Abstract
Seasonal epidemics of cholera in Bangladesh are self-limited in nature, presumably due to phage predation of the causative Vibrio cholerae during the late stage of an epidemic, when cholera patients excrete large quantities of phage in their stools. To further understand the mechanisms involved, we studied the effect of phage on the infectivity and survival of V. cholerae shed in stools. The 50% infectious dose of stool vibrios in infant mice was ∼10-fold higher when the stools contained a phage (1.8 × 103 to 5.7 × 106 PFU/ml) than when stools did not contain a detectable phage. In competition assays in mice using a reference strain and phage-negative cholera stools, the infectivity of biofilm-like clumped cells was 3.9- to 115.9-fold higher than that of the corresponding planktonic cells. However, the difference in infectivity of these two cell populations in phage-positive stools was significantly less than that in phage-negative stools (P = 0.0006). Coculture of a phage and V. cholerae or dilutions of phage-positive cholera stools in nutrient medium, but not in environmental water, caused rapid emergence of phage-resistant derivatives of the bacteria, and these derivatives lost their O1 antigen. In cholera stools and in intestinal contents of mice prechallenged with a mixture of V. cholerae and phage, the bacteria remained completely phage susceptible, suggesting that the intestinal environment did not favor the emergence of phage-resistant derivatives that lost the O1 antigen. Our results indicate that phages lead to the collapse of epidemics by modulating the required infectious dose of the bacteria. Furthermore, the dominance of phage-resistant variants due to the bactericidal selective mechanism occurs rarely in natural settings, and the emerging variants are thus unable to sustain the ongoing epidemic.
Toxigenic Vibrio cholerae strains belonging to the O1 and O139 serogroups cause cholera, a waterborne acute diarrheal disease that occurs frequently as epidemics in many developing countries (7, 11). The occurrence of epidemics is known to coincide with increased prevalence of the causative V. cholerae strain in the aquatic environment (12), and populations lacking adequate safe drinking water and sanitation are the ones mostly affected. Epidemics of cholera occur regularly in the Ganges Delta region of Bangladesh and India, with two epidemics occurring every year (7).
V. cholerae belongs to a group of organisms whose major habitats are aquatic ecosystems (2, 3, 7, 11, 12). A variety of physical and biological parameters are likely to influence the survival and abundance of V. cholerae as a species in the environment, but these factors do not exclusively modulate the prevalence of toxigenic V. cholerae O1 and O139 strains. In contrast, bacteriophages in the environment have recently been found to modulate the abundance of toxigenic V. cholerae in water samples and the incidence rates of cholera (5, 8, 10). Phages may also play a role in the emergence of pathogenic clones and may be involved in territorialism between different strains of V. cholerae (5). For example, the emergence and dominance of V. cholerae O139 in Bangladesh and India during 1992 to 1993 may have involved phages both as a means of horizontal gene transfer and as a selective mechanism (17).
We have recently shown that cholera patients excrete large quantities of lytic phages in their stools during the late stage of an epidemic, coinciding with the period of increased phage prevalence in the surface water (8). These and other findings suggested that in vivo phage amplification in cholera victims and contamination of surface water by stools of cholera patients may contribute to increased concentrations of the phage in water (6, 8). Thus, biological control of V. cholerae by phage amplified in cholera victims and subsequent phage predation in the environment likely influences the temporal dynamics of cholera epidemics. To further understand the mechanism associated with this phenomenon, we studied the effect of phage on the infectivity of V. cholerae excreted in stools of cholera patients and the dose of the pathogen required to cause an infection in the mouse model. Furthermore, we monitored the emergence of phage-resistant derivatives of the parent epidemic strain under both in vivo and in vitro conditions, to understand whether these derivatives are likely to replace the original strain as a cause of subsequent cholera epidemics.
MATERIALS AND METHODS
Clinical specimens.
The International Centre for Diarrheal Disease Research in Bangladesh (ICDDR,B) maintains a surveillance system at its Dhaka hospital in which data from every 50th patient presenting to the hospital are collected; cholera stool samples used in the study were obtained from these patients. Specimens from these patients were also used to obtain clinical V. cholerae strains and phages as required for the studies described. Protocols for studies involving human specimens were reviewed and approved by the Research Review Committee and Ethical Review Committee at the ICDDR,B in Bangladesh and the Institutional Review Boards at Harvard Medical School and Massachusetts General Hospital.
Assays in cholera stools.
Freshly collected cholera stools from patients who did not receive antibiotics before reporting to the hospital were examined by dark-field microscopy to ascertain the presence of typical vibrio-shaped organisms. Dilutions of the stools in phosphate-buffered saline (PBS) were cultured immediately to calculate the number of V. cholerae organisms present in the stools, as well as to isolate the strain for further analysis. Since many of the stool samples were likely to contain phage particles to which the bacteria were susceptible, an aliquot of the stools was centrifuged to precipitate the cells and resuspended in fresh PBS, thus reducing the phage content before the dilutions were plated and the number of CFU was estimated. The presence of phage in the stools and the phage susceptibilities of the V. cholerae strains in the same stool were also quantified by plaque assays (5) and a modified plating technique as described below. In addition, the stool specimens were used for a variety of assays including infectivity assays in mice and for study of the dynamics of phage and bacterial growth in nutrient medium and filter-sterilized environmental water.
