Abstract
G1-specific transcription in the budding yeast Saccharomyces cerevisiae depends upon SBF and MBF. Whereas inactivation of SBF-regulated genes during the G1/S transition depends upon mitotic B-type cyclins, inactivation of MBF has been reported to involve multiple regulators, Nrm1 and Stb1. Nrm1 is a transcriptional corepressor that inactivates MBF-regulated transcription via negative feedback as cells exit G1 phase. Cln/cyclin-dependent kinase (CDK)-dependent inactivation of Stb1, identified via its interaction with the histone deacetylase (HDAC) component Sin3, has also been reported to inactivate MBF-regulated transcription. This report shows that Stb1 is a stable component of both SBF and MBF that binds G1-specific promoters via Swi6 during G1 phase. It is important for the growth of cells in which SBF or MBF is inactive. Although dissociation of Stb1 from promoters as cells exit G1 correlates with Stb1 phosphorylation, phosphorylation is only partially dependent upon Cln1/2 and is not involved in transcription inactivation. Inactivation depends upon Nrm1 and Clb/CDK activity. Stb1 inactivation dampens maximal transcriptional induction during late G1 phase and also derepresses gene expression in G1-phase cells prior to Cln3-dependent transcriptional activation. The repression during G1 also depends upon Sin3. We speculate that the interaction between Stb1 and Sin3 regulates the Sin3/HDAC complex at G1-specific promoters.
Periodic expression of large families of genes during the cell cycle is one of the primary cellular mechanisms imposing an orderly progression of events during the cell cycle. The most extensive analysis of the cell cycle regulatory network has been carried out in the budding yeast Saccharomyces cerevisiae. In budding yeast, transcriptional activation of more than 200 G1-specific genes is the earliest indicator of cell cycle initiation, often referred to as “Start” (reviewed in reference 30). The G1-specific gene family encodes the components of the cellular machinery required for subsequent events associated with cell cycle initiation. In budding yeast the regulation of G1-S genes is mediated by two transcription factors, MBF (Mlu1 cell cycle box binding factor) and SBF (Swi4 cell cycle box binding factor), which are functionally analogous to the mammalian E2F transcription factors (5). SBF and MBF each contain a Swi6 subunit and a distinct DNA-binding subunit, Swi4 and Mbp1, respectively. Each heterodimeric factor binds to a specific DNA sequence found in the promoters of G1-specific genes. Although some genes are influenced by both factors, the regulation of most genes depends upon one of the two factors, the identity of which is correlated with the frequency of the specific binding motif (17, 25, 26).
SBF regulates genes encoding proteins involved in the initiation of the G1-S transition and progression into S phase, whereas MBF is primarily involved in the regulation of genes involved in DNA replication and repair. SBF and MBF are bound to G1-specific promoters prior to G1-specific transcription activation (6, 15, 19). Activation of G1-specific transcription depends on Cln3-associated cyclin-dependent kinase (CDK) activity (11, 27, 28). Whereas both SBF and MBF are dependent on Cln3/CDK for activation, they are distinctly regulated (1, 9, 10, 20, 23). SBF is required for transcriptional activation during G1 phase, but MBF restricts the expression of its targets to the G1 phase by repressing transcription outside of G1 (2, 9, 20). Consequently, inactivation of SBF results in constitutively low expression of SBF-specific targets, whereas inactivation of MBF results in constitutively high expression of its specific targets (9, 19). Although there seems to be little overlap, there is significant redundancy in SBF and MBF promoter binding when the other factor is inactivated (9).
The timing of activation of SBF-dependent transcription during G1 phase depends upon Whi5, an SBF-specific transcriptional repressor (10). Whi5 is inactivated, during late G1 phase, via phosphorylation by Cln3/CDK (7, 10). Although it has been reported that Whi5 binds synthetic promoters containing MCB elements via MBF (7), we find it specifically binds to the SBF complex at SBF-regulated promoters to repress expression from SBF-dependent promoters in vivo (10). Interestingly, in the absence of MBF a significant number of G1-specific genes can be bound by SBF and visa versa (2, 9), which, at least at some promoters, results in Whi5-dependent repression in G1 phase (9). The mechanism of Cln3/CDK-dependent transcriptional activation of MBF targets remains unknown but may involve additional regulatory proteins similar to Whi5.
Inactivation of SBF-dependent transcription upon exit from G1 phase depends largely upon Clb2/CDK activity (1, 9, 20, 23). In contrast, the repression of MBF genes as cells exit G1 phase has been reported to involve several proteins. Nrm1, encoded by an MBF-dependent gene, promotes the timely repression via negative feedback during the G1-S transition (9). In its absence, the delayed repression that is observed most likely depends upon Clb/CDK. Finally, Cln/CDK-dependent phosphorylation of Stb1 was reported to promote dissociation from Swi6 and repress MBF targets (8).
Stb1 was originally identified via its interaction with the histone deacetylase (HDAC) complex component Sin3 by two-hybrid analysis (18). It was later found to interact with Swi6 and to be a target of Cln-associated CDK involved in the timing of G1-specific transcription activation (16). It was proposed that Cln/CDK-dependent phosphorylation of Stb1 activates G1-specific transcription at Start, much like Whi5 (7, 16). However, a subsequent study suggested that Stb1 is required specifically to regulate the expression of MBF, but not SBF, target genes (8). That study suggested that phosphorylation of Stb1 by Cln/CDK inhibits its ability to associate with Swi6, thereby inactivating MBF-dependent transcription.
In the interest of understanding the role of Stb1 in transcriptional repression of MBF-regulated genes, we investigated its role in G1-specific transcription. We found that endogenous Stb1 forms a complex with both the SBF and the MBF transcription factors. Furthermore, Stb1 associates with SBF- and MBF-regulated promoters via Swi6 during the G1 phase of the cell cycle. We find that, although phosphorylation of Stb1 correlates with its disassociation from G1-specific promoters and transcriptional inactivation, Stb1 does not play a role in the timing of transcriptional inactivation. In addition, phosphorylation of Stb1 is not entirely dependent upon Cln1 and Cln2. Inactivation of Stb1 elevates the level of G1-specific transcripts in G1 phase prior to Cln3-dependent transcriptional activation and affects the maximal transcriptional activation of G1-specific genes, predominantly via MBF, such that the overall induction of G1-specific genes is dampened. Finally, full repression of G1-specific transcripts during G1 prior to transcriptional activation requires both Stb1 and the HDAC complex component Sin3. We conclude that timely inactivation of G1-specific genes does not depend on Stb1 but rather on Clb/CDK activity and accumulation of Nrm1. We speculate that Stb1 is required for efficient modulation of G1-specific transcription during the G1 phase by modulating Sin3/HDAC activity at G1-specific promoters.
MATERIALS AND METHODS
Strains and DNAs.
