1Δ::hisGc | This study |
| NGY6 | 3236a | vps15Δ::kanMX4 trp1Δ::hisGc | This study |
| NGY7 | 5149a | vps34Δ::kanMX4 trp1Δ::hisGc | This study |
| JBY46 | BY4741a | RIM20-GFP::HIS3MX6 | 9 |
| JBY207 | JBY46 | RIM20-GFP-HIS3MX6 rim8::kanMX4 | 9 |
| JBY208 | JBY46 | RIM20-GFP-HIS3MX6 rim9::kanMX4 | 9 |
| JBY209 | JBY46 | RIM20-GFP-HIS3MX6 rim13::kanMX4 | 9 |
| JBY210 | JBY46 | rim20::kanMX4 | 9 |
| JBY211 | JBY46 | RIM20-GFP-HIS3MX6 rim21::kanMX4 | 9 |
| JBY212 | JBY46 | RIM20-GFP-HIS3MX6 rim101::kanMX4 | 9 |
| JBY213 | JBY46 | RIM20-GFP-HIS3MX6 dfg16::kanMX4 | 9 |
| JBY115 | BY4741a | RIM20-GFP::HIS3MX6 vps4Δ::URA3 | 9 |
| JBY197 | BY4741a | RIM20-GFP::HIS3MX6 vps23Δ::kanMX4 | 9 |
| JBY198 | BY4741a | RIM20-GFP::HIS3MX6 vps28Δ::kanMX4 | 9 |
| JBY200 | BY4741a | RIM20-GFP::HIS3MX6 vps25Δ::kanMX4 | 9 |
| JBY201 | BY4741a | RIM20-GFP::HIS3MX6 vps36Δ::kanMX4 | 9 |
| JBY202 | BY4741a | RIM20-GFP::HIS3MX6 snf8Δ::kanMX4 | 9 |
| JBY203 | BY4741a | RIM20-GFP::HIS3MX6 vps20Δ::kanMX4 | 9 |
| JBY204 | BY4741a | RIM20-GFP::HIS3MX6 snf7Δ::kanMX4 | 9 |
| JBY133 | JBY115 | RIM20-GFP::HIS3MX6 vps4Δ::URA3 vps23Δ::kanMX4 | 9 |
| JBY176 | JBY115 | RIM20-GFP::HIS3MX6 vps4Δ::URA3 vps28Δ::kanMX4 | 9 |
| JBY136 | JBY115 | RIM20-GFP::HIS3MX6 vps4Δ::URA3 vps25Δ::kanMX4 | 9 |
| JBY182 | JBY115 | RIM20-GFP::HIS3MX6 vps4Δ::URA3 vps36Δ::kanMX4 | 9 |
| JBY185 | JBY115 | RIM20-GFP::HIS3MX6 vps4Δ::URA3 snf8Δ::kanMX4 | 9 |
| JBY139 | JBY115 | RIM20-GFP::HIS3MX6 vps4Δ::URA3 vps20Δ::kanMX4 | 9 |
| JBY142 | JBY115 | RIM20-GFP::HIS3MX6 vps4Δ::URA3 snf7Δ::kanMX4 | 9 |
| FZY709 | JBY115 | RIM20-GFP::HIS3MX6 vps4Δ::ura3 | This study |
| FZY711 | JBY133 | RIM20-GFP::HIS3MX6 vps4Δ::ura3 vps23Δ::kanMX4 | This study |
| FZY713 | JBY176 | RIM20-GFP::HIS3MX6 vps4Δ::ura3 vps28Δ::kanMX4 | This study |
| FZY715 | JBY142 | RIM20-GFP::HIS3MX6 vps4Δ::ura3 snf7Δ::kanMX4 | This study |
| HQY1232 | BY4741a | pep4::HIS3 | This study |
| FZY688 | 249a | gcn4Δ::kanMX4 pep4::HIS3 | This study |
| HQY1230 | 1580a | snf7Δ::kanMX4 pep4::HIS3 | This study |
| FZY694 | 5149a | vps34Δ::kanMX4 pep4::HIS3 | This study |
| FZY707 | 5588a | vps4Δ::kanMX4 pep4::HIS3 | This study |
| FZY693 | 3236a | vps15Δ::kanMX4 pep4::HIS3 | This study |
| FZY700 | 3682a | pep7Δ::kanMX4 pep4::HIS3 | This study |
| FZY690 | 1812a | pep12Δ::kanMX4 pep4::HIS3 | This study |
| FZY659 | 1580a | snf7Δ::kanMX4 gcn4Δ::hisGc | This study |
| FZY661 | 2826a | snf8Δ::kanMX4 gcn4Δ::hisGc | This study |
| NGY9 | 3236a | vps15Δ::kanMX4 gcn4Δ::hisGc | This study |
| NGY10 | 5149a | vps34Δ::kanMX4 gcn4Δ::hisGc | This study |
| 1044a | BY4741a | gal4Δ::kanMX4 | Research Genetics |
| RH144-3D | NA | MATaura3 leu2 his4 bar1-1 | 48 |
| RH268-1C | RH144-3D | MATaura3 leu2 his4 bar1-1 end4-1 | 48 |
| LCY14 | RH144-3D | MATaura3 leu2 his4 bar1-1 vps1Δ::LEU2 | 48 |
| LCY19 | RH144-3D | MATaura3 leu2 his4 bar1-1 end4-1 vps1Δ::LEU2 | 48 |
| BJ3511 | MATapep4::HIS3 ura3-52 (HIS GAL) | 33 | |
| ALHWT | NA | MATaleu2 his3 ura3 trp1 ade8 | 39 |
| ALH715 | ALHWT | MATaleu2 his3 ura3 trp1 ade8 wsc1::ADE8 wsc3::TRP1 | 39 |
| NGY13 | ALHWT | MATaleu2 his3 ura3 trp1 ade8 pep7::kanMX4 | This study |
| NGY14 | ALH715 | MATaleu2 his3 ura3 trp1 ade8 wsc1::ADE8 wsc3::TRP1 pep7::kanMX4 | This study |
| FZY721 | 5149a | vps34Δ::kanMX4 snf7Δ::HIS3 | This study |
| FZY724 | 5149a | vps34Δ::kanMX4 vps20Δ::HIS3 | This study |
| FZY727 | 2362a | vam3Δ::kanMX4 snf7Δ::HIS3 | This study |
| RSY249 | NA | MATahis4-619 ura3-52 sec18-1 | R. Schekman |
| RSY272 | NA | MATasec18-1 his4-619 ura3-52 sec18-1 | R. Schekman |
Purchased from Research Genetics.
HIS3* designates the HIS3 allele from Saccharomyces kluyveri.
hisG sequences from Salmonella enterica serovar Typhimurium.
NA, not applicable.
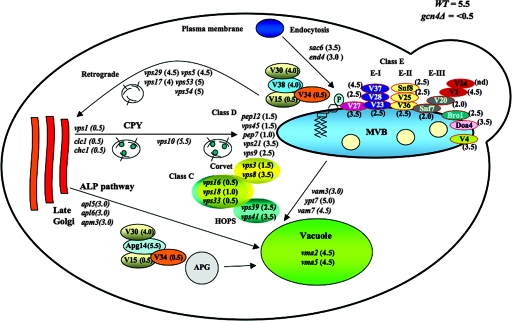
FIG. 2.
Summary of the functions of Vps factors in vesicular trafficking and the SMs/Gcn− phenotypes conferred by vps deletions. Functions of Vps factors in vesicular trafficking are depicted (see text for details), and SMs phenotypes of the corresponding deletion mutants are given in parentheses, as in Table 4, where WT cells have an SMs score of 5.5 and gcn4Δ cells are scored as <0.5.
TABLE 4.
SM sensitivity of vps mutants
| Deletiona | Alias(es) | Class or function(s)b | Relative SM resistancec |
|---|---|---|---|
| None (WT) | NAh | NA | 5.5 |
| gcn4Δ | AAS3, ARG9 | Transcriptional activator | <0.5 |
| vps10Δ | PEP1, VPT1 | A Vps, transport receptor for carboxypeptidase Y | 5.5 |
| vps29Δ | PEP11 | A Vps, retromer complex for retrograde MVB-to-Golgi transport | 4.5 |
| vps30Δ | APG6, VPT30, ATG6 | A Vps, associated with Vps34/Vps15 in PtdIns[3] kinase complex II for PtdIns[3]P synthesis at endosomes, supports retromer recruitment | 4.0 |
| vps38Δ | VPL17 | A Vps, associated with Vps34, Vps15, and Vps30 in PtdIns[3] kinase complex II for PtdIns[3]P synthesis at endosomes, supports retromer recruitment | 4.0 |
| vam7Δ | VPS43 | B Vps, SNARE complex for vesicle fusion at vacuole | 4.5 |
| vps5Δ | GRD2, PEP10, VPT5 | B Vps, retromer complex for MVB-to-Golgi transport | 4.5 |
| vps17Δ | PEP21 | B Vps, retromer complex for MVB-to-Golgi transport | 4.0 |
| vam6Δf | CVT4, VPL18, VPL22, VPS39 | B Vps, component of HOPS tethering complex for vesicle fusion at vacuole | 2.5 |
| vps41Δ | CVT8, FET2, SVL2, VAM2, VPL20 | B Vps, supports Ap-3 in ALP pathway from Golgi complex to vacuole, component of HOPS tethering complex for vesicle fusion at vacuole | 3.5 |
| vps53Δ | NA | B Vps, Golgi-associated retrograde complex for vesicle fusion at Golgi apparatus | 5.0 |
| vps54Δ | LUV1, CGP1, TCS3 | B Vps, Golgi-associated retrograde complex for vesicle fusion at Golgi apparatus | 5.0 |
| vps16Δ | SVL6, VAM9, VPT16 | C Vps, component of class C Vps tethering complex shared between HOPS and CORVET complexes for vesicle fusion at vacuole and MVB, respectively | 0.5 |
| vps18Δ | VAM8, PEP3, VPT18 | C Vps, component of class C Vps tethering complex shared between HOPS and CORVET complexes for vesicle fusion at vacuole and MVB, respectively | 1.0 |
| vps33Δ | CLS14, MET27, PEP14, SLP21, VAM5, VPL25, VPT33 | C Vps, SM protein, component of class C Vps tethering complex shared between HOPS and CORVET complexes for vesicle fusion at vacuole and MVB, respectively | 0.5 |
| vps3Δ | PEP6, VPL3, VPT17 | D Vps, component of CORVET tethering complex for vesicle fusion at MVB | 1.5 |
| pep12Δ | VPL6, VPS6, VPT13 | D Vps, QA-SNARE, in multiple SNARE complexes, vesicle fusion at MVB from Golgi complex, EE, vacuole | 1.5 |
| vps8Δ | FUN15, VPT8 | Dd Vps, component of CORVET tethering complex for vesicle fusion at MVB | 3.5 |
| vps9Δ | VPL31, VPT9 | D Vps, Vps21 (Rab) GEF, vesicle fusion at MVB from Golgi apparatus | 2.5 |
| pep7Δ | VAC1, VPL21, VPS19, VPT19 | D Vps, Vps21 (Rab) effector, vesicle fusion at MVB from Golgi apparatus | 1.0 |
| vps21Δ | VPS12, VPT12, YPT21, YPT51 | Rab GTPase, vesicle fusion at MVB from Golgi apparatus | 3.5 |
| vps34Δ | END12, PEP15, VPL7, VPT29 | D Vps, PtdIns[3] kinase associated with Vps15, in complex II synthesizes PtdIns[3]P at endosomes, supports ESCRT-0 and retromer function at MVB | 0.5 |
| vps15Δ | GRD8, VAC4, VPL19 | D Vps, protein kinase associated with Vps34, in complex II stimulates PtdIns[3]P synthesis at endosomes, supports ESCRT-0 and retromer function at MVB | 0.5 |
| vps45Δ | STT10, VPL28 | D Vps, SM protein, binds QA-SNARE Pep12, vesicle fusion at MVB from Golgi apparatus | 1.5 |
| vps27Δ | GRD11, SSV17, VPL23, VPT27, DID7 | E Vps, ESCRT-0, MVB-to-vacuole transport | 3.5 |
| vps37Δ | SRN2, SRN10 | E Vps, ESCRT-I, MVB-to-vacuole transport | 4.5 |
| vps23Δ | STP22 | E Vps, ESCRT-I, MVB-to-vacuole transport | 2.5 |
| vps28Δ | VPT28 | E Vps, ESCRT-I, MVB-to-vacuole transport | 2.5 |
| mvb12Δ | NA | E Vps, ESCRT-I subunit required to stabilize oligomers of the ESCRT-I core complex | 5 |
| vps36Δ | GRD12, VAC3, VPL11 | E Vps, ESCRT-II, MVB-to-vacuole transport | 2.5 |
| snf8Δ | VPS22 | E Vps, ESCRT-II, MVB-to-vacuole transport | 2.5 |
| vps25Δ | VPT25 | E Vps, ESCRT-II, MVB-to-vacuole transport | 2.5 |
| vps20Δ | CHM6, VPT20, VPL10 | E VPS, ESCRT-III, MVB-to-vacuole transport | 2.0 |
| snf7Δ | DID1, VPS32, RNS4 | E Vps, ESCRT-III, MVB-to-vacuole transport | 2.0 |
| vps2Δ | DID4, GRD7, REN1, VPL2, VPT14, CHM2 | E Vps, ESCRT-III, MVB-to-vacuole transport | 4.5 |
| did2Δ | FTI1, CHM1, VPS46 | E Vps, ESCRT-III associated, MVB-to-vacuole transport | 4.5 |
| bro1Δ | LPF2, VPS31, ASI6, NPI3 | E Vps, ESCRT-III associated, MVB-to-vacuole transport | 2.5 |
| vta1Δ | NA | E Vps, ESCRT-III disassembly, MVB-to-vacuole transport | 5.0 |
| vps60Δ | MOS10, CHM5 | E Vps, ESCRT-III disassembly, MVB-to-vacuole transport | 5.0 |
| vps4Δ | CSC1, END13, GRD13, VPL4, VPT10, DID6 | E Vps, AAA-ATPase, ESCRT-III disassembly, MVB-to-vacuole transport | 3.5 |
| doa4Δe | DOS1, MUT4, NPI2, SSV7, UBP4 | Deubiquitinates MVB cargo during MVB-to-vacuole transport | 3.5 |
| vps1Δg | GRD1, LAM1, SPO15, VPL1, VPT26 | F Vps dynamin-related GTPase, Golgi-to-MVB transport | 0.5 |
| chc1Δg | SWA5 | Clathrin HC, Golgi complex to MVB, also promotes ESCRT-0 in MVB-to-vacuole transport, enhances endocytosis | 0.5 |
| clc1Δ | SCD4 | Clathrin LC, Golgi complex to MVB, also promotes ESCRT-0 in MVB-to-vacuole transport, enhances endocytosis | 0.5 |
| ypt7Δ | AST4, VAM4 | Rab GTPase for vesicle fusion at vacuole via HOPS complex | 5.0 |
