Abstract
Yersinia enterocolitica is an enteric pathogen that exploits diverse means to survive in the human host. Upon Y. enterocolitica entry into the human host, bacteria sense and respond to variety of signals, one of which is the temperature. Temperature in particular has a profound impact on Y. enterocolitica gene expression, as most of its virulence factors are expressed exclusively at 37°C. These include two outer membrane proteins, YadA and Ail, that function as adhesins and complement resistance (CR) factors. Both YadA and Ail bind the functionally active complement alternative pathway regulator factor H (FH). In this study, we characterized regions on both proteins involved in CR and the interaction with FH. Twenty-eight mutants having short (7 to 41 amino acids) internal deletions within the neck and stalk of YadA and two complement-sensitive site-directed Ail mutants were constructed to map the CR and FH binding regions of YadA and Ail. Functional analysis of the YadA mutants revealed that the stalk of YadA is required for both CR and FH binding and that FH appears to target several conformational and discontinuous sites of the YadA stalk. On the other hand, the complement-sensitive Ail mutants were not affected in FH binding. Our results also suggested that Ail- and YadA-mediated CR does not depend solely on FH binding.
The complement system is the first line of immune defense against invading pathogens that directly activate the lectin pathway or the alternative pathway (AP) cascades in the human host. To survive, the pathogens have developed strategies to prevent deleterious consequences of complement activation. One of these strategies involves the acquisition of the host AP regulator factor H (FH). FH consists of 20 repetitive units, named short consensus repeats (SCRs), of ca. 60 amino acids each (46). The binding of FH is beneficial for microbes, as FH acts as a cofactor for the factor I (FI)-mediated cleavage of C3b, interferes with the association of factor B with C3b, and contributes to the dissociation of preformed AP C3 convertase C3bBb (23, 38, 55). Several pathogens were previously reported to take advantage of FH protective properties, including Streptococcus pyogenes (8, 18), group B streptococci (1), Borrelia sp. (16, 45), and Neisseria gonorrhoeae (35, 42). Also, Yersinia enterocolitica, an enteropathogen resistant to complement (2, 15, 26, 27, 39, 41, 48), has been shown to bind FH from human serum (3, 9, 44).
The Y. enterocolitica outer membrane proteins YadA and Ail confer serum resistance (4, 5, 9, 40, 52) and mediate the binding of FH and the classical and lectin pathway inhibitor C4b-binding protein (C4bp) to bacteria (3, 21). YadA functions as the major FH and C4bp receptor, while Ail can bind only the regulators when not blocked by the lipopolysaccharide O antigen and outer core (3, 21). Furthermore, our results showed that YadA appears to bind throughout the entire polypeptide chain of FH, while Ail targets SCRs 6 and 7. Both YadA-bound FH and Ail-bound FH, however, were found fully functional as cofactors in the FI-mediated cleavage of C3b (3).
YadA is encoded on a 70-kb virulence plasmid (pYV) (14, 53, 57) and expressed exclusively at 37°C (24, 50). YadA is a homotrimeric autotransporter protein with a monomer size of about 44 kDa that forms a lollipop-shaped structure on the bacterial surface (see Fig. 2A). From the N terminus to the C terminus, YadA contains a head, a neck, and a coiled-coil stalk domain, followed by a translocation and membrane-anchoring unit (17, 22). It was previously suggested that YadA serum resistance determinants are located within the stalk (43).
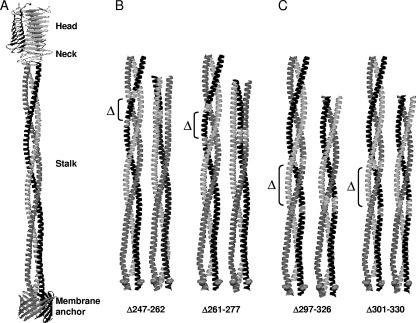
FIG. 2.
Effect of deletions on the structural model of the YadA stalk. (A) Ribbon presentation of the full-length wild-type YadA structure. (Modified from reference 22 with permission of the publisher.) (B and C) Comparison of the trimeric coiled-coil stalks of the wild type and four deletion mutants. Two of the mutants shown were predicted to alter the periodicity of the coiled-coil (B), and the other two were predicted to retain the wild-type periodicity of the stalk (C). The deleted regions in each stalk pair are indicated for the wild-type stalks.
The membrane anchor of YadA is a 12-stranded β-barrel with the pore closed by the C-terminal part of the stalk, a left-handed coiled-coil segment of four heptads (residues 368 to 395 of the Y. enterocolitica O:3 YadA sequence [GenBank accession no. CAA32086]) (22). The stalk continues into a right-handed coiled coil (residues 214 to 367) formed of nine 15-residue repeats (pentadecads) and a 19-residue segment separating the right- and left-handed parts (17, 22). The neck region (residues 192 to 213) comprises three safety pin-like elements (36) and connects the stalk to the head domain (residues 26 to 191). The head domain is a tight cylindrical trimeric structure formed by left-handed β-rolls (36); it binds to extracellular matrix proteins and mediates bacterial autoagglutination (reviewed in reference 11). The neck and head are dispensable for the YadA-mediated complement resistance (CR) (43).