All of the budding yeast strains used in the present study were derived from 15Daub (MATa ade1 leu2-3,112 his2 trp1-1 ura3Δns bar1Δ). The yeast strains used here are presented in Table 1. The strategy of Rigaut et al. (22) was used to append a tandem affinity purification (TAP) tag to SWI6 at the endogenous loci. The PCR method of Longtine et al. (21) was used to disrupt STB1, MBP1, SWI4, and SWI6, and 13-myc-tagged STB1 and NRM1, at the carboxy terminus. Six-myc-tagged alleles of SWI6 were constructed by plasmid integration.
TABLE 1.
Yeast strains in this study
| Yeast strain | Genotypea | Source or reference |
|---|---|---|
| CWY231 | ade1 leu2-3,112 his2 trp1-1 ura3Δns bar1Δ | 14 (strain 15 Daub) |
| CWY1682 | mbp1::LEU2 | 10 |
| CWY1683 | swi4::KANr | 10 |
| CWY1410 | SWI6-6xmyc::URA3 | 10 |
| CWY1559 | NRM1-13xmyc::KANr | 10 |
| CWY1678 | STB1-13xmyc::URA3 | This study |
| CWY1894 | STB1-13xmyc::URA3 swi6::HIS2 | This study |
| CWY1442 | SWI6-TAP::KANr | 10 |
| CWY1696 | SWI6-TAP::KANrSTB1-13xmyc::URA3 | This study |
| CWY1697 | SWI6-TAP::KANrSTB1-13xmyc::URA3 mbp1::LEU2 | This study |
| CWY1990 | SWI6-TAP::KANrSTB1-13xmyc::URA3 swi4::TRP1 | This study |
| CWY1922 | cln1Δ cln2xs STB1-13xmyc::URA3 | This study |
| CWY1925 | cln1Δ cln2xs NRM1-13xmyc::URA3 | This study |
| CWY905 | cln1Δ cln2xs leu2::GAL-CLN3::LEU2 | 27 |
| CWY1959 | cln1Δ cln2xs leu2::GAL-CLN3::LEU2 stb1::URA3 | This study |
| CWY908 | cln1Δ cln2xs leu2::GAL-CLN3::LEU2 sic1::URA3 | 27 |
| CWY1688 | stb1::URA3 | This study |
| CWY1685 | mbp1::LEU2 stb1::URA3 | This study |
| CWY1687 | swi4::KANrstb1::URA3 | This study |
| CWY1975 | sin3::KANr | This study |
| CWY1976 | sin3::KANrstb1::URA3 | This study |
KANr, kanamycin resistance.
Coimmunoprecipitation.
Immunoprecipitations were carried out using TAP purification buffers (3). Immunoprecipitated proteins were resolved by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis.
Cell synchronization.
Mating pheromone arrest synchrony experiments were carried out as described previously (27). For the experimental results shown in Fig. 4C, the strains also carried GAL-CLN3 and were grown on yeast extract-peptone (YEP)-galactose to conditionally enhance synchrony prior to arrest by mating pheromone (2.5 h) and release in YEP-glucose.
FIG. 4.
cln1Δ cln2Δ mutation results in prolonged binding of Stb1 to the CLN2 promoter. (A) Wild-type and cln1Δ cln2Δ cells were arrested by alpha-factor and released to allow cells to synchronously progress through the cell cycle. (Top) Samples were obtained at 15-min intervals; the budding indices and mRNA levels (quantitative RT-PCR; expressed as the percentage of highest level [100%] in wild-type cells after normalization of all values to the ACT1 mRNA) were determined. (Middle) In addition, Stb1 protein levels were analyzed in WCE probed with anti-myc and with anti-PSTAIRE antibody to detect Cdc28 protein level. (Bottom) ChIP of CLN2 (SBF-dependent gene), RNR1 (MBF-dependent gene), and ACT1 (MBF/SBF-independent gene) promoter DNA by Stb1-myc in cells from the same time course. ChIP of untagged genes (no tag) and WCE are shown as negative and positive controls. (B) Same as in panel A except that Nrm1-myc protein levels and binding to the RNR1 promoter were analyzed. (C) Wild-type cells and cln1Δ cln2Δ cells, either with or without an stb1Δ or sic1Δ mutation, all carrying GAL-CLN3, were grown on galactose media. Cells were arrested by alpha-factor for 2.5 h and released on glucose medium to progress synchronously through the cell cycle as described in the text. Samples were taken at 10-min intervals, and budding (right) and mRNA levels (quantitative RT-PCR; expressed as the percentage of the highest level [100%] in wild-type cells after normalization of all values to the ACT1 mRNA) were analyzed.
Real-time PCR and RT-PCR.
Total RNA was isolated by using an RNeasy Plus kit (Qiagen). The iQ Sybr Green Supermix (Bio-Rad) was used for quantitative PCR on chromatin immunoprecipitation (ChIP) samples, and the iScript One-Step RT-PCR kit with Sybr green (Bio-Rad) was used for reverse transcription-PCR (RT-PCR) experiments. Reactions were run on a Chromo-4 real-time PCR detector (Bio-Rad) using standard PCR and RT-PCR conditions. The data were analyzed by using MJ Opticon monitor analysis software 3.0.
ChIP analysis.
ChIP was performed as described previously (12).
Other methods.
Cell size analysis was performed using a Coulter Z2 particle cell analyzer (Beckman-Coulter). The cell size distribution was analyzed using Z2 AccuComp software (Beckman-Coulter).
RESULTS
Stb1 is a component of both SBF and MBF.
To elucidate the mechanism by which G1-specific transcription is regulated, we sought to identify proteins that physically interact with the SBF and MBF transcription factors. We recently reported mass spectrometry-based MudPIT (multi-dimensional protein interaction technology) (31) analysis of affinity-purified Swi4, Swi6, or Mbp1 complexes, which led to the identification of Whi5 as an SBF-specific inhibitor and Nrm1 as an MBF-specific corepressor (9, 10). Stb1, which was previously shown to interact with Swi6 (16), was identified in that screen as an interactor of both SBF and MBF (Swi6, Swi4, and Mbp1 fractions; data not shown). The coverage of Stb1 by specific peptides was greater in the Mbp1 fraction than in the Swi6 and Swi4 fractions, a finding consistent with the proposed specificity for MBF.
To confirm that Stb1 interacts with both SBF and MBF, we performed coimmunoprecipitation analysis. Anti-myc immune complexes prepared from cells expressing myc-tagged Stb1 and TAP-tagged Swi6, revealed an interaction (Fig. 1A). To determine whether the interaction with Swi6 depends on either Swi4 or Mbp1, the DNA-binding components of SBF and MBF, respectively, the experiment was carried out with either swi4Δ or mbp1Δ mutant strains. Neither inactivation of SWI4 nor of MBP1 abolishes the interaction between Stb1 and Swi6, indicating that Stb1 interacts with the Swi6 component of both transcription factors (Fig. 1A).
FIG. 1.
