Abstract
The Moraxella catarrhalis ubiquitous surface proteins (UspAs) are autotransporter molecules reported to interact with a variety of different host proteins and to affect processes ranging from serum resistance to cellular adhesion. The role of UspA1 as an adhesin has been confirmed with a number of different human cell types and is mediated by binding to eukaryotic proteins including carcinoembryonic antigen-related cellular adhesion molecules (CEACAMs), fibronectin, and laminin. A distinct difference in the ability of prototypical M. catarrhalis strains to adhere to CEACAM-expressing cell lines prompted us to perform strain-specific structure-function analyses of UspA1 proteins. In this study, we characterized CEACAM binding by a diverse set of UspA1 proteins and showed that 3 out of 10 UspA1 proteins were incapable of binding CEACAM. This difference resulted from the absence of a distinct CEACAM binding motif in nonadhering strains. Our sequence analysis also revealed a single M. catarrhalis isolate that lacked the fibronectin-binding motif and was defective in adherence to Chang conjunctival epithelial cells. These results clearly demonstrate that UspA1-associated adhesive functions are not universally conserved. Instead, UspA1 proteins must be considered as variants with the potential to confer both different cell tropisms and host cell responses.
Moraxella catarrhalis is a gram-negative, unencapsulated bacterium whose natural existence is generally restricted to the respiratory tract of humans. Previously considered to be a commensal organism, it is now recognized as an important mucosal surface pathogen responsible for causing many cases of acute otitis media in children (9, 26). M. catarrhalis is also an important cause of exacerbations of chronic obstructive pulmonary disease (COPD) in adults (28, 37). These facts have led to increased efforts to identify bacterial factors that contribute to mucosal colonization and disease pathogenesis (16). Of these, the interaction between the surface-exposed proteins of M. catarrhalis and their receptors in the respiratory tract has become an important focus of research to define vaccine targets (23, 27) and novel therapeutics to treat infection caused by this organism (12).
A particular interest concerning M. catarrhalis interactions with its human host is the ubiquitous surface proteins (UspAs). The UspA proteins are subdivided into three basic groups based on conserved amino acids motifs within the N and C termini of these macromolecules: UspA1 (∼88 kDa) (2, 7), UspA2 (∼62 kDa) (2, 7), and the hybrid UspA2H (∼92 kDa) (18). The UspA proteins are homologues of the YadA protein of Yersinia sp. (14, 17) and, as such, are predicted to be autotransporter proteins (11) that are present on the bacterial cell surface. Each protein consists of three distinct structural domains (14, 17). The N-terminal region is considered to form a β-sheet-based globular head, while the C-terminal region is predicted to form a membrane-spanning β-barrel structure (17). The stalk region that connects the head to the membrane-anchoring domains consists of a coiled-coil structure (6, 17). Although predicted to be structurally conserved, UspA1 and UspA2 proteins are distinct in that they are made up of a variable assortment of polypeptide modules that alter the size and composition of both the stalk and globular regions of each different protein (4a).
UspA1 and UspA2 have been shown to interact with a variety of human-derived targets normally found on certain eukaryotic cell surfaces or in the extracellular matrix, including fibronectin (42), laminin (41), and carcinoembryonic antigen (CEA)-related cellular adhesion molecule 1 (CEACAM1) (13). Each binding function contributes to M. catarrhalis interactions with different cell types to various degrees (13, 18, 24). In addition to its role as an adhesin, UspA1 was previously reported to confer serum resistance by interacting with both C3 (32) and C4b (31), components of the complement cascade. Most recently, UspA1 was shown to be associated with the development of apoptosis in epithelial cells exposed to M. catarrhalis (30) and to be involved in the invasion of human epithelial cells in vitro (39, 40). Remarkably, the interaction with each of these host proteins has been localized to a different region of UspA1, suggesting a potential for this bacterial macromolecule to simultaneously coordinate multiple ligands. However, the studies that have attributed each individual phenotype to UspA1 have typically been performed using different strains, and each binding activity is frequently considered in isolation, making it difficult to reconcile sometimes contradictory observations regarding UspA1 function. This prompted us to perform a sequence-based functional analysis with a diverse set of M. catarrhalis isolates, thereby allowing us to demonstrate a previously unappreciated link between the variability in UspA1 primary amino acid sequence and cellular tropism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The M. catarrhalis wild-type strains and mutants employed in this study are listed in Table 1. The UspA1 mutants used in this study were previously described (2, 3, 18) and were designated ΔUspA1 in the present study because they contain an internal deletion in the UspA1 open reading frame. For the preparation of whole-cell lysates, M. catarrhalis strains were grown at 37°C on brain heart infusion (BHI) agar (Difco/Becton Dickinson, Sparks, MD) in an atmosphere containing 5% CO2 or in broth. Recombinant Escherichia coli DH5α strains containing pACYC184-based recombinant plasmids were grown on Luria-Bertani (LB) medium containing chloramphenicol (10 μg/ml) at 37°C. E. coli strain EPI300 cells (Epicentre Biotechnologies, Madison, WI) expressing recombinant UspA1 proteins were grown at 37°C on LB medium with standard antibiotic supplementation and in LB medium with chloramphenicol and CopyControl induction solution (Epicentre) as required.
TABLE 1.
