Abstract
Nuclear receptors activate or repress target genes depending on the recruitment of coactivators or corepressors. The corepressor RIP140 and the PPAR coactivator 1α (PGC-1α) both play key roles in the regulated transcription of genes involved in energy homeostasis. We investigated the roles of RIP140 and PGC-1α in controlling the expression of CIDEA, an important regulatory factor in adipose cell function and obesity. Ectopically expressed CIDEA surrounded lipid droplets in brown adipocytes and induced the formation of lipid droplets in nonadipogenic cell lines. The expression and promoter activity of CIDEA was repressed by RIP140 and induced by PGC-1α, mediated through the binding of estrogen-related receptor α and NRF-1 to their cognate binding sites. Importantly, we demonstrate that RIP140 interacts directly with PGC-1α and suppresses its activity. The direct antagonism of PGC-1α by RIP140 provides a mechanism for regulating target gene transcription via nuclear receptor-dependent and -independent pathways.
Adipose tissue plays a key role in the development of obesity and diabetes, functioning both as an energy store and as a major endocrine organ to regulate whole-body energy balance. Energy homeostasis is maintained by the regulated expression of gene networks in response to nutritional status, physical activity, temperature, or other changes in environmental conditions (14). A number of transcription factors, including nuclear receptors (NRs) such as peroxisome proliferator-activated receptors (PPARs) and estrogen-related receptors (ERRs), are responsible for regulating transcription from many metabolic gene networks (12, 15). Their activity depends on factors which recruit chromatin remodeling enzymes. PPAR coactivator 1α (PGC-1α), which promotes the activity of several transcription factors, including PPARs, ERRs, nuclear respiratory factors (NRFs), and FoxO transcription factors, plays an important role in mitochondrial biogenesis and respiration (20, 35, 42, 44, 47, 48). Conversely, RIP140 (4) is a corepressor for NRs that suppresses catabolic signaling pathways in white adipose tissue (WAT) (30, 40) and muscle (49) while, in contrast, promoting lipogenesis in liver (19). Overall, the changes in metabolic gene expression found in RIP140-null mice are accompanied by a reduction in triglyceride stores in WAT, protection from hepatic steatosis, resistance to diet-induced obesity, and increased glucose clearance and insulin sensitivity (30, 40).
Inspection of the metabolic genes that are upregulated in the absence of RIP140 (30, 40, 49) suggests that they may also be targets for transcriptional control by PGC-1α. Notable among these is the gene encoding uncoupling protein 1 (Ucp1) (5, 11), which is normally restricted to expression in brown adipose tissue (BAT) (3, 29). Other potential target genes that are highly expressed in BAT include those encoding carnitine palmitoyltransferase 1b, cytochrome c oxidase, subunit VIIIb (Cox8b), PPARα, somatic cytochrome c (Cycs), and cell death-inducing DFFA-like effector A (CIDEA) (5, 30). We have focused on CIDEA, since it plays an important role in metabolism in addition to its reported role in programmed cell death (21). In murine BAT, it is a regulator of Ucp1 function (56), and in human WAT, it was found to be a negative regulator of lipolysis in human white adipocytes (39). Interestingly, its expression was decreased in mouse and human WAT following diet-induced obesity (17, 36), and it was the CIDEA gene that exhibited the greatest increase in expression in human adipose tissue following weight loss (9). A polymorphism in the CIDEA gene has also been found to be associated with obesity (8).
To investigate the function of CIDEA, we have expressed it ectopically and observed the appearance of cytoplasmic lipid droplets. To study transcriptional regulation of the CIDEA gene, we generated immortalized adipocytes that can be induced to differentiate into brown adipocytes. We found that CIDEA transcription is stimulated by ERRα and NRF-1 and demonstrate that both transcription factors are targets for activation by PGC-1α and repression by RIP140. Remarkably, RIP140 antagonizes PGC-1α activity, and these two cofactors interact directly. We postulate that PGC-1α and RIP140 play key roles in determining the activities of ERRα and NRF-1 and thereby regulate transcription from metabolic genes such as the CIDEA gene.
MATERIALS AND METHODS
Cell culture.
The IMBAT-1 cell line was generated by the culture of cells prepared from interscapular BAT deposits of the mouse line (H-2Kb-tsA58) (22) expressing a gamma interferon-inducible thermolabile simian virus 40 large T antigen, as described previously (11). They were grown in the presence of 2 ng/ml gamma interferon (Chemicon) at 33°C and differentiated at 37°C (11). Differentiated cells were stained using Oil Red O (30).
Transfections and plasmids.
The CIDEA coding region was PCR amplified from the I.M.A.G.E. clone ID 1397772 provided by Geneservice Ltd. (www.geneservice.co.uk) and cloned into the vector pcDNA3.1D/V5-His-TOPO or pcDNA3.1/NT-GFP-TOPO (Invitrogen). CIDEA expression was monitored by immunostaining with anti-V5 antibody (Invitrogen) diluted 1/300 in phosphate-buffered saline and with the secondary antibody AF568 (Invitrogen), an anti-mouse antibody conjugate, diluted 1/300 in the presence of 2.5 μg/ml BODIPY 493/503 (Invitrogen). Triglyceride accumulation was examined by Oil Red O staining (30). Stable cell lines expressing green fluorescent protein (GFP)-CIDEA or GFP alone were generated by Lipofectamine 2000 (Invitrogen) transfection of 293 cells with pcDNA3.1/NT-GFP-CIDEA or pcDNA3.1/NT-GFP followed by selection with G418.
Reporter assays were carried out in 96-well plates with 20 ng of reporter gene and 5 ng of pRL-CMV, in the absence or presence of ERRα (20 ng), PGC-1α (10 ng), pSG-NRF-1 (10 ng), pEF-RIP140, pSUPER-SiRIP140 (11), or pSUPER-Scambled (pSUPER-Scam) using FuGene6 (Roche). The 4×NRF-1/Cycs/luc promoter construct (16) and the ligand-binding domain (LBD) of ERRα fused to the Gal4 DNA binding domain (DBD) (the Gal-ERRα-LBD fusion protein) (23) have been described previously.
Adenoviral infection.
Adenoviruses expressing CIDEA or GFP were constructed and prepared with the pAdEasy system (Stratagene) by following the manufacturer's instructions. IMBAT-1 adipocytes were infected with adenovirus vectors expressing CIDEA, RIP140, PGC-1α, GFP, small interfering RNA (siRNA) against RIP140, or a nontargeting random sequence (37). Equal levels of infection were confirmed by fluorescence microscopy for the coexpressed GFP. Cells infected with adenovirus to express GFP or CIDEA were methanol fixed and immunostained with anti-V5 antibody (1/200; QED Bioscience), followed by a Cy3-conjugated anti-goat antibody (1/400; Jackson ImmunoResearch Labs) and visualized by confocal microscopy.
Immunoprecipitation.
Cells were lysed in 50 mM Tris (pH 8), 250 mM NaCl, 1 mM EDTA, and 0.5% NP-40 with protease inhibitor cocktail (Roche). Anti-PGC-1α antibody (sc-13067; Santa Cruz Biotechnology) or anti-V5 antibody (Invitrogen) was incubated with lysate and protein AG plus agarose beads (Santa Cruz Biotechnology).
Western blotting.