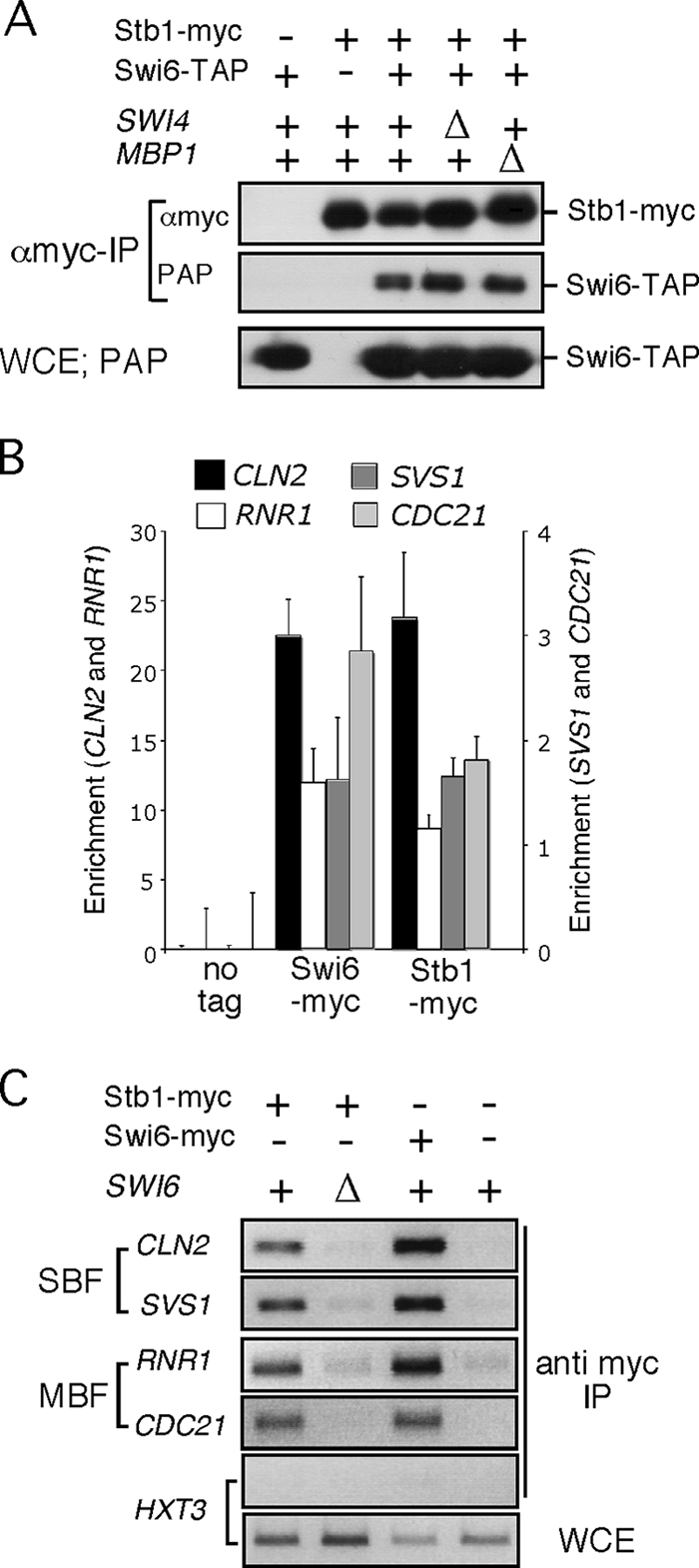
Stb1 binds via Swi6 to G1-specific promoters. (A) Extracts were prepared from wild type, swi4Δ or mbp1Δ strains carrying both STB1-myc13 and SWI6-TAP or from wild type carrying either STB1-myc13 or SWI6-TAP. Whole-cell extract (WCE) from asynchronous cultures were probed with PAP (PAP-IP; peroxidase-conjugated anti-peroxidase immunoglobulin G) to detect Swi6-TAP. Anti-myc immune complexes (αmyc-IP) were probed with anti-myc to detect Stb1-myc13 or PAP to detect Swi6-TAP. (B) Quantitative PCR of chromatin-immunoprecipitated RNR1 and CDC21 (MBF-dependent gene) and CLN2 and SVS1 (SBF-dependent gene) promoter DNA in an untagged strain (no tag) or wild-type cells carrying Swi6-myc or Stb1-myc. Bars represent the specific signal derived from the indicated promoter DNA detected by quantitative PCR in the relevant ChIP analysis. Efficiency of immunoprecipitation was determined by calculating immunoprecipitated “target/ACT1” DNA. The average value from three independent experiments, each sample run in triplicate, is presented with the standard error. (C) ChIP of RNR1 and CDC21 (MBF-dependent gene) and CLN2 and SVS1 (SBF-dependent gene) and HXT3 (SBF/MBF-independent gene) promoter DNA from wild-type and swi6Δ cells by Stb1-myc immunoprecipitation and wild-type cells by Swi6-myc immunoprecipitation. The results of ChIP analyses of untagged strain (no tag) and HXT3, which does not bind SBF or MBF, are provided as negative controls. Amplification of DNA from a WCE is shown as a positive control.
Stb1 binds via Swi6 to G1-specific promoters.
Whereas Swi4 and Mbp1 both bind a specific subset of G1-specific promoters, Swi6 binds to both SBF and MBF promoters. To determine whether Stb1 associates with SBF and MBF target genes, the binding of epitope-tagged Stb1 to promoters of RNR1 and CDC21 (MBF-specific) and CLN2 and SVS1 (SBF-specific) was assessed by quantitative ChIP analysis (Fig. 1B). CDC21 and SVS1 are exclusively regulated by MBF or SBF, respectively, whereas RNR1 and CLN2 are influenced by both factors (9). ChIP analysis using quantitative PCR shows that Stb1 binds to both MBF- and SBF-regulated promoters, as does Swi6. To establish whether binding of Stb1 to G1-specific promoters depends on Swi6, the ChIP experiment was repeated with wild-type and swi6Δ mutant cells. Binding of Stb1 to both SBF and MBF promoters was lost in extracts from swi6Δ mutant cells demonstrating that the interaction is dependent upon Swi6 (Fig. 1C). We conclude that Stb1 binds to SBF and MBF promoters via the common component of these transcription factors, Swi6.
Inactivation of Stb1 strongly affects cell size in the absence of either SBF or MBF.