Ail is a 17-kDa protein encoded chromosomally (33, 34). It belongs to a family of proteins predicted to form eight outer membrane-spanning amphipatic β-strands and four short extracellular loops (51, 54). Critical regions for CR of serotype O:8 Ail have been mapped to the C-terminal end of loop 2 and the N-terminal end of loop 3 (32).
In this report, we aimed to characterize YadA and Ail regions mediating serum resistance and FH binding. We show that FH targets several structural motifs of the YadA stalk and that this is biologically significant. The Ail region(s) involved in FH binding, however, could not be precisely mapped.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. For bactericidal assays, collagen affinity blotting, verification of expression of YadA deletion mutants, and adsorption of goat anti-human FH antiserum, bacteria were grown to stationary phase overnight in 5 ml of MedECa (4) at 37°C without shaking. For the examination of bacterial ability to bind serum FH, bacterial cultures grown overnight were diluted 1:20 in fresh medium and incubated for 3 h at 37°C without shaking to obtain bacteria in exponential phase of growth. When appropriate, antibiotics were added to the growth medium at the following concentrations: kanamycin at 100 μg/ml in agar plates and 20 μg/ml in broth and chloramphenicol at 20 μg/ml.
TABLE 1.
Bacteria and plasmids used in this work
| Bacterial strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Y. enterocolitica | ||
| 6471/76-c (YeO3-c) | Virulence plasmid cured derivative of YeO3 | 47 |
| YeO3-c-Ail | Δail::Km-GenBlock; derivative of YeO3-c; Kmr | 4 |
| YeO3-c-Ail-OCR | Spontaneous OC mutant derivative of YeO3-c-Ail-R; Kmr | 4 |
| E. coli | ||
| JM109 | reaA1 Δlac-pro endA1 gyrA96 thi-1 hsdR17 supE44 relA1 F′ traD36 proAB+lacIqZΔM15 | 56 |
| DH10B | F−mcrA Δ(mrr-hsd RMS-mcrBC), φ80lacZΔM15 ΔlacX74 deoR recA1 endA1 araΔ139Δ(ara leu)7697 galU galK λ−rpsL nupG λ−tonA | Life Technologies |
| HB101/pRK2013 | Triparental conjugation helper strain; Kmr | 12 |
| Plasmids | ||
| pYMS4505 | yadA gene of YeO3 cloned as a 1,735-bp PCR fragment (nucleotides 478-2213)a into the BamHI site of pTM100; Clmr; the yadA gene is transcribed by the constitutive tet promoter of pTM100 | 10 |
| pYMS4505:ΔNECK | Deletion of bp 1194-1271 of the yadA gene in pYMS 4505; expresses YadA with a 26-aa deletion (aa 193-218) | This work |
| pYMS4505:A | Deletion of bp 1257-1301 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 214-228) | This work |
| pYMS4505:S | Deletion of bp 1281-1325 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 222-236) | This work |
| pYMS4505:Δ#1 | Deletion of bp 1311-1355 of the yadA gene in pYMS 4505; expresses YadA with a 16-aa deletion (aa 231-246) | This work |
| pYMS4505:B | Deletion of bp 1347-1391 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 244-258) | This work |
| pYMS4505:Δ#2 | Deletion of bp 1356-1403 of the yadA gene in pYMS 4505; expresses YadA with a 16-aa deletion (aa 247-262) | This work |
| pYMS4505:C | Deletion of bp 1359-1403 of the yadA gene in pYMS 4505; does not express YadA due to a frameshift mutation introduced during construction | This work |
| pYMS4505:D | Deletion of bp 1371-1415 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 252-266) | This work |
| pYMS4505:E | Deletion of bp 1383-1427 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 256-270) | This work |
| pYMS4505:Δ#3+6 | Deletion of bp 1398-1448 of the yadA gene in pYMS 4505; expresses YadA with a 17-aa deletion (aa 261-277) | This work |
| pYMS4505:F | Deletion of bp 1392-1436 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 259-273) | This work |
| pYMS4505:Δ#3 | Deletion of bp 1404-1448 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 263-277) | This work |
| pYMS4505:H | Deletion of bp 1416-1460 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 267-281) | This work |