Determination of ID50s in mice.
To determine the 50% infectious dose (ID50) of V. cholerae strains, 5-day-old Swiss Albino mice in groups of five were inoculated intragastrically with dilutions of cholera stools or laboratory-grown V. cholerae, as well as with a mixture of V. cholerae and phage whenever appropriate. Aliquots of these samples were also cultured as described above to determine the cell count, and plaque assays were conducted to determine the concentration of phage. Mice were euthanized at 24 h, and the small intestines were homogenized in PBS. The homogenate was centrifuged, and the pellet was resuspended in fresh PBS to eliminate possible phage particles (derived from phage-positive samples) which could negatively influence the cell count. Dilutions of the suspension were then plated on taurocholate tellurite gelatin agar plates (16) containing streptomycin (70 μg/ml) to determine the number of CFU. The percentage of infected mice was plotted against the input dose (see Fig. 2) to determine the ID50 as described previously (1, 21).
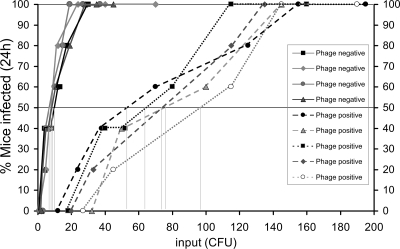
FIG. 2.
Determination of ID50 of V. cholerae excreted in phage-positive and phage-negative cholera stools. Groups of five mice were infected with V. cholerae O1 by using dilutions of stools from cholera patients coexcreting both phage and V. cholerae (broken lines) or from patients excreting V. cholerae without phage (solid lines). The doses used are shown on the x axis, and the percentage of mice infected after 24 h is shown on the y axis. The ID50 for each sample, i.e., the dose at which 50% of the challenged mice would show amplification of the strain in vivo, was determined graphically and is indicated by the vertical lines.
Competition assay in infant mice.
Stools from cholera patients were fractionated according to the size of suspended particles by centrifugation, initially at 2,000 × g to precipitate cells bound to debris, while the supernatant contained mostly planktonic cells. The precipitate was washed in PBS (pH 7.2) to remove any loosely adhering cells and finally resuspended in fresh PBS. Relative infectivity of V. cholerae present in different fractions of stools was determined by competition assays using methods described previously (6, 15). Briefly, the abilities of different populations of V. cholerae cells to colonize infant mice in competition with a reference strain were determined. The reference strain used was DSM-V984 (15), which is LacZ negative and hence could be differentiated from the test strains when grown on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-containing plates. Competition assays were conducted by mixing the reference strain grown in LB medium to log phase with stool bacteria in a ratio of 1:10 (vol/vol). A total of 50 μl of a 1,000-fold dilution of this mixture, containing 104 to 105 CFU, was administered to 3- to 5-day-old Swiss Albino mice. An in vitro analysis of the inoculum was also done to determine the precise ratio of test strain to reference strain. After 24 h, mice were euthanized, the small intestines were removed and homogenized, and serial dilutions of the homogenates were plated on GA (1% peptone, 1% NaCl, 3% gelatin, 1.5% agar) medium supplemented with X-Gal to allow for subsequent enumeration of LacZ-positive and LacZ-negative V. cholerae colonies to determine the ratio of the test and reference strains. The competitive index or infectivity of each population of cells was calculated by dividing the output ratios by the input ratio of the test and reference strains. The infectivity of the test strain was considered to be more than that of the reference strain when the competitive index was greater than 1. Statistical comparison of the infectivities between any two different populations of cells was done using statistical software (Sigmastat for Windows, version 2.03; Jandel Scientific, San Rafael, CA). Differences were considered to be significant when two-tailed P values were ≤0.05.
Estimation and analysis of phage.
Suitable recipient V. cholerae strains including isolates from recent cholera patients were used to estimate phage concentration by plaque assay as described previously (5). Briefly, logarithmic-phase cells (500 μl) of each host bacterial strain in nutrient broth (Difco, Detroit, MI) were mixed with 3.5-ml aliquots of soft agar (nutrient broth containing 0.8% Bacto agar; Difco), and the mixtures were overlaid on nutrient agar plates. For detection and quantification of phage in cholera stools or intestinal extracts of mice, aliquots of the samples were prefiltered through 0.22-μm-pore-size filters (Millipore Corporation, Bedford, MA) and dilutions of this filtrate were inoculated on plates. A sample was scored positive for phage when a plaque was observed on the bacterial lawn. Plaques were counted to estimate the concentration of phage particles in the sample. When appropriate, phages from representative plaques were further purified and saved for subsequent analysis. Nucleic acids were prepared from representative phage isolates using previously described methods (5) and were analyzed for restriction endonuclease cleavage patterns of the phage genomes using standard procedures (14).
Dynamics of phage and bacterial growth.