M. catarrhalis strains used in this study
| Strain | Adult or child | Disease | Location of isolation | Geographical location | GenBank accession no. for UspA | Reference, source |
|---|---|---|---|---|---|---|
| ATCC 25238 | Adult | Unknown | Unclear; nasophayrnx (Neisseria isolate) | University of Rochester | AAD43465 | 7, ATCC |
| O35E | Child | Otitis media | Tympanocentesis-derived isolate | Dallas, TX | AAB96359 | 2, John D. Nelson |
| O35E.118 | 19 | |||||
| O35EΔUspA1 | 2 | |||||
| O12E | Child | Otitis media | Tympanocentesis-derived isolate | Dallas, TX | AAF40118 | 18, John D. Nelson |
| O12EΔUspA1 | 18 | |||||
| V1145 | Child | Healthy | Nasopharynx | North Carolina | ABY64882 | 3, Frederick Henderson |
| V1145 ΔUspA1 | 3 | |||||
| V1156 | Child | Healthy | Nasopharynx | North Carolina | ACC44785 | 3, Frederick Henderson |
| V1156 ΔUspA1 | 3 | |||||
| V1118 | Child | Healthy | Nasopharynx | North Carolina | ACB58308 | 3, Frederick Henderson |
| 7169 | Child | Otitis media | Tympanocentesis-derived isolate | Buffalo, NY | ACB59385 | 21, Anthony Campagnari |
| 7169 ΔUspA1 | 3 | |||||
| TTA37 | Adult | COPD | Transtracheal aspirate | Tennessee | AAF40122 | 18, Steven Berk |
| FIN2344 | Child | Unclear | Unknown | Finland | ACC44784 | 3, Merja Helminen |
| FIN2344 ΔUspA1 | 3 | |||||
| ATCC 43617 | Adult | COPD | Transtracheal aspirate from coal miner with chronic bronchitis | Belgium | ACB58310 | 45, ATCC |
General DNA methods.
M. catarrhalis genomic DNA was isolated from agar plate-grown cells using the Easy-DNA kit (Invitrogen, Carlsbad, CA). The preparation of plasmid DNA and the purification of PCR products were performed using kits manufactured by Qiagen (Santa Clarita, CA). Nucleotide sequence data obtained from automated sequencing systems were analyzed using the MacVector analysis package (version 6.5; Oxford Molecular Group, Campbell, CA). Mult-Align (8) was used to generate sequence alignments.
DNA cloning.
Plasmid pELU1-10G, which encodes the uspA1 gene from M. catarrhalis O35E, was described previously (18). To clone uspA1 genes into the CopyControl vector (Epicentre), PCR was used to amplify DNA encoding the entire uspA1 open reading frame and approximately 40 to 200 nucleotides of DNA 5′ from the uspA1 translational start codon using bacterial chromosomal DNA as the template for PCR. This PCR product was then cloned into the Blunt Cloning-Ready pCC1 vector (Epicentre) to generate recombinant plasmids expressing each different UspA1 protein. Each cloned uspA1 gene was fully sequenced and confirmed to be identical to that in the chromosome of the relevant M. catarrhalis strain. A kanamycin resistance cassette was cloned into pCC1 to generate the pCC-Kan negative control plasmid. These CopyControl-based plasmids were electroporated into Electrocompetent TransforMax E. coli EPI300 as described for the Epicentre CopyControl cDNA, gene, and PCR cloning kit.
Measurement of bacterial attachment to human cells.
Attachment of wild-type and mutant M. catarrhalis strains and recombinant E. coli strains expressing M. catarrhalis UspA1 proteins to Chang human conjunctival epithelial cells (ATCC CCL20.2) and to A549 human lung epithelial cells (ATCC CCL-185) was measured by using a modification of a previously described attachment assay (1). It must be noted that these Chang cells, purchased from the American Type Culture Collection (ATCC), were accompanied by an ATCC notice that this line has been contaminated with HeLa cells. Briefly, tissue culture medium was removed from human cells grown overnight in 24-well plates and replaced with 0.5 ml of fresh tissue culture medium to which 25 μl of M. catarrhalis cells (∼106 CFU) or recombinant E. coli cells (∼106 CFU) was then added. These bacterial suspensions were obtained by harvesting growth from agar plates that had been grown overnight and suspending these cells in either BHI (for M. catarrhalis) or LB (for E. coli) medium to a density of approximately 300 Klett units as measured in a Klett-Summerson colorimeter (VWR Scientific, West Chester, PA). The tissue culture plates were then subjected to centrifugation at 200 × g for 5 min and incubated at 37°C in an atmosphere of 95% air-5% CO2 for 30 min or 1 h, after which the wells were washed five times with BHI or LB medium. The human cells and attached bacteria were removed from the wells by treatment with trypsin (0.05% to 0.25%), subjected to vigorous mixing, and plated onto either BHI or LB agar to determine the number of attached bacteria.
Additional bacterial attachment experiments with Chang cells examined the effect of specific antibodies or serum components on attachment. To determine the effect of CEACAM antibodies on bacterial attachment, tissue culture medium was removed from Chang cells grown overnight in 24-well plates and replaced with 1 ml of fresh tissue culture medium containing 50 μl of rabbit antiserum to CEACAM (catalog number A0115; Dako North America, Carpinteria, CA), normal rabbit serum (catalog number X0903; Dako), or phosphate-buffered saline (PBS). These two antisera had been previously dialyzed overnight against PBS to remove azide. These plates were subjected to centrifugation at 200 × g for 5 min and then incubated at 37°C for 1 h. Next, the medium was removed from each well and replaced with fresh tissue culture medium to which 25 μl of recombinant E. coli cells containing plasmid pACYC184 or pELU1-10G (suspended to 250 Klett units) was then added, and the attachment assay was carried out as described above except that attachment was allowed to proceed for 2 h.
To determine the effect of serum components on attachment, Chang cell monolayers grown in the presence of 10% (vol/vol) fetal calf serum (FCS) were washed once with serum-free tissue culture medium, and tissue culture medium with or without 10% FCS was then added to each well. Recombinant E. coli cells containing M. catarrhalis uspA1 genes cloned into the CopyControl vector were induced for 3 h with CopyControl induction solution and adjusted to a density of 300 Klett units, 25-μl portions of this suspension were then added to the wells, and the attachment assay was performed as described above, with a 1-h incubation period.