| sac6Δf,g | ABP67 | Actin filament bundling protein essential for endocytic vesicle formation in receptor-mediated endocytosis | 3.5 |
| sla2Δg | END4, MOP2 | Liquid phase and receptor-mediated endocytosis | 3.0 |
| apg14Δ | CVT12, ATG14 | Involved in macroautophagy as component of PtdIns[3] kinase complex I with Vps30, Vps15, and Vps34 | 5.5 |
| vma2Δf,g | VAT2 | Vacuolar membrane H+-ATPase subunit 5 | 4.5 |
| vma5Δ | CSL5, VAT3 | Vacuolar membrane H+-ATPase subunit 5 | 4.5 |
| vam3Δ | PTH1 | Syntaxin-related protein required for vacuolar assembly | 3.0 |
| apl5Δ | YKS4 | Delta adaptin-like subunit of the clathrin-associated protein complex | 3.0 |
| apl6Δ | YKS5 | β3-like subunit of the yeast AP-3 complex; | 3.0 |
| apm3Δ | YKS6 | Mu3-like subunit of the clathrin-associated protein complex (AP-3) | 3.0 |
| lsb6Δ | NA | Type II phosphatidylinositol 4-kinase | 5.0 |
| slm4Δ | EGO3, NIR1, GSE1 | Component of the GSE complex, required for sorting of general amino acid permease Gap1 from MVB to plasma membrane | 4.5 |
| meh1Δ | EGO1, GSE2 | Component of the GSE complex, required for sorting of general amino acid permease Gap1 from MVB to plasma membrane | 4.5 |
| gtr1Δ | NA | Component of the GSE complex, required for sorting of general amino acid permease Gap1 from MVB to plasma membrane | 4.5 |
| gtr2Δ | NA | Component of the GSE complex, required for sorting of general amino acid permease Gap1 from MVB to plasma membrane | 4.5 |
| ltv1Δ | YKL2 | Component of the GSE complex, required for sorting of general amino acid permease Gap1 from MVB to plasma membrane | 4.0 |
| kex2Δg | QDS1, VMA45, SRB1 | Calcium-dependent serine protease involved in activation of proproteins of secretory pathway | 5.0 |
All strains were obtained from Research Genetics and are isogenic to BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ).
Resistance to SM was measured as described for Fig. 1C, and relative growth was assigned by considering both colony size, the maximum dilution where colonies were visible, and the number of days required to observe visible colonies.
vps8Δ is listed in both the class A and class D mutants (8).
doa4Δ was not classified as a vps mutant by Bowers et al., but it is grouped with the class E mutants based on its function in the ESCRT pathway.
Homozygous diploid deletion mutant; the haploid mutant was unavailable.
Demonstration of equivalent SMs phenotypes in haploid versus homozygous diploid mutants was not possible with strains available from Research Genetics.
NA, not applicable.
Deletion of GCN4 using plasmid pHQ1240 was conducted and verified as described previously (71). The snf7Δ::kanMX, vps15Δ::KanMX, vps34Δ::kanMX, vps2Δ::KanMX, and vps4Δ::KanMX alleles were isolated from the chromosomal DNA of strains 1580, 3236, 5149, 4850, and 5588, respectively, by PCR amplification and introduced into H1486 (65) to produce the G418-resistant strains FZY512, NGY12, NGY11, FZY720, and FZY718, respectively. The pep7Δ::KanMX allele was PCR amplified from the chromosomal DNA of strain 3682 and introduced into ALHWT and ALH715 (39) to produce NGY13 and NGY14, respectively. The strains JBY115, JBY133, JBY176, and JBY142 were subjected to a mock transformation and plated on 5-fluoroorotic acid medium to isolate their ura3 derivatives. The pep4Δ::HIS3 allele was amplified from chromosomal DNA of BJ3511 and introduced into BY4741, 249, 1580, 5588, 5149, 3236, 3682, 1812, and 2826 to produce their pep4::HIS3 derivatives. The trp1Δ::hisG allele was introduced into BY4741, 1580, 3682, 1812, 3236, and 5149 using pNKY1009. The snf7Δ::HIS3 and vps20Δ::HIS3 cassettes were PCR amplified from plasmid pFA6a-HIS3MX6 (40) and introduced into 5149 to produce FZY721 and FZY724, respectively. The snf7Δ::HIS3 cassette amplified as above was introduced into 1580 to produce FZY727.
Biochemical methods.
The reporter gene assays were performed as described previously (62). For Western analysis, whole-cell extracts (WCEs) were prepared with tricholoracetic acid, as described previously (55), and analyzed with antibodies against Gcd6 (12), Gcn4 (affinity purified as described below), or lexA (Abcam, Inc.). Northern analysis was conducted as described previously (37). Chromatin immunoprecipitation (ChIP) assays were conducted as described previously (62, 72) using the primers in Table 2 and antibodies against Gcn4 (described above) and Rpb3 (Neoclone).
Affinity purification of Gcn4 antibodies. (i) Purification of His6-Gcn4.
A transformant of Escherichia coli strain BL21(DE3) carrying pCD377-2/p1934 encoding His6-Gcn4 was grown to saturation overnight, diluted (1:50) into four 1-liter volumes of LB plus ampicillin in four 4-liter flasks, grown for 3 h, and induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG; to 1 mM) and incubating for another 3 h. Cells were harvested by centrifugation and His6-Gcn4 purified with Ni-nitrilotriacetic acid (NTA) resin following the vendor's instructions (Qiagen).
(ii) Preparation of Gcn4 affinity column.
Purified His6-Gcn4 (∼30 mg) was dialyzed overnight against coupling buffer (0.5 M NaCl, 0.1 M NaHCO3 [pH 8.3]). A 1.5-g aliquot of CNBr-activated Sepharose 4B (Pharmacia) was washed with 200 ml ice-cold 1 mM HCl followed by 40 ml coupling buffer. Dialyzed His6-Gcn4 was mixed with the HCl-washed resin and mixed on a nutator at room temperature for 3 h (for coupling). The resin was collected by centrifugation, resuspended in 10 ml of 200 mM glycine (pH 8.0), and mixed at room temperature for 2 h (for blocking). After blocking, the resin was washed twice with 10 volumes of coupling buffer and twice with phosphate-buffered saline (PBS), and the His6-Gcn4-coupled resin was stored under PBS at 4°C. The coupling efficiency was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the His6-Gcn4 levels in the supernatant before and after coupling.
(iii) Affinity purification of Gcn4 antibody.
A 10-ml aliquot of anti-Gcn4 polyclonal antiserum (HL2871) raised in rabbits against recombinant full-length Gcn4 (Covance) was mixed with 2 volumes of PBS, added to 6 ml of the His6-Gcn4-coupled resin, and mixed in the cold for 2 h. The resin was poured into a column (1.2-cm diameter), washed with 10 bed volumes of PBS, 10 volumes of 2× PBS, 0.05% Tween 20, and 5 volumes of PBS. Affinity-purified Gcn4 antibodies were eluted with 3 volumes of 0.1 M diethylamine (pH 11.5), collecting 1-ml fractions, which were mixed immediately with an equal volume of 1 M Tris-HCl (pH 7.0) to neutralize the pH. Fractions with an A280 greater than 0.05 were pooled and concentrated ∼15-fold with a Centriprep (Amicon). The concentrated antibody was dialyzed against PBS in the cold, divided into 50-μl aliquots, and stored at −80°C.
Pulse-chase analysis of Gcn4 synthesis and degradation rates.
For the pulse-chase analysis, modifications of a protocol described previously (38) were employed, as follows. Cells were cultured in synthetic complete (SC) medium lacking isoleucine and valine (SC-Ile/Val) to an optical density at 600 nm of 0.6 to 0.8, harvested by centrifugation, washed once with SC-Ile/Val lacking methionine (SC-Ile/Val-Met), resuspended in 0.5 ml of SC-Ile/Val-Met, transferred to a 1.5-ml screw-cap tube containing SM (at a final concentration of 1.0 μg/ml), and incubated for 15 min at 30°C. One mCi of [35S]methionine/cysteine labeling mix was added, and cells were incubated another 15 min before harvesting in a microcentrifuge and resuspending in 5 ml prewarmed SC-Ile/Val containing 10 mM methionine and 10 mM cysteine. A 1-ml aliquot was removed immediately (for the 0-min chase) and the remainder was incubated for 20 min, taking 1-ml aliquots at the appropriate times of chase, and each aliquot was added to 170 μl of 1.85 M NaOH, 7.4% 2-mercaptoethanol in a 1.5-ml screw-cap tube and placed on ice for 10 min. After adding 70 μl 100% trichloroacetic acid (TCA) and incubating on ice for 10 min, the extracts were centrifuged at 4°C for 5 min and the pellets were washed with ice-cold acetone and dried in a SpeedVac centrifuge. The dried pellets were resuspended in 120 μl of 2.5% SDS, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride by vortexing, and the suspensions were heated to >90°C for 1 min and cleared by centrifugation. Incorporation of label was measured by combining 2 μl of extract with 20 μl bovine serum albumin (BSA; 10 mg/ml) and 1 ml 5% TCA, incubating on ice for 15 min, collecting the precipitate on Whatman GF/C glass fiber filters, and measuring the radioactivity by scintillation counting. Aliquots of extract containing equal amounts of radioactivity (1 × 107 cpm) were combined with 1 ml of immunoprecipitation (IP) buffer (50 mM Na-HEPES [pH 7.5], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride) containing 1 mg/ml BSA and 1 μl affinity-purified anti-Gcn4 antibody and mixed by rotating at 4°C for 2 h. Twenty μl of a 50% slurry of protein A-agarose beads pretreated with IP buffer containing BSA (1 mg/ml) was added, and mixing continued for 2 h. The beads were washed with cold IP buffer containing 0.1% SDS (500 μl; three times), resuspended in loading buffer, heated, and resolved by SDS-PAGE using 4 to 20% gels. The gel was dried and subjected to autoradiography, and the 35S-labeled Gcn4 was quantified by phosphorimaging analysis.