Adipocytes were lysed in Laemmli loading buffer, electrophoresed on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, and transferred to a polyvinylidene difluoride membrane. Membranes were probed with specific antibodies and bands and visualized with SuperSignal West Pico substrate (Pierce). Primary antibodies were incubated at the following dilutions: for CIDEA (AB16922; Chemicon), 1 in 1,000; RIP140 (6D7 generated against residues 301 to 478), 1 in 500; and GAPDH (glyceraldehyde-3-phosphate dehydrogenase; MAB374; Chemicon), 1 in 5,000.
Expression analysis.
Total RNA was isolated from cells using TRIzol (Invitrogen), and cDNA was prepared (5). Expression levels of genes were determined by using specific primers with Sybr green reagent or TaqMan probes and correlated to that for the L19 ribosomal coding gene. Primer sequences may be obtained on request.
GST assays.
Expression vectors were transcribed and translated in vitro in the presence of [35S]methionine in reticulocyte lysate (Promega). Glutathione S-transferase (GST) fusion proteins were induced, purified, bound to glutathione-Sepharose (Amersham Biosciences), and incubated with translated proteins, as described previously (6). After being washed, the samples were separated by SDS-10% PAGE. Gels were fixed and dried, and the 35S-labeled proteins were visualized by autoradiography. The expression of GST-RIP140 and GST-PGC-1α fragments was confirmed by Coomassie staining (see Fig. S1 in the supplemental material) (27).
EMSA.
Electrophoretic mobility shift assay (EMSA) studies were carried out as described previously (38). The labeled oligonucleotide probe corresponded to the NRF-1 binding site (CTCGGTACCGTTTGCGCACGAAGG), in the CIDEA promoter (indicated by underlining).
ChIP assay.
Differentiated IMBAT-1 cells were cross-linked (1% formaldehyde for 15 min), lysed, sonicated, and immunoprecipitated with protein A/G PLUS-agarose (Santa Cruz Biotechnology) using antibodies raised against RIP140 (raised against residues 587 to 843), PGC-1α (sc-13067; Santa Cruz Biotechnology), NRF-1 (sc-721; Santa Cruz Biotechnology), or normal mouse immunoglobulin G (IgG) (Santa Cruz Biotechnology). DNA fragments were purified with a QIAquick PCR purification kit (Qiagen) and used for templates in PCRs. Cells were synchronized with α-amanitin (2.5 μM for 2 h), followed by removal prior to fixation for the sequential chromatin immunoprecipitation (re-ChIP). Chromatin was eluted with dithiothreitol (10 mM) from the first antibody-protein A/G PLUS-agarose complex.
RESULTS
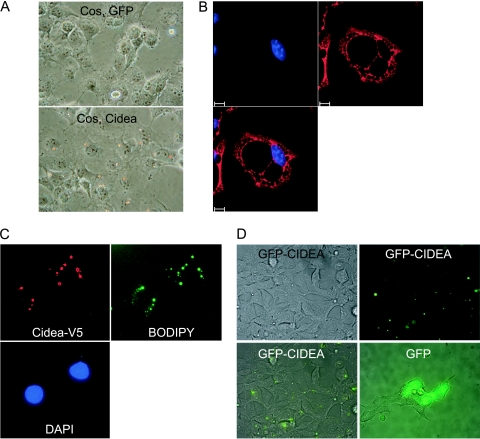
The function of CIDEA was examined by expressing the gene in a number of cell lines, and we noted the appearance of spherical structures that stained with Oil Red O, indicating that they were lipid droplets (Fig. 1A, C, and D). The response was observed not only in preadipocyte lines (3T3-L1) but also in Cos, 293, MDA-MB-231, and human fetal mesenchymal stem cells (Fig. 1 and data not shown). Immunocytochemical staining in differentiated brown adipocytes showed the cytoplasmic presence of exogenous CIDEA in proximity to the surfaces of lipid droplets (Fig. 1B). Localization was further studied in Cos cells transiently transfected with CIDEA-V5, which induced the appearance of spherical structures that stained positively with the neutral lipid stain BODIPY and colocalized with immunostain for the V5 epitope (Fig. 1C). Lipid droplets were also induced by the presence of the GFP-CIDEA fusion protein but not by GFP alone (Fig. 1D). Fluorescence microscopy of viable cells detected GFP-CIDEA in discrete cytoplasmic regions that, when overlaid with a phase-contrast image, colocalized to the lipid droplets, whereas GFP was distributed diffusely throughout the cell (Fig. 1D). We investigated whether ectopic CIDEA expression caused induction of PPARγ expression, a key regulator of the adipogenic process, but there was no increase as judged by Western blotting (see Fig. S2A in the supplemental material), indicating that the accumulation of lipid droplets is not due to initiation of the adipogenic program.
FIG. 1.
CIDEA-dependent induction of lipid droplet accumulation. (A) Oil Red O stained Cos cells infected with adenovirus to express GFP or CIDEA. (B) Confocal microscopy of differentiated IMBAT-1 cells immunostained for V5 and DAPI (4′,6-diamidino-2-phenylindole) following adenoviral infection to express CIDEA-V5. Bars, 10 μm. (C) Cos cells transiently transfected with pcDNA3.1-CIDEA-V5 were immunostained with anti-V5 antibody and costained with BODIPY 493/503. (D) Stable cell lines expressing GFP or GFP were visualized by phase-contrast (upper left) or fluorescence (upper right) microscopy, and images were overlaid (lower panels).
With the knowledge that CIDEA promotes the formation of lipid droplets and inhibits catabolic processes, including lipolysis and respiratory uncoupling, we tested its ability to modify total cellular oxygen consumption. The level of oxygen consumed by cells expressing CIDEA was considerably less than that detected in control cells expressing GFP (see Fig. S2C in the supplemental material). These data show that CIDEA expression in adipocytes causes a metabolic shift leading to reduced oxygen consumption and lipid droplet formation in both adipogenic and nonadipogenic cell lines.
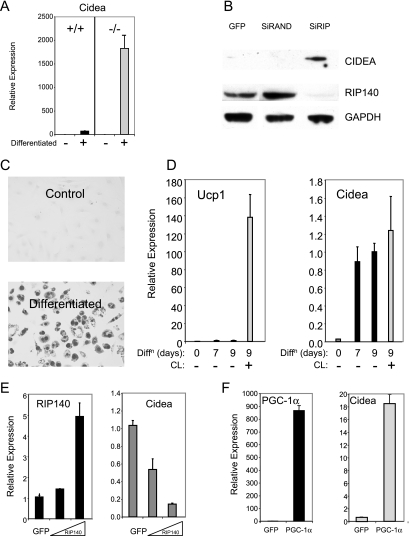
Quantitative mRNA analysis of adipocytes differentiated from primary mouse embryo fibroblasts (Fig. 2A) confirmed that CIDEA expression was elevated in cells lacking RIP140. Since BAT is a major site of CIDEA gene expression (50), we utilized the IMBAT-1 cell line expressing an inducible temperature-sensitive version of simian virus 40 T antigen to study its regulation (22). Knockdown of endogenous RIP140 levels in IMBAT-1 adipocytes by adenoviral infection of siRNA targeting RIP140 resulted in elevated levels of CIDEA protein that were not found in the presence of a nontargeting sequence (Fig. 2B). We characterized the IMBAT-1 cells which undergo adipogenesis, which is manifested morphologically by the appearance of lipid droplets staining positive with Oil Red O (Fig. 2C), coincident with the induction of mRNA for aP2 and PPARγ (see Fig. S3A in the supplemental material). The differentiated IMBAT-1 cell line retained functional responses characteristic of BAT. For example, Ucp1 expression was induced 5-fold after adipocyte differentiation and was further elevated, more than 100-fold, by the selective β3 agonist CL316,243 (Fig. 2D). CIDEA levels also increased with differentiation; however, no additional increase was observed with CL316,243. We have found that many genes involved in catabolic pathways in adipocytes are upregulated in the absence of RIP140 (5, 40) and noted that a high proportion are also targets for PGC-1α. Both RIP140 and PGC-1α levels were induced following differentiation, although only PGC-1α expression was increased with CL316,243 (see Fig. S3B in the supplemental material).