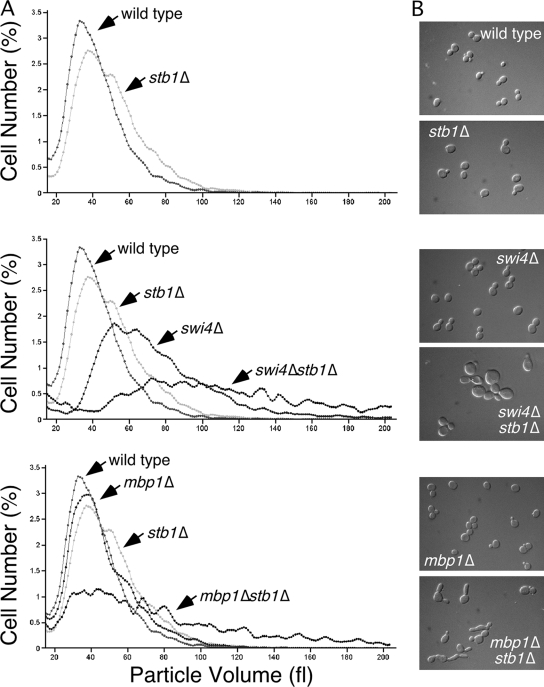
Based on the association of Stb1 to both SBF- and MBF-dependent promoters, we speculated that Stb1 might affect both SBF- and MBF-dependent transcription. Therefore, we examined their genetic interactions with Stb1. Inactivation of both Swi4 and Mbp1 leads to lethality, suggesting that inactivation of SBF renders cells dependent on MBF and vise versa. We therefore reasoned that cells should be more sensitive to a defect in the expression of SBF or MBF targets if the other transcription factor has been inactivated. While stb1Δ cells are slightly larger than wild-type cells, inactivation of Stb1 in a swi4Δ or mbp1Δ mutant results in a dramatic increase in cell size, a finding consistent with the hypothesis that Stb1 regulates both SBF and MBF (Fig. 2A). However, there are significant differences in the effect on cell size in stb1Δ mutants depending on the status of MBF and SBF (Fig. 2B). Inactivation of Stb1 in mbp1Δ cells mainly increases the heterogeneity of cells, with some cells still born small, whereas inactivation of Stb1 in swi4Δ cells significantly increases cell size at birth (Fig. 2A and B). In addition, whereas inactivation of Stb1 in swi4Δ mutant cells leads to large round cells, when combined with mbp1Δ the cells are noticeably elongated, suggesting a hyperaccumulation of G1 cyclins or a deficiency of B cyclins. Although this phenotype has not been explored further, we favor the latter hypothesis. Together, these observations suggest that the effect of Stb1 on G1-specific transcription is exerted via both SBF and MBF.
FIG. 2.
The increased cell size of stb1Δ depends on SBF and MBF. (A) The cell volume distribution (cell number/particle volume) of log-phase cultures of each of the indicated strains grown in yeast extract-peptone-dextrose as determined by a Coulter Z2 particle analyzer. (B) The indicated strains were grown to mid-log phase in rich medium, viewed, and photographed using Nomarski optics.
Stb1 is associated with G1-specific promoters during G1 phase.
To evaluate the relationship between Stb1 binding to G1-specific targets and G1-specific transcriptional regulation, we analyzed the timing of Stb1 association with the RNR1 (MBF-specific) and CLN2 and SVS1 (SBF-specific) promoters in cells synchronized by release from G1 arrest by mating pheromone. Stb1 binding to G1-specific promoters occurs throughout the G1 phase of the cell cycle, decreasing at the time of bud emergence (Fig. 3, lower panel). This is similar to Swi6 binding to SBF-dependent promoters (9, 10). Unlike SBF targets, Swi6 does not dissociate from MBF targets during the G1/S transition (9). Nevertheless, Stb1 association with these promoters is also lost as cells progress into S phase (Fig. 3, lower panel). Overall, association of Stb1 with SBF- and MBF-dependent promoters during G1 phase coincides with transcriptional activation, and its subsequent dissociation correlates with transcriptional inactivation.
FIG. 3.
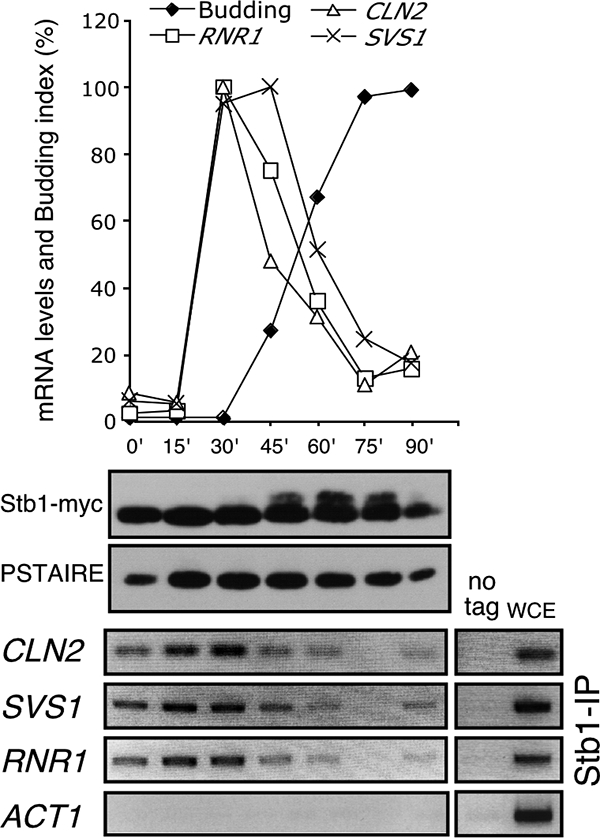
Stb1 binds to G1-specific promoter during G1 phase. Cells were arrested by alpha-factor and released to allow cells to synchronously progress through the cell cycle. (Top) Samples were taken at 15-min intervals, the budding index was determined, and mRNA was analyzed by quantitative RT-PCR. The transcript levels are expressed as the percentage of the highest level (100%) after normalization of all values to the ACT1 mRNA. (Middle) Stb1 protein levels were determined in WCE probed with anti-myc to detect Stb1-myc in cells from the same time course and with anti-PSTAIRE antibody to detect Cdc28 protein level. (Bottom) ChIP of RNR1 (MBF-dependent gene), CLN2 and SVS1 (SBF-dependent genes), and ACT1 (MBF/SBF-independent gene) promoter DNA by Stb1-myc in cells from the same time course. ChIP of untagged genes (no tag) and WCE are shown as negative and positive controls.
Stb1 hyperphosphorylation is associated with release from G1-specific promoters during the G1-S transition.
To determine whether the status of the Stb1 protein changes in these cells, we examined the mobility of the Stb1 protein in the same population of synchronized cells used for the ChIP analysis of Stb1 (Fig. 3, middle panel). Whereas Stb1-myc migrates as a single species during the G1 phase, a slower-migrating form of Stb1-myc appears and becomes increasingly apparent as cells progress into the budded phase of the cell cycle. Previous studies have shown that these slower-migrating forms of Stb1 are a consequence of phosphorylation (16). The timing of phosphorylation of Stb1 is similar to that observed in earlier studies. Phosphorylation of Stb1 correlates with disassociation of Stb1 from G1-specific promoters (Fig. 3). These observations are consistent with previous in vitro data showing that Stb1 is a direct target for phosphorylation by Cln-associated CDK and that Cln-dependent phosphorylation inhibits the interaction between Stb1 and Swi6 (8, 16). We conclude that Stb1 phosphorylation correlates with its dissociation from SBF- and MBF-dependent promoters in vivo during exit from G1 phase.
Inactivation of CLN1 and CLN2 reduces Stb1 phosphorylation and prolongs its association with G1-specific promoters.