Live-cell imaging by fluorescence microscopy.
To stain cellular DNA, 4′,6-diamidino-2-phenylindole (DAPI; Sigma) was applied to cells as a 1/400 dilution of a 1-mg/ml aqueous solution after the prescribed period of Ile/Val starvation with SM (0.5 μg/ml). After a 5-min incubation, cells were washed and transferred to fresh medium containing SM and examined. Distribution of Gcn4-GFP in living cells was analyzed with an oil immersion, 100×/1.4 numerical aperture objective using the Olympus Cell R detection and analyzing system based on the motorized Olympus IX-71 inverted microscope, a Hammamatsu Orca/ER digital camera, and the following highly specific mirror units: (i) enhanced green fluorescent protein (EGFP) filter block U-MGFPHQ, excitation maximum at 488 nm, emission maximum at 507 nm; (ii) a DAPI filter block U-MNUA2, excitation maximum at 440 nm, emission maximum at 500 to 520 nm. The Cell R system enabled us to obtain up to nine optical sections through the yeast cell. Nomarski (differential interference contrast) optics were used to record transmission images.
RESULTS
Deletions of multiple VPS genes confer sensitivity to sulfometuron.
We screened the entire library of viable haploid deletion mutants produced by the Saccharomyces Genome Deletion Project (19) for SM sensitivity and identified a large number of vps mutants with Gcn− phenotypes (Fig. 1C and Table 4). Most of the mutants showing the strongest SM sensitivity are class C or D vps mutants, which have defects in vesicle fusion at the MVB. This includes mutants lacking the Q-SNARE Pep12/Vps6, the “SM” proteins Vps33 and Vps45, Rab GTPase Vps21, and Rab effector Pep7/Vps19 (8). It also includes Vps18, Vps16, Vps3, and Vps8, which reside with Vps33 in the tethering complex CORVET, which is thought to link the membranes prior to vesicle fusion (49) (summarized in Fig. 2). Vesicle fusion at the MVB depends on synthesis of phosphotidyl inositol-3 phosphate (PtdIns[3]P) in the membrane by lipid kinase Vps34 and associated protein kinase Vps15. Vesicle budding at the Golgi apparatus involves the clathrin coat, and Golgi complex-to-endosome trafficking requires the dynamin-related GTPase Vps1 (a class F factor) (8). Deletion mutants lacking each of these latter proteins also exhibit strong SMs phenotypes (Fig. 1C, Table 4, and Fig. 2), suggesting that defects in vesicular transport from the Golgi apparatus to MVB impair the GAAC.
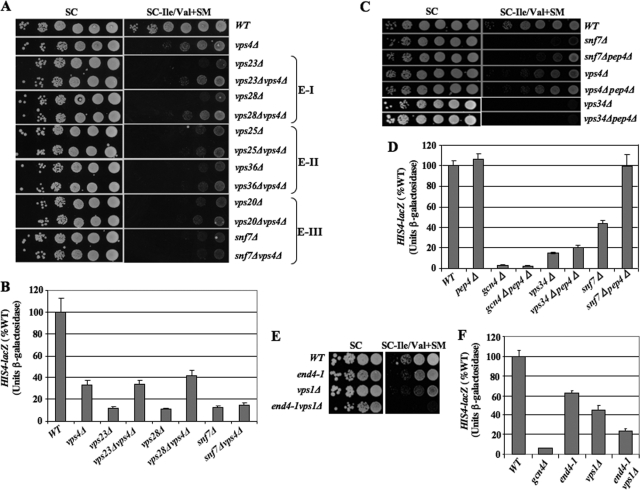
Class E mutants lack subunits of the ESCRT complexes, which bind ubiquitinated cargo proteins on the MVB outer membrane for delivery to ILVs and subsequent transport to the vacuole lumen. Vps27 binds ubiquitin moieties of cargo proteins, membrane-associated PtdIns[3]P, and clathrin and helps recruit E-I components Vps37, Vps28, Vps23, and Mvb12. E-I activates the E-II heterotrimer Vps25, Vps36, and Snf8/Vps22, which in turn recruits the E-III subunits Snf7, Vps20, Vps2, and Vps24. The Vps2-Vps24 subcomplex then recruits the AAA-ATPase Vps4, which functions to deliver the cargo proteins, deubiquitinated by Doa4, into ILVs for transport to the vacuole (Fig. 1B). We found that many of the class E vps mutants exhibited SMs phenotypes, albeit less severe than for most class C/D mutants (Fig. 1C, Table 4, and Fig. 2). Surprisingly, vps2Δ and vps4Δ, defective for the last step of the ESCRT pathway, have significantly weaker SMs phenotypes compared to snf7Δ, snf8Δ, and several other mutants lacking subunits of E-I, E-II, or E-III (Fig. 1C and 2). This suggests that the functions of E-I, E-II, and E-III in associating with cargo proteins on the MVB outer membrane are more important than the Vps4-dependent delivery of cargo via ILVs to the vacuole lumen to achieve a robust GAAC response. We pursue this unexpected finding below in greater detail.
To verify that class E mutants are less SM sensitive than class C/D vps mutants, we measured the doubling times of selected mutants in liquid SC medium lacking Ile and Val and containing SM at 0.5 μg/ml. We found that the class D mutants pep7Δ, pep12Δ, vps15Δ, and vps34Δ had doubling times of between 8.5 and 13.5 h, comparable to that of the gcn4Δ strain (8.9 h). By contrast, the class E mutants snf7Δ and vps4Δ had doubling times of 7.2 h and 6.2 h, respectively, intermediate between those of the class D mutants and the isogenic WT strain (4.2 h) (data not shown).
Vps proteins are required for efficient PIC assembly by chromatin-bound Gcn4.
The abundance on the cell surface of certain plasma membrane proteins is downregulated by endocytosis and vesicular transport to the MVB and vacuole, including the general amino acid permease (Gap1) (57). We considered the possibility that the SMs phenotype of vps mutants could result from elevated uptake or retention of SM, owing to defective regulation of a permease (like Gap1) to thereby produce an exaggerated level of Ile/Val starvation that can't be counteracted by induction of Gcn4 and its target genes. We took several approaches to eliminate this possibility and to demonstrate that transcriptional activation by Gcn4 is truly impaired in vps mutants. For these studies, we focused on vps15Δ, vps34Δ, pep12Δ, and pep7Δ as exemplars of class D mutants with strong SMs phenotypes and the class E mutants snf7Δ and snf8Δ.
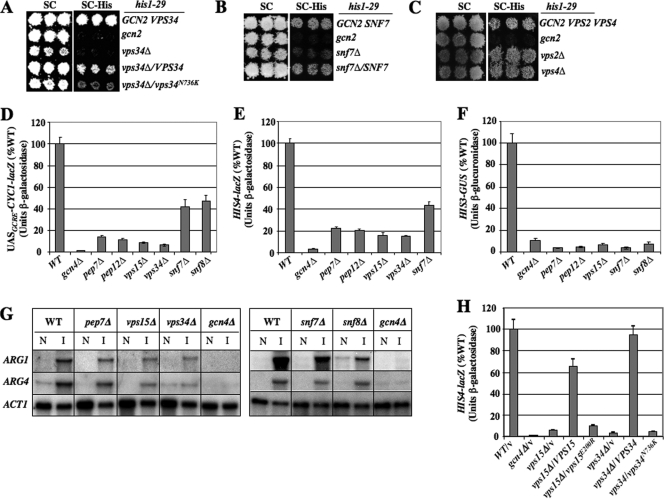
First, we asked whether vps34Δ and snf7Δ impair Gcn4 induction of the HIS1 gene in response to histidine starvation of his1-29 cells on medium lacking histidine. Because his1-29 reduces, but doesn't abolish, activity of the first enzyme of histidine biosynthesis, his1-29 cells can grow without exogenous histidine, due to transcriptional activation of his1-29 by Gcn4. Hence, mutations that impair Gcn4 induction, like gcn2Δ, render his1-29 cells unable to grow without histidine (67). As shown in Fig. 3A, vps34Δ resembles gcn2Δ by conferring a histidine requirement in his1-29 cells, and the His− phenotype of the vps34Δ his1-29 double mutant is reduced by plasmid-borne VPS34 but not by catalytically defective Vps34-N736K (61). Similar results were obtained by combining vps15Δ or snf7Δ with his1-29 (Fig. 3B and data not shown), although as expected, the His− phenotype of snf7Δ his1-29 cells is less severe than that of vps34Δ his1-29 cells (cf. Fig. 3A and B). By contrast, vps2Δ and vps4Δ did not produce a His− phenotype in the his1-29 background (Fig. 3C), in accordance with their weak SMs phenotypes (Fig. 1C and Table 4). The fact that vps34Δ, vps15Δ, and snf7Δ all impair the Gcn4-mediated response to histidine deficiency imposed without an inhibitor of histidine biosynthesis in the medium argues against a contribution of altered permease regulation to the phenotypes of these mutants. By exacerbating the histidine requirement of his1-29 cells and conferring SM sensitivity in otherwise-WT cells (Fig. 1C), these vps mutations confer key phenotypes of bone fide Gcn− mutants (26, 67).
FIG. 3.
Class D and E vps mutations impair transcriptional activation by Gcn4. (A) his1-29 strains H1486 (GCN2 VPS34), H1485 (gcn2-508), and NGY11 (vps34Δ), all transformed with empty vector (top three rows), and vps34Δ strain NGY11 transformed with either VPS34 plasmid pNG11 (fourth row) or vps34-N736K plasmid pNG12 (fifth row), were grown on SC-Ura and replica plated to SC-His-Ura at 30°C. (B) The same experiment as in panel A, except with his1-29 strains H1486, H1485, and snf7Δ strain FZY512 transformed with empty vector (top three rows) and FZY512 transformed with SNF7 plasmid pJT4 (bottom row). (C) The same experiment as in panel A, except with his1-29 strains H1486, H1485, FZY720 (vps2Δ), and FZY718 (vps4Δ) transformed with empty vector. (D to F) Transformants of vpsΔ mutants described in Fig. 1C harboring episomal Gcn4-dependent reporters were grown under inducing conditions (SC-Ile/Val-Ura containing 0.5 μg/ml SM), and β-galactosidase or β-glucuronidase activities were assayed in WCEs. Means and standard errors from three or more cultures are plotted as percentages of WT values. (D) UASGCRE-CYC1-lacZ reporter plasmid pHYC2; (E) HIS4-lacZ reporter plasmid p367; (F) HIS3-GUS reporter plasmid pKN7. (G) Total RNA isolated from the indicated strains grown under the same inducing conditions (I) or noninducing (N) conditions (SC-Ile/Val) was subjected to Northern analysis using probes for ARG1, ARG4, and ACT1 mRNA. (H) Transformants of strains described in Fig. 1C with empty vector (v) or sc plasmids with the indicated VPS15 or VPS34 alleles were analyzed as for panel E.
We demonstrated next that the vps mutations impair activation of Gcn4-dependent transcriptional reporters. The UASGCRE-CYC1-lacZ reporter, containing Gcn4 binding sites (UASGCRE) upstream of the CYC1 promoter, is induced by Gcn4 in cells treated with SM (62). Induction of this reporter is diminished to ∼10% of WT in the class D vps mutants and to ∼40% of WT in snf7Δ and snf8Δ cells (Fig. 3D). Similar results were obtained for HIS4-lacZ and HIS3-GUS reporters harboring the native HIS4 and HIS3 5′ noncoding regions (Fig. 3E and F). We also observed decreased induction of two mRNAs produced by the arginine biosynthetic genes ARG1 and ARG4, relative to the nontarget gene ACT1, although to a lesser extent than observed for the reporter constructs in the class D mutants (Fig. 3G). As discussed below, Mitchell and colleagues previously reported that mRNAs of 12 different Gcn4 target genes are downregulated in snf7Δ cells at elevated pH (7), providing independent evidence for diminished Gcn4 function in class E mutants. Importantly, the impaired induction of HIS4-lacZ expression in the vps15Δ and vps34Δ mutants was complemented by plasmid-borne VPS15 or VPS34, but not by alleles encoding catalytically inactive forms of these kinases (61) (Fig. 3H), thus confirming that the kinase activities of both proteins are required to stimulate GAAC.