FIG. 2.
CIDEA expression is regulated by RIP140 and PGC-1α in the brown fat cell line IMBAT-1. (A) Results from real-time PCR analysis of mRNAs for CIDEA. Primary cultures of mouse embryo fibroblasts prepared from wild-type (+/+) or RIP140-null (−/−) tissue were untreated (−) or differentiated (+) for 6 days. Data are expressed relative to results for untreated wild-type cells. (B) Immunoblot for CIDEA, RIP140, and GAPDH in IMBAT-1 cells following adenovirus-mediated expression of GFP, scrambled small inhibitory RNA (SiRAND), or RIP140-targeting small inhibitory RNA (SiRIP). (C) IMBAT-1 cells, untreated or differentiated, were stained with Oil Red O. (D) The expression of Ucp1 and CIDEA in IMBAT-1 cells that were untreated, differentiated, or treated with CL316,243 for 5 h was analyzed by real-time PCR. Diffn, differentiated; CL, CL316, 243. (E and F) CIDEA gene expression in differentiated IMBAT-1 cells was assessed by real-time PCR following adenovirus-mediated expression of RIP140 (E) or PGC-1α (F).
Since RIP140 and PGC-1α seem to regulate transcription from a number of common gene targets, we investigated their functional interplay. In preliminary experiments, we determined whether genes whose expression was upregulated in the absence of RIP140 were repressed or activated by exogenous expression of the corepressor or coactivator, respectively. In this way, we were able to confirm that the targeted repression of CIDEA by RIP140 was recapitulated in adipocytes by adenovirus-mediated RIP140 expression (Fig. 2E). Furthermore, the expression of Cox8b and Cycs, but not aP2, was repressed by RIP140 (see Fig. S4 in the supplemental material). Conversely, expression of the transcriptional coregulator PGC-1α increased the expression of the same set of genes, including the CIDEA gene (Fig. 2F; see Fig. S4 in the supplemental material).
To investigate the regulation of the CIDEA promoter, we identified potential regulatory elements in the 5′-end flanking region following rapid amplification of cDNA ends to determine the start sites of transcription in brown adipocytes. From the 5′ rapid amplification of cDNA ends, we could identify several clustered transcription start sites in the first exon. CpG islands were present at the transcription start sites, and potential binding sites were identified for NRs (see Fig. S5 in the supplemental material).
Given that CIDEA expression was increased by PGC-1α (Fig. 2C), we investigated the ability of PPARs to stimulate its expression in differentiated adipocytes. CIDEA levels were not affected by PPARα or -δ ligands and increased slightly with a PPARγ agonist (see Fig. S6 in the supplemental material). Furthermore, the CIDEA promoter was unaffected by any of the PPAR ligands, even in the presence of PGC-1α, whereas a synthetic PPAR response element-containing reporter construct was potently induced (data not shown).
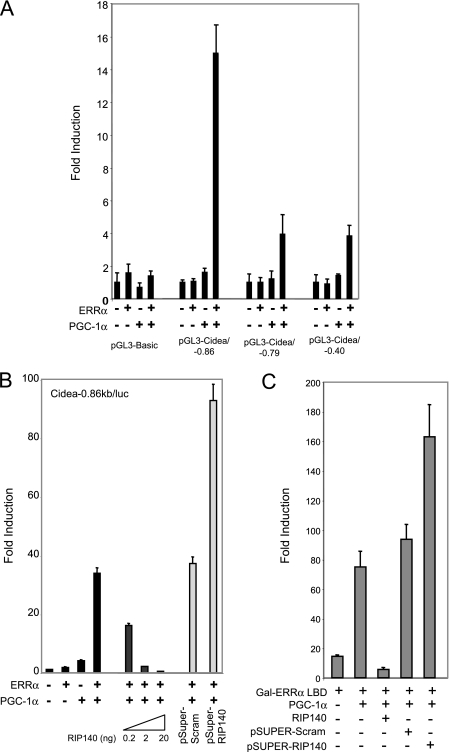
PGC-1α is a coactivator for a number of other NRs, including ERRα and transcription factors such as Foxo1A, SREBP, and NRF-1 (34, 42, 47). We found that transcription from the CIDEA promoter was stimulated by ERRα expression in the presence of PGC-1α (Fig. 3A). Deletion analysis indicated that DNA sequences between −861 and −793 were responsive to ERRα/PGC-1α, and inspection of this sequence revealed a perfect match to the consensus ERR response element (ERRE), TGAAGGTCA at −856. The expression of RIP140 inhibited ERRα/PGC-1α activation of the CIDEA promoter in a dose-dependent manner (Fig. 3B). Furthermore, siRNA-mediated knockdown of RIP140 elevated ERRα/PGC-1α-dependent reporter gene activity (Fig. 3B), indicating that endogenous RIP140 inhibited CIDEA promoter activity. These observations indicate that both cofactors are able to target the CIDEA promoter, possibly via recruitment by ERRα.
FIG. 3.
CIDEA promoter activity is activated by ERRα under the control of PGC-1α and RIP140. (A and B) Transient transfection of Cos cells with CIDEA promoter constructs in the presence of ERRα and/or PGC-1α (A) and in the absence and presence of RIP140 or pSuper-RIP140 (B). (C) Transient transfection of Cos cells with pGal-DBD×5/luc and Gal-ERRα-LBD in the absence and presence of PGC-1α, RIP140, pSuper-Scrambled, or pSuper-RIP140.
To investigate the mechanism by which RIP140 repressed ERRα activity, we analyzed its effect on the transcriptional activity of the Gal-ERRα-LBD fusion protein in transient-transfection assays. The activity of the Gal-ERRα-LBD fusion protein, which is dependent on the presence of PGC-1α, was inhibited by RIP140 (Fig. 3C). Furthermore, siRNA-mediated knockdown of RIP140 enhanced the ability of Gal-ERRα-LBD/PGC-1α to induce reporter gene expression. Therefore, overexpression of RIP140 or the depletion of its endogenous levels regulates PGC-1α-dependent coactivation of ERRα.