| pYMS4505:I | Deletion of bp 1428-1472 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 271-285) | This work |
| pYMS4505:Δ#4 | Deletion of bp 1449-1493 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 278-292) | This work |
| pYMS4505:J | Deletion of bp 1482-1571 of the yadA gene in pYMS 4505; expresses YadA with a 30-aa deletion (aa 289-318) | This work |
| pYMS4505:Δ#5 | Deletion of bp 1494-1538 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 293-307) | This work |
| pYMS4505:Δ#5-6 | Deletion of bp 1494-1583 of the yadA gene in pYMS 4505; expresses YadA with a 30-aa deletion (aa 293-322) | This work |
| pYMS4505:K | Deletion of bp 1506-1595 of the yadA gene in pYMS 4505; expresses YadA with a 30-aa deletion (aa 297-326) | This work |
| pYMS4505:L | Deletion of bp 1518-1607 of the yadA gene in pYMS 4505; expresses YadA with a 30-aa deletion (aa 301-330) | This work |
| pYMS4505:Δ#6 | Deletion of bp 1539-1583 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 308-322) | This work |
| pYMS4505:Δ#6+3 | Deletion of bp 1539-1586 of the yadA gene in pYMS 4505; expresses YadA with a 16-aa deletion (aa 308-323) | This work |
| pYMS4505:M | Deletion of bp 1572-1616 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 319-333) | This work |
| pYMS4505:Δ#7-9 | Deletion of bp 1584-1706 of the yadA gene in pYMS 4505; expresses YadA with a 41-aa deletion (aa 323-363) | This work |
| pYMS4505:N | Deletion of bp 1617-1661 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 334-348) | This work |
| pYMS4505:O | Deletion of bp 1662-1706 of the yadA gene in pYMS 4505; expresses YadA with a 15-aa deletion (aa 349-363) | This work |
| pYMS4505:P | Deletion of bp 1707-1727 of the yadA gene in pYMS 4505; expresses YadA with a 7-aa deletion (aa 364-370) | This work |
| pYMS4505:Δ1-3 | Deletion of bp 1707-1772 of the yadA gene in pYMS 4505; does not express surface-located YadA; 22-aa deletion (aa 364-385) | This work |
| pYMS4505:Q | Deletion of bp 1728-1748 of the yadA gene in pYMS 4505; expresses YadA with a 7-aa deletion (aa 371-377) | This work |
| pYMS4505:R | Deletion of bp 1749-1769 of the yadA gene in pYMS 4505; expresses YadA with a 7-aa deletion (aa 378-384) | This work |
| pTM100 | Mobilizable derivative of pACYC184 | 31 |
| pTM100-ail | ail gene cloned in a 1,711-bp PCR fragment (nucleotides 1490-3201)b into EcoRV site of pTM100; Clmr | This work |
| pTM100-ail1 | CAC at bp 2459-2461 substituted for GCA in pTM100-ail; expresses Ail with His65Ala substitution (loop 2) | This work |
| pTM100-ail2 | GATCTT at bp 2465-2470 substituted for GGAGGT in pTM100-ail; expresses very low levels of Ail with Asp67Gly and Leu68Gly substitutions (loop 2) | This work |
| pTM100-ail3 | GATCTT at bp 2465-2470 substituted for GCACGA in pTM100-ail; expresses very low levels of Ail with Asp67Ala and Leu68Arg substitutions (loop 2) | This work |
| pTM100-ail4 | TCA at bp 2564-2566 substituted for GAT in pTM100-ail; expresses Ail with Ser100 substituted for Asp (loop 3) | This work |
In all yadA mutants, nucleotide positions refer to the numbering of the 2,551-bp sequence containing the yadA gene of Y. enterocolitica O:3 under GenBank accession no. X13882.
In all ail mutants, nucleotide positions refer to the numbering of the 3,865-bp sequence containing the ail gene of Y. enterocolitica O:3 under GenBank accession no. AJ605740.
Cloning of ail.
The ail gene was PCR amplified using genomic DNA from YeO3-c as a template and primers ail-9 and ail-12 (Table 2). A PCR product of 1,711 bp was gel purified, ligated into EcoRV-digested pTM100, and transformed into Escherichia coli JM109 cells. The transformant carrying pTM100 with the ail gene cloned in the correct orientation was named pTM100-ail. The expression of Ail by JM109/pTM100-ail was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously (33).
TABLE 2.