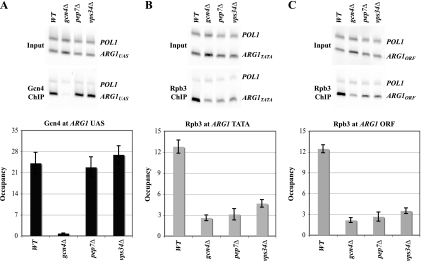
To probe the molecular mechanism of transcriptional attenuation in vps mutants, we conducted chromatin immunoprecipitation assays to measure binding of Pol II and Gcn4 at ARG1. SM treatment of WT cells leads to rapid increases in Gcn4 binding to the UAS (Fig. 4A, WT), recruitment of Pol II subunit Rpb3 to the promoter TATA element (Fig. 4B, WT), and increased Pol II occupancy at the 3′ end of the coding sequences (Fig. 4C, WT), all in agreement with previous findings (21). Importantly, Gcn4 occupancy of the UAS was unaffected by class D mutations pep7Δ and vps34Δ (Fig. 4A), indicating that induction of Gcn4 and its nuclear localization and UAS binding activity occur at levels sufficient for high-level Gcn4 occupancy of the ARG1 UAS in these mutants. By contrast, pep7Δ and vps34Δ elicit strong reductions in Pol II (Rpb3) occupancies at TATA and 3′ ORF sequences, comparable to the effect of deleting Gcn4 itself (Fig. 4B and C). Nearly identical results were obtained for class D mutants pep12Δ and vps15Δ (data not shown). The reduced Pol II occupancy in the ORF provides direct evidence that Gcn4-dependent transcription of ARG1 is diminished, and the reduced TATA occupancy by Pol II implies that Gcn4-stimulated PIC assembly is attenuated in class D mutants.
FIG. 4.
Class D vps mutations impair PIC assembly by chromatin-bound Gcn4. Strains described in Fig. 1C were induced with SM (0.5 μg/ml) for 30 min and treated with formaldehyde. Extracted, sonicated chromatin was immunoprecipitated with antibodies against Gcn4 (A) or Rpb3 (B and C). DNA extracted from the immunoprecipitates (ChIP) or starting chromatin (input) was PCR amplified in the presence of [33P]dATP with primers for the ARG1 UAS (A), TATA (B), or 3′ ORF sequences (C) and separately with primers for the POL1 ORF (as a nonspecific control). PCR products were quantified by phosphorimaging, and the ratios of ChIP-to-input signals for ARG1 were normalized for the corresponding ratios for POL1, to yield occupancy values. Averages and standard errors from two PCR amplifications for each of two independent immunoprecipitates from two independent cultures are plotted.
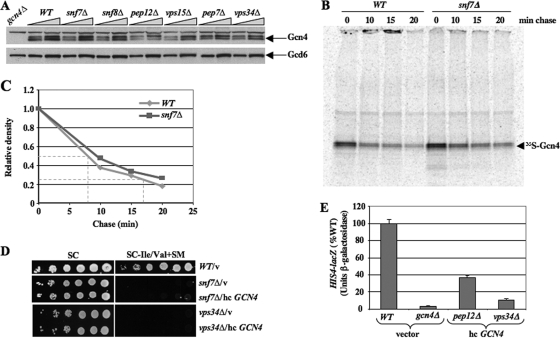
We attempted to conduct a ChIP analysis in snf7Δ and snf8Δ cells but found that Gcn4 and Rpb3 are degraded in chromatin extracts from these strains (data not shown). Accordingly, we asked whether the activation defect in these mutants results from reduced Gcn4 expression levels. Western analysis of Gcn4 in WCEs prepared under denaturing conditions (by TCA extraction) revealed no significant differences in the induced levels of Gcn4 (relative to Gcd6) among WT, snf7Δ, and snf8Δ cells (Fig. 5A). We also analyzed the rate of synthesis and turnover of Gcn4 in snf7Δ cells by pulse-chase analysis with [35S]methionine/cysteine. We observed no differences in the extent of 35S labeling of Gcn4 during the 15-min pulse, or of its rate of degradation during the chase, between WT and snf7Δ cells (Fig. 5B and C), thus indicating that snf7Δ does not alter Gcn4 synthesis or degradation in vivo.
FIG. 5.
Deletion of SNF7 does not alter Gcn4 synthesis or stability. (A) Western blot analysis of strains described in Fig. 1C induced with 1 μg/ml SM for 30 min. WCEs were extracted under denaturing conditions and analyzed with anti-Gcn4 and anti-Gcd6 antibodies. Aliquots differing by twofold were loaded in adjacent lanes. (B and C) WT and snf7Δ strains were grown in SC-Ile/Val, resuspended in SC-Ile/Val-Met with SM (1 μg/ml) for 15 min, and labeled in the same medium with [35S]methionine/cysteine for 15 min. Cells were resuspended in SC-Ile/Val (containing both Met and Cys at 10 mM), and aliquots were removed immediately (0 min) and at the indicated times (chase). Aliquots of WCEs containing 1 × 107 cpm were immunoprecipitated with anti-Gcn4 antibodies, immunocomplexes were collected with protein A-agarose beads and resolved by SDS-PAGE, and the [35S]Gcn4 bands were quantified by phosphorimaging (C). (D) The indicated strains transformed with vector or hc GCN4 plasmid pHQ1377 were tested for SM sensitivity, as in Fig. 1C. (E) The indicated strains transformed with vector or hc GCN4 plasmid pHQ1377 were analyzed for HIS4-lacZ expression as in Fig. 3E.
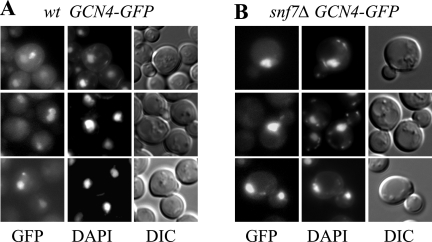
We asked next whether the snf7Δ mutation impairs nuclear localization of a Gcn4-GFP fusion. This fusion is expressed from a high-copy-number (hc) plasmid under the native GCN4 promoter and complements the SMs phenotype of a gcn4Δ strain indistinguishably from hc GCN4+ (data not shown). Although overexpressing Gcn4-GFP was required to visualize its fluorescence, this approach should be valid, because neither hc GCN4 nor the hc GCN4-GFP construct suppresses the SMs phenotype of a snf7Δ mutant (Fig. 5D and data not shown). As expected, we observed extensive colocalization of Gcn4-GFP with nuclei in WT cells induced with SM (Fig. 6A, cf. GFP with DAPI). Importantly, nuclear localization of Gcn4-GFP is evident in the majority of snf7Δ cells under the same conditions (Fig. 6B and data not shown). Thus, Snf7 is not required for nuclear localization of Gcn4. The normal expression and nuclear localization of Gcn4 in snf7Δ cells is consistent with the possibility that class E mutations, like class D mutations, impair the ability of UAS-bound Gcn4 to activate transcription.
FIG. 6.
Deletion of SNF7 does not reduce nuclear localization of Gcn4. WT (A) and snf7Δ (B) strains described in Fig. 1C containing episomal GCN4-GFP (pHQ1483) were cultured in SC-Ile/Val and treated with SM (0.5 μg/ml) for 2 h. Cells were stained with DAPI for 5 min and visualized by using the Olympus Cell R fluorescence microscopy system. The images represent single optical layers of the optically sectioned cells.
In the Western analysis described above, we observed moderate (≤50%) reductions in Gcn4 abundance in class D vps mutants (Fig. 5A and data not shown). However, the ChIP data in Fig. 4 indicated strong UAS occupancies by Gcn4 in these strains. Hence, the reduced cellular levels of Gcn4 cannot explain the activation defects of class D mutants. Supporting this conclusion, introducing additional copies of GCN4 on a high-copy-number plasmid does not suppress the SMs phenotype of vps34Δ or other class D mutants (Fig. 5D and data not shown). Introducing hc GCN4 also does not rescue activation of the HIS4-lacZ reporter (Fig. 5E), even though it restores near-WT levels of Gcn4 (relative to Gcd6) in these mutants (data not shown).
vps mutations reduce the functions of Gcn4, Gal4, and VP16 activation domains.
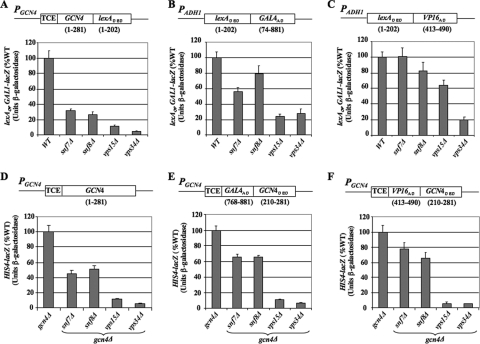
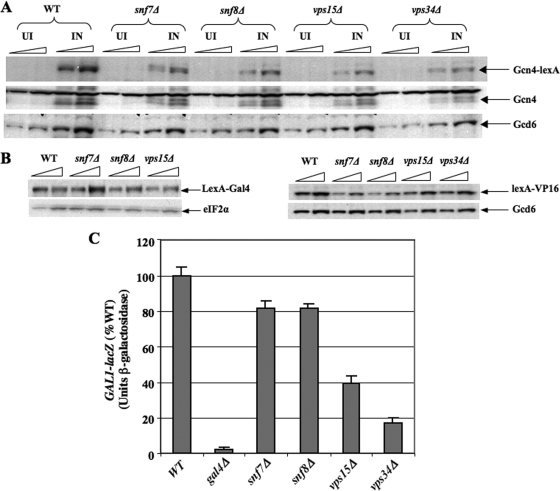
The results of our ChIP analysis suggested that the activation function, not DNA binding activity, of Gcn4 is impaired in class D vps mutants. To support this conclusion and extend it to class E mutants, we examined the ability of a Gcn4-lexA fusion to activate transcription of a lacZ reporter containing lexA binding sites as the only UAS upstream of the GAL1 promoter. The Gcn4-lexA fusion was expressed under the GCN4 promoter and TCE. This lexAop-GAL1-lacZ reporter is strongly induced by Gcn4-lexA on SM treatment of WT cells (≈50-fold) (data not shown), and this induction is reduced by factors of 3 to 4 in snf7Δ and snf8Δ mutants and factors of 10 to 20 in vps15Δ and vps34Δ mutants (Fig. 7A), similar to the effects of these mutations on activation by native Gcn4 (Fig. 3D to F). Western analysis revealed <2-fold reductions in the steady-state level of Gcn4-lexA in snf7Δ and snf8Δ cells and ≈2.5-fold reductions in vps15Δ and vps34Δ cells, compared to WT (Fig. 8A). Thus, it seems unlikely that the impaired activation by Gcn4-lexA in these mutants results merely from diminished expression of this hybrid activator. These data support our conclusion that the activation function, rather than DNA binding activity, of Gcn4 is enhanced by Vps factors.
FIG. 7.
vps mutations impair transcriptional activation by several activation domains. (A to C) Expression of lexAOP-GAL1-lacZ reporter (pSH18-34) was measured in the indicated strains (described in Fig. 1C) containing activator fusions depicted schematically and encoded by the low-copy-number (lc) plasmid pTXZA-GCN4-LexA (A), hc plasmid pSH17-4 (B), and sc plasmid pDB198 (C). Cells were grown in SC-Ile/Val-Ura-Trp with 1 μg/ml SM (A), SC-Ura-His (B), or SC-Ura-Trp (C). (D to F) Cells were grown in SC-Ile/val-Ura-Leu with 1 μg/ml SM, and expression of the HIS4-lacZ reporter on p367 was measured in the gcn4Δ strains FZY659 (snf7Δ), FZY661 (snf8Δ), NGY9 (vps15Δ), and NGY10 (vps34Δ) expressing activators encoded by hc plasmids pHQ1377 (D), pNG13 (E), and pNG14 (F).