Western blot and overlay analyses.
Bacterial whole-cell lysates (2) were boiled and resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 12.5% (wt/vol) polyacrylamide separating gels, transferred onto Immobilon-P membranes (Millipore, Bedford, MA), and probed with the appropriate antibody. UspA1 was detected using monoclonal antibody (MAb) 24B5 (7). Whole-cell lysates of Chang and A549 cells were prepared from confluent monolayers grown in 24-well tissue culture plates. These monolayers were trypsinized, suspended in PBS, and then solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis digestion buffer. Rabbit antiserum to CEACAM (described above) and mouse antiserum to glyceraldehyde-3-phosphate dehydrogenase were used as the primary antibodies. CEACAMs were detected using CEA-specific rabbit polyclonal antisera (Dako, Glostrup, Denmark). In overlay experiments, the blots were probed with soluble CEACAM5 (Fitzgerald, Concord, MA) or soluble CEACAM1 (36), followed by the CEA-specific antiserum, which was then detected using secondary antiserum. The secondary antibodies were goat anti-mouse immunoglobulin G or anti-rabbit immunoglobulin G conjugated to either Bodipy or Cy5 fluorophores (Jackson ImmunoResearch, West Grove, PA). Antigen-antibody complexes were visualized by immunofluorescence using a Typhoon 9400 instrument (Amersham Biosciences).
CEACAM-binding assays.
Cultures of E. coli cells expressing M. catarrhalis UspA1 proteins were grown overnight in LB chloramphenicol (12.5 μg/ml) and Copycontrol induction solution (Epicentre) for 5 h. Cells were washed with PBS-Mg-Ca (10 mM MgCl2 and 5 mM CaCl2). For bacterial dot blots, bacterial density was calculated by measuring the optical density at 550 nm and then diluting to 106 bacteria/ml before spotting onto nitrocellulose. Cells were dried overnight at room temperature, and the membranes were blocked in PBS containing 4% skim milk. These membranes were probed with soluble CEACAM5, which was subsequently detected using CEA-specific antiserum and horseradish peroxidase (HRP)-coupled secondary antibodies. For enzyme-linked immunosorbent assays (ELISAs), Nunc plates were coated with heat-killed bacteria and dried at 37°C overnight. Wells were blocked using PBS containing 4% skim milk, followed by incubation with soluble CEACAM5 or CEACAM1. CEACAM was detected as described above for the dot blot experiments except that the HRP-based colorimetric detection system (Sure Blue) was used to measure CEACAM binding by detection at 650 nm.
RESULTS
UspA1 proteins differ in cellular adherence through CEACAM.
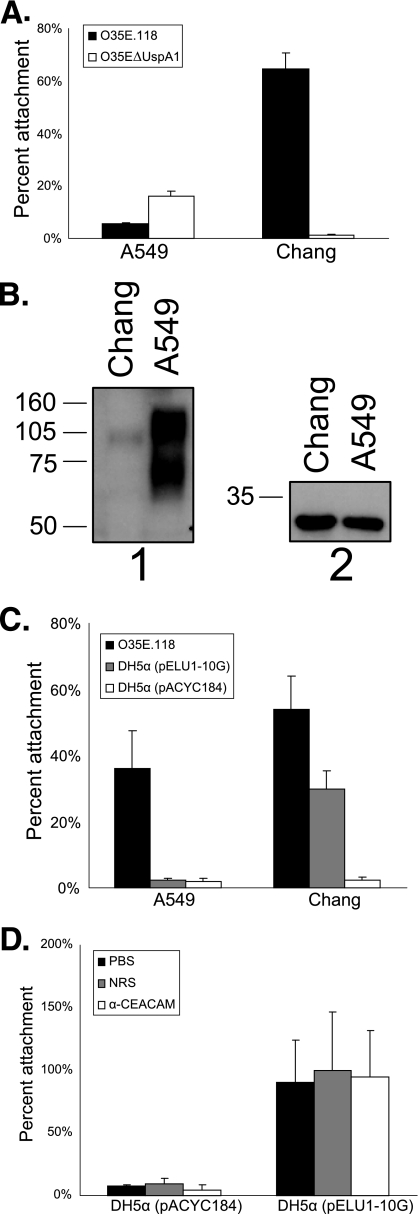
Recent work clearly demonstrates that the UspA1 protein from M. catarrhalis strain ATCC 25238 mediates bacterial binding to CEACAM1 (13). However, the cellular tropism of prototypical M. catarrhalis strain O35E did not correlate with CEACAM expression. Specifically, UspA1-expressing M. catarrhalis strain O35E demonstrated effective binding to Chang conjunctival epithelial cells (Fig. 1A), which express relatively little CEACAM (Fig. 1B), while demonstrating little association with A549 respiratory epithelial cells (Fig. 1A) (P < 0.05) that express relatively large quantities of several different CEACAMs (Fig. 1B) (43, 44). An isogenic UspA1-deficient strain of M. catarrhalis O35E (O35EΔUspA1) did not adhere to Chang cells, allowing us to attribute 035E adherence to UspA1 expression, yet the uspA1 mutant binding to the A549 cells was greater than that of the wild-type strain (P < 0.05) (Fig. 1A), suggesting that another adhesin(s) contributed to this interaction. This prompted us to express the UspA1 protein (UspA1O35E) from M. catarrhalis O35E in E. coli so that we could characterize host cellular binding by UspA1 in the absence of other M. catarrhalis adhesins.
FIG. 1.