Oligonucleotide primers used in inverse PCR with template plasmids pYMS4505 and pTM100-ail to mutagenize the yadA and ail genes, respectivelya
| Primer | Sequence (5′-3′)b | 5′-3′ positions | Purpose |
|---|---|---|---|
| yadA-1193r | AAGGCTTTCATGACCAATGG | 1193-1974 | Deletion of aa 193-218 of YadA |
| yadA-1272f | GAAAAAACACAGGAAAATACAAATAAAAG | 1272-1300 | |
| yadA-1310r | CTCAGCTGATCTTTTATTTGTATTTTCCTG | 1310-1281 | Deletion of aa 231-246 of YadA |
| yadA-1356f | GTGCTAGGGATCGCAAATAACTATAC | 1356-1381 | |
| yadA-1355r | GCTAGAAGACTTGTTGTCTGCATACG | 1355-1330 | Deletion of aa 247-262 of YadA |
| yadA-1404f | TTGGAAAATGCGCGTAAAGAGGC | 1404-1426 | |
| yadA-1403r | TGTTTCAGCACTTTTACTATCAGT | 1403-1380 | Deletion of aa 263-277 of YadA |
| yadA-1449f | TTGAATATGGCAAAAGCACACTC | 1449-1471 | |
| yadA-1448r | AACATCTTTAGACTGAGCAAAAGC | 1448-1425 | Deletion of aa 278-292 or 293-307 of YadA |
| yadA-1494f | TTAGAAACTGCTGAAGAACATGC | 1494-1516 | |
| yadA-1493r | AGTTGTTCTAGCAACACTATTTGAG | 1493-1469 | |
| yadA-1538r | AGTTGTTCTGGCAACACTATTTGC | 1538-1515 | Deletion of aa 308-322 of YadA |
| yadA-1584f | TTAGCAAGCGCTAATGTGTATGC | 1584-1606 | |
| yadA-1583r | CGCCTCAGCTGATTTTTTATTTGC | 1583-1560 | Deletion of aa 323-363 of YadA |
| yadA-1707f | ACAGATCATAAATTCCGTCAACTTGAC | 1707-1733 | |
| yadA-1256r | TTGCGCGACATTCACTG | 1256-1240 | Deletion of aa 214-228 of YadA |
| yadA-1302f | TCAGCTGAGCTGTTAGCA | 1302-1319 | |
| yadA-1346r | CTTGTTGTCTGCATACGCAT | 1346-1327 | Deletion of aa 244-258 of YadA |
| yadA-1392f | AGTGCTGAAACATTGGAAAAT | 1392-1412 | |
| yadA-1358r | CACGCTAGAAGACTTGTTGTCTGC | 1358-1335 | Deletion of aa 248-262 of YadA |
| yadA-1404f | TTGGAAAATGCGCGTAAAGAGGC | 1404-1426 | |
| yadA-1370r | TGCGATCCCTAGCACGCT | 1370-1353 | Deletion of aa 252-266 of YadA |
| yadA-1416f | CGTAAAGAGGCTTTTGCTCAGTCTAAA | 1416-1442 | |
| yadA-1382r | AGTATAGTTATTTGCGATCCCTA | 1382-1360 | Deletion of aa 256-270 of YadA |
| yadA-1428f | TTTGCTCAGTCTAAAGATGTT | 1428-1448 | |
| yadA-1391r | TTTACTATCAGTATAGTTATTTGCGAT | 1391-1365 | Deletion of aa 259-273 of YadA |
| yadA-1437f | TCTAAAGATGTTTTGAATATGGCA | 1437-1460 | |
| yadA-1415r | CGCATTTTCCAATGTTTCAGCA | 1415-1394 | Deletion of aa 267-281 of YadA |
| yadA-1461f | AAAGCACACTCAAATAGTGTTGC | 1461-1483 | |
| yadA-1427r | AGCCTCTTTACGCGCAT | 1427-1411 | Deletion of aa 271-285 of YadA |
| yadA-1473f | AATAGTGTTGCTAGAACAACTTTAGAAAC | 1473-1501 | |
| yadA-1481r | AACACTATTTGAGTGTGCTTTTGC | 1481-1458 | Deletion of aa 289-318 of YadA |
| yadA-1572f | TCAGCTGAGGCGTTAGC | 1572-1588 | |
| yadA-1505r | AGCAGTTTCTAAAGTTGTTCTAGCA | 1505-1481 | Deletion of aa 297-326 of YadA |
| yadA-1596f | AATGTGTATGCAGACAGCAAGT | 1596-1617 | |
| yadA-1517r | TGCATGTTCTTCAGCAGTTTCTAA | 1517-1494 | Deletion of aa 301-330 of YadA |
| yadA-1608f | GACAGCAAGTCTTCTCACACAC | 1608-1629 | |
| yadA-1571r | TTTTTTATTTGCATGTTCTTCAGCAGT | 1571-1545 | Deletion of aa 319-333 of YadA |
| yadA-1617f | TCTTCTCACACACTAAAAACTGCA | 1617-1640 | |
| yadA-1616r | CTTGCTGTCTGCATACACATTAG | 1616-1594 | Deletion of aa 334-348 of YadA |
| yadA-1662f | GTAAGTAATTCGACTAAGAAAGCAATCC | 1662-1689 | |
| yadA-1661r | AGTCACATCGGTATAGCTATTTGCA | 1661-1637 | Deletion of aa 349-363 of YadA |
| yadA-1707f | ACAGATCATAAATTCCGTCAACTTGAC | 1707-1733 | |