FIG. 8.
vps mutations have modest effects on expression of hybrid activators and impaired transcriptional activation by Gal4 in class D mutants. (A) Western analysis of TCA-extracted WCEs was conducted on cells grown identically to those analyzed for β-galactosidase activity in Fig. 7A, probing with antibodies against LexA, Gcn4, and Gcd6, as indicated. Aliquots differing by twofold were loaded in adjacent lanes. (B) Western analysis of the LexA-Gal4 (left) or LexA-VP16 (right) proteins conducted on aliquots of cells of the indicated genotype that were analyzed for β-galactosidase activity in Fig. 7. (C) The GAL1-lacZ reporter containing the native GAL1 promoter on plasmid pCGS286 was assayed in strains of the indicated genotypes described in Fig. 1C, plus isogenic gal4Δ strain 1044, after growing the cells in SC-Ura with 2% raffinose as carbon source and then adding galactose to 2% and incubating for 6 h.
We also analyzed activation by proteins containing the Gcn4 DBD fused to the ADs of Gal4 or herpesvirus VP16, expressed in gcn4Δ cells under the GCN4 promoter and TCE. Activation of HIS4-lacZ by both hybrid activators was strongly reduced by the vps15Δ and vps34Δ mutations, comparable to their effects on activation by native Gcn4 expressed from a hc plasmid (Fig. 7, cf. E and F with D). Activation by the hybrid activators was also reduced in snf7Δ and snf8Δ cells (Fig. 7E and F), but not to the same extent observed for native Gcn4 (Fig. 7D).
Finally, we examined the completely heterologous hybrid activators containing the LexA DBD and Gal4 or VP16 ADs. Activation of the lexAop-GAL1-lacZ reporter by the LexADBD-Gal4AD hybrid activator was modestly impaired by snf7Δ and snf8Δ (factor of ≤2) but substantially reduced by vps15Δ and vps34Δ (factor of ∼4) (Fig. 7B). Activation of the same reporter by the LexADBD-VP16AD activator was not significantly affected by snf7Δ and snf8Δ, weakly impaired by vps15Δ (by ∼40%), and markedly reduced by vps34Δ (∼5-fold) (Fig. 7C). Western analysis indicated only small (<2-fold) reductions in the levels of these hybrid activators, which cannot account for their reduced functions in the vps15Δ and vps34Δ mutants (Fig. 8B). We went on to analyze induction of a GAL1-lacZ reporter by native Gal4 in galactose medium and observed activation defects in the vps mutants (Fig. 8C) similar to those shown above for hybrid activators harboring the Gal4 AD (Fig. 7B and E). Together, these data suggest that the Gcn4 activation domain is not unique in requiring class D and class E Vps factors for full activity; however, the degree of dependence on Vps functions, especially class E proteins, is greater for Gcn4 than for the Gal4 or VP16 ADs.
The Gcn− phenotype of vps mutants does not involve the ASR pathway.
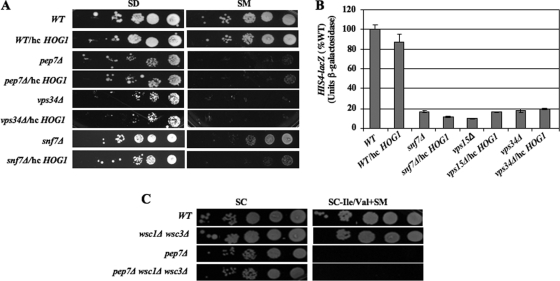
Certain secretory-defective (sec) mutants impaired for vesicular transport impair nuclear import of various proteins in the “arrest of secretion response” (ASR) (43). Thus, we considered whether the Gcn− phenotypes of vps mutants might involve the ASR. Inconsistent with this idea, the vps18 and vps45 mutants have strong SMs phenotypes (Table 4) but were shown previously not to elicit the ASR (43). In addition, our ChIP and Gcn4-GFP imaging data imply that nuclear entry of Gcn4 is not impaired in Gcn− vps mutants. To rule out further the involvement of the ASR, we asked whether the Gcn− phenotypes of vps mutants are diminished by overexpressing the mitogen-activated protein kinase Hog1, shown previously to suppress the ASR (43). Overexpressing Hog1 from an hc plasmid does not suppress the SMs phenotypes of class D mutants pep7Δ, pep12Δ, vps15Δ, and vps34Δ or class E mutant snf7Δ (Fig. 9A and data not shown). Consistently, hc HOG1 does not rescue activation of the HIS4-lacZ reporter in these strains (Fig. 9B and data not shown).
FIG. 9.
The Gcn− phenotypes of vps mutants do not involve the ASR. (A) Strains of the indicated genotypes described in Fig. 1C were transformed with vector or hc HOG1 plasmid pRS424-HOG1 and tested for SM sensitivity as in Fig. 1C, except using synthetic medium with minimal supplements (SD). (B) HIS4-lacZ expression was measured in transformants of strains from panel A and strain 3236, harboring p367, as described for Fig. 3E. (C) Strains ALHWT, ALH715, NGY13, and NGY14 were tested for SM sensitivity at 0.5 μg/ml as described for Fig. 1C.
It was shown previously that the plasma membrane protein Wsc1, involved in signal transduction in response to changes in cell wall integrity, is required for a strong ASR (43), and that deleting WSC1 and WSC3 simultaneously abrogates the repression of ribosomal protein genes provoked by a block to the secretory pathway (39). By contrast, we found that the double mutation wsc1Δ wsc3Δ does not ameliorate the SMs phenotype of class D mutant pep7Δ (Fig. 9C). We conclude that the ASR does not make an important contribution to diminished activation by Gcn4 in the vps mutants described above and that attenuation of GAAC in these mutants is functionally distinct from the downregulation of ribosomal protein genes in response to secretory defects.
The Gcn− phenotype of snf7Δ cells likely involves PrA-dependent degradation of MVB cargo.
We showed above that snf7Δ mutants have a more severe SMs phenotype than do vps2Δ or vps4Δ strains (Fig. 1C), even though all of these mutants are defective for protein trafficking via ILVs from the MVB to vacuole. This paradox is reminiscent of the specialized function carried out by Snf7 in the response to alkaline pH in the RIM pathway. Transcriptional repressor Rim101 is proteolytically activated at the MVB in a manner requiring Snf7 and other ESCRT proteins, but independently of E-III subunits Vps24 and Vps2, the AAA-ATPase Vps4, Bro1, and Doa4 (69). Moreover, the requirements for all ESCRT proteins except Snf7 in processing Rim101 were suppressed by vps4Δ (9), which was attributed to the fact that Snf7 accumulates on MVB membranes in cells lacking Vps4 due to the impaired recycling of E-III complexes from the MVB (4). This allows Snf7 to recruit Rim20 (or Bro1) to the MVB independent of other ESCRT factors in the vps4Δ background (9), which implies that the stimulatory functions of E-I and E-II factors in promoting Snf7 assemblies at the MVB are bypassed in vps4Δ cells. At alkaline pH, Snf7 recruits Rim20 to the MVB in place of Bro1, and Rim20 then recruits Rim101 and, most likely, the Rim101-activating protease (9, 68). Thus, Snf7 has a regulatory role in the RIM pathway distinct from protein trafficking by providing a platform for Rim101 processing on the MVB outer membrane, and the other ESCRT proteins support this auxiliary function primarily by enhancing Snf7 recruitment to the MVB (7, 9, 27).
We found that none of the deletion mutants specifically impaired for Rim101 processing (rim8, −9, −13, −20, −21, or −101 and dfg16) (7, 69) exhibit SMs phenotypes (data not shown). Moreover, bro1Δ confers a strong SMs phenotype (Table 4) but, as mentioned above, is dispensable for the RIM pathway. Hence, the functions of ESCRT proteins in GAAC appear to be unrelated to Rim101 processing. It is intriguing, however, that vps4Δ reduces the SM sensitivity conferred by E-I mutations vps23Δ and vps28Δ in double mutants to yield the less severe SMs/Gcn− phenotype seen in vps4Δ single mutants (Fig. 10A). By contrast, vps4Δ has only a small effect on the SMs/Gcn− phenotypes of E-II mutations vps36Δ, vps25Δ, and snf8Δ and no effect on the E-III mutants vps20Δ and snf7Δ (Fig. 10A and data not shown). Similarly, vps4Δ impairs transcriptional activation of HIS4-lacZ by Gcn4 to a lesser extent than do vps23Δ and vps28Δ. Furthermore, vps4Δ suppresses the stronger activation defects conferred by these E-I mutations to yield a level of HIS4-lacZ expression in the double mutants comparable to the vps4Δ single mutant, whereas vps4Δ does not increase HIS4-lacZ expression in snf7Δ cells (Fig. 10B).
FIG. 10.
Evidence that the strongest Gcn− phenotypes in class E mutants result from failure to recruit the Snf7/Vps20 E-III subunits and protect MVB cargo from PrA-dependent degradation. (A) Mutants derived from JBY46 of the indicated genotype (described in Table 4) were tested for SM sensitivity as in Fig. 1C. (B) HIS4-lacZ expression was measured in p367 transformants of strains JBY46, FZY709, JBY197, FZY711, JBY198, FZY713, JBY204, and FZY715, as for Fig. 3E. (C) Strains BY4741, 1580, HQY1230, 5588, FZY707, 5149, and FZY694 were tested for SM sensitivity as in Fig. 1C. (D) Strains BY4741, HQY1232, 249, FZY688, 5149, FZY694, 1580, and HQY1230 were analyzed for HIS4-lacZ expression from p367, as for Fig. 3E. (E and F) Strains RH144-3D, RH268-1C, LCY14, and LCY19 were tested for SM sensitivity (E) and HIS4-lacZ expression from p367 (F) as in Fig. 1C and 3E, respectively, except cells were cultured at 21°C.
As noted above, there is evidence that vps4Δ reduces the requirement for E-I and E-II factors in assembling Snf7/Vps20 E-III subcomplexes at the MVB. Consistent with this, it was shown that overexpressing the E-II complex partially suppresses the MVB sorting phenotype of E-I mutants (3). Accordingly, the partial suppression of the Gcn− phenotypes of E-I mutants by vps4Δ shown in Fig. 10A and B implies that the functions of E-I factors in enhancing the assembly of E-II and E-III complexes at the MVB is more critical for GAAC than is delivery of MVB cargo to the vacuole lumen. Given that E-II complexes also stimulate E-III assembly, it appears that impairment of GAAC is greatest in the class E mutants that not only disrupt protein sorting but also fail to assemble the Snf7/Vps20 E-III subcomplex on endosomal membranes. Hence, although the RIM pathway is not involved in GAAC, the two regulatory systems share a greater requirement for Snf7 on the MVB outer membrane than for Vps4 function in cargo delivery via ILVs.
We wished to explore further why GAAC is impaired more severely by the failure to assemble the Snf7/Vps20 E-III subcomplex at the MVB than by merely disrupting protein trafficking to the vacuole lumen by vps4Δ. Emr and colleagues have shown that vps4Δ and other ESCRT mutants that accumulate Snf7/Vps20 on endosomal membranes differ from snf7Δ and vps20Δ with regard to proteolysis of the MVB cargo vacuolar proteinase Cps1, a portion of which accumulates in the class E compartment in ESCRT mutants. (The remainder of Cps1 is mislocalized to the limiting membrane of the vacuole in class E mutants.) Removing Snf7/Vps20 from endosomal membranes increases the rate of proteolytic maturation of pro-Cps1, possibly by eliminating steric hindrance of the maturing protease by direct binding of Snf7/Vps20 to pro-Cps1 (3). Hence, the more severe SMs phenotype of snf7Δ might result from proteolytic cleavage of Cps1, or other MVB cargo, that accumulates in the class E compartment and is not protected by association with Snf7/Vps20 in the manner that occurs in vps4Δ cells. To test this, we examined the effect of deleting PEP4 on the activation defect in snf7Δ cells, as PEP4-encoded PrA is essential for maturation of multiple vacuolar hydrolyases (33) and PrA is known to accumulate in the class E compartment along with PrB and other cargo that reaches the vacuole via the endosome in class E mutants (3, 8, 54).