Cellular tropism of the M. catarrhalis O35E UspA1 protein. (A) Attachment of wild-type strain O35E.118 and mutant strain O35EΔUspA1 to A549 and Chang cells in vitro using a 30-min incubation period for attachment. (B) Expression of CEACAMs by Chang and A549 cells determined by Western blot analysis. Whole-cell lysates of these two human cells lines were probed with antibody to CEACAM (panel 1) or glyceraldehyde-3-phosphate dehydrogenase (panel 2), followed by HRP-conjugated secondary antibody. (C) Attachment of wild-type M. catarrhalis O35E.118 and recombinant E. coli DH5α strains containing either pELU1-10G, which expresses the O35E UspA1 or the empty pACYC184 vector to A549 and Chang cells using a 30-min incubation period for attachment. (D) Effect of rabbit antibody to CEACAM (αCEACAM) and normal rabbit serum (NRS) on the binding of the two recombinant E. coli strains to Chang cells. Representative experiments are shown (A, C, and D). Statistical analysis of attachment data involved the use of one-way or two-way ANOVA with a Bonferroni posttest.
Consistent with the phenotype of the parental M. catarrhalis strains, Chang cell binding of E. coli cells expressing UspA1O35E was much greater than that of A549 cells (Fig. 1C) (P < 0.05), suggesting that CEACAM receptors are not the primary determinants of A549 binding by this UspA1 variant. Next, we determined the contribution of CEACAM to adherence by performing attachment assays in the presence of a CEACAM-specific polyclonal antiserum, which effectively blocks the binding of M. catarrhalis ATCC 25238 (13). There was no effect of the CEACAM antiserum on Chang cell adherence conferred by UspA1O35E (P > 0.05) (Fig. 1D), suggesting that CEACAM binding was not essential for adherence to this cell line.
CEACAM binding by UspA1 protein variants.
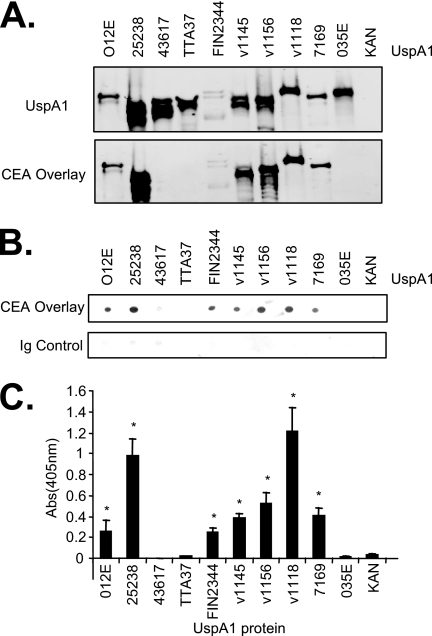
Previous studies confirmed that UspA1 proteins are expressed in recombinant E. coli strains and that their function reflects that in the parental M. catarrhalis strain (18). In an attempt to compare CEACAM binding by a diverse set of UspA1 proteins, we expressed a total of 10 UspA1 proteins in E. coli (Table 1). To confirm UspA1 expression, bacterial lysates were probed with UspA1-specific antibodies. Consistent with data from previous reports (25), individual UspA1 proteins were expressed at various levels (Fig. 2A). In addition, all of these cloned UspA1 proteins could be detected on the surfaces of their respective recombinant E. coli strains when these strains were probed with UspA1-specific MAb 24B5 by flow cytometry (data not shown).
FIG. 2.
CEACAM binding by recombinant E. coli expressing UspA1 variants. (A) E. coli whole-cell lysates were Western blotted and then probed to detect UspA1 protein (top) with MAb 24B5, and soluble CEACAM5 binding (bottom) was detected with anti-CEACAM (Dako). CEACAM5 binding was associated with UspA1 in 7 of 10 strains, while no binding was evident in the other three strains. (B) E. coli cells expressing the indicated UspA1 variants were spotted onto nitrocellulose and then incubated with soluble CEACAM5, followed by antibodies to detect bound CEACAM5. Ig, immunoglobulin. (C) E. coli cells expressing the indicated UspA1 variants were adsorbed to ELISA plates and then incubated with soluble CEACAM5, which was then detected using specific antibodies.
It was previously demonstrated that denatured UspA1 bound to purified soluble CEACAM (13). The blots containing the lysates were therefore reprobed using soluble CEACAM5. Seven of 10 UspA1 proteins tested revealed CEACAM5 binding, while the other three (UspA1ATCC 43617, UspA1TTA37, and UspA1O35E) did not (Fig. 2A). To confirm that the binding in the Western blot overlay assays reflected the phenotype of UspA1 in the recombinant bacterial membrane, the E. coli strains were spotted onto nitrocellulose (Fig. 2B) and adsorbed onto wells of an ELISA plate (Fig. 2C). In each case, binding reflected that of the overlay assays, with the same seven UspA1 variants conferring CEACAM5 binding in each assay (compare Fig. 2A, B, and C). These results suggest that CEACAM binding is not an inherent property of all UspA1 variants, as certain strains express UspA1 variants that do not bind CEACAMs.