| yadA-1706r | GTATTGATTCGATTCACGGATTGCTTTC | 1706-1679 | Deletion of aa 364-370 of YadA |
| yadA-1728f | CTTGACAACCGGTTAGATAAACTTGAC | 1728-1754 | |
| yadA-1727r | TTGACGGAATTTATGATCTGTGTATTGA | 1727-1700 | Deletion of aa 371-377 of YadA |
| yadA-1749f | CTTGACACACGAGTTGACAAAG | 1749-1770 | |
| yadA-1748r | TTTATCTAACCGGTTGTCAAGTTGA | 1748-1724 | Deletion of aa 378-384 of YadA |
| yadA-1770f | GGTTTAGCCAGTTCAGCC | 1770-1787 | |
| yadA-1280r | TGTTTTTTCAATTTCTTTCTTTAATTGC | 1280-1253 | Deletion of aa 222-236 of YadA |
| yadA-1326f | AATGCGTATGCAGACAAC | 1326-1343 | |
| yadA-1706r | GTATTGATTCGATTCACGGATTGCTTTC | 1706-1679 | Deletion of aa 364-385 of YadA |
| yadA-1773f | TTAGCCAGTTCAGCCGCTTTAAAC | 1773-1796 | |
| ail-9 | AAGCGCTTGAGGTCAAAGCACAA | 1490-1512 | Cloning of ail gene |
| ail-12 | TTTTCCGGCAGCGGCCCGG | 3201-3183 | |
| H65Ar | ACCAAACTTATTACTGCCATAGAAGAAATCGTAT | 2458-2424 | His65Ala substitution in loop 2 of Ail |
| H65Af | GCAGGTGATCTTGATTACTATTCAGTAACAATGGGGC | 2459-2494 | |
| D67G L68Gr | ACCGTGACCAAACTTATTACTGCCATAGAAG | 2464-2433 | Asp67Gly, Leu68Gly, Asp67Ala, and Leu68Arg substitutions in loop 2 of Ail |
| D67G L68Gf | GGAGGTGATTACTATTCAGTAACAATGGGGCCATCTTTC | 2465-2503 | |
| D67A L68Rf | GCACGAGATTACTATTCAGTAACAATGGGGCCATCTTTC | 2465-2503 | |
| S100r | CTTAACCTTTCCGTGAGCAGCAC | 2563-2540 | Ser100Asp substitution in loop 3 of Ail |
| S100Df | GATTCTGTATTTGATGGGTCAGTCAGTACAAGTAAG | 2564-2599 |
Mutagenesis of yadA and ail.
Site-directed mutagenesis of the yadA and ail genes was carried out by inverse PCR with pYMS4505 and pTM100-ail as PCR templates, respectively (7). The primers used are listed in Table 2. PCR fragments were DpnI digested, phosphorylated, ligated, and electroporated into E. coli DH10B (yadA mutagenesis) and JM109 (ail mutagenesis) cells. The recovered plasmids were analyzed by PCR and sequencing to confirm the desired mutations. Triparental conjugation with the helper strain E. coli HB101/pRK2013 was used to mobilize the plasmids carrying (i) the mutated yadA of Y. enterocolitica strains YeO3-c and YeO3-c-Ail and (ii) ail genes of Y. enterocolitica strain YeO3-c-Ail-OCR, as described previously (4).
Antibodies and antisera.
The antibodies used were horseradish peroxidase (HRP)-conjugated rabbit anti-mouse immunoglobulin G (IgG) (catalog no. P260; Dako), HRP-conjugated swine anti-rabbit IgG (catalog no. P217; Dako), HRP-conjugated rabbit anti-goat IgG (catalog no. P449; Dako), mouse anti-collagen type I (catalog no. C2456; Sigma), rabbit anti-human C3c (catalog no. A0062; Dako), rabbit anti-human C3d (catalog no. A0063; Dako), and goat antiserum against human FH (catalog no. A312; Quidel). The latter antiserum cross-reacted with Y. enterocolitica O:3 proteins, including YadA. The anti-YadA antibodies were efficiently removed by 1 h of adsorption on ice with YadA-expressing bacteria, YeO3 and YeO3-R2, as shown elsewhere previously (3). The preadsorbed antiserum was used for the FH detection in all the experiments performed unless otherwise indicated. The YadA-specific monoclonal antibodies (MAbs) 2A9, 2G12, and 3G12 raised against Tx-114-isolated YadA were described previously (49).
SDS-PAGE, immunoblotting, dot blotting, and affinity blotting.