The dynamics of growth of V. cholerae and a lytic phage was studied in a mixed culture of the phage and the bacteria in LB medium as well as in filter-sterilized environmental water. The water samples were collected in Dhaka during an epidemic of cholera in September 2007. Before the experiments were set up, aliquots of water samples were filtered through cheesecloth, followed by 0.45- and 0.22-μm-pore-size membranes (Millipore). The LB medium or water was inoculated with a mixture of laboratory-grown bacteria and phage (to a concentration of 104 bacteria and 104 phage particles per ml) or dilutions of phage-positive cholera stools. Samples were removed hourly and analyzed for the presence of phage and the proportions of phage-susceptible and phage-resistant V. cholerae as described below.
Monitoring the emergence of phage-resistant V. cholerae.
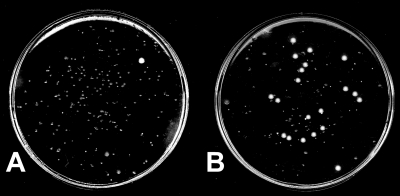
To determine the emergence of phage-resistant V. cholerae in cultures of the bacteria in the presence of a phage or in phage-positive biological specimens, we developed a modified plating technique. Dilutions of the culture or biological specimens (cholera stools or intestinal extracts of animals) were spread on a nylon membrane placed on an LB agar plate and incubated for ∼6 h till small colonies became visible. The nylon membrane was then removed and placed upside down on a sampling plate (i.e., LB agar plate preinoculated with ∼108 phage particles) to obtain an impression of the colonies. The membrane was transferred back onto the original plate, and the sampling plate, with the impression of the colonies, was incubated at 37°C for 16 h. The phage-sensitive colonies did not grow any further, whereas the phage-resistant colonies continued to grow (Fig. 1). This method allowed us to estimate the total number of CFU as well as the number of phage-resistant CFU. Representative phage-resistant colonies from the plate were picked and further verified by a conventional soft agar plating method described previously (5).
FIG. 1.
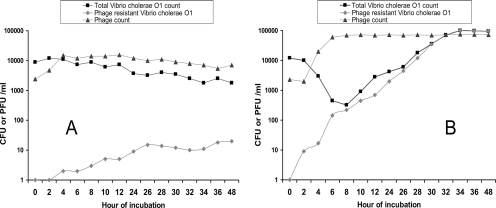
Plate assay for estimation of the emergence of phage-resistant V. cholerae in cultures containing both phage and V. cholerae. Dilutions of the culture were spread on a nylon membrane placed on an LB agar plate and incubated for ∼6 h till small colonies became visible. The membrane was removed and transiently placed upside down on an LB agar plate preinoculated with ∼108 phage particles to obtain an impression of the small colonies. The plate was then incubated at 37°C for 16 h. The small colonies are phage sensitive, whereas the large colonies are resistant to the phage. Plates derived from mixed cultures of V. cholerae O1 and JSF4 phage in filtered environmental water at a 10−3 dilution (A) and in LB at a 10−6 dilution (B) are shown.
RESULTS
V. cholerae and phage in cholera stools.
We determined the concentration of V. cholerae and phage particles excreted in the stools of cholera patients by analyzing representative stool samples collected from successive cholera epidemics in Dhaka, Bangladesh, during 2004 to 2007. In agreement with a previous report (8), many of the cholera patients were found to excrete high titers of phage (1.8 × 103 to 5.7 × 107 PFU/ml) in their stools, particularly during the late stages of the epidemics. Interestingly, despite the presence of the phage, all stools also contained high concentrations of V. cholerae (1.2 × 105 to 2.4 × 107 CFU/ml) (Table 1).
TABLE 1.
Concentrations of Vibrio cholerae O1 cells and phage excreted in the stools of cholera patients
| Date | V. cholerae concn (CFU/ml) | Phage concn (PFU/ml)b | Identity of phagea |
|---|---|---|---|
| 2 August 2004 | 4.5 × 105 | 2.7 × 104 | JSF4 |
| 10 August 2004 | 2.7 × 105 | 1.8 × 103 | JSF4 |
| 6 August 2004 | 5.7 × 106 | 2.0 × 104 | JSF4 |
| 16 August 2004 | 3.3 × 105 | 8.1 × 105 | JSF4 |
| 18 September 2005 | 3.7 × 105 | 3.5 × 104 | JSF4 |
| 25 September 2005 | 3.2 × 106 | 7.8 × 105 | JSF4 |
| 26 September 2005 | 1.2 × 105 | 1.1 × 104 | JSF4 |
| 4 August 2006 | 3.0 × 105 | 4.5 × 104 | JSF4 |
| 16 May 2007 | 2.0 × 105 | 2.2 × 105 | JSF4 |
| 11 June 2007 | 3.0 × 106 | 5.7 × 107 | JSF4 |
| 4 July 2007 | 2.3 × 106 | 3.0 × 106 | JSF4 |
| 7 August 2007 | 1.4 × 106 | 2.0 × 105 | JSF4 and JSF6 |
| 19 August 2007 | 2.4 × 107 | 1.5 × 104 | JSF4 |
| 25 September 2007 | 3.4 × 105 | 3.8 × 106 | JSF4 |
| 7 August 2004 | 5.0 × 105 | <10 | |
| 9 August 2004 | 3.4 × 106 | <10 | |
| 10 August 2004 | 1.6 × 107 | <10 | |
| 16 August 2006 | 3.4 × 105 | <10 | |
| 9 May 2007 | 3.4 × 106 | <10 | |
| 16 May 2007 | 8.0 × 106 | <10 | |
| 22 May 2007 | 6.2 × 106 | <10 | |
| 11 June 2007 | 2.4 × 106 | <10 | |
| 8 July 2007 | 6.0 × 108 | <10 | |
| 27 August 2007 | 1.6 × 106 | <10 | |
| 4 September 2007 | 6.4 × 106 | <10 | |
| 30 September 2007 | 8.3 × 106 | <10 | |
| 30 October 2007 | 8.0 × 106 | <10 |
The identity of each phage isolate was established by comparing its host specificity as well as the restriction pattern of its genome with that of previously described phages (5).