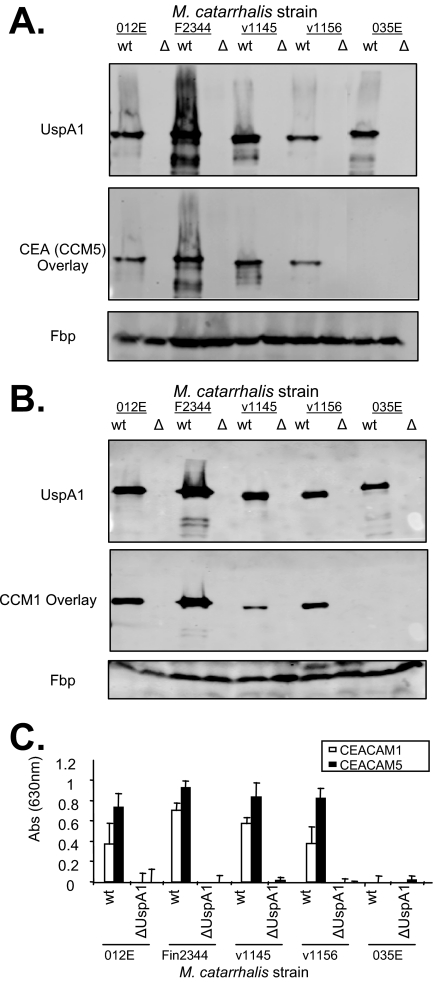
In order to confirm that the CEACAM-binding phenotype of UspA1 expressed by E. coli reflected that of the parental M. catarrhalis strains, we used a panel of UspA1-deficient M. catarrhalis strains (1, 3, 18). Lysates derived from the wild-type and UspA1-deleted strains were probed with MAb to the UspA1 protein to confirm the expected UspA1 expression (Fig. 3A and B, top). These blots were subsequently reprobed with soluble CEACAM1 or CEACAM5 to assess the CEACAM-binding activity of blotted proteins. CEACAM binding colocalized with the UspA1 variants of M. catarrhalis strains O12E, FIN2344, V1145, and V1156 but not with that from strain O35E (Fig. 3A and B, middle). To confirm that overlay-based experiments reflect the phenotypes of the intact M. catarrhalis strains, wild-type and mutant M. catarrhalis strain binding to soluble CEACAM1 and CEACAM5 was assessed using the ELISA-based assay (Fig. 3C). While different variants of the adhesins from other pathogenic bacteria may be specific for either CEACAM1 and/or CEACAM5 (10), the M. catarrhalis UspA1 proteins appear to bind either both CEACAMs or neither one. Altogether, these results demonstrate that CEACAM binding is conferred by certain UspA1 protein variants and that no other CEACAM-binding adhesins are expressed by M. catarrhalis that are detectable in this assay system.
FIG. 3.
M. catarrhalis wild-type (wt) or UspA1-deficient (Δ) strain binding to CEACAMs. M. catarrhalis whole-cell lysates were separated by electrophoresis and were immunoblotted with MAb 24B5 to detect UspA1 (A and B, top) or binding to soluble CEACAM5 (A, bottom) or CEACAM1 (B, bottom). UspA proteins were detected using Cy5-conjugated secondary antibodies, while bound CEACAM was using Bodipy-conjugated antisera. M. catarrhalis ferric binding (Fbp) was probed using mouse anti-Fbp antiserum as a loading control (bottom). (C) M. catarrhalis strains were adsorbed into ELISA plates and then incubated with soluble CEACAM1 or CEACAM5. The bound CEACAMs were then detected using CEACAM cross-specific antisera (Dako CEA), followed by HRP-conjugated goat anti-rabbit immunoglobulin. Abs, absorbance.
Sequence analysis of CEACAM binding in UspA1 proteins.
To reveal the basis of the differences in CEACAM binding for UspA1, the UspA1 amino acid sequences for each of the 10 strains tested were aligned. Previous work by Hill et al. (12) localized CEACAM1 binding to a 141-amino-acid polypeptide fragment derived from the C-terminal stalk region of UspA1ATCC 25238. Peptides derived from this region are able to bind CEACAM1 and block UspA1-dependent M. catarrhalis binding to CEACAM1-expressing cells (12). Our sequence alignments show that each UspA1 variant that lacks CEACAM-binding activity is missing a portion of the CEACAM-binding region (Fig. 4A). Consistent with the loss of these sequences not being an isolated event, the size of the deletion varied among the three proteins. Of these, UspA1TTA37 had the smallest deletion, lacking only 33 amino acids of the sequence corresponding to the CEACAM-binding region described previously by Hill et al. (12).
FIG. 4.
UspA1 protein sequence alignments. (A) Alignment of variant sequences spanning the CEACAM-binding region of UspA1, defined by Hill et al. (12), which is delineated with a horizontal black line above the sequences. CEACAM binding by M. catarrhalis and/or recombinant E. coli strains, as determined in the aforementioned studies, is indicated. +, CEACAM binding; −, does not bind CEACAM. (B) Alignment of UspA1 variant sequences spanning the region reported to contribute to fibronectin and Chang cell binding (42). Relevant peptides from the UspA1 and UspA2 proteins of M. catarrhalis strain Bc5, which was originally used to define the fibronectin-binding sequences, are included for comparison with the sequences considered in this study. (C) Attachment of recombinant E. coli cells expressing UspA1O35E or UspA1ATCC 43617 to Chang conjunctival epithelial cells in the presence or absence of FCS. The negative control is E. coli containing the pCC1-Kan construct (Kan).
Fibronectin and Chang cell binding.
In addition to CEACAM binding, various UspA1 proteins have been shown to bind extracellular matrix protein fibronectin (24). In particular, a 153-amino-acid peptide derived from the UspA1 protein from M. catarrhalis strain Bc5 was shown to be sufficient to mediate fibronectin binding and could inhibit UspA1-dependent binding to Chang cells (42). The minimal sequence capable of binding fibronectin spans a region containing a so-called “NINNY” repeat flanked on either side by a “VEEG” repeat (Fig. 4B), which was conserved between the UspA1 and UspA2 proteins expressed by strain Bc5 (42). Whether or not this entire region was necessary for fibronectin binding has remained undetermined. In order to understand whether the presence of these fibronectin-binding motifs correlated with CEACAM binding, we compared our UspA1 sequences (Fig. 4B). Based on this analysis, we observed that UspA1ATCC 43617 was unique in that it lacked the complete VEEG-NINNY-VEEG repeat. While the NINNY repeat is considered to represent the core of the fibronectin-binding sequence, the flanking regions were suggested to play a role in binding. The ordered VEEG-NINNY-VEEG sequence is highly conserved among UspA1 proteins but is less well conserved in UspA2 proteins. Subsequently, UspA2 proteins lacking portions of this ordered repeat were recognized to be deficient in adherence to fibronectin and in Chang cell binding (18, 24). Based upon these analyses, we predicted that UspA1ATCC 43617 would be unable to bind to Chang cells.