Bacteria were grown overnight at 37°C in 5 ml of MedECa. Whole-cell lysates were prepared from 5 ml of bacterial suspension with the optical density at 600 nm adjusted to 0.5. The pelleted bacteria were resuspended into 50 μl of phosphate-buffered saline (PBS), 200 μl of Laemmli buffer was then added, and the samples were heated at 95°C for 5 to 10 min. The samples (10 μl) were subjected to electrophoresis in 8 or 10% polyacrylamide gels and transferred onto nitrocellulose membranes, or 1 μl was spotted directly onto the membrane for dot blotting. After blocking with 5% skim milk-PBS, the membranes were incubated overnight at +4°C with appropriate primary antibody diluted in blocking solution. The YadA protein was visualized using Coomassie staining or detected with MAbs 3G12-15, 2A9, and 2G12 or with 1:2,000-diluted nonadsorbed polyclonal goat anti-FH antiserum. After washes with PBS, primary antibodies were detected with HRP-conjugated rabbit anti-mouse (1:2,000) or rabbit anti-goat (1:2,000) Ig. Peroxidase activity was detected using an electrochemiluminescence system. Quantitative analysis of YadA expression by the mutants was performed using Coomassie-stained gels and NIH ImageJ software. The loading differences between the lanes were corrected using the intensities of a distinct nonvariable bacterial protein band.
The same samples (10 μl) were used for collagen affinity blotting as described previously (10). Membranes were incubated with 20 μg/ml of type I collagen (catalog no. C7774; Sigma) overnight at +4°C, and bound collagen was detected with anti-collagen type I MAb.
Serum bactericidal assay.
Normal human serum (NHS) was obtained on two occasions from the same healthy human donors who were devoid of anti-Yersinia antibodies, and the sera were pooled (pools 1 and 2). The ability of Y. enterocolitica O:3 strains to survive in the pooled 66.7% NHS-EGTA-Mg serum or in heat-inactivated (56°C for 30 min) serum (HIS) was performed as described previously (4). The survival percentage was calculated by taking the bacterial counts obtained with bacteria incubated in HIS as 100%. The killing experiment was repeated for each strain at least three times, each time in duplicate, starting from independent cultures. For the calculation of statistical significances between the deletion mutants and wild-type YadA, the percentages of survival values for each experiment were used. To allow comparison between separate experiments, the results of the tested strains were adjusted to the wild-type survival percentage, which was set to 100.
Binding of serum FH to bacteria and cofactor assay for C3b inactivation.
The FH binding experiments by immunoblotting and enzyme-linked immunosorbent assay (ELISA) as well as the cofactor assay performed to analyze FI-mediated cleavage of C3b to iC3b were performed as described in detail elsewhere previously (3). The bacterial pellets from each cofactor assay sample were additionally subjected to 8% SDS-PAGE and immunoblotting using preadsorbed goat antiserum against human FH for detection (1:2,000).
Binding of truncated recombinant FH constructs to bacteria.
Bacteria were grown to logarithmic phase and washed three times with Veronal-buffered saline (VBS). The bacteria (109) were incubated with shaking (850 rpm) at 37°C for 30 min in 200 μl of VBS alone or containing SCRs 1 to 5, 1 to 6, 1 to 7, 8 to 11, 11 to 15, or 8 to 20 in equimolar (0.3 μM) concentrations. Following incubation, bacteria were washed three times with VBS, and bacterial pellets were suspended in 30 μl of VBS and mixed with Laemmli buffer. As all the recombinant FH constructs are recognized by the goat anti-FH antiserum (3), the samples were subjected to 12% SDS-PAGE and immunoblotting using goat antiserum against human FH for the detection of the truncated FH constructs.
Modeling of the stalk coiled-coil structure.
The stalk models for the YadA deletion mutants were generated with a modified version of BeammotifcCC (37), in which the supercoil radius can be specified individually for each residue (22). The basic periodicity used to model the entire stalk (residues 214 to 396) was 15-15-15-15-15-15-15-15-15-19-7-7-7-7; from this, we subtracted the deletion to be modeled and adjusted the local periodicity. The adjustments were based on the observation that insertions and deletions can be accommodated into coiled coils without disrupting the helices (25) as long as the local sequence can be resolved into combinations of 3- and 4-residue segments, each of which starts with a hydrophobic residue or, when multiple segments of 4 succeed each other, with a small residue (Ala, Ser, Thr, and Val). In all 28 deletion mutants that could be expressed, the effect of the deletions could be resolved in this way. We were particularly intrigued by deletion at residues 323 to 363 (Δ323-363), which we found could be modeled best by assuming that a sequence element of 23 residues (4-3-4-4-4-4) would form at the deletion site. While this may seem unusual, stalks that include this periodicity are found in other trimeric autotransporter adhesins, and, in the present case, all the resulting segments of 4 residues started with Ala, Ser, or Thr, while the single segment of 3 started with Leu. Thus, even though some deletions seemed incompatible with coiled-coil periodicity at first glance, they actually yielded models with favorable core interactions. Coordinates for the models can be obtained upon request.