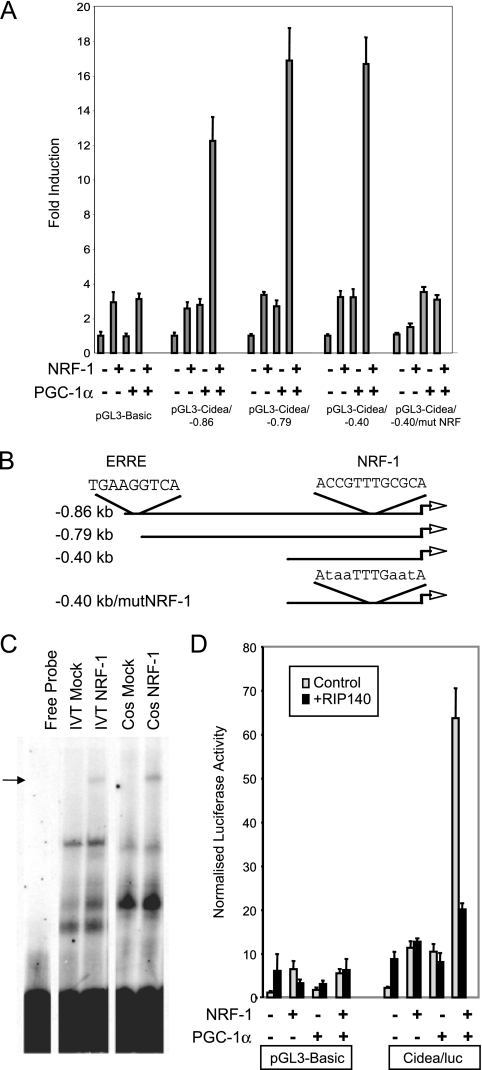
Given that many of the RIP140-repressed metabolic genes are targets for NRF-1, we tested the ability of this transcription factor to stimulate transcription from the CIDEA promoter. NRF-1, in the presence of PGC-1α, induced reporter activity from a series of promoter fragments, with the smallest fragment tested (0.40 kb 5′ of the start codon) maintaining full reporter activity (Fig. 4A). Within this promoter sequence, we identified a putative binding site for NRF-1 (Fig. 4B) that formed a complex with NRF-1 in an electrophoretic mobility shift assay (Fig. 4C) and was necessary for NRF-1-dependent reporter gene activation (Fig. 4A). Conversely, activation of the CIDEA promoter by NRF-1/PGC-1α was inhibited by cotransfection with RIP140 (Fig. 4D), which is surprising since we were unable to demonstrate an interaction between RIP140 and NRF-1 in GST pull-down assays (data not shown). We concluded that RIP140 represses NRF-1 activity by an indirect mechanism.
FIG. 4.
CIDEA promoter activity is activated by NRF-1 under the control of PGC-1α and RIP140. (A) Transient transfection of Cos cells with wild-type and NRF-1 binding site mutants CIDEA promoter constructs in the presence of NRF-1 and/or PGC-1α. (B) Schematic diagram of the CIDEA promoter showing the binding sites for ERRα and NRF-1 (wild-type and site-directed mutants). (C) 32P-labeled oligonucleotide corresponding to the NRF-1 binding site was incubated with mock or NRF-1 in vitro translation (TNT T7 polymerase quick-coupled transcription/translation system; Promega) or Cos cell nuclear protein extract that was mock infected or transfected with NRF-1e. (D) pGL3-CIDEA/−1.3kb/luc was cotransfected with NRF-1 and/or PGC-1α in the absence and presence of RIP140.
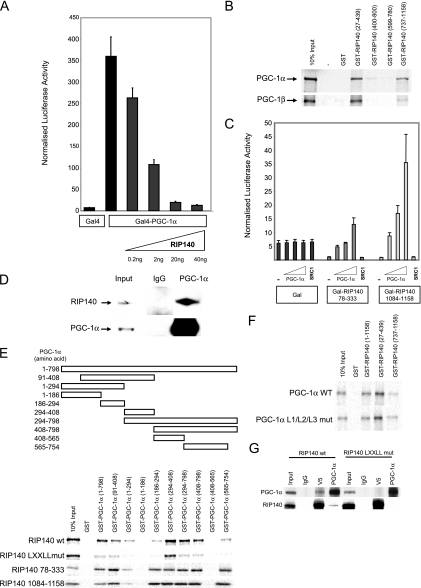
The requirement of PGC-1α for the activation of ERRα and NRF-1 makes it a potential target for RIP140 action. We tested the ability of RIP140 to repress PGC-1α directly by examining the activities of Gal4-DBD fusion proteins. RIP140 suppressed the activity of Gal4-PGC-1α in a dose-dependent manner (Fig. 5A), whereas the activity due to Gal4-SRC-1, which served as a negative control, was unaffected (see Fig. S7 in the supplemental material). Moreover, we were able to demonstrate a direct interaction between the two cofactors in a GST pull-down assay in which 35S-labeled PGC-1α bound to N- and C-terminal fragments of RIP140 (amino acids 1 to 439 and 737 to 1158) (Fig. 5B). 35S-labeled PGC-1β also interacted with GST-RIP140 (Fig. 5B).
FIG. 5.
RIP140 directly interacts with PGC-1α. (A) pGal-DBD×5/luc was cotransfected with Gal-PGC-1α in Cos cells in the presence of increasing amounts of pEF-RIP140. (B and F) RIP140 fragments fused to GST and GST alone used as a control were immobilized on glutathione beads and incubated with 35S-labeled PGC-1α, PGC-1β, or PGC-1α with mutated leucine-rich motifs 1 to 3. Bound proteins were eluted and resolved by SDS-10% PAGE, dried, and exposed to autoradiography. (C) Transient transfection of Cos cells with pGal-DBD/luc and Gal4 or Gal-RIP140 fragments in the absence and presence of PGC-1α (0.2, 2, and 20 ng) or SRC1e (20 ng). (D) Immunoblot for RIP140 and PGC-1α following coimmunoprecipitation with anti-PGC-1α antibody from differentiated IMBAT-1 cell lysate. (E) PGC-1α fragments fused to GST and GST alone used as a control were immobilized on glutathione beads and incubated with 35S-labeled RIP140 wild type (wt), LXXLL mutant, or amino acids 78 to 333 or 1084 to 1158 fused to Gal4. (G) Immunoblot for RIP140 and PGC-1α following coimmunoprecipitation of protein lysate from 293 cells transfected with PGC-1α and V5-RIP140 wild type or LXXLL mutant. Lysates were immunoprecipitated with antibodies raised against PGC-1α, V5 epitope, or a control (normal IgG).
We next investigated the interaction of RIP140 and PGC-1α in intact cells by testing the ability of PGC-1α, via its endogenous transcriptional activation function, to reverse the repressive action of Gal-RIP140 (6) on a luciferase reporter gene. Gal-RIP140 repression domain 1 (RD1; amino acids 78 to 333) and RD4 (amino acids 1084 to 1158) fusion proteins repressed reporter gene activity compared to the effect of Gal4-DBD alone. The repression by these Gal-RIP140 fusion proteins was relieved by PGC-1α coexpression, whereas the activity of Gal4-DBD alone was unaffected (Fig. 5C). In contrast, the p160 coactivator SRC1e did not reverse the repression of these Gal-RIP140 fragments, indicating selectivity for PGC-1α. By using coimmunoprecipitation followed by Western blotting, we were able to demonstrate that the endogenous proteins associate in differentiated IMBAT-1 (Fig. 5D).