Plaque count of <10 means that no plaque was detected on plating of 100-μl aliquots of specimen. In theory, the concentration of phage should be <10 PFU/ml.
The phages isolated from all stools during the epidemics were identical to a previously characterized phage, JSF4 (5). One patient, however, excreted two types of phages which were previously designated as JSF4 and JSF6 (Table 1). The V. cholerae strains isolated from phage-positive stools were completely susceptible to lysis by the phage from the same stools. We used a modified plaque assay (see Materials and Methods for details) to determine the frequency of isolating possible phage-resistant derivatives of the V. cholerae strain in each stool. In this assay, one could examine a large number of V. cholerae cells in stools for phage resistance on a single plate (Fig. 1). However, we failed to isolate any JSF4 phage-resistant V. cholerae directly from stools, and we estimate that the presence of phage-resistant cells, if any, in cholera stools would be extremely weak (frequency, <10−6). However, as described below, phage-resistant derivatives of the parent V. cholerae strain emerged rapidly when diluted stools were grown in LB medium but not in filtered environmental water or under in vivo conditions in infant mice.
Phages can modulate the infectivity of V. cholerae excreted in cholera stools.
We investigated whether the presence of phage in the stools of cholera patients would alter the dose of the bacteria in the stools required to cause a productive infection. The assay to determine the infectious dose was conducted by inoculating infant mice with various dilutions of each cholera stool (and hence various doses of V. cholerae) and determining the ID50, the dose at which 50% of the challenged mice would show amplification of the strain in vivo.
We analyzed the infectious doses of V. cholerae in five different phage-positive cholera stools and compared them with those in phage-negative cholera stool samples (Fig. 2). The observed ID50s of V. cholerae in stools containing a phage were 54 to 97 CFU (mean, 73 CFU), which were ∼10-fold higher than the ID50s of V. cholerae in stools that did not contain a detectable phage (ID50 = 5 to 12 CFU; mean, 8.7 CFU). Remarkably, the ID50 of V. cholerae in a stool that contained a low count of phage (∼103 PFU/ml) was still higher than that of phage-negative stools, although the dilutions (103 to 105) of stool used in the assay should further reduce the phage concentration to a minimum level (≤1 particle/ml). This observation can be explained by assuming that phage particles had already adhered to a proportion of V. cholerae cells, and hence, despite the dilution of stools, a considerable number of phage particles were inoculated into the mice. A recent study showed that the motility of V. cholerae cells in phage-positive cholera stools was less than that in phage-negative stools (18). We presume that the reduced motility could be due to the adherence of phage particles to the bacterial cells.
One caveat to this assay, however, was that the number of infectious V. cholerae cells in stools containing a phage (or in mixed cultures of phage and V. cholerae), as determined by plate count, is likely to be an underestimate, since the phage would kill a proportion of the V. cholerae cells during the assay procedure. Although we washed the bacterial cells to reduce the phage content before plating, it is likely that phages already adsorbed by the cells could still negatively influence the count of viable cells. This would mean that the real ID50 of V. cholerae cells in stools containing a phage would be even higher than that observed in our assays. On the other hand, the negative influence of phage on the estimation of V. cholerae cells would also apply when plating homogenates of the mouse intestines to determine colonization, and this may compensate for the effect of possible underestimation of input counts of V. cholerae.