In order to determine whether the absence of one VEEG repeat would affect fibronectin-based Chang cell binding, we compared Chang cell binding by recombinant E. coli bacteria expressing UspA1ATCC 43617 to that of the prototypical UspA1O35E variant. While UspA1O35E expression conferred binding to Chang cells, bacteria expressing UspA1ATCC 43617 mirrored that of the background control E. coli strain (Fig. 4C). The adherence of the UspA1O35E bacteria was further increased in the presence of serum, consistent with the ability of serum components such as fibronectin to facilitate binding (42). These results indicate that the first VEEG repeat is essential for UspA1-dependent binding to Chang epithelial cells and that naturally occurring variants of UspA1 can lack this binding function.
DISCUSSION
The surface protein UspA1 has been shown to play a primary role in M. catarrhalis attachment to a variety of different cell types (1, 13, 24). This association appears to result from its ability to bind host-derived molecules including fibronectin (42), laminin (41), and members of the CEACAM family of intercellular adhesion molecules (13). While not considered in this study, UspA1 also interacts with components of both the classical and alternative complement pathways by interacting with both C3 (32) and C4b (31).
The diversity of functions attributed to UspA1 is remarkable. However, the various binding activities have been largely considered to be independent of each other and often with different strains of M. catarrhalis. While it was generally assumed that each function would be conserved among UspA1 variants, the current study was prompted by our finding that prototypical M. catarrhalis strain O35E does not bind to certain CEACAM-expressing cell lines. Since the sequence of variant UspA1 proteins is frequently unavailable, we cloned and sequenced several additional uspA1 genes to obtain a total of 10 genes encoding different UspA1 proteins. Three of the 10 UspA1 variants tested did not bind to CEACAMs (Fig. 2), and each of these variants lacked a portion of the sequence that has been shown to confer CEACAM1 binding (12). While M. catarrhalis expresses a number of different adhesins (4, 5, 15, 18, 20, 22, 33, 34), our UspA1-deficient mutants clearly demonstrated that UspA1 was the only adhesin with the potential to confer CEACAM binding.
The lack of fibronectin binding exhibited by the UspA1 protein from M. catarrhalis ATCC 43617 (Fig. 4C) is also a clear demonstration of the variant nature of individual UspA1 proteins. Fibronectin binding has been shown to mediate the adherence of M. catarrhalis to Chang cells and, unlike CEACAM binding, has been associated with both UspA1 and UspA2 (42). The amino acid sequence considered to contribute to fibronectin binding is present in UspA1, UspA2, and the naturally occurring chimera UspA2H; however, its association with each UspA protein is strain specific. The fact that both UspA1 and UspA2 can confer fibronectin-mediated binding to Chang cells makes it difficult to test the relative contribution of each one in the context of M. catarrhalis. However, our nucleotide sequence analyses indicated that UspA1 from strain ATCC 43617 contained a deletion within the region that was previously shown to contribute to fibronectin binding (42), and our binding assays demonstrated that this variant could not adhere to Chang epithelial cells.
The existence of three different M. catarrhalis isolates (O35E, TTA37, and ATCC 43617) that are unable to bind CEACAM receptors makes it intriguing to speculate how this phenotype may contribute to M. catarrhalis colonization of the nasopharynx or the production of disease in the respiratory tract. M. catarrhalis can be found as a member of the commensal flora in healthy individuals or associated with distinct diseases in either children (i.e., otitis media) or adults (i.e., exacerbations of COPD). The specific location (nasopharynx, trachea, or middle ear) and manner (swab, tympanocentesis, or aspirate) in which M. catarrhalis isolates are obtained must be considered when strain-specific differences in phenotype are analyzed. It is interesting in this regard that two of the three strains that lack the CEACAM-binding ability were isolated from adult tracheal aspirates.
While much remains to be learned about how each UspA1-associated function contributes to colonization or disease production (or both), it is clear that the ability to bind CEACAM and fibronectin is not a universally conserved function among M. catarrhalis strains. Sequence analyses and functional assays must be performed before the spectrum of potential functions can be deduced. We must also consider that other functions, including complement binding, may also vary among M. catarrhalis isolates (3). It is therefore enticing to consider that the ability of UspA1 to engage different combinations of host receptors and serum components will elicit different cellular responses (29, 35, 38) and that the different combination of activities may define the colonization ability and virulence potential of each strain.
Acknowledgments
This work was supported by funding from the Canadian Institutes for Health Research grant no. MOP-15499 to S.D.G.-O. and U.S. Public Health Service grant no. AI36344 to E.J.H. S.D.G.-O. is supported by New Investigator Awards from the Canadian Institutes of Health Research and is a recipient of the Province of Ontario Premier's Research Excellence Award.
We thank John Nelson, Anthony Campagnari, David Goldblatt, Richard Wallace, Steven Berk, Merja Helminen, and Frederick Henderson for providing the wild-type isolates of M. catarrhalis and Anthony Schryvers for the FpbA-specific antiserum used in this study.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 4 August 2008.