Interestingly, deleting PEP4 partially suppresses the SMs phenotype of the snf7Δ strain but has little effect on the weaker SMs phenotype of vps4Δ cells and no effect on the stronger SMs phenotypes of vps15Δ, vps34Δ, pep7Δ, or pep12Δ mutants (Fig. 10C and data not shown). Consistent with this, pep4Δ suppresses the defective transcriptional activation of HIS4-lacZ expression in snf7Δ cells, but not in vps34Δ or vps4Δ cells (Fig. 10D and data not shown). (The extent to which pep4Δ suppresses the HIS4-lacZ activation defect in snf7Δ cells varied among different experiments and was often incomplete, consistent with its partial suppression of the SMs phenotype in snf7Δ cells [Fig. 10C].) These results support the idea that PrA-dependent proteolysis of MVB cargo in the class E compartment of snf7Δ cells contributes to its stronger Gcn− phenotype, compared to the simple impairment of protein trafficking in vps4Δ cells.
Evidence that trafficking from the Golgi complex to the MVB is critical for GAAC.
As many Vps proteins are required for delivery of cargo proteins to the MVB from the plasma membrane by endocytosis, we asked whether impairment of receptor-mediated endocytosis alone elicits a Gcn− phenotype. At odds with this possibility, deleting SAC6 does not produce a strong SMs phenotype (Table 4 and Fig. 2), whereas Sac6 is required for endocytosis (34). It is also noteworthy that vps1Δ confers a strong SMs phenotype (Table 4), as this dynamin-related GTPase is thought to be required for vesicular trafficking from the Golgi complex to the late endosome in the CPY pathway but is not crucial for endocytosis (8). Indeed, in vps1Δ mutants, certain Golgi and vacuolar membrane proteins reach the vacuole circuitously by mislocalization to the plasma membrane followed by transport to the MVB via endocytosis. This alternative pathway is impaired by disrupting endocytosis with the end4-1 Ts− mutation, so that all detectable Golgi complex-to-vacuole trafficking is blocked at 36°C and growth is strongly impaired above 21°C in a vps1Δ end4-1 double mutant (48). Remarkably, we found that both the SMs phenotype and defective activation of HIS4-lacZ expression in a vps1Δ single mutant are exacerbated by end4-1 at 21°C (Fig. 10E and F). This provides strong evidence that a defect in Golgi complex-to-MVB trafficking is involved in downregulating GAAC, as GAAC is reduced in vps1Δ cells lacking conventional Golgi complex-to-MVB transport but diminished further in the vps1Δ end4-1 double mutant, where the alternative endocytic pathway for Golgi complex-to-MVB transport is additionally impaired.
DISCUSSION
In this report we show that transcriptional activation by Gcn4 is attenuated in vivo by numerous mutations that disrupt vesicular protein trafficking at the late endosome/MVB. Our analysis of class D mutants pep12Δ, pep7Δ, vps15Δ, and vps34Δ, all defective for vesicular transport from the Golgi complex to MVB, revealed marked defects in induction of reporters and mRNAs representing various Gcn4 target genes and a decreased association of Pol II with ARG1 coding sequences. Importantly, UAS occupancy of Gcn4 is not reduced at ARG1, indicating that Gcn4 enters the nucleus and binds to target sequences in chromatin at near-WT levels in these mutants. However, Pol II occupancy of the ARG1 promoter is lower in all four class D mutants, indicating that UAS-bound Gcn4 cannot efficiently stimulate PIC assembly. This shows that transcription initiation, rather than mRNA stability, is impaired in the class D vps mutants. Our finding that activation by the hybrid protein Gcn4-lexA is strongly impaired in vps15Δ and vps34Δ strains supports the idea that the activation function, not DNA binding activity, of Gcn4 is being attenuated. These mutations also diminish the functions of hybrid activators containing Gal4 or VP16 ADs, and of native Gal4, although not to the same extent observed for the Gcn4 AD. Most class E vps mutants, defective for vesicular transport from MVB to vacuole, also confer SMs phenotypes but ones that are less severe than for the class C/D mutants with the strongest Gcn− phenotypes. Analysis of snf7Δ and snf8Δ mutants revealed impaired induction of Gcn4-dependent mRNAs and reporters, despite WT levels of Gcn4 protein and nuclear localization of Gcn4-GFP, and also reduced activation by Gcn4-lexA. Thus, these class E vps mutations most likely also diminish transcriptional activation by chromatin-bound Gcn4.
As summarized in Fig. 2, the vps mutants with the strongest Gcn− phenotypes are class C/D mutants lacking factors required for vesicle fusion at the MVB, including lipid kinase Vps34 and its associated protein kinase, Vps15, Q-SNARE Pep12, SM protein Vps45, Rab effector Pep7/Vps19, and four of six components of the CORVET tethering complex, Vps16, Vps18, Vps33, and Vps3 (8, 49). (Our deletion library lacks the vps11Δ mutant for the final CORVET subunit.) Deletions of Vps1 and clathrin subunits also confer strong Gcn− phenotypes, consistent with their roles in Golgi complex-to-MVB trafficking (8). The fact that eliminating the Rab GTPase (Vps21) and its GEF (Vps9) results in weaker Gcn− phenotypes than deletions of other factors involved in Golgi complex-to-MVB transport (Fig. 2) might be explained by the existence of other related Rab GTPases in yeast, Ypt52 and Ypt53, which could partially substitute for Vps21 (60).
Interestingly, most vps mutations that specifically impair vesicular transport to the vacuole do not provoke strong Gcn− phenotypes (Fig. 2). Thus, deletions eliminating SNARE component Vam7 and Rab GTPase Ypt7, both involved in vesicle fusion at the vacuole, produce weaker Gcn− phenotypes than do pep12Δ, vps21Δ, and vps9Δ, which remove a SNARE component, a Rab GTPase, and GEF, respectively, working at the MVB (Fig. 2) (8). Furthermore, eliminating Vps39/Vam6, the Ypt7 exchange factor of the HOPS tethering complex, confers a weaker Gcn− phenotype than does eliminating the analogous subunit of the CORVET tethering complex, Vps3 (Fig. 2). This is significant because the HOPS complex functions in vesicle fusion primarily at the vacuole, while CORVET operates mostly at the MVB (49). The fact that deleting CORVET-specific Rab effector VPS8 confers a relatively weak Gcn− phenotype might be explained by the fact that the analogous HOPS subunit, Vps41, can replace Vps8 in the intermediate i-CORVET complex in vps8Δ cells (49). Together, these comparisons suggest that GAAC is more strongly attenuated by defects in vesicular transport to the MVB than to the vacuole. This inference is supported by the fact that eliminating Vps4, which impairs proper trafficking to the vacuole from the MVB, has a relatively weak Gcn− phenotype (Fig. 2). Deleting subunits of the vacuolar ATPase, including Vma2 and Vma5, also does not confer SM sensitivity (Fig. 2), ruling out the possibility that a defect in vacuolar acidification is responsible for the Gcn− phenotype in vps mutants.
The strongest overall Gcn− phenotype was observed with deletion of the PtdIns[3] kinase Vps34, which resides with its activating protein kinase Vps15 in several different complexes. One complex containing Vps30 and Vps38 supports PtdIns[3]P formation at endosomes and is important for retrograde trafficking from MVB to the Golgi apparatus by the retromer complex, a process that serves to retrieve components of the CPY pathway. A second complex contains Vps30 and Apg14 and mediates transport of cargo directly to the vacuole for degradation in macroautophagy (8, 36). Whereas vps30Δ produces a moderate SMs phenotype, vps38Δ and apg14Δ produce little or no SM sensitivity, respectively (Fig. 2). Thus, these two complexes cannot account for the critical role of Vps34 in GAAC. This is not surprising, since neither complex is critical for vesicular transport from the Golgi complex to MVB. Importantly, it was shown that vps34Δ, but not vps30Δ, impairs PrA and PrB trafficking to the vacuole, implying that Vps34 functions in vesicular transport outside of the Vps30-containing complexes. Consistent with this, deleting VPS30 or VPS38 produced only a ≈20% reduction, and deletion of APG14 had no effect, on Vps34 kinase activity (36). Presumably, additional complexes containing Vps34 would have to be disrupted to obtain the strong Gcn− phenotype of the vps34Δ mutant.
One concern could be that the Gcn− phenotype of vps34Δ results from the fact that numerous vesicular trafficking events are impaired in this mutant. However, the sec18-1 mutation also impacts multiple intracellular transport steps (reference 10 and references therein), yet we found that incubation of temperature-sensitive sec18-1 cells at the restrictive temperature for 3 h had little (<10%) effect on HIS4-lacZ induction by SM (data not shown). We also found that deletion of LSB6, encoding a nonessential type II phosphatidylinositol 4-kinase (24), did not confer SM sensitivity. In addition, pep7Δ, which eliminates a Vps21 effector of vesicle fusion at the MVB, produces a defect in transcriptional activation by UAS-bound Gcn4 comparable to that observed for vps34Δ. Hence, it appears that Vps34 functions in conjunction with other class C/D Vps factors in maintaining a robust GAAC response.
The class E vps mutants with the strongest Gcn− phenotypes affect subunits of the ESCRT complexes that sort monoubiquitinated cargo proteins on the MVB membrane for delivery to ILVs. These include mutants lacking subunits of E-I (vps23Δ and vps28Δ), all three subunits of E-II (snf8Δ, vps36Δ, and vps25Δ), and the Vps20/Snf7 components of the E-III complex. These mutants have in common a failure to recruit the Snf7/Vps20 E-III subunits to the MVB outer membrane. The fact that eliminating Vps4 produces weaker SMs phenotypes than was observed for snf7Δ and snf8Δ mutants shows that blocking proper delivery of MVB cargo to the vacuole lumen (as in vps4Δ cells) is not sufficient for the more-pronounced Gcn− defects conferred by snf7Δ and snf8Δ. It is known that inactivating Vps4 leads to accumulation of Snf7/Vps20 on the MVB membrane, owing to impaired recycling of E-III subunits to the cytoplasm (4, 9). Hence, our finding that vps4Δ suppresses the stronger SMs of the E-I mutations vps23Δ and vps28Δ but fails to suppress snf7Δ and vps20Δ mutations implies that defective recruitment of Snf7/Vps20 (and possibly E-II) underlies the stronger Gcn− phenotypes of E-I mutations vps23Δ and vps28Δ compared to vps4Δ. Together, these results indicate that eliminating the Snf7/Vps20 E-III subunits from the MVB outer membrane is required, and not merely disrupting vesicular transport from the MVB, for the strongest Gcn− phenotypes displayed in class E mutants.
We ruled out the possibility that the particularly strong requirement for Snf7/Vps20 in GAAC involved the RIM pathway (9), wherein Snf7 provides a platform on the MVB outer membrane for processing and activation of transcription factor Rim101. Indeed, Mitchell and colleagues had suggested previously that Snf7, Vps20, and Vps25 (E-II) operate outside of the RIM pathway in a signal transduction response to nutrient starvation. Among the mRNAs that showed reductions in snf7Δ but not rim21Δ cells at pH 8 in their study (7), we identified 12 known Gcn4 target genes involved in amino acid, vitamin, or nucleotide biosynthesis (ASN1, ARG1, ARG3, ARG5, ARG6, ARG8, HIS4, MET6, GLN1, BIO4, ADE4, and ADE17) (45). Consonant with our findings, they reported that vps20Δ and vps25Δ, but not vps4Δ, reduced the mRNA level of the Gcn4 target gene ASN1 (7). These previous results support our conclusion that efficient activation by Gcn4 is more dependent on endosomal recruitment of Snf7/Vps20 than on Vps4-dependent protein trafficking to the vacuole. Barwell et al. (7) identified many other mRNAs downregulated specifically in snf7Δ cells which are not Gcn4 targets, consistent with the idea that activators besides Gcn4 are attenuated in this mutant. This last conclusion is also supported by the fact that snf7Δ and snf8Δ mutations were first identified on the basis of diminished SUC2 mRNA expression (Snf− phenotype) (63, 70).