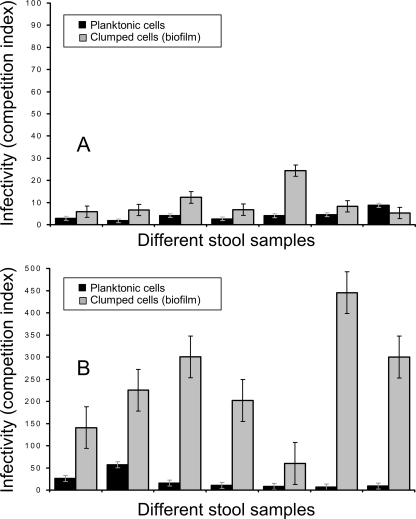
Cholera stools contain at least two morphologically different populations of cells, which include planktonic and clumped biofilm-like cells (6), and a previous study showed that clumped cells were apparently more infectious. We, therefore, separated the two fractions of cells by low-speed centrifugation. The determination of ID50 was done using the planktonic cells from phage-positive and phage-negative stools. However, we used the competition assay in infant mice to compare the infectivities of both planktonic and clumped cells derived from phage-positive and phage-negative stools as described previously (6). The relative infectivities of these stool vibrios are presented in Fig. 3. Comparison of the apparent infectivities of planktonic and clumped stool vibrios using the reference strain, DSM-V984, grown to log phase showed that the mean infectivities of the biofilm-like clumped cells from different phage-negative stools were 3.9- to 115.9-fold higher than those for the corresponding planktonic cells. This difference in infectivity between the two populations of cells was also statistically significant (P = 0.0041). However, the difference in infectivity of these two cell populations in phage-positive stools was significantly less than that in phage-negative stools (P = 0.0006) (Fig. 3).
FIG. 3.
Mean competitive indices of V. cholerae O1 present in different fractions of phage-positive and phage-negative cholera stool samples, shown in panels A and B, respectively, against a reference strain grown to exponential phase. The mean infectivities of clumped cell fractions were significantly higher than those of the planktonic cell fractions in phage-negative cholera stools (P = 0.0041). However, the difference in infectivity of these two cell populations in phage-positive stools was significantly less than that in phage-negative stools (P = 0.0006).
As pointed out previously (6), the apparently higher infectivity of clumped cells results from underestimation of input colony counts during competition assays with the “reference” laboratory-grown strain to determine the competitive index. In phage-positive stools, the clumped cells also appeared to carry phage particles despite washing, and between 103 and 105 PFU/ml of infectious phage could be recovered from the challenged mice (data not shown). Presumably the phage particles adhered to the cell clumps, and on inoculation of mice, the phage acted on the V. cholerae cells in vivo.
Effect of phage on the infectivity of laboratory-grown V. cholerae.
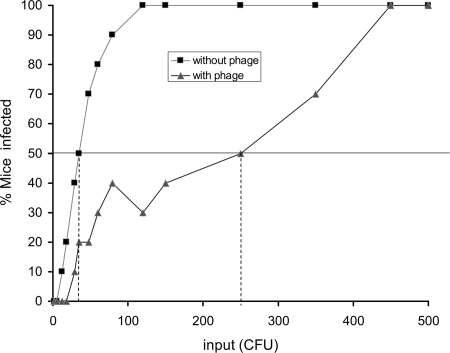
To further verify the findings from the assays in human cholera stools, we also studied the effect of phage on the infectious dose of V. cholerae strains grown under laboratory conditions using the infant mouse model. V. cholerae O1 El Tor strain C6706 was grown to log phase, and various dilutions of the culture were inoculated in mice with or without a phage previously designated JSF4 (5). The mean ID50 of the strain was 32 CFU in the absence of the phage but 250 CFU in the presence of 104 phage particles coinoculated with the V. cholerae strain (Fig. 4). This ID50 was higher than that of V. cholerae excreted in stools (Fig. 2). The apparently lower ID50 for stool vibrios could be due to the presence in the planktonic cell fraction of small clumps of cells that could not be efficiently separated and thus caused an underestimation of the input count. However, stools from cholera patients contain complex mixtures of cells, presumably with various transcriptional programs that remain to be fully characterized.
FIG. 4.
ID50 of a laboratory-grown V. cholerae O1 strain in the presence and absence of a lytic phage, JSF4. Groups of five mice were infected with strain C6706 together with a lytic phage, JSF4, and without phage. The doses used are shown on the x axis, and the percentage of mice infected after 24 h is shown on the y axis. The ID50 for each group was determined graphically and is indicated by the vertical broken lines.
Dynamics of phage-bacterial growth and emergence of phage-resistant bacteria.
To assess whether the selection pressure of a lytic phage leads to rapid emergence of phage-resistant derivatives of the susceptible bacterium, we examined the effect of phage on the appearance of phage-resistant V. cholerae both under in vitro conditions and in the mouse model by using either cholera stools or laboratory-grown bacterial and phage preparations. The proportions of phage-resistant and phage-sensitive bacteria in the samples were determined using the plate assay described above.
When phage-positive cholera stools were diluted and grown in LB medium, initially the number of total V. cholerae cells rapidly declined, and within 6 h, the bacterial count was reduced to less than 1/10 of the original inoculum (Fig. 5B). Thereafter, phage-resistant derivatives of the V. cholerae strain began to be detected. After overnight incubation, the culture was found to contain mostly phage-resistant derivatives of the parent phage-susceptible strain (Fig. 5B). Interestingly the phage-resistant colonies were found to have lost their O1 antigen and hence did not agglutinate with the O1-specific antiserum. While a proportion of these colonies (51 of 145 colonies tested) were rough and agglutinated in normal saline, there were also phage-resistant derivatives that formed smooth colonies, like the parents, but did not agglutinate with the O1 antiserum, suggesting possible serotype conversion. These preliminary findings indicated that the O antigen of V. cholerae O1 may be a possible receptor in JSF4 phage infection. Further studies are under way to conduct molecular characterization of the phage-resistant derivatives. Similar results were observed when laboratory-grown V. cholerae O1 strains (which are sensitive to phage JSF4) were grown together with phage JSF4 in LB medium (data not shown).