GST pull-downs, using PGC-1α fusion proteins, were performed to map the regions necessary for interaction with RIP140. Full-length 35S-RIP140 interacted most strongly with PGC-1α amino acids 294 to 408 and, to a lesser extent, with amino acids 186 to 294 and the C-terminal amino acids 565 to 754 (Fig. 5E). Truncated fragments of RIP140 encompassing amino acids 78 to 333 and 1084 to 1158 showed similar levels of strong binding to amino acids 186 to 294, 294 to 408, and 565 to 754 of PGC-1α (Fig. 5E). Thus, there are a number of distinct sites that facilitate the interaction between RIP140 and PGC-1α. Mutation of all the LXXLL motifs within RIP140 resulted in reduced interaction with PGC-1α fragments (Fig. 5E). Conversely, mutation of the three leucine-rich motifs with PGC-1α did not affect its ability to interact with RIP140 (Fig. 5F). The LXXLL motifs were required for in vivo protein interaction, as there was no interaction between transfected PGC-1α and the RIP140 LXXLL mutant, whereas wild-type RIP140 coimmunoprecipitated with PGC-1α (Fig. 5G).
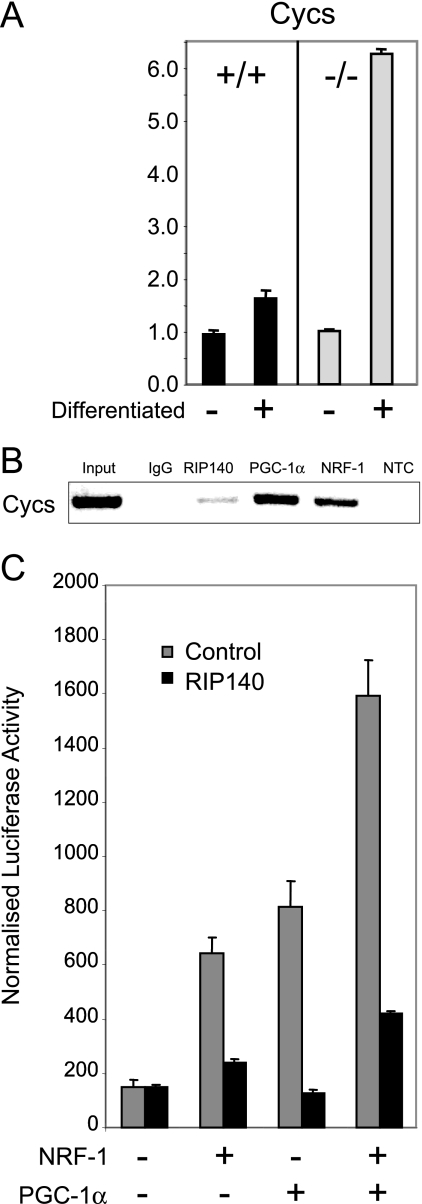
The association of RIP140 and PGC-1α, which exhibit opposing transcriptional functions, was unexpected. To investigate this further, we examined the Cycs gene since its expression was elevated in RIP140-depleted adipocytes (Fig. 6A) and it is a known NRF-1 target (18). In ChIP assays, we demonstrated that NRF-1 and the coregulators RIP140 and PGC-1α were present on the Cycs promoter in the proximity of an NRF-1 binding site (Fig. 6B). Furthermore, we found that the ability of NRF-1, together with PGC-1α, to stimulate transcription from a reporter containing four tandem Cycs NRF-1 binding sites upstream from a truncated Cycs promoter was repressed by RIP140 (Fig. 6C). Thus, it appears likely that the targeting of NRF-1-regulated genes, such as the CIDEA and Cycs genes, by RIP140 may represent a key determinant of gene expression in metabolic cells.
FIG. 6.
RIP140 and PGC-1α associate with the NRF-1 binding site on the Cycs promoter. (A) Results from real-time PCR analysis of mRNAs for Cycs. Primary cultures of mouse embryo fibroblasts prepared from wild-type (+/+) or RIP140-null (−/−) tissue were left untreated (−) or were differentiated (+) for 6 days. Data are expressed relative to results for untreated wild-type cells. (B) Chromatin prepared from differentiated IMBAT-1 cells was immunoprecipitated with antibodies raised against RIP140, PGC-1α, NRF-1, or IgG (control), as indicated. Immunoprecipitated products were PCR amplified with primers designed to flank the NRF-1 binding site in the Cycs promoter. NTC, nontemplate control. (C) Cos cells were cotransfected with 4×NRF-1/Cycs/luc and expression vectors for NRF-1 and PGC-1α, in the absence and presence of RIP140, as indicated.
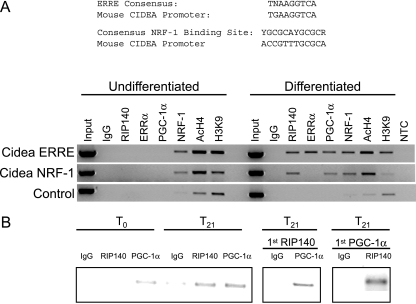
Finally, to investigate the recruitment of transcription factors to the CIDEA promoter, we performed ChIP assays on undifferentiated and differentiated IMBAT-1 cells. ERRα was recruited to the ERRE after differentiation, whereas NRF-1 binding was detected in the vicinity of the NRF-1 and ERR binding sites both before and after differentiation (Fig. 7A). Interestingly, the binding of RIP140 and PGC-1α, in proximity to the ERRE and NRF-1 binding sites, was also differentiation dependent. We next analyzed histone modifications characteristic of transcriptional activation and repression. General acetylation of H4 (AcH4) was detected at the CIDEA ERRE and NRF-1 binding sites before and after differentiation (Fig. 7A). In contrast, methylation of histone H3 at lysine 9 (H3K9), a mark characteristic of repressed gene promoters, was lower in differentiated cells than in undifferentiated cells.
FIG. 7.
ChIP assays for transcription factor recruitment to CIDEA promoter. (A) Chromatin prepared from undifferentiated or differentiated IMBAT-1 cells was immunoprecipitated with antibodies raised against RIP140, ERRα, PGC-1α, NRF-1, AcH4, methylated H3K9, or IgG (control) as indicated. Immunoprecipitated products were PCR amplified with primers designed to flank the ERRE or NRF-1 binding site in the CIDEA promoter or an upstream control region. The ERRE and NRF-1 binding sites in the CIDEA promoter are indicated (top). NTC, nontemplate control. (B) Chromatin was prepared from differentiated IMBAT-1 cells at 0 (T0) and 21 (T21) min following α-amanatin synchronization, and products were immunoprecipitated with antibodies raised against RIP140, PGC-1α, or IgG. A re-ChIP assay was performed by initial immunoprecipitation with the first antibody, followed by elution with dithiothreitol (10 mM) and immunoprecipitation with IgG (control) or the second antibody and PCR amplification with primers flanking the CIDEA ERRE.
We tested whether RIP140 and PGC-1α colocalized on the CIDEA promoter by performing re-ChIP experiments. In differentiated IMBAT-1 cells, we detected recruitment of RIP140 and PGC-1α to the CIDEA promoter (Fig. 7B). The re-ChIP assay was performed by initial immunoprecipitation with an antibody raised against RIP140, followed by elution and immunoprecipitation with an antibody raised against PGC-1α. We detected an enrichment of CIDEA promoter DNA with RIP140/PGC-1α antibodies but not with IgG controls (Fig. 7B), indicating that these two coregulator molecules are present at the same time on the promoter. Similar results were observed for the reciprocal experiment in which chromatin was immunoprecipitated with anti-PGC-1α followed by anti-RIP140 antibody (Fig. 7B). Therefore, the action of RIP140 in metabolic tissues to regulate CIDEA gene expression is directed via NR-dependent and -independent mechanisms and involves direct interactions between both RIP140 and PGC-1α.