REFERENCES
- 1.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. L. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect. Immun. 663113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 654367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attia, A. S., S. Ram, P. A. Rice, and E. J. Hansen. 2006. Binding of vitronectin by the Moraxella catarrhalis UspA2 protein interferes with late stages of the complement cascade. Infect. Immun. 741597-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balder, R., J. Hassel, S. Lipski, and E. R. Lafontaine. 2007. Moraxella catarrhalis strain O35E expresses two filamentous hemagglutinin-like proteins that mediate adherence to human epithelial cells. Infect. Immun. 752765-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Brooks, M. J., J. L. Sedillo, N. Wagner, C. A. Laurence, W. Wang, A. S. Attia, E. J. Hansen, and S. D. Gray-Owen. 2008. Modular arrangement of allelic variants explains the divergence in Moraxella catarrhalis UspA protein function. Infect. Immun. 765330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullard, B., S. L. Lipski, and E. R. Lafontaine. 2005. Hag directly mediates the adherence of Moraxella catarrhalis to human middle ear cells. Infect. Immun. 735127-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conners, R., D. J. Hill, E. Borodina, C. Agnew, S. J. Daniell, N. M. Burton, R. B. Sessions, A. R. Clarke, L. E. Catto, D. Lammie, T. Wess, R. L. Brady, and M. Virji. 2008. The Moraxella adhesin UspA1 binds to its human CEACAM1 receptor by a deformable trimeric coiled-coil. EMBO J. 271779-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. J. Hasemann, C. Aebi, F. W. Henderson, G. H. J. McCracken, and E. J. Hansen. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 1814026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1610881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enright, M. C., and H. McKenzie. 1997. Moraxella (Branhamella) catarrhalis—clinical and molecular aspects of a rediscovered pathogen. J. Med. Microbiol. 46360-371. [DOI] [PubMed] [Google Scholar]
- 10.Gray-Owen, S. D., D. R. Lorenzen, A. Haude, T. F. Meyer, and C. Dehio. 1997. Differential Opa specificities for CD66 receptors influence tissue interactions and cellular response to Neisseria gonorrhoeae. Mol. Microbiol. 26971-980. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill, D. J., A. M. Edwards, H. A. Rowe, and M. Virji. 2005. Carcinoembryonic antigen-related cell adhesion molecule (CEACAM)-binding recombinant polypeptide confers protection against infection by respiratory and urogenital pathogens. Mol. Microbiol. 551515-1527. [DOI] [PubMed] [Google Scholar]
- 13.Hill, D. J., and M. Virji. 2003. A novel cell-binding mechanism of Moraxella catarrhalis ubiquitous surface protein UspA: specific targeting of the N-domain of carcinoembryonic antigen-related cell adhesion molecules by UspA1. Mol. Microbiol. 48117-129. [DOI] [PubMed] [Google Scholar]
- 14.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 195989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holm, M. M., S. L. Vanlerberg, I. M. Foley, D. D. Sledjeski, and E. R. Lafontaine. 2004. The Moraxella catarrhalis porin-like outer membrane protein CD is an adhesin for human lung cells. Infect. Immun. 721906-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2547-559. [DOI] [PubMed] [Google Scholar]
- 17.Koretke, K. K., P. Szczesny, M. Gruber, and A. N. Lupas. 2006. Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J. Struct. Biol. 155154-161. [DOI] [PubMed] [Google Scholar]
- 18.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 1821364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 1831540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipski, S. L., C. Akimana, J. M. Timpe, R. M. Wooten, and E. R. Lafontaine. 2007. The Moraxella catarrhalis autotransporter McaP is a conserved surface protein that mediates adherence to human epithelial cells through its N-terminal passenger domain. Infect. Immun. 75314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luke, N. R., and A. A. Campagnari. 1999. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect. Immun. 675815-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luke, N. R., J. A. Jurcisek, L. O. Bakaletz, and A. A. Campagnari. 2007. The expression of type IV pili by Moraxella catarrhalis contributes to biofilm formation and adherence to respiratory epithelial cells, abstr. D-168. Abstr. 107th Gen. Meet. Am. Soc. Microbiol.
- 23.McMichael, J. C. 2000. Vaccines for Moraxella catarrhalis. Vaccine 19(Suppl. 1)S101-S107. [DOI] [PubMed] [Google Scholar]
- 24.McMichael, J. C., M. J. Fiske, R. A. Fredenburg, D. N. Chakravarti, K. R. VanDerMeid, V. Barniak, J. Caplan, E. Bortell, S. Baker, R. Arumugham, and D. Chen. 1998. Isolation and characterization of two proteins from Moraxella catarrhalis that bear a common epitope. Infect. Immun. 664374-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier, P. S., R. Troller, N. Heiniger, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2005. Moraxella catarrhalis strains with reduced expression of the UspA outer membrane proteins belong to a distinct subpopulation. Vaccine 232000-2008. [DOI] [PubMed] [Google Scholar]
- 26.Murphy, T. F. 2005. Moraxella (Branhamella) catarrhalis and other gram-negative cocci, p. 2529. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Mandell, Douglas, and Bennett's principles and practice of infectious disease. Elsevier Inc., Philadelphia, PA.