Statistical methods.
Statistical analyses were performed using the two-sample t test; a P value of <0.05 was considered to be statistically significant.
RESULTS
Deletion mapping of YadA functions and MAb epitopes.
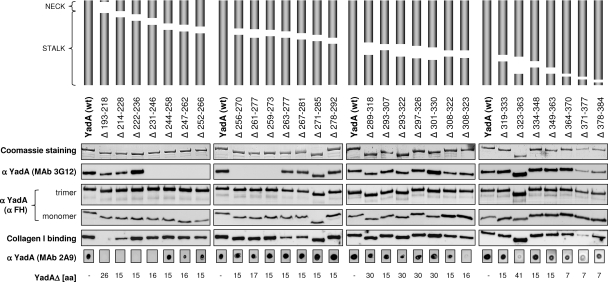
Since YadA appeared to be the major FH binding determinant of Y. enterocolitica O:3 (3), we aimed to identify YadA regions involved in CR and FH binding. To this end, we engineered 30 overlapping in-frame deletions to the yadA gene of plasmid pYMS4505. The deletions mapped within the neck and stalk regions of YadA (Fig. 1). The head domain was not mutated since it was previously reported not to play a role in CR (43). Two of the constructed mutants did not express YadA. One of them carried a deletion of 22 residues overlapping the junction between the right-handed and left-handed stalks (Δ364-385), while the other (pYMS4505:C) had an out-of-frame deletion within the stalk-coding region. The latter served as a control in all the experiments. The other 28 mutants expressed trimeric YadA as shown by Coomassie-stained SDS-PAGE and by immunoblottings and affinity blottings (Fig. 1). Quantitative analyses revealed no significant differences between the YadA expression levels of these mutants (data not shown).
FIG. 1.
Characteristics of YadA deletion mutants. A schematic representation of the in-frame deletions generated within the YadA neck and stalk is shown at the top. Whole-cell lysates of the strains were subjected to SDS-PAGE, and the gels were either stained with Coomassie blue (top row) or processed for immunoblotting or affinity blotting (four middle rows). Parts of the gels or immunoblots with the trimeric YadA bands and, in the case of nonadsorbed polyclonal anti-FH antiserum (that cross-reacts with YadA) (see Materials and Methods), also the monomeric band are shown. An aliquot of the lysates was analyzed for MAb 2A9 reactivity by dot blotting (bottom row). Affinity blotting was used to test the ability of YadA deletion mutants to bind type I collagen. The bound collagen was detected using type I collagen-specific MAb. wt, wild type.
To elucidate the impact of the generated deletions on the stalk structure, we modeled the stalks of the 28 YadA deletion mutants (for details, see Materials and Methods). According to the modeling, most deletions would not alter the structure of the rest of the stalk, while a few would change the local periodicity and thus the sense and degree of supercoiling (Fig. 2).
Several structure-related observations could be made based on the analyses of these YadA mutants. Firstly, the epitopes of MAbs 3G12-15, 2A9, and 2G12 could be mapped to amino acids 236 to 263, 193 to 244, and 193 to 214 of YadA, respectively (Fig. 1 and data not shown). All the epitopes were distinct, in close proximity to each other at the N-terminal end of the YadA stalk. Interestingly, the reduced signal for MAb 2A9, observed with the C-terminal deletions Δ308-323, Δ323-363, and Δ371-377 (Fig. 1), suggested a possible long-distance effect on the conformation of the N-terminal epitope. Secondly, all the oligomerized YadA forms as well as YadA monomers were detected by the cross-reacting YadA-specific antibodies present in the unadsorbed polyclonal anti-FH antiserum (see Materials and Methods) (Fig. 1). Thirdly, all the mutants were tested for the ability to bind collagen type I to demonstrate that the introduced mutations did not affect the integrity of the head domain. All but the neck deletion mutant (Δ193-218) were able to bind collagen as expected (Fig. 1), confirming that the constructed mutants had an intact head domain. The neck deletion covers the neck region and a few N-terminal amino acids of the stalk; this result corroborates the previously reported observation that the neck region is also necessary for the collagen binding ability of YadA (43).
Serum resistance profiles of the YadA deletion mutants.