The previous observation that Cln/CDK can phosphorylate Stb1 and prevent its interaction with Swi6 led to the hypothesis that Cln1/2-associated CDK promotes dissociation from promoters, leading to transcriptional inactivation. To evaluate the relationship between Cln1/2-CDK-dependent phosphorylation and Stb1 promoter binding, we compared the mobility and promoter binding of the Stb1 protein in a synchronized population of cln1Δ cln2Δ mutant cells with that in wild-type cells. cln1Δ cln2Δ cells, and wild-type cells expressing myc-tagged Stb1 were synchronized by mating pheromone arrest and release. Mating pheromone arrested cells grow larger than the minimum cell size required for cell cycle initiation, thereby minimizing the cell cycle delay observed in cln1Δ cln2Δ mutants. The accumulation of low mobility forms of Stb1 was less robust and delayed in the cln1Δ cln2Δ mutants relative to wild-type cells (Fig. 4A). This indicates that the majority, but not all, of the cell cycle-dependent phosphorylation of Stb1 depends on Cln1/2-associated CDK activity.
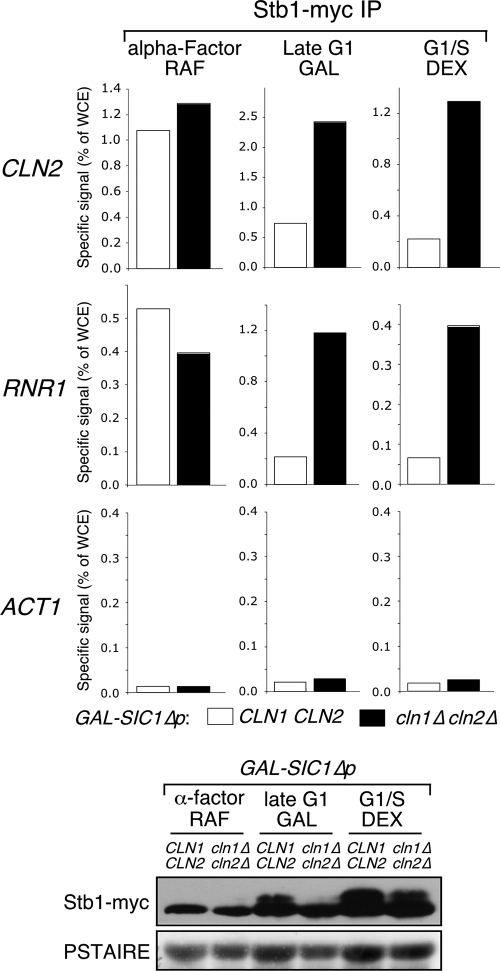
Consistent with the hypothesis that phosphorylation leads to dissociation of Stb1 from promoters, we observed an extended interval of Stb1 binding to CLN2 and RNR1 promoters in the cln1Δ cln2Δ mutants (Fig. 4A). To establish whether Cln1/2-CDK-dependent phosphorylation is sufficient to dissociate Stb1 from G1-specific promoters, wild-type and cln1Δ cln2Δ cells were synchronized using mating pheromone arrest and released from the arrest concurrent with induction of expression of a hyperstable form of the Clb/CDK inhibitor Sic1 from the GAL promoter (GAL-SIC1ΔP). Stb1 associated with G1-specific promoters in both wild-type and cln1Δ cln2Δ during the mating pheromone arrest. However, whereas Stb1 disassociates from promoters in the Sic1ΔP-expressing wild-type cells, arresting during late G1 phase, it remains bound to promoters in similarly treated cln1Δ cln2Δ cells (Fig. 5). This correlates with the accumulation of low-mobility forms of Stb1 in wild-type cells in which B-type cyclins are inhibited by Sic1 but not in cln1Δ cln2Δ mutant cells in which B-type cyclins are similarly inhibited. In cln1Δ cln2Δ cells that have progressed through G1 phase in the absence of exogenous Sic1ΔP, phosphorylation of Stb1 is decreased and promoter binding is prolonged relative to wild-type cells. We conclude that, whereas Clb-dependent phosphorylation of Stb1 may contribute to its dissociation from promoters, Cln1/2-CDK-dependent phosphorylation of Stb1 is sufficient to promote dissociation. This experiment provides evidence that phosphorylation of Stb1 is dependent upon the G1 cyclins Cln1 and Cln2 in vivo and is consistent with previous in vitro observations showing that Cln/CDK-dependent phosphorylation of Stb1 prevents Swi6 binding (8). There is no evidence that Stb1 is phosphorylated by Cln3/CDK in vivo.
FIG. 5.
Cln1/2-CDK-dependent phosphorylation of Stb1 is sufficient to release Stb1 from promoters. Wild-type (CLN1 CLN2) and cln1Δ cln2Δ cells carrying ura3:YIpGAL1-SIC1ΔP:URA3 were grown in raffinose medium (RAF) and synchronized by alpha-factor and subsequently released into galactose (GAL) or glucose (DEX) media. Quantitative PCR (percentage of the WCE signal) of chromatin-immunoprecipitated CLN2 (SBF-dependent gene), RNR1 (MBF-dependent gene), and ACT1 (MBF/SBF-independent gene) promoter DNA by Stb1-myc in cells arrested by alpha-factor or released from the arrest for 120 min in galactose (budded late G1 arrest with maximal SBF-dependent transcription [see reference 9]) or 75 min in glucose (late G1/early S phase with maximum budding and minimum G1-specific transcription; see Fig. 4). Whole-cell protein extract probed with anti-myc was used to detect Stb1-myc protein levels and mobility. The same extract probed with anti-PSTAIRE antibody was used to detect Cdc28 protein level for the loading control (bottom panel).
Absence of Cln1/2-associated CDK activity, not cell cycle delay, extends Stb1 promoter binding but does not affect the timely inactivation of MBF targets.
The persistence of Stb1 at MBF target promoters in cln1Δ cln2Δ mutants appears to be consistent with the conclusions of Costanzo et al. (8) that Cln1/2-CDK-dependent dissociation of Stb1 is required for inactivation of MBF targets. However, we observed no effect of inactivating Stb1 on the timing of transcriptional inactivation of RNR1 in cells synchronized by release from mating pheromone arrest (Fig. 4C). This is consistent with the finding that Nrm1 accumulates and binds to MBF promoters in cln1Δ cln2Δ mutants with the same kinetics as in wild-type cells (Fig. 4B). Binding of Nrm1 correlates with and is required for inactivation of MBF-regulated transcription at the G1-S transition (9). We conclude that, although Stb1 persists at MBF promoters, it has no effect on the effectiveness of transcription inactivation as cells enter S phase. Furthermore, Stb1 does not influence the binding or activity of Nrm1 at MBF promoters.
Increase of G1-specific transcript levels and persistence of SBF-dependent gene expression in cln1Δ cln2Δ cells depends, in large part, on low Clb/CDK activity.