If endosomal Snf7 does not affect GAAC in the context of the RIM pathway, then how can we explain the stronger Gcn− phenotype of snf7Δ versus vps4Δ? It was shown previously that recruitment of Snf7/Vps20 to endosomal membranes protects the MVB cargo Cps1 from rapid proteolytic processing in the aberrant endosome of class E mutants (3). Interestingly, we found that the Gcn− phenotype of snf7Δ is diminished by inactivation of PrA (pep4Δ). Hence, the more severe Gcn− phenotype of class E mutants which fail to recruit (or lack) Snf7/Vps20 might result partly from rapid PrA-dependent proteolysis of MVB cargo in the class E compartment. Other mislocalized vacuolar proteases, like PrB, could participate with PrA in this aberrant degradation process. If the destruction of MVB cargo in the late endosome is the key defect responsible for the Gcn− phenotype of snf7Δ cells, then perhaps the severe depletion of cargo proteins in the MVB that occurs in class C/D mutants, owing to their defects in vesicle fusion, is responsible for their even stronger Gcn− phenotypes. In this view, depletion of intact MVB cargo proteins in the late endosome represents a key molecular condition, or signal, responsible for attenuating transcriptional activation by Gcn4 in all vps mutants with Gcn− phenotypes. As depicted schematically in Fig. 11, this aberration would increase in severity in the progression from (i) vps4Δ to (ii) snf7Δ pep4Δ to (iii) snf7Δ to (iv) vps34Δ mutants (Fig. 11). This model predicts that the stronger Gcn− phenotypes of class C/D mutations should be epistatic to the weaker Gcn− phenotypes of class E mutations, since the former evoke a more profound reduction in MVB cargo at the late endosome. Consistent with this prediction, we found that vps34Δ vps20Δ and vps34Δ snf7Δ double mutants exhibited the more severe SMs Gcn− phenotype characteristic of the vps34Δ single mutant (data not shown).
FIG. 11.
Hypothetical model for coupling activation by Gcn4 to the efficiency of vesicular trafficking from the Golgi complex to MVB. The model proposes that vps mutations attenuate transcriptional activation by Gcn4 to different extents according to their effect in reducing levels of MVB cargo proteins in the MVB. (A) In WT cells, MVB cargo proteins (black and gray shapes) interact with the ESCRT factors, including Snf8 (E-II) and Snf7 (E-III), on the limiting membrane of the MVB and are delivered to ILVs in the MVB via Vps4. The MVB cargo is transported to the vacuole lumen and, in the case of endocytic cargo, is degraded by vacuolar proteinases (purple shapes), such as PrA and PrB, which also reach the vacuole through the late endosome. Activation by Gcn4 during amino acid limitation is efficient. (B) vps34Δ cells have very low levels of cargo proteins at the MVB, owing to profound defects in vesicular fusion at the MVB and, accordingly, exhibit the strongest inhibition of transcriptional activation by Gcn4. (C) snf7Δ cells have a less severe activation defect because cargo proteins still reach the MVB, even though they are not delivered to ILVs. The cargo falls below normal levels owing to proteolysis by PrA (and possibly other vacuolar hydrolases) that accumulate in the class E compartment of this and other class E vps mutants (54). Thus, inactivation of PrA (pep4Δ) reduces the proteolysis of MVB cargo and diminishes the signal for downregulating Gcn4 in snf7Δ cells. (The situation in the snf7Δ pep4Δ mutant is not depicted here.) (D) vps4Δ cells exhibit the smallest activation defect because cargo proteins accumulating in the class E compartment are relatively protected from proteolysis by interaction with E-II and E-III complexes on the MVB membrane (3). See text for further details.
Many of the known MVB cargo proteins are cell surface transmembrane receptors or permeases, whereas others (like Cps1) are transmembrane proteins in the vacuole with biosynthetic functions (35). Interestingly, the general amino acid permease Gap1 is an MVB cargo that is downregulated at the plasma membrane in amino acid-replete cells by ubiquitination and trafficking to the MVB en route to the vacuolar lumen for degradation (57). However, we found that deletion of GAP1, or eliminating subunits of the GSE complex required for sorting Gap1 from the MVB to plasma membrane (18), does not confer SM sensitivity (Table 4 and data not shown). This indicates that proper trafficking of Gap1 is not required to maintain a functional GAAC response. Furthermore, the stronger Gcn− phenotype we observed for vps C/D mutants versus sac6Δ cells suggests that a broad reduction in receptor-mediated endocytosis of cell surface proteins is not sufficient for a severe inhibition of Gcn4 function. This suggests that the MVB cargo proteins most relevant to the GAAC are biosynthetic vacuolar proteins delivered from the Golgi complex, rather than cell surface membrane proteins transported to the MVB by endocytosis (Fig. 2).
This last conclusion fits with our finding that vps1Δ, which specifically impairs Golgi complex-to-MVB transport, provokes a stronger Gcn− phenotype than does impairing endocytosis by the sac6Δ or end4-1 mutations. An alternative pathway for Golgi complex-to-endosome trafficking operates in vps1Δ cells wherein Golgi cargo is mislocalized to the plasma membrane and then transported to the MVB by endocytosis. This backup pathway is impaired by disrupting endocytosis with the end4-1 mutation in the vps1Δ background (48). Our finding that the Gcn− phenotype in vps1Δ cells is exacerbated by end4-1 mirrors the previously established effects of combining these two mutations in disrupting Golgi complex-to-endosome transport (48). Thus, this finding further supports the notion that a defect in Golgi complex-to-MVB transport is the key malfunction involved in downregulating GAAC in vps mutants. However, since end4-1 impairs endocytosis, we cannot exclude the possibility that the stronger Gcn− phenotype of the vps1Δ end4-1 double mutant results from simultaneously impairing delivery of MVB cargo from the plasma membrane and the Golgi complex.
Conditional mutations affecting different stages of the secretory pathway, and also the drug chlorpromazine, which deforms membrane properties of internal organelles, elicit increased eIF2α phosphorylation, with a strong inhibition of general protein synthesis (14). Interestingly, Gcn4 is induced but does not transcriptionally activate HIS4-lacZ in chlorpromazine-treated cells (13), the phenotype observed here in vps mutants. These drastic impairments of the secretory pathway also elicit the ASR, which inhibits nuclear import (43) and probably mediates the decreased transcription of ribosomal protein genes, rDNA (39, 46), and certain Gcn4 targets (13). However, vps18Δ and vps45Δ confer strong Gcn− phenotypes but do not elicit the ASR (43). Moreover, overexpressing Hog1 or deleting WSC genes, which suppress the ASR (42), does not affect the Gcn− phenotype of vps mutants. Finally, we showed that nuclear import of Gcn4 and its occupancy of the ARG1 UAS were unaffected in Gcn− vps mutants and found that inactivation of Sec18 (which elicits the ASR) does not impair activation of HIS4-lacZ transcription by Gcn4. Hence, the attenuation of Gcn4 function in vps mutants is distinct from the ASR and the downregulation of ribosomal protein genes in response to severe disruptions of the secretory pathway.
One way to account for our findings is to propose that a signal transduction pathway operates in yeast to attenuate transcriptional activation by Gcn4 and certain other activators in response to impaired vesicular trafficking at the MVB. It may make sense to have a signaling mechanism to prevent Gcn4 from increasing production of amino acid substrates for new protein synthesis in the face of toxic compounds or conditions that disrupt protein trafficking. A precedent for this idea is the reduction in protein synthesis that occurs on activation of the mammalian eIF2α kinase PERK by endoplasmic reticulum stress, to prevent endoplasmic reticulum overload (56). And in yeast, it was shown that general nitrogen starvation blocks the translational induction of Gcn4 by phosphorylated eIF2α during amino acid limitation, presumably because induction of amino acid biosynthetic genes by Gcn4 is detrimental during nitrogen starvation (22). Rather than blocking translational induction of Gcn4, the disruption of endosomal trafficking in vps mutants reduces the ability of induced Gcn4 to stimulate transcription. The fact that at least some activation domains besides Gcn4's are affected by vps mutations suggests that a transcriptional coactivator could be downregulated by endosome dysfunction. Targeting a coactivator that is critical for Gcn4 and other stress-responsive activators, but not universally required by all activators at all promoters, e.g., SAGA, could eliminate competition for general transcription factors at genes that don't require the targeted coactivator and which encode proteins that ameliorate the consequences of endosome dysfunction.
Rather than positing a signaling pathway that couples Gcn4 activity to MVB function, it could be proposed instead that an unknown regulator of Gcn4 is activated at the late endosome, analogous to the regulation of Rim101 at this organelle, or is dependent on MVB integrity for its proper function. In this view, the vps mutations with Gcn− phenotypes impair this hypothetical regulator by altering MVB structure or the makeup of resident cargo proteins. In mammalian cells, there is evidence that Vps34 mediates the stimulatory effects of amino acids on the TOR pathway, a central regulator of gene expression and protein synthesis in response to nutrients in yeast and mammals (47). Tor and several of its effectors are localized to intracellular membranes and vesicles, leading to suggestions that the regulation of TOR signaling occurs on membranes (5). A simple model that vps34Δ impairs Gcn4 function by attenuating TOR activity cannot be ruled out, but it seems unlikely considering that Gcn4 is induced translationally (11) and contributes to activation of certain target genes by Gln3 when TOR is inhibited by rapamycin (64). Nevertheless, it is possible that another regulator of transcriptional activation by Gcn4 and certain other activators functions from late endosomal membranes and is affected by disruption of vesicular trafficking at this organelle in vps mutants.
It was shown previously that inactivation of Kex2, an endoprotease in the Golgi complex that processes the precursors of α-factor and M1 killer toxin (17), suppresses temperature-sensitive mutations in the largest subunit of Pol II, Rpb1 (41). Hence, it was possible that Kex2 would process a cargo protein in the Golgi complex that is transported to the MVB and has an impact on GAAC; however, we found that the kex2Δ mutant does not exhibit an SMs/Gcn− phenotype (Table 4).
While this work was under review, Puria et al. reported that a number of vps C/D mutants, and to a lesser extent vps E mutants (including vps20), impair transcriptional activation by Gln3 with a poor nitrogen source (proline) (51). Nuclear localization of Gln3 was reduced in class C mutants, which could account at least partly for their defects in Gln3-mediated transcriptional activation. It was proposed that Golgi complex-endosome trafficking is an obligate step in routing of Gln3 to the nucleus during growth in a poor nitrogen source. These authors also obtained evidence that Gln3 is associated with Golgi apparatus- or endosome-related vesicles and speculated that Gln3 is sequestered in cytoplasmic vesicles that can't fuse with the endosome in the vps mutants, reducing its nuclear localization. However, it seems possible that activation by the residual nucleus-localized Gln3, which they observed in a class D mutant (vps45Δ), was attenuated in the manner we have described here for Gcn4. Unlike the case of Gln3, nuclear localization of Gcn4 is constitutive in WT cells (50), and we found that Gcn4 is not excluded from the nucleus in class C, D, or E vps mutants. Thus, it appears that transcriptional activation can be downregulated at more than one stage for different activators in response to defective vesicular transport at the late endosome.
We have discussed the involvement of Vps factors in modulating transcriptional activation by Gcn4 and Gln3, the central role of endosomal Snf7 in the RIM pathway, and evidence implicating human Vps34 in TOR signaling. In addition, it was shown that the α-subunit of the G protein involved in pheromone signaling in yeast (Gpa1) is present at endosomes and interacts with Vps34 and Vps15 to stimulate PtdIns[3]P synthesis in a manner that enhances mating (61). Thus, there is a growing body of evidence that endosomal membranes serve as an important intracellular platform for a variety of signaling and regulatory mechanisms in eukaryotic cells.
Acknowledgments
We thank Jacob Boysen, Aaron Mitchell, and Tom Stevens for generous gifts of strains and many helpful suggestions, Krishnamurthy Natarajan, Marion Carlson, Stan Fields, Susan Michaelis, George Santangelo, Michael Gustin, Hans Ronne, Stefan Bjorklund, Roger Brent, Tom Stevens, Scott Emr, Tom Donahue, Ray Deshaies, Jon Warner, Peter Novick, Randy Schekman, and Mark Hochstrasser for strains or plasmids, Scott Emr for antibodies, and S. D. Kohlwein and H. Wolinski for help with microscopy. We thank Alex Strunnikov, Jinsheng Dong, and Caroline Philpott for valuable advice and technical assistance and Aaron Mitchell, Jacob Boysen, and Chhabi Govind for critical comments on the manuscript.