DISCUSSION
CIDEA is a member of a family of three closely related proteins that also includes CIDEB (21) and fat-specific protein 27 (FSP27/CIDEC) (21), all of which display a high degree of homology at the N- and C-terminal regions of the proteins. CIDEA has been identified as a key factor in the metabolic function of adipose cells in view of its ability to regulate Ucp1 activity during adaptive thermogenesis (56) and its relationship to adiposity in mice and humans (8, 9, 17, 36, 39). Recent studies provide evidence that the CIDE family of proteins is important in the control of metabolic processes that contribute to lipid storage. FSP27, a protein linked with terminal differentiation of fat cells (10, 26), has been detected in association with lipid droplets of 3T3-L1 adipocytes (2) and has the capacity to induce lipid droplet formation (45). Similarly, we have found that CIDEA is localized surrounding lipid droplets in adipocytes and that its expression in nonadipogenic cell lines induced the accumulation of lipid droplets, an observation that was recently reported by Puri and coworkers (46). CIDEB is highly expressed in the liver, where it seems to regulate lipid metabolism and is implicated in hepatic steatosis (33). Similarly, CIDEA is expressed in normal adult mouse liver at older ages and positively correlates with liver steatosis in a mouse model of obesity-induced type 2 diabetes (24).
The expression of CIDEA in liver is stimulated by PPARα and PPARγ (52), but in adipose tissue their effects are minimal (52), and, consistent with these data, we observed negligible effects of PPAR ligands in IMBAT-1 adipocytes. We have found functional binding sites for ERRα and NRF-1 in the CIDEA promoter and determined that both factors stimulate CIDEA expression in brown adipocytes, as judged by promoter analysis using reporter genes and ChIP assays. Recent studies have indicated that ERRα and NRF-1 promote mitochondrial biogenesis and the oxidative capacity necessary for providing the energy utilized for adaptive thermogenesis by acting directly at genes important for mitochondrial function (20, 25, 48, 55). The ability of RIP140 to repress ERRα-dependent CIDEA expression is not unexpected given our previous results that show the corepressor inhibits ERRα activation of target genes, including Ucp1 (11, 40). However, the finding that NRF-1 is a target for repression was rather surprising, since we were unable to demonstrate a direct interaction with the corepressor. The relative contributions of these transcription factors to promoter activity are difficult to assess, but since CIDEA expression is maintained in BAT from ERRα knockout mice (51), it is conceivable that NRF-1 plays a crucial function. In any event, the ability of RIP140 to block the activity of NRF-1, in addition to many NRs, provides an explanation for the marked sensitivity of CIDEA gene expression to inhibition.
Energy homeostasis is controlled by the regulated transcription of gene networks in response to environmental factors and as such depends on a balance between activating and repressive signals. Previous work has established that PGC-1α activates and RIP140 represses genes involved in mitochondrial biogenesis and respiration (28, 30, 40, 43, 49). The possibility that PGC-1α and RIP140 regulate transcription from overlapping sets of target genes is emerging. RIP140 and PGC-1α are capable of interacting directly with ligand-activated NRs (32, 44), and thus, their mutual antagonism could be explained by the abilities of these two cofactors to compete for binding. However, ChIP assays indicate that both RIP140 and PGC-1α can bind simultaneously to the CIDEA promoter in the vicinity of the binding sites for ERRα and NRF-1. Due to the proximity of these sites to one another, we cannot investigate the occupancies of the individual sites, and one possibility is that the two factors bind independently, at least to the ERRα binding sites. However, given the abilities of PGC-1α and RIP140 to interact, the recruitment of RIP140 to either site could be mediated via its direct interaction with PGC-1α. Moreover, the abilities of the two cofactors to interact directly provides a mechanism for RIP140 to function as a corepressor for transcription factors, such as NRF-1, that do not appear to bind to RIP140 but do depend on PGC-1α for their activity. The interaction could be important for the function of all members of the PGC-1 family, as binding was also detected between the corepressor and PGC-1β, a close homolog of PGC-1α. It is conceivable that the interaction is transient, at least at ERRα target genes, to allow the exchange of PGC-1α with RIP140, since both cofactors can bind to the LBD. On the other hand, both cofactors may form stable ternary complexes with ERRα or NRF-1, although additional studies will be required to distinguish the relative functional importance of these different mechanisms. Finally, it is worth mentioning that RIP140 and PGC-1α have been found to colocalize in nuclear foci (31), and it is conceivable that this involves a direct interaction.
We have mapped the interacting regions to RD1 and RD4 of RIP140, which were both recruited to two regions of PGC-1α at amino acids 186 to 408 and 565 to 754. Interestingly, the region of amino acids 186 to 408 is known to inhibit transcriptional activation (41), and so it is conceivable that, in addition to the p160 myb binding protein (13), the recruitment of RIP140 may contribute to repression. The interaction between the cofactors was dependent on intact LXXLL motifs in RIP140 but not in PGC-1α. These motifs, which are well characterized in their role for cofactor recruitment to the LBD of NRs, have also been implicated in the interaction of the TRAP220 cofactor with PGC-1α (53) and may therefore represent an important motif for coregulator binding to PGC-1α.
The recruitment of RIP140 and PGC-1α to the CIDEA promoter in brown adipocytes was differentiation dependent and coincident with induction of CIDEA mRNA expression and reduction in the repressing chromatin mark H3K9. It is possible that the coexistence of histone marks associated with both active (AcH4) and inactive (H3K9) chromatin at the CIDEA promoter, in preadipocytes, has a role in priming the gene ready for transcriptional activation. Such bivalency has been reported for lineage-specific gene expression in embryonic stem cells (1). However, further work is necessary to fully characterize the epigenetic changes induced by the recruitment of RIP140 and PGC-1α to their target promoters.
The observation that RIP140 can directly repress the activity of PGC-1α raises the question of whether the corepressor inhibits transcription from all or a subset of PGC-1α target genes and, in particular, ERRα and NRF-1 target genes. We have found that numerous ERRα target genes are subject to repression by RIP140 in adipocytes (40) and skeletal muscle (49). Similarly, we have noted that the NRF-1 Cycs target gene, whose activation is promoted by PGC-1α (25, 54), is subject to repression by RIP140. Further investigations to identify the full repertoire of genes targeted by these coregulators and their transcription factors will require global expression analysis after overexpression of PGC-1α with ERRα or NRF-1 in the presence and absence of RIP140.
The identification of a direct functional link between key activators such as PGC-1α and RIP140 raises the possibility that important regulatory steps in many metabolic processes may depend on the relative levels of these cofactors. Our studies indicate that the interplay between RIP140 and PGC-1α plays a critical role in the determination of CIDEA expression and may underpin the transcriptional regulation of many metabolic genes that control energy homeostasis in metabolic tissues.
Supplementary Material
Acknowledgments
We thank R. Scarpulla, A. Kakizuka, and B. Herzog for their gifts of plasmids, Hongwu Chen for RIP140 antibody, Greg Brook for assistance with confocal microscopy, and the members of the Molecular Endocrinology Group for help and advice.
This work was supported by Wellcome Trust program grants 061930/Z/00/Z and 079200/Z/06/Z and BBSRC grant BB/C504327/1.