Inactivation of Cln1 and Cln2 results in increased levels of G1-specific transcription (11, 27) (Fig. 4). Whereas Cln/CDK-dependent phosphorylation of Stb1 has been proposed to inactivate MBF-dependent transcription (8), repression of G1-specific gene expression also depends on Clb/CDK activity (1, 9, 20, 23) and Nrm1 (9). Cln-dependent phosphorylation of the Clb/CDK specific inhibitor Sic1 targets it for degradation by SCFcdc4 (29). It has been suggested that the hyperinduction of G1-specific transcription observed in cln1Δ cln2Δ cells might, therefore, depend on low Clb/CDK levels (27). To determine whether the increase in G1-specific transcript levels and their extended duration in the cln1Δ cln2Δ mutant cells results from an inability to inactivate Stb1 or to activate Clb/CDK, we compared G1 transcript levels in wild-type, cln1Δ cln2Δ, cln1Δ cln2Δ stb1Δ, and cln1Δ cln2Δ sic1Δ cells synchronized by mating pheromone arrest and release (Fig. 4C). In cln1Δ cln2Δ cells the MBF-regulated transcript RNR1 and the SBF target CLN2 are significantly elevated compared to wild-type cells. However, MBF transcript RNR1 is still inactivated in a timely manner, due to the activity of Nrm1 (Fig. 4B) (9), whereas repression of the SBF target CLN2 exhibits a 10-min delay. Because comparable elevated transcript levels are observed in the cln1Δ cln2Δ and cln1Δ cln2Δ stb1Δ cells, we conclude that neither the increase of G1-specific transcript levels nor timely inactivation of SBF-dependent gene expression in cln1Δ cln2Δ cells depends upon Stb1. In contrast, inactivation of the Clb/CDK specific inhibitor Sic1 in cln1Δ cln2Δ cells significantly lowers G1-specific transcript levels and largely restores the timely repression of the SBF target CLN2. Strikingly, whereas timely inactivation of transcription depends on Nrm1 the peak expression level of the MBF target RNR1 is restored to its wild-type status in cln1Δ cln2Δ sic1Δ mutants. Therefore, both the increase of G1-specific transcript levels and the delay in transcriptional repression of SBF targets observed in cln1Δ cln2Δ cells can largely be explained by the delayed activation of Clb/CDK associated with the delay in Sic1 proteolysis. Based on these results, we conclude that Nrm1 and Clb/CDK, but not Stb1, mediate inactivation of G1-specific transcription during the G1-S transition.
Stb1 is required for repression of G1-specific transcripts in G1 phase prior to Start and peak expression of MBF-dependent transcripts.
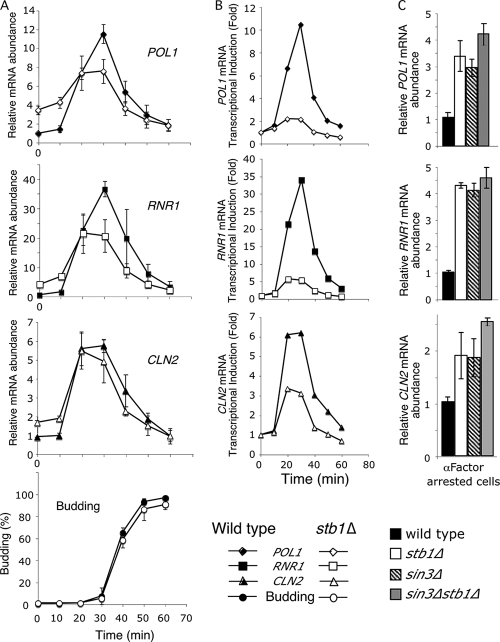
Because binding of Stb1 to G1-specific promoters coincides with G1-specific transcription but its disassociation has no role in inactivation, we sought to determine whether Stb1 is involved in the regulation of this wave of cell cycle-regulated transcription during G1 (Fig. 6). Wild-type and stb1Δ cells were synchronized by release from alpha-factor arrest and gene expression monitored upon release into the cell cycle. In wild-type and stb1Δ cells both the MBF-regulated transcripts (RNR1, POL1, and CDC21) and the SBF targets (CLN2 and SVS1), as determined by quantitative RT-PCR, are activated after 10 min and reached a maximum level at 20 to 30 min after release from mating pheromone arrest, which coincides with the time of bud emergence (Fig. 6A and data not shown). Whereas timely activation and inactivation was not affected in stb1Δ cells compared to the wild type, a significant increase in transcript levels was observed in cells arrested in G1 phase by mating pheromone and immediately after release (0- and 10-min time points; Fig. 6A and C). Previous results obtained from cells synchronized by centrifugal elutriation also show that G1-specific transcriptional induction is dampened in stb1Δ cells (16).
FIG. 6.
Stb1 is required for repression of G1-specific genes in G1 phase and for attainment of the wild-type levels of MBF gene expression. (A) mRNA levels (quantitative RT-PCR; expressed as the increase over lowest level detected in wild-type cells after normalization to ACT1 mRNA) in alpha-factor synchronized wild-type and stb1Δ cells for the indicated time points. At the bottom of panel A, the budding index of wild type and stb1Δ cells is presented as an indicator of synchrony. (B) Data from one of the time courses used in panel A expressed as the mRNA induction in wild-type and stb1Δ cells compared to levels detected in alpha-factor arrested cells from these strains. (C) Bar graph representation of mRNA levels detected in alpha-factor arrested wild-type, stb1Δ, sin3Δ, and sin3Δstb1Δ cells expressed as the induction over wild-type levels. In panels A and C, the average values from three independent experiments, each run in triplicate, are presented with the standard errors.
Previous studies show that Stb1 is required for MBF transcriptional activity but not SBF-dependent transcription (8). Consistent with these observations deletion of Stb1 results in a significant reduction of the peak level of transcript accumulation from MBF-regulated genes but not from SBF-regulated genes (Fig. 6A). Overall, inactivation of Stb1 results in a dampening of transcriptional induction of G1-specific genes (Fig. 6B). We conclude that Stb1 is required for repression of G1-specific genes in G1 phase prior to Cln3-depdendent transcriptional activation at Start and for attainment of the wild-type levels of MBF gene expression. Inactivation of CLN1 and CLN2 is often associated with transcriptional derepression in large G1-arrested cells, although the basis for this effect is unknown (see Fig. 4C).
Stb1 and Sin3 are required for transcriptional repression prior to Cln-dependent transcriptional activation.
The effect of inactivation of Stb1 on G1-specific genes is largely on transcriptional repression prior to Cln3-dependent activation of transcription (Fig. 6C). Stb1 was originally identified via its interaction with the HDAC complex component Sin3 in a two-hybrid screen (18). Sin3 is commonly associated with transcriptional repression, but a role in G1-specific transcription regulation has not been established. Interestingly, inactivation of Sin3 has the same effect on early expression of both SBF and MBF targets. Furthermore, sin3Δ appears to be epistatic to stb1Δ in that the double mutant exhibits a similar level of transcriptional derepression as the individual mutants (Fig. 6C). These results suggest that Stb1 and Sin3 participate in the same pathway.