This work was supported in part by the Intramural Research Program of the NIH. J.H. was supported by LC545 and Institutional Research Concept no. AV0Z50200510.
Footnotes
Published ahead of print on 15 September 2008.
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst, M. 2005. A protein's final ESCRT. Traffic 62-9. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3271-282. [DOI] [PubMed] [Google Scholar]
- 4.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 172982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backer, J. M. 2008. The regulation and function of class III PI3Ks: novel roles for Vps34. Biochem. J. 4101-17. [DOI] [PubMed] [Google Scholar]
- 6.Balciunas, D., M. Hallberg, S. Bjorklund, and H. Ronne. 2003. Functional interactions within yeast mediator and evidence of differential subunit modifications. J. Biol. Chem. 2783831-3839. [DOI] [PubMed] [Google Scholar]
- 7.Barwell, K. J., J. H. Boysen, W. Xu, and A. P. Mitchell. 2005. Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 4890-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowers, K., and T. H. Stevens. 2005. Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1744438-454. [DOI] [PubMed] [Google Scholar]
- 9.Boysen, J. H., and A. P. Mitchell. 2006. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol. Biol. Cell 171344-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burd, C. G., M. Peterson, C. R. Cowles, and S. D. Emr. 1997. A novel Sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol. Biol. Cell 81089-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherkasova, V. A., and A. G. Hinnebusch. 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2α kinase GCN2. Genes Dev. 17859-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cigan, A. M., M. Foiani, E. M. Hannig, and A. G. Hinnebusch. 1991. Complex formation by positive and negative translational regulators of GCN4. Mol. Cell. Biol. 113217-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Filippi, L., M. Fournier, E. Cameroni, P. Linder, C. De Virgilio, M. Foti, and O. Deloche. 2007. Membrane stress is coupled to a rapid translational control of gene expression in chlorpromazine-treated cells. Curr. Genet. 52171-185. [DOI] [PubMed] [Google Scholar]
- 14.Deloche, O., J. de la Cruz, D. Kressler, M. Doere, and P. Linder. 2004. A membrane transport defect leads to a rapid attenuation of translation initiation in Saccharomyces cerevisiae. Mol. Cell 13357-366. [DOI] [PubMed] [Google Scholar]
- 15.Donahue, T. F., and A. M. Cigan. 1988. Genetic selection for mutations that reduce or abolish ribosomal recognition of the HIS4 translational initiator region. Mol. Cell. Biol. 82955-2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drysdale, C. M., E. Dueñas, B. M. Jackson, U. Reusser, G. H. Braus, and A. G. Hinnebusch. 1995. The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol. Cell. Biol. 151220-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuller, R. S., A. Brake, and J. Thorner. 1989. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl. Acad. Sci. USA 861434-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, M., and C. A. Kaiser. 2006. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 8657-667. [DOI] [PubMed] [Google Scholar]
- 19.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418387-391. [DOI] [PubMed] [Google Scholar]
- 20.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74527-534. [DOI] [PubMed] [Google Scholar]
- 21.Govind, C. K., S. Yoon, H. Qiu, S. Govind, and A. G. Hinnebusch. 2005. Simultaneous recruitment of coactivators by Gcn4p stimulates multiple steps of transcription in vivo. Mol. Cell. Biol. 255626-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grundmann, O., H. U. Mosch, and G. H. Braus. 2001. Repression of GCN4 mRNA translation by nitrogen starvation in Saccharomyces cerevisiae. J. Biol. Chem. 27625661-25671. [DOI] [PubMed] [Google Scholar]
- 23.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75791-803. [DOI] [PubMed] [Google Scholar]
- 24.Han, G. S., A. Audhya, D. J. Markley, S. D. Emr, and G. M. Carman. 2002. The Saccharomyces cerevisiae LSB6 gene encodes phosphatidylinositol 4-kinase activity. J. Biol. Chem. 27747709-47718. [DOI] [PubMed] [Google Scholar]
- 25.Hanes, S. D., and R. Brent. 1989. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell 571275-1283. [DOI] [PubMed] [Google Scholar]
- 26.Harashima, S., E. M. Hannig, and A. G. Hinnebusch. 1987. Interactions between positive and negative regulators of GCN4 controlling gene expression and entry into the yeast cell cycle. Genetics 117409-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi, M., T. Fukuzawa, H. Sorimachi, and T. Maeda. 2005. Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol. Cell. Biol. 259478-9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinnebusch, A. G. 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae, p. 319-414. In J. R. Broach, E. W. Jones, and J. R. Pringle (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 29.Hinnebusch, A. G. 2005. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59407-450. [DOI] [PubMed] [Google Scholar]
- 30.Hinnebusch, A. G., G. Lucchini, and G. R. Fink. 1985. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene CYC1 of yeast. Proc. Natl. Acad. Sci. USA 82498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinnebusch, A. G., and K. Natarajan. 2002. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 122-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia, M. H., R. A. Larossa, J. M. Lee, A. Rafalski, E. Derose, G. Gonye, and Z. Xue. 2000. Global expression profiling of yeast treated with an inhibitor of amino acid biosynthesis, sulfometuron methyl. Physiol. Genomics 383-92. [DOI] [PubMed] [Google Scholar]
- 33.Jones, E. W. 1991. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 194428-453. [DOI] [PubMed] [Google Scholar]
- 34.Kaksonen, M., C. P. Toret, and D. G. Drubin. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell 123305-320. [DOI] [PubMed] [Google Scholar]
- 35.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3893-905. [DOI] [PubMed] [Google Scholar]
- 36.Kihara, A., T. Noda, N. Ishihara, and Y. Ohsumi. 2001. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, S. J., M. J. Swanson, H. Qiu, C. K. Govind, and A. G. Hinnebusch. 2005. Activator Gcn4p and Cyc8p/Tup1p are interdependent for promoter occupancy at ARG1 in vivo. Mol. Cell. Biol. 2511171-11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kornitzer, D. 2002. Monitoring protein degradation. Methods Enzymol. 351639-647. [DOI] [PubMed] [Google Scholar]
- 39.Li, Y., R. D. Moir, I. K. Sethy-Coraci, J. R. Warner, and I. M. Willis. 2000. Repression of ribosome and tRNA synthesis in secretion-defective cells is signaled by a novel branch of the cell integrity pathway. Mol. Cell. Biol. 203843-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 41.Martin, C., and R. A. Young. 1989. KEX2 mutations suppress RNA polymerase II mutants and alter the temperature range of yeast cell growth. Mol. Cell. Biol. 92341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nanduri, J., S. Mitra, C. Andrei, Y. Liu, Y. Yu, M. Hitomi, and A. M. Tartakoff. 1999. An unexpected link between the secretory path and the organization of the nucleus. J. Biol. Chem. 27433785-33789. [DOI] [PubMed] [Google Scholar]
- 43.Nanduri, J., and A. M. Tartakoff. 2001. The arrest of secretion response in yeast: signaling from the secretory path to the nucleus via Wsc proteins and Pkc1p. Mol. Cell 8281-289. [DOI] [PubMed] [Google Scholar]
- 44.Natarajan, K., B. M. Jackson, E. Rhee, and A. G. Hinnebusch. 1998. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol. Cell 2683-692. [DOI] [PubMed] [Google Scholar]
- 45.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 214347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nierras, C. R., and J. R. Warner. 1999. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 27413235-13241. [DOI] [PubMed] [Google Scholar]
- 47.Nobukuni, T., M. Joaquin, M. Roccio, S. G. Dann, S. Y. Kim, P. Gulati, M. P. Byfield, J. M. Backer, F. Natt, J. L. Bos, F. J. Zwartkruis, and G. Thomas. 2005. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA 10214238-14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nothwehr, S. F., E. Conibear, and T. H. Stevens. 1995. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J. Cell Biol. 12935-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peplowska, K., D. F. Markgraf, C. W. Ostrowicz, G. Bange, and C. Ungermann. 2007. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev. Cell 12739-750. [DOI] [PubMed] [Google Scholar]
- 50.Pries, R., K. Bomeke, S. Irniger, O. Grundmann, and G. H. Braus. 2002. Amino acid-dependent Gcn4p stability regulation occurs exclusively in the yeast nucleus. Eukaryot. Cell 1663-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puria, R., S. A. Zurita-Martinez, and M. E. Cardenas. 2008. Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1057194-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu, H., C. Hu, S. Yoon, K. Natarajan, M. J. Swanson, and A. G. Hinnebusch. 2004. An array of coactivators is required for optimal recruitment of TATA binding protein and RNA polymerase II by promoter-bound Gcn4p. Mol. Cell. Biol. 244104-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu, H., C. Hu, F. Zhang, G. J. Hwang, M. J. Swanson, C. Boonchird, and A. G. Hinnebusch. 2005. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Mol. Cell. Biol. 253461-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raymond, C. K., I. Howald-Stevenson, C. A. Vater, and T. H. Stevens. 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 31389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reid, G. A., and G. Schatz. 1982. Import of proteins into mitochondria. J. Biol. Chem. 25713062-13067. [PubMed] [Google Scholar]
- 56.Ron, D., and H. P. Harding. 2000. PERK and translational control by stress in the endoplasmic reticulum, p. 547-560. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 57.Rubio-Texeira, M., and C. A. Kaiser. 2006. Amino acids regulate retrieval of the yeast general amino acid permease from the vacuolar targeting pathway. Mol. Biol. Cell 173031-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rue, S. M., S. Mattei, S. Saksena, and S. D. Emr. 2008. Novel ist1-did2 complex functions at a late step in multivesicular body sorting. Mol. Biol. Cell 19475-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 12219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singer-Kruger, B., H. Stenmark, A. Dusterhoft, P. Philippsen, J. S. Yoo, D. Gallwitz, and M. Zerial. 1994. Role of three rab5-like GTPases, Ypt51p, Ypt52p, and Ypt53p, in the endocytic and vacuolar protein sorting pathways of yeast. J. Cell Biol. 125283-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Slessareva, J. E., S. M. Routt, B. Temple, V. A. Bankaitis, and H. G. Dohlman. 2006. Activation of the phosphatidylinositol 3-kinase Vps34 by a G protein alpha subunit at the endosome. Cell 126191-203. [DOI] [PubMed] [Google Scholar]
- 62.Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S.-J. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 232800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tu, J., L. G. Vallier, and M. Carlson. 1993. Molecular and genetic analysis of the SNF7 gene in Saccharomyces cerevisiae. Genetics 13517-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valenzuela, L., C. Aranda, and A. Gonzalez. 2001. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J. Bacteriol. 1832331-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wek, R. C., J. F. Cannon, T. E. Dever, and A. G. Hinnebusch. 1992. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2α kinase GCN2. Mol. Cell. Biol. 125700-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. E. Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. J. Hegemann, T., M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-McDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 67.Wolfner, M., D. Yep, F. Messenguy, and G. R. Fink. 1975. Integration of amino acid biosynthesis into the cell cycle of Saccharomyces cerevisiae. J. Mol. Biol. 96273-290. [DOI] [PubMed] [Google Scholar]
- 68.Xu, W., and A. P. Mitchell. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 1836917-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu, W., F. J. Smith, Jr., R. Subaran, and A. P. Mitchell. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 155528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeghiayan, P., J. Tu, L. G. Vallier, and M. Carlson. 1995. Molecular analysis of the SNF8 gene of Saccharomyces cerevisiae. Yeast 11219-224. [DOI] [PubMed] [Google Scholar]
- 71.Yoon, S., H. Qiu, M. J. Swanson, and A. G. Hinnebusch. 2003. Recruitment of SWI/SNF by Gcn4p does not require Snf2p or Gcn5p but depends strongly on SWI/SNF integrity, SRB mediator, and SAGA. Mol. Cell. Biol. 238829-8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, F., L. Sumibcay, A. G. Hinnebusch, and M. J. Swanson. 2004. A triad of subunits from the Gal11/tail domain of Srb mediator is an in vivo target of transcriptional activator Gcn4p. Mol. Cell. Biol. 246871-6886. [DOI] [PMC free article] [PubMed] [Google Scholar]