Footnotes
Published ahead of print on 15 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Azuara, V., P. Perry, S. Sauer, M. Spivakov, H. F. Jorgensen, R. M. John, M. Gouti, M. Casanova, G. Warnes, M. Merkenschlager, and A. G. Fisher. 2006. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 8532-538. [DOI] [PubMed] [Google Scholar]
- 2.Brasaemle, D. L., G. Dolios, L. Shapiro, and R. Wang. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J. Biol. Chem. 27946835-46842. [DOI] [PubMed] [Google Scholar]
- 3.Cao, W., K. W. Daniel, J. Robidoux, P. Puigserver, A. V. Medvedev, X. Bai, L. M. Floering, B. M. Spiegelman, and S. Collins. 2004. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol. Cell. Biol. 243057-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavailles, V., S. Dauvois, F. L'Horset, G. Lopez, S. Hoare, P. J. Kushner, and M. G. Parker. 1995. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 143741-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christian, M., E. Kiskinis, D. Debevec, G. Leonardsson, R. White, and M. G. Parker. 2005. RIP140-targeted repression of gene expression in adipocytes. Mol. Cell. Biol. 259383-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christian, M., J. M. Tullet, and M. G. Parker. 2004. Characterization of four autonomous repression domains in the corepressor receptor interacting protein 140. J. Biol. Chem. 27915645-15651. [DOI] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Dahlman, I., M. Kaaman, H. Jiao, J. Kere, M. Laakso, and P. Arner. 2005. The CIDEA gene V115F polymorphism is associated with obesity in Swedish subjects. Diabetes 543032-3034. [DOI] [PubMed] [Google Scholar]
- 9.Dahlman, I., K. Linder, E. Arvidsson Nordstrom, I. Andersson, J. Liden, C. Verdich, T. I. Sorensen, and P. Arner. 2005. Changes in adipose tissue gene expression with energy-restricted diets in obese women. Am. J. Clin. Nutr. 811275-1285. [DOI] [PubMed] [Google Scholar]
- 10.Danesch, U., W. Hoeck, and G. M. Ringold. 1992. Cloning and transcriptional regulation of a novel adipocyte-specific gene, FSP27. CAAT-enhancer-binding protein (C/EBP) and C/EBP-like proteins interact with sequences required for differentiation-dependent expression. J. Biol. Chem. 2677185-7193. [PubMed] [Google Scholar]
- 11.Debevec, D., M. Christian, D. Morganstein, A. Seth, M. Parker, and R. White. 2007. Receptor interacting protein 140 regulates expression of uncoupling protein 1 in adipocytes through specific peroxisome proliferator activated receptor isoforms and estrogen-related receptor α. Mol. Endocrinol. 211581-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desvergne, B., L. Michalik, and W. Wahli. 2006. Transcriptional regulation of metabolism. Physiol. Rev. 86465-514. [DOI] [PubMed] [Google Scholar]
- 13.Fan, M., J. Rhee, J. St-Pierre, C. Handschin, P. Puigserver, J. Lin, S. Jaeger, H. Erdjument-Bromage, P. Tempst, and B. M. Spiegelman. 2004. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: modulation by p38 MAPK. Genes Dev. 18278-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feige, J. N., and J. Auwerx. 2007. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 17292-301. [DOI] [PubMed] [Google Scholar]
- 15.Giguere, V. 2002. To ERR in the estrogen pathway. Trends Endocrinol. Metab. 13220-225. [DOI] [PubMed] [Google Scholar]
- 16.Gugneja, S., C.-M. A. Virbasius, and R. C. Scarpulla. 1996. Nuclear respiratory factors 1 and 2 utilize similar glutamine-containing clusters of hydrophobic residues to activate transcription. Mol. Cell. Biol. 165708-5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gummesson, A., M. Jernas, P. A. Svensson, I. Larsson, C. A. Glad, E. Schele, L. Gripeteg, K. Sjoholm, T. C. Lystig, L. Sjostrom, B. Carlsson, B. Fagerberg, and L. M. Carlsson. 2007. Relations of adipose tissue CIDEA gene expression to basal metabolic rate, energy restriction, and obesity: population-based and dietary intervention studies. J. Clin. Endocrinol. Metab. 924759-4765. [DOI] [PubMed] [Google Scholar]
- 18.Herzig, R. P., S. Scacco, and R. C. Scarpulla. 2000. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J. Biol. Chem. 27513134-13141. [DOI] [PubMed] [Google Scholar]
- 19.Herzog, B., M. Hallberg, A. Seth, A. Woods, R. White, and M. G. Parker. 2007. The nuclear receptor cofactor, receptor-interacting protein 140, is required for the regulation of hepatic lipid and glucose metabolism by liver X receptor. Mol. Endocrinol. 212687-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huss, J. M., I. P. Torra, B. Staels, V. Giguère, and D. P. Kelly. 2004. Estrogen-related receptor α directs peroxisome proliferator-activated receptor α signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 249079-9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inohara, N., T. Koseki, S. Chen, X. Wu, and G. Nunez. 1998. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 172526-2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jat, P. S., M. D. Noble, P. Ataliotis, Y. Tanaka, N. Yannoutsos, L. Larsen, and D. Kioussis. 1991. Direct derivation of conditionally immortal cell lines from an H-2Kb-tsA58 transgenic mouse. Proc. Natl. Acad. Sci. USA 885096-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamei, Y., H. Ohizumi, Y. Fujitani, T. Nemoto, T. Tanaka, N. Takahashi, T. Kawada, M. Miyoshi, O. Ezaki, and A. Kakizuka. 2003. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. USA 10012378-12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelder, B., K. Boyce, A. Kriete, R. Clark, D. E. Berryman, S. Nagatomi, E. O. List, M. Braughler, and J. J. Kopchick. 2007. CIDE-A is expressed in liver of old mice and in type 2 diabetic mouse liver exhibiting steatosis. Comp. Hepatol. 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelly, D. P., and R. C. Scarpulla. 2004. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 18357-368. [DOI] [PubMed] [Google Scholar]
- 26.Kim, J. Y., K. Liu, S. Zhou, K. Tillison, Y. Wu, and C. M. Smas. 2008. Assessment of fat-specific protein 27 in the adipocyte lineage suggests a dual role for FSP27 in adipocyte metabolism and cell death. Am. J. Physiol. Endocrinol. Metab. 294E654-E667. [DOI] [PubMed] [Google Scholar]
- 27.Kiskinis, E., M. Hallberg, M. Christian, M. Olofsson, S. M. Dilworth, R. White, and M. G. Parker. 2007. RIP140 directs histone and DNA methylation to silence Ucp1 expression in white adipocytes. EMBO J. 264831-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koves, T. R., P. Li, J. An, T. Akimoto, D. Slentz, O. Ilkayeva, G. L. Dohm, Z. Yan, C. B. Newgard, and D. M. Muoio. 2005. Peroxisome proliferator-activated receptor-γ co-activator 1α-mediated metabolic remodeling of skeletal myocytes mimics exercise training and reverses lipid-induced mitochondrial inefficiency. J. Biol. Chem. 28033588-33598. [DOI] [PubMed] [Google Scholar]