DISCUSSION
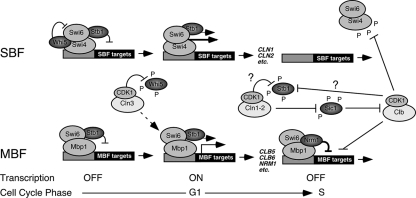
A recent model for Stb1-dependent regulation of G1-specific transcription predicts that Stb1 functions to activate G1-specific transcription and that Cln/CDK-dependent phosphorylation of Stb1, which promotes the dissociation of Stb1-Swi6, is required for inactivation of MBF-specific transcription as cells exit G1 phase (8, 16). We show here that Stb1 plays a role in modulating G1-specific transcription by binding to both SBF and MBF via the common component Swi6. Stb1 associates with SBF- and MBF-dependent promoters throughout G1 phase and dissociates coincident with transcriptional inactivation (Fig. 7). Whereas Stb1 dissociates from MBF at promoters, it dissociates from SBF targets at the same time as the SBF complex. In agreement with published observations, we found that dissociation from G1-specific promoters is associated with an increase in phosphorylated Stb1, but this depends only in part upon the accumulation of Cln1/2 CDK during late G1 phase. However, whereas inactivation of Cln1 and Cln2 prolongs the binding of Stb1 to promoters, the increase of G1-specific gene expression observed in cln1Δ cln2Δ mutants does not depend on Stb1. Instead, both the high magnitude of expression and the delayed repression of SBF-dependent gene expression depend upon the extended period of low Clb/CDK activity. We conclude that Stb1 is not required for activation of G1-specific transcription (also see reference 8), nor is it required for timely inactivation during exit from G1 phase as previously suggested (16). However, Stb1 does appear to play a role at both SBF and MBF promoters for the maintenance of transcriptional repression prior to transcriptional activation during G1 phase and for attainment of maximal transcription of some G1-specific targets during G1 (Fig. 7).
FIG. 7.
Stb1 modulates G1-specific transcription during G1. Model depicting G1-specific transcriptional regulation in budding yeasts. In G1 phase, prior to Cln3-dependent activation, transcriptional repression is maintained by a complex containing Stb1/SBF/Whi5 and Stb1/MBF bound to target promoters. Cln3/CDK relieves transcriptional repression by inactivating Whi5 via phosphorylation, thereby activating SBF-specific transcription and by antagonizing the repressive activity of Stb1/MBF via a mechanism that has yet to be established. During the G1/S transition, Stb1 can be phosphorylated by Cln/CDK or Clb/CDK and released from promoters. Transcription of MBF targets is inactivated by binding of the MBF-associated corepressor Nrm1, whereas the transcription of SBF targets is inactivated by Clb/CDK-dependent phosphorylation of SBF components. Inactivation of MBF targets is also reinforced by Clb/CDK.
Whereas Nrm1 and Whi5 act as transcriptional repressors that confine transcription to the G1 phase, Stb1 seems to play a modulatory role in both transcriptional repression and activation. The mechanism by which Stb1 represses transcription prior to Cln3/CDK activation and activates MBF-specific genes during late G1 phase remains unknown. The identification of Stb1 as an interactor of the transcriptional regulator Sin3 (18) may provide some insight into Stb1-dependent transcriptional repression in G1 phase. Sin3 has been associated with transcriptional repression of a large number of genes (reviewed in reference 24), but a role in G1-specific transcription regulation has not been established. We now demonstrate that inactivation of Stb1 or Sin3 has a similar effect on expression from G1-specific promoters prior to transcriptional induction during G1 phase. Furthermore, the effect on transcription indicates that both Sin3 and Stb1 participate in the pathway leading to the repression of G1-specific transcription. In addition, our preliminary results show that Sin3 associates with both SBF and MBF-dependent promoters (data not shown). However, we find that the binding of Sin3 is not dependent upon Stb1. These data are consistent with a model in which Stb1 regulates the activity of Sin3 (presumably in the context of the Rpd3 HDAC complex) without affecting its binding. It remains to be established whether Stb1 or other transcriptional repressors, including Whi5 and Nrm1, regulate the Sin3/HDAC complex to G1-specific promoters and whether Sin3 participates in the regulation of G1-specific gene expression.
The Stb1 protein contains 5 perfect and 12 minimal putative CDK sites. Previous studies have shown that Stb1 is a direct target for phosphorylation by Cln1/2-associated CDK in vitro (16). Furthermore, in vitro phosphorylation of Stb1 by Cln-CDK regulates its ability to associate with Swi6 (8). In addition, our data show that a Cln1/2-CDK-dependent shift of Stb1 in vivo correlates with disassociation from G1-specific promoters (Fig. 5). However, we also show that phosphorylation of Stb1 in vivo depends only in part upon Cln1/2-CDK and that the effect of Cln1 and Cln2 on the level and duration of G1-specific transcription is independent of Stb1. It seems likely that the residual phosphorylation of Stb1 depends upon Clb/CDK. Whether Stb1 phosphorylation is required for its release from the transcription complex awaits the analysis of CDK-dependent phosphorylation sites.
Although it has been reported that Stb1 binds and activates synthetic promoters containing MCB elements but not synthetic promoters containing SCB's (8), we show that it binds endogenous MBF and SBF target promoters in vivo and regulates their transcription (Fig. 7). Our data, obtained using mating pheromone synchronized cells, agree with previous results obtained using cells synchronized by centrifugal elutriation showing that G1-specific transcriptional induction is dampened in stb1Δ cells (16). Consistent with these findings, we observed an additive cell size defect in double mutants of stb1Δ and either swi4Δ or mbp1Δ (Fig. 3). In contrast, Ho et al. (16) reported that a swi4Δ stb1Δ has no obvious additive phenotype, whereas Costanzo et al. (8) found the same double mutant to be lethal. Whether these differences are a result of differences in background remains to be established. Nevertheless, all studies show that the interaction of Stb1 with Swi6 is important for the role of Stb1 in transcriptional regulation of G1-specific genes.
Inactivation of Stb1 has a greater effect on the expression of MBF-regulated genes than those regulated by SBF. That difference might be explained by the differences in the manner by which SBF and MBF regulate gene expression. Although SBF and MBF evoke identical pattern of cell cycle-regulated transcription, SBF acts as a transcriptional activator, whereas MBF acts as a transcriptional repressor (2, 9, 20). However, until it is known how Stb1 mediates transcriptional regulation, we can only speculate as to why the two classes of promoters are affected differently.
The increase in G1-specific transcript levels observed in cln1Δ cln2Δ cells has been previously proposed to result from the low Clb/CDK activity that occurs as a consequence of the persistence of the Clb/CDK specific inhibitor Sic1 in those mutants (27). Here we show that inactivating the Clb/CDK inhibitor in cln1Δ cln2Δ mutants lowers the maximal transcript levels of SBF genes and restores the level of MBF transcripts to that observed in wild-type cells. Conversely, inactivating Clb/CDK results in persistence of SBF-dependent gene expression (1, 9). These observations are consistent with a negative-feedback loop in which Cln1-2/CDK inactivates Sic1, thereby activating Clb/CDK, which not only affects the timing of transcriptional inactivation of SBF-dependent transcription but also restricts the maximal expression of G1-specific genes (Fig. 7).