FIG. 5.
Dynamics of phage and bacterial growth, when phage-positive cholera stools were diluted in filtered environmental water (A) and LB medium (B) and incubated at 30°C. Each data point represents the average value derived from three cholera stools.
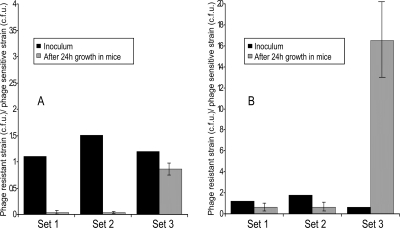
We compared the infectivities of a phage-susceptible V. cholerae O1 strain and its O-antigen-negative phage-resistant derivatives by inoculating a mixture of the parent strain and its derivatives in infant mice with or without the phage. As a control, we also included a V. cholerae O139 strain, which is resistant to phage JSF4, used in the study. The input ratios of the mutant and parent strains and their respective output ratios after 24 h of incubation in mice (Fig. 6) suggested that the O-antigen-negative mutants were impaired in colonization, both in the absence and in the presence of the phage. However, since the mutants were resistant to JSF4 phage, the mutants appeared to compete better with the phage-sensitive strain in the presence of the phage (Fig. 6). On the other hand, the control V. cholerae O139 strain outcompeted the phage-sensitive O1 strain in the presence of the phage and the mean output ratio was ∼25-fold higher than the input ratio of the strains.
FIG. 6.
Competition assays in infant mice for colonization by a JSF4 phage-sensitive V. cholerae O1 strain and its O1-antigen-negative phage-resistant derivatives. Assays were done in the absence (A) and presence (B) of the phage. The first two sets of strains (set 1 and set 2) represent mixtures of the parent V. cholerae O1 strain with two different O-antigen-negative phage-resistant isolates. Set 3 represents a mixture of the phage-sensitive V. cholerae O1 strain with a V. cholerae O139 strain that is resistant to phage JSF4. Each mixture of strains was assayed in five different mice.
It may be mentioned that, in a previous study, phage-resistant derivatives of an O1 El Tor strain resistant to a temperate phage, K139, which uses the O antigen of V. cholerae O1 as its receptor, were investigated according to their lipopolysaccharide (LPS) pattern, and interestingly, different groups of altered LPS structures were found. These included O-antigen-negative strains, where one mutant was characterized for the loss of O antigen due to transposition of IS1004 into wbeW, encoding a putative glycosyltransferase. Another class identified was rough LPS (R-LPS) mutants comprising an altered core oligosaccharide with no O antigen attached. In this same study, phage-resistant isolates with intact O antigen but with altered core oligosaccharide and additionally affected in galactose catabolism were also identified; these strains had mutations in the galU gene (20, 22).
In contrast to the dynamics of V. cholerae and phage growth in LB medium, when the stool was inoculated in filter-sterilized environmental water, the phage-resistant derivatives did not substantially enrich and the total bacterial population declined (Fig. 5A). However, the phage counts remained stable in water for more than 2 months. Furthermore, when infant mice were inoculated with a mixture of phage and V. cholerae in the infectivity assay, the intestinal contents recovered after 24 h contained both V. cholerae and phage. However, only 3 of 108 CFU of V. cholerae screened showed a phage-resistant phenotype, suggesting that phage-resistant derivatives appeared at a low frequency (3 × 10−8). Thus, although in nutrient medium phage-susceptible cells are rapidly replaced by resistant derivatives of the epidemic V. cholerae strain, it appears that in the aquatic environment, phage-resistant derivatives do not emerge substantially to replace the phage-susceptible cells. Similarly, the mouse intestinal environment did not appear to favor emergence of phage-resistant derivatives. Notably, V. cholerae strains isolated from stools of cholera patients were also found to remain completely susceptible to the phage isolated from the same stools.
DISCUSSION
We predict that the results described herein will have profound significance in understanding how phages modulate the dynamics of cholera epidemics, causing the epidemics to be self-limited in nature, and why phage-resistant derivatives of the epidemic V. cholerae strain do not emerge rapidly under bactericidal selection and continue to sustain an ongoing epidemic. The ID50 of V. cholerae cells in stools as well as that of laboratory-grown cells was found to be higher in the presence of a phage, suggesting that at least 10-fold-more cells would be required to cause a productive infection under the conditions in which cholera patients excrete phages in their stools. The sustainability of an epidemic depends on amplification of the causative strain in each cholera victim and waterborne spread of the pathogen to infect more individuals. Hence, the increase of the required infectious dose due to the deleterious effect of coingesting phage with V. cholerae would therefore lead to a reduced number of patients. Furthermore, in the face of phage predation, the environment would fail to support a heavy load of viable V. cholerae, and hence the epidemic would collapse.