- 29.Kozak, L. P., and M. E. Harper. 2000. Mitochondrial uncoupling proteins in energy expenditure. Annu. Rev. Nutr. 20339-363. [DOI] [PubMed] [Google Scholar]
- 30.Leonardsson, G., J. H. Steel, M. Christian, V. Pocock, S. Milligan, J. Bell, P. W. So, G. Medina-Gomez, A. Vidal-Puig, R. White, and M. G. Parker. 2004. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. USA 1018437-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lerin, C., J. T. Rodgers, D. E. Kalume, S.-H. Kim, A. Pandey, and P. Puigserver. 2006. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab. 3429-438. [DOI] [PubMed] [Google Scholar]
- 32.L'Horset, F., S. Dauvois, D. M. Heery, V. Cavailles, and M. G. Parker. 1996. RIP-140 interacts with multiple nuclear receptors by means of two distinct sites. Mol. Cell. Biol. 166029-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, J. Z., J. Ye, B. Xue, J. Qi, J. Zhang, Z. Zhou, Q. Li, Z. Wen, and P. Li. 2007. Cideb regulates diet-induced obesity, liver steatosis, and insulin sensitivity by controlling lipogenesis and fatty acid oxidation. Diabetes 562523-2532. [DOI] [PubMed] [Google Scholar]
- 34.Lin, J., R. Yang, P. T. Tarr, P.-H. Wu, C. Handschin, S. Li, W. Yang, L. Pei, M. Uldry, P. Tontonoz, C. B. Newgard, and B. M. Spiegelman. 2005. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1β coactivation of SREBP. Cell 120261-273. [DOI] [PubMed] [Google Scholar]
- 35.Mootha, V. K., C. Handschin, D. Arlow, X. Xie, J. St. Pierre, S. Sihag, W. Yang, D. Altshuler, P. Puigserver, N. Patterson, P. J. Willy, I. G. Schulman, R. A. Heyman, E. S. Lander, and B. M. Spiegelman. 2004. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA 1016570-6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moraes, R. C., A. Blondet, K. Birkenkamp-Demtroeder, J. Tirard, T. F. Orntoft, A. Gertler, P. Durand, D. Naville, and M. Begeot. 2003. Study of the alteration of gene expression in adipose tissue of diet-induced obese mice by microarray and reverse transcription-polymerase chain reaction analyses. Endocrinology 1444773-4782. [DOI] [PubMed] [Google Scholar]
- 37.Morganstein, D. L., M. Christian, J. J. O. Turner, M. G. Parker, and R. White. 2008. Conditionally immortalized white preadipocytes: a novel adipocyte model. J. Lipid Res. 49679-685. [DOI] [PubMed] [Google Scholar]
- 38.Nichol, D., M. Christian, J. H. Steel, R. White, and M. G. Parker. 2006. RIP140 expression is stimulated by estrogen-related receptor α during adipogenesis. J. Biol. Chem. 28132140-32147. [DOI] [PubMed] [Google Scholar]
- 39.Nordstrom, E. A., M. Ryden, E. C. Backlund, I. Dahlman, M. Kaaman, L. Blomqvist, B. Cannon, J. Nedergaard, and P. Arner. 2005. A human-specific role of cell death-inducing DFFA (DNA fragmentation factor-α)-like effector A (CIDEA) in adipocyte lipolysis and obesity. Diabetes 541726-1734. [DOI] [PubMed] [Google Scholar]
- 40.Powelka, A. M., A. Seth, J. V. Virbasius, E. Kiskinis, S. M. Nicoloro, A. Guilherme, X. Tang, J. Straubhaar, A. D. Cherniack, M. G. Parker, and M. P. Czech. 2006. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J. Clin. Investig. 116125-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puigserver, P., G. Adelmant, Z. Wu, M. Fan, J. Xu, B. O'Malley, and B. M. Spiegelman. 1999. Activation of PPARgamma coactivator-1 through transcription factor docking. Science 2861368-1371. [DOI] [PubMed] [Google Scholar]
- 42.Puigserver, P., J. Rhee, J. Donovan, C. J. Walkey, J. C. Yoon, F. Oriente, Y. Kitamura, J. Altomonte, H. Dong, D. Accili, and B. M. Spiegelman. 2003. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 423550-555. [DOI] [PubMed] [Google Scholar]
- 43.Puigserver, P., and B. M. Spiegelman. 2003. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha): transcriptional coactivator and metabolic regulator. Endocr. Rev. 2478-90. [DOI] [PubMed] [Google Scholar]
- 44.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92829-839. [DOI] [PubMed] [Google Scholar]
- 45.Puri, V., S. Konda, S. Ranjit, M. Aouadi, A. Chawla, M. Chouinard, A. Chakladar, and M. P. Czech. 2007. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J. Biol. Chem. 28234213-34218. [DOI] [PubMed] [Google Scholar]
- 46.Puri, V., S. Ranjit, S. Konda, S. M. Nicoloro, J. Straubhaar, A. Chawla, M. Chouinard, C. Lin, A. Burkart, S. Corvera, R. A. Perugini, and M. P. Czech. 2008. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc. Natl. Acad. Sci. USA 1057833-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scarpulla, R. C. 2006. Nuclear control of respiratory gene expression in mammalian cells. J. Cell. Biochem. 97673-683. [DOI] [PubMed] [Google Scholar]
- 48.Schreiber, S. N., R. Emter, M. B. Hock, D. Knutti, J. Cardenas, M. Podvinec, E. J. Oakeley, and A. Kralli. 2004. The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. USA 1016472-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seth, A., J. H. Steel, D. Nichol, V. Pocock, M. K. Kumaran, A. Fritah, M. Mobberley, T. A. Ryder, A. Rowlerson, J. Scott, M. Poutanen, R. White, and M. Parker. 2007. The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab. 6236-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unami, A., Y. Shinohara, K. Kajimoto, and Y. Baba. 2004. Comparison of gene expression profiles between white and brown adipose tissues of rat by microarray analysis. Biochem. Pharmacol. 67555-564. [DOI] [PubMed] [Google Scholar]
- 51.Villena, J. A., M. B. Hock, W. Y. Chang, J. E. Barcas, V. Giguere, and A. Kralli. 2007. Orphan nuclear receptor estrogen-related receptor α is essential for adaptive thermogenesis. Proc. Natl. Acad. Sci. USA 1041418-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Viswakarma, N., S. Yu, S. Naik, P. Kashireddy, K. Matsumoto, J. Sarkar, S. Surapureddi, Y. Jia, M. S. Rao, and J. K. Reddy. 2007. Transcriptional regulation of Cidea, mitochondrial cell death-inducing DNA fragmentation factor α-like effector A, in mouse liver by peroxisome proliferator-activated receptor α and γ. J. Biol. Chem. 28218613-18624. [DOI] [PubMed] [Google Scholar]
- 53.Wallberg, A. E., S. Yamamura, S. Malik, B. M. Spiegelman, and R. G. Roeder. 2003. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Mol. Cell 121137-1149. [DOI] [PubMed] [Google Scholar]
- 54.Wright, D. C., D.-H. Han, P. M. Garcia-Roves, P. C. Geiger, T. E. Jones, and J. O. Holloszy. 2007. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1α expression. J. Biol. Chem. 282194-199. [DOI] [PubMed] [Google Scholar]
- 55.Wu, Z., P. Puigserver, U. Andersson, C. Zhang, G. Adelmant, V. Mootha, A. Troy, S. Cinti, B. Lowell, R. C. Scarpulla, and B. M. Spiegelman. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98115-124. [DOI] [PubMed] [Google Scholar]
- 56.Zhou, Z., S. Yon Toh, Z. Chen, K. Guo, C. P. Ng, S. Ponniah, S.-C. Lin, W. Hong, and P. Li. 2003. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat. Genet. 3549-56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.