- 27.Murphy, T. F. 2005. Vaccine development for non-typeable Haemophilus influenzae and Moraxella catarrhalis: progress and challenges. Expert Rev. Vaccines 4843-853. [DOI] [PubMed] [Google Scholar]
- 28.Murphy, T. F., A. L. Brauer, B. J. Grant, and S. Sethi. 2005. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am. J. Respir. Crit. Care Med. 172195-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.N′Guessan, P. D., B. Temmesfeld-Wollbruck, J. Zahlten, J. Eitel, S. Zabel, B. Schmeck, B. Opitz, S. Hippenstiel, N. Suttorp, and H. Slevogt. 2007. Moraxella catarrhalis induces ERK- and NF-kappaB-dependent COX-2 and prostaglandin E2 in lung epithelium. Eur. Respir. J. 30443-451. [DOI] [PubMed] [Google Scholar]
- 30.N′Guessan, P. D., M. Vigelahn, S. Bachmann, S. Zabel, B. Opitz, B. Schmeck, S. Hippenstiel, J. Zweigner, K. Riesbeck, B. B. Singer, N. Suttorp, and H. Slevogt. 2007. The UspA1 protein of Moraxella catarrhalis induces CEACAM1-dependent apoptosis in alveolar epithelial cells. J. Infect. Dis. 1951651-1660. [DOI] [PubMed] [Google Scholar]
- 31.Nordstrom, T., A. M. Blom, A. Forsgren, and K. Riesbeck. 2004. The emerging pathogen Moraxella catarrhalis interacts with complement inhibitor C4b binding protein through ubiquitous surface proteins A1 and A2. J. Immunol. 1734598-4606. [DOI] [PubMed] [Google Scholar]
- 32.Nordstrom, T., A. M. Blom, T. T. Tan, A. Forsgren, and K. Riesbeck. 2005. Ionic binding of C3 to the human pathogen Moraxella catarrhalis is a unique mechanism for combating innate immunity. J. Immunol. 1753628-3636. [DOI] [PubMed] [Google Scholar]
- 33.Plamondon, P., N. R. Luke, and A. A. Campagnari. 2007. Identification of a novel two-partner secretion locus in Moraxella catarrhalis. Infect. Immun. 752929-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reddy, M. S., T. F. Murphy, H. S. Faden, and J. M. Bernstein. 1997. Middle ear mucin glycoprotein; purification and interaction with nontypeable Haemophilus influenzae and Moraxella catarrhalis. Otolaryngol. Head Neck Surg. 116175-180. [DOI] [PubMed] [Google Scholar]
- 35.Rosseau, S., K. Wiechmann, S. Moderer, J. Selhorst, K. Mayer, M. Krull, A. Hocke, H. Slevogt, W. Seeger, N. Suttorp, J. Seybold, and J. Lohmeyer. 2005. Moraxella catarrhalis-infected alveolar epithelium induced monocyte recruitment and oxidative burst. Am. J. Respir. Cell Mol. Biol. 32157-166. [DOI] [PubMed] [Google Scholar]
- 36.Schumann, D., J. Huang, P. E. Clarke, J. Kirshner, S. W. Tsai, V. N. Schumaker, and J. E. Shively. 2004. Characterization of recombinant soluble carcinoembryonic antigen cell adhesion molecule 1. Biochem. Biophys. Res. Commun. 318227-233. [DOI] [PubMed] [Google Scholar]
- 37.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slevogt, H., B. Schmeck, C. Jonatat, J. Zahlten, W. Beermann, L. van, V. B. Opitz, S. Dietel, P. D. N′Guessan, S. Hippenstiel, N. Suttorp, and J. Seybold. 2006. Moraxella catarrhalis induces inflammatory response of bronchial epithelial cells via MAPK and NF-kappaB activation and histone deacetylase activity reduction. Am. J. Physiol. Lung Cell. Mol. Physiol. 290L818-L826. [DOI] [PubMed] [Google Scholar]
- 39.Slevogt, H., J. Seybold, K. N. Tiwari, A. C. Hocke, C. Jonatat, S. Dietel, S. Hippenstiel, B. B. Singer, S. Bachmann, N. Suttorp, and B. Opitz. 2007. Moraxella catarrhalis is internalized in respiratory epithelial cells by a trigger-like mechanism and initiates a TLR2- and partly NOD1-dependent inflammatory immune response. Cell. Microbiol. 9694-707. [DOI] [PubMed] [Google Scholar]
- 40.Spaniol, V., N. Heiniger, R. Troller, and C. Aebi. 2007. Outer membrane protein UspA1 and lipooligosaccharide are involved in invasion of human epithelial cells by Moraxella catarrhalis. Microbes Infect. 103-11. [DOI] [PubMed] [Google Scholar]
- 41.Tan, T. T., A. Forsgren, and K. Riesbeck. 2006. The respiratory pathogen Moraxella catarrhalis binds to laminin via ubiquitous surface proteins A1 and A2. J. Infect. Dis. 194493-497. [DOI] [PubMed] [Google Scholar]
- 42.Tan, T. T., T. Nordstrom, A. Forsgren, and K. Riesbeck. 2005. The respiratory pathogen Moraxella catarrhalis adheres to epithelial cells by interacting with fibronectin through ubiquitous surface proteins A1 and A2. J. Infect. Dis. 1921029-1038. [DOI] [PubMed] [Google Scholar]
- 43.Virji, M., D. Evans, A. Hadfield, F. Grunert, A. M. Teixeira, and S. M. Watt. 1999. Critical determinants of host receptor targeting by Neisseria meningitidis and Neisseria gonorrhoeae: identification of Opa adhesiotopes on the N-domain of CD66 molecules. Mol. Microbiol. 34538-551. [DOI] [PubMed] [Google Scholar]
- 44.Virji, M., K. Makepeace, D. J. P. Ferguson, and S. Watt. 1996. Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol. Microbiol. 22941-950. [DOI] [PubMed] [Google Scholar]
- 45.Wallace, R. J., Jr., V. A. Steingrube, D. R. Nash, D. G. Hollis, C. Flanagan, B. A. Brown, A. Labidi, and R. E. Weaver. 1989. BRO β-lactamases of Branhamella catarrhalis and Moraxella subgenus Moraxella, including evidence for chromosomal β-lactamase transfer by conjugation in B. catarrhalis, M. nonliquefaciens, and M. lacunata. Antimicrob. Agents Chemother. 331845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]