The transmissibility of cholera, particularly during an epidemic, may depend on the physiological state and morphological forms of V. cholerae cells excreted by cholera patients (6, 18). We demonstrated previously that stools from cholera patients contain a heterogeneous mixture of biofilm-like aggregates and free-swimming planktonic cells of V. cholerae (6). Estimation of relative infectivities of these different forms of V. cholerae cells suggested that the enhanced infectivity of V. cholerae shed in human stools is largely due to the presence of clumps of cells that disperse in vivo, providing a high dose of the pathogen. Furthermore, the clumped cells were found to better survive the hypotonic conditions of environmental water than the planktonic cells. In the present study, we found that the presence of phage in cholera stools affected the infectivity of both the planktonic and clumped cell populations. Moreover, even the clumped cell population in cholera stools that contained a lytic phage did not show a drastically higher infectivity than that of the planktonic cells (Fig. 3), as would be expected in the absence of a phage (6).
As in cholera stools, V. cholerae strains in the environment often exist as aggregates of cells. We previously described these cells as conditionally viable environmental cells (CVEC), which resist cultivation by conventional techniques (6, 9). Further studies suggested that CVEC could be derived from cholera patients, since V. cholerae shed in stools of cholera patients, when inoculated in environmental water samples in the laboratory, exhibited characteristics similar to those of CVEC. Human victims ingesting clumps of CVEC in the water might get a fully infectious dose of the pathogen and might further amplify the strain to begin or sustain an epidemic due to that strain. In the present study, the difference in infectivity of clumped and planktonic cell populations in phage-positive stools was significantly less than that in phage-negative stools (P = 0.0006). Based on this observation, we predict that toward the end of an epidemic when the environmental phage concentration is known to be high, even CVEC would fail to sustain the epidemic, presumably due to predation of the strain when the clumped cells in CVEC disperse in vivo.
The recent recognition that phage predation may play a role in controlling cholera epidemics (5, 8) also suggests that changes in this organism may be driven by factors outside the human host. The emergence of new serogroups (e.g., O139) and the displacement of old biotypes (e.g., the classical by the El Tor) may be driven by environmental forces that include phages, propensity to form biofilms on aquatic matter, and even interaction with marine animals (4, 8, 13). Our method for quantitative estimation of the emergence of phage-resistant bacterial cells provided an opportunity to understand why more-frequent replacement of an epidemic clone by its phage-resistant derivative does not happen, as would be expected in a bactericidal selective mechanism. Investigation with laboratory-grown V. cholerae and phage in mixed culture suggested that growth of phage and V. cholerae allows generation of phage-resistant derivatives that rapidly dominate (Fig. 5). Interestingly, V. cholerae cells excreted in phage-positive cholera stools were completely susceptible to the phage. This observation suggested that possible phage-resistant derivatives may not compete well with the parent strain under in vivo conditions. In this study we have demonstrated convincingly that phage-resistant mutants of V. cholerae O1 that lose their O1 antigen are outcompeted by their parent strain in the mouse model of infection (Fig. 6).
It may be mentioned that in a previous study, phage-resistant mutants of V. cholerae O1 that were O antigen negative or altered in their LPS structure, including the galU and R-LPS mutants (20, 22), were also found to be defective in colonization in infant mice. It was suggested that galU and R-LPS mutants were more sensitive to short-chain organic acids, cationic antimicrobial peptides, the complement system, and bile salts as well as other hydrophobic agents. O-antigen-negative strains were found to be sensitive to complement and cationic peptides, but they displayed significant resistance to bile salts and short-chain organic acids. In another study, loss of LPS O side chain or capsular polysaccharide in V. cholerae O139 strains resulted in a significant reduction in colonization of the infant mouse small intestine, indicating that the presence of both LPS O side chain and capsular polysaccharide is important during the colonization process (19).
Our epidemiological observation suggests that the clone of V. cholerae causing recent cholera epidemics in Bangladesh has remained largely unchanged for several years in terms of its phage sensitivity. Similarly, the phage designated JSF4 has persisted through several epidemic seasons, since its first detection by us in 2003. Thus, although phages are likely to select phage-resistant clones leading to the emergence of new genetic variants, our results suggest that this change does not happen frequently under natural settings. Clearly, in filtered environmental water, the emergence of phage-resistant derivatives occurred at a low frequency compared to that in nutrient medium (Fig. 5). The water samples used in the study were collected during an epidemic of cholera, when expectedly the physicochemical conditions and dissolved nutrients in the water should be conducive to the growth of V. cholerae. Our data suggest that even during the period of the epidemic, the overall growth of V. cholerae cells and emergence of phage-resistant mutants occurred at a lower rate in water than in LB medium under laboratory conditions. Taken together, these findings explain why cholera epidemics continue to be self-limiting in nature. Presumably, when the balance of phage and bacteria tips in favor of the phage, there is a dramatic decline of numbers of bacteria, leading to the collapse of the epidemic. Moreover, phage-resistant derivatives of the epidemic strain do not emerge rapidly enough under natural conditions to be able to sustain the epidemic.