Transcriptional repression is a critically important mechanism for the maintenance of cellular homeostasis. The present study highlights the diversity of mechanisms that cells use to maintain appropriate repression of periodically expressed genes during the cell cycle. Despite limited conservation of the primary sequence, the G1-specific transcription system seems to be conserved from yeast to humans (5). Regulation of G1-specific transcription in mammals depends on the E2F family of transcription factors and their regulators, the pocket proteins (4). Like Whi5 and Nrm1, pocket proteins appear to implement the repression of G1-specific genes in collaboration with transcriptional activators and repressors. Like the regulation invoked by Stb1 and Sin3, which we presume to be mediated by histone deacetylation, G1-specific transcriptional regulators recruit HDACs to their target promoters (13). However, the details of that regulation are only poorly understood. Maintenance of transcriptional repression of G1-specific genes is likely to be important for the maintenance of quiescence during nutrient limitation and cell cycle arrest by physiological signals. Further investigation of the mechanisms governing transcriptional repression will be important in unraveling this complex regulatory system.
Acknowledgments
We thank M. Guaderrama for technical support, J. Yates III and W. H. McDonald for mass spectrometry analysis that led to the identification of Stb1 as an SBF and MBF interacting protein, and members of the TSRI Cell Cycle Group for helpful discussion.
This study was supported by USPHS grant GM059441 to C.W.
Footnotes
Published ahead of print on 15 September 2008.
REFERENCES
- 1.Amon, A., M. Tyers, B. Futcher, and K. Nasmyth. 1993. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell 74993-1007. [DOI] [PubMed] [Google Scholar]
- 2.Bean, J. M., E. D. Siggia, and F. R. Cross. 2005. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics 17149-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates III, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107537-548. [DOI] [PubMed] [Google Scholar]
- 4.Cobrinik, D. 2005. Pocket proteins and cell cycle control. Oncogene 242796-2809. [DOI] [PubMed] [Google Scholar]
- 5.Cooper, K. 2006. Rb, whi it's not just for metazoans anymore. Oncogene 255228-5232. [DOI] [PubMed] [Google Scholar]
- 6.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 71213-1220. [DOI] [PubMed] [Google Scholar]
- 7.Costanzo, M., J. L. Nishikawa, X. Tang, J. S. Millman, O. Schub, K. Breitkreuz, D. Dewar, I. Rupes, B. Andrews, and M. Tyers. 2004. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 117899-913. [DOI] [PubMed] [Google Scholar]
- 8.Costanzo, M., O. Schub, and B. Andrews. 2003. G1 transcription factors are differentially regulated in Saccharomyces cerevisiae by the Swi6-binding protein Stb1. Mol. Cell. Biol. 235064-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bruin, R. A., T. I. Kalashnikova, C. Chahwan, W. H. McDonald, J. Wohlschlegel, J. Yates III, P. Russell, and C. Wittenberg. 2006. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol. Cell 23483-496. [DOI] [PubMed] [Google Scholar]
- 10.de Bruin, R. A., W. H. McDonald, T. I. Kalashnikova, J. Yates III, and C. Wittenberg. 2004. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 117887-898. [DOI] [PubMed] [Google Scholar]
- 11.Dirick, L., T. Bohm, and K. Nasmyth. 1995. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. EMBO J. 144803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flick, K. M., N. Spielewoy, T. I. Kalashnikova, M. Guaderrama, Q. Zhu, H. C. Chang, and C. Wittenberg. 2003. Grr1-dependent inactivation of Mth1 mediates glucose-induced dissociation of Rgt1 from HXT gene promoters. Mol. Biol. Cell 143230-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frolov, M. V., and N. J. Dyson. 2004. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 1172173-2181. [DOI] [PubMed] [Google Scholar]
- 14.Hadwiger, J. A., C. Wittenberg, H. E. Richardson, M. de Barros Lopes, and S. I. Reed. 1989. A family of cyclin homologs that control the G1 phase in yeast. Proc. Natl. Acad. Sci. USA 866255-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrington, L. A., and B. J. Andrews. 1996. Binding to the yeast SwI4,6-dependent cell cycle box, CACGAAA, is cell cycle regulated in vivo. Nucleic Acids Res. 24558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho, Y., M. Costanzo, L. Moore, R. Kobayashi, and B. J. Andrews. 1999. Regulation of transcription at the Saccharomyces cerevisiae start transition by Stb1, a Swi6-binding protein. Mol. Cell. Biol. 195267-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409533-538. [DOI] [PubMed] [Google Scholar]
- 18.Kasten, M. M., and D. J. Stillman. 1997. Identification of the Saccharomyces cerevisiae genes STB1-STB5 encoding Sin3p binding proteins. Mol. Gen. Genet. 256376-386. [DOI] [PubMed] [Google Scholar]
- 19.Koch, C., T. Moll, M. Neuberg, H. Ahorn, and K. Nasmyth. 1993. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 2611551-1557. [DOI] [PubMed] [Google Scholar]
- 20.Koch, C., A. Schleiffer, G. Ammerer, and K. Nasmyth. 1996. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at Start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 10129-141. [DOI] [PubMed] [Google Scholar]
- 21.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14953-961. [DOI] [PubMed] [Google Scholar]
- 22.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 171030-1032. [DOI] [PubMed] [Google Scholar]
- 23.Siegmund, R. F., and K. A. Nasmyth. 1996. The Saccharomyces cerevisiae Start-specific transcription factor Swi4 interacts through the ankyrin repeats with the mitotic Clb2/Cdc28 kinase and through its conserved carboxy terminus with Swi6. Mol. Cell. Biol. 162647-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silverstein, R. A., and K. Ekwall. 2005. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 471-17. [DOI] [PubMed] [Google Scholar]
- 25.Simon, I., J. Barnett, N. Hannett, C. T. Harbison, N. J. Rinaldi, T. L. Volkert, J. J. Wyrick, J. Zeitlinger, D. K. Gifford, T. S. Jaakkola, and R. A. Young. 2001. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell 106697-708. [DOI] [PubMed] [Google Scholar]
- 26.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 93273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuart, D., and C. Wittenberg. 1995. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 92780-2794. [DOI] [PubMed] [Google Scholar]
- 28.Tyers, M., G. Tokiwa, and B. Futcher. 1993. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. EMBO J. 121955-1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma, R., R. S. Annan, M. J. Huddleston, S. A. Carr, G. Reynard, and R. J. Deshaies. 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278455-460. [DOI] [PubMed] [Google Scholar]
- 30.Wittenberg, C., and S. I. Reed. 2005. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene 242746-2755. [DOI] [PubMed] [Google Scholar]
- 31.Wolters, D. A., M. P. Washburn, and J. R. Yates III. 2001. An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 735683-5690. [DOI] [PubMed] [Google Scholar]