Since the same V. cholerae strain often reemerges and causes a subsequent epidemic during the next cholera season, it is obvious that a proportion of the epidemic V. cholerae cells survive the phage attack. Therefore, the ability to survive phage attacks in the environment may be an important aspect of the epidemic cycle of cholera. The metabolic status of V. cholerae cells and the optimum expression of phage receptors can presumably influence the susceptibility of cells to phage infection. Understanding the complex interactions of many different factors, including environmental factors, the lytic phages, and the role of the human host, may lead to the development of a model that can more fully explain the dynamics of cholera epidemics.
Acknowledgments
This research was funded in part by National Institutes of Health grants 2RO1-GM068851-5, AI070963-01A1, and U01-AI58935 under different subagreements between the Harvard Medical School, Massachusetts General Hospital, and the ICDDR,B. The ICDDR,B is supported by countries and agencies which share its concern for the health problems of developing countries.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 15 September 2008.
REFERENCES
- 1.Butler, S. M., E. J. Nelson, N. Chowdhury, S. M. Faruque, S. B. Calderwood, and A. Camilli. 2006. Cholera stool bacteria repress chemotaxis to increase infectivity. Mol. Microbiol. 60417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colwell, R. R. 2004. Infectious disease and environment: cholera as a paradigm for waterborne disease. Int. Microbiol. 7285-289. [PubMed] [Google Scholar]
- 3.Colwell, R. R., and A. Huq. 1994. Vibrios in the environment: viable but non-culturable Vibrio cholerae, p. 117-133. In I. K. Wachsmuth, P. A. Blake, and O. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. ASM Press, Washington, DC.
- 4.Colwell, R. R., and W. M. Spira. 1992. The ecology of Vibrio cholerae, p. 107-127. In D. Barua and W. B. Greenough III (ed.), Cholera. Plenum Medical Book Co., New York, NY.
- 5.Faruque, S. M., I. B. Naser, M. J. Islam, A. S. G. Faruque, A. N. Ghosh, G. B. Nair, D. A. Sack, and J. J. Mekalanos. 2005. Seasonal epidemics of cholera inversely correlate with the prevalence of environmental cholera phages. Proc. Natl. Acad. Sci. USA 1021702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faruque, S. M., K. Biswas, S. N. Udden, Q. S. Ahmad, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. USA 1036350-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 621301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque, S. M., M. J. Islam, Q. S. Ahmad, A. S. G. Faruque, D. A. Sack, G. B. Nair, and J. J. Mekalanos. 2005. Self-limiting nature of seasonal cholera epidemics: role of host-mediated amplification of phage. Proc. Natl. Acad. Sci. USA 1026119-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faruque, S. M., M. J. Islam, Q. S. Ahmad, K. Biswas, A. S. G. Faruque, G. B. Nair, R. B. Sack, D. A. Sack, and J. J. Mekalanos. 2006. An improved technique for isolation of environmental Vibrio cholerae with epidemic potential: monitoring the emergence of a multiple-antibiotic-resistant epidemic strain in Bangladesh. J. Infect. Dis. 1931029-1036. [DOI] [PubMed] [Google Scholar]
- 10.Jensen, M. A., S. M. Faruque, J. J. Mekalanos, and B. R. Levin. 2006. Modeling the role of bacteriophage in the control of cholera outbreaks. Proc. Natl. Acad. Sci. USA 1034652-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaper, J. B., J. G. Morris, and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 848-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan, M. U., M. Shahidullah, M. S. Haque, and W. U. Ahmed. 1984. Presence of vibrios in surface water and their relation with cholera in a community. Trop. Geogr. Med. 36335-340. [PubMed] [Google Scholar]
- 13.Kirn, T. J., B. A. Jude, and R. K. Taylor. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438863-866. [DOI] [PubMed] [Google Scholar]
- 14.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 15.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monsur, K. A. 1961. A highly selective gelatine-taurocholoate-tellurite medium for the isolation of Vibrio cholerae. Trans. R. Soc. Trop. Med. Hyg. 55440-442. [DOI] [PubMed] [Google Scholar]
- 17.Mooi, F. R., and E. M. Bik. 1997. The evolution of epidemic Vibrio cholerae strains. Trends Microbiol. 5161-165. [DOI] [PubMed] [Google Scholar]
- 18.Nelson, E. J., A. Chowdhury, J. B. Harris, Y. A. Begum, F. Chowdhury, A. I. Khan, R. C. Larocque, A. L. Bishop, E. T. Ryan, A. Camilli, F. Qadri, and S. B. Calderwood. 2007. Complexity of rice-water stool from patients with V. cholerae plays a role in the transmission of infectious diarrhea. Proc. Natl. Acad. Sci. USA 10419091-19096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nesper, J., S. Schild, C. M. Lauriano, A. Kraiss, K. E. Klose, and J. Reidl. 2002. Role of Vibrio cholerae O139 surface polysaccharides in intestinal colonization. Infect. Immun. 705990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 22.Reidl, J., and J. J. Mekalanos. 1995. Characterization of Vibrio cholerae bacteriophage K139 and use of a novel mini transposon to identify a phage-encoded virulence factor. Mol. Microbiol. 18685-701. [DOI] [PubMed] [Google Scholar]