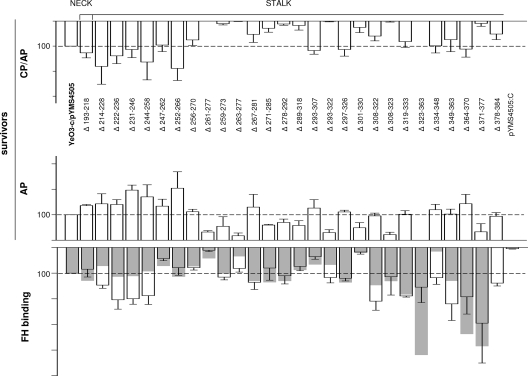
In order to map the YadA regions involved in CR, the YadA deletion mutants were all expressed in a virulence plasmid-cured strain, YeO3-c, and tested for the ability to survive under conditions of normal and EGTA-Mg-treated serum and classical pathway (CP)/AP- and AP-mediated killing, respectively (Fig. 3). For a number of mutants, the deletions did not apparently affect the CR. However, deletions within the neck domain (Δ193-218) or within the N-terminal end of the stalk (Δ214-228, Δ222-236, Δ231-246, Δ244-258, Δ247-262, and Δ252-266) resulted in increased CR (Fig. 3). The differences in the survival in EGTA-Mg-treated serum between these strains and YeO3-c/pYMS4505 were statistically significant (P values of between 0.0001 and 0.037). The only exception was Δ247-262, for which the difference did not reach significance (P = 0.066). On the other hand, several deletions within the rest of the right-handed part of the stalk (Δ261-277, Δ259-273, Δ263-277, Δ271-285, Δ278-292, Δ289-318, Δ293-322, Δ301-330, Δ308-323, and Δ323-363) resulted in reduced CR. The differences in survival in EGTA-Mg-treated serum between these strains and YeO3-c/pYMS4505 were also statistically significant (P values of between 0.0001 and 0.042). Finally, the deletion of the middle heptad in the left-handed part of the stalk (Δ371-377), but not the flanking heptads (Δ364-370 and Δ378-384), strongly affected CR (P = 0.0014 for EGTA-Mg serum).
FIG. 3.
Deletion mapping of YadA for CR determinants. The resistance of the mutants to CP/AP- and AP-mediated killing is presented for the 2-h time point. For the same strains, the FH binding from HIS was determined by ELISA (for each strain, the transparent columns with standard deviation bars represent the average FH binding for three independent duplicate experiments) and by immunoblotting (superimposed gray columns) (quantitation of the FH band intensities was performed with the Typhoon 9400 Image Quant analyzer). Survival and FH binding values of the strain expressing wild-type YadA (YeO3-c/pYMS4505) were set to 100, and the survival percentage and FH binding of the mutants are expressed relative to those of the wild type. For the calculation of statistical significances between the deletion mutants and wild-type YadA, the actual survival percentages for each experiment were used. The average YeO3-c/pYMS4505 survival value for AP killing in serum pool 1 was 76.1 ± 22.1, and that in pool 2 was 103.7 ± 27.7; the value for the CP/AP killing in serum pool 1 was 49.4 ± 17.4, and that in pool 2 was 73.3 ± 7.2. In ELISA, the average optical density at 492 nm value for strain YeO3-c/pYMS4505 was 0.47 ± 0.19 (range, 0.27 to 0.92). The strain with a frameshift mutation in the yadA gene (YeO3-c/pYMS4505:C), unable to express YadA, was included as a negative control.
Serum resistance versus FH binding phenotype of the YadA deletion mutants.
To examine if the decrease in resistance to AP killing was due to reduced FH binding, the mutants were further analyzed by immunoblotting and ELISA for the ability to bind this AP regulator from HIS (Fig. 3). Of note, the heat treatment of serum does not denature FH, as even after 20 min of incubation at 95°C, FH displays cofactor activity for the FI-mediated cleavage of C3b (20). Not surprisingly, all the AP-resistant mutants were able to bind FH (Fig. 3).
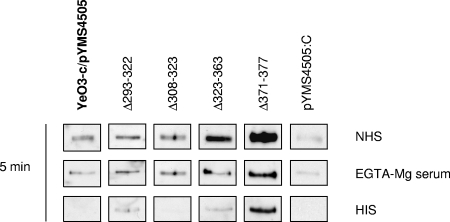
On the other hand, the FH binding ability had not been lost by most of the AP-sensitive mutants; instead, four of them displayed wild-type levels of FH binding (Δ293-322, Δ308-323, and Δ323-363) or even significantly higher levels (P = 0.013 forΔ371-377 by ELISA). Furthermore, when tested for the ability to bind FH from NHS or EGTA-Mg-treated serum during a short 5-min incubation, these four sensitive strains again displayed FH binding at wild-type levels or higher (Fig. 4). Interestingly, in this 5-min experiment, bacteria acquired much more FH from NHS or EGTA-Mg-treated serum than from HIS, indicating that the binding is enhanced by complement activation (Fig. 4).
FIG. 4.
Comparison of FH binding to bacteria expressing wild-type YadA to that of bacteria expressing mutated YadA during a 5-min incubation at 37°C in NHS, EGTA-Mg-treated serum, or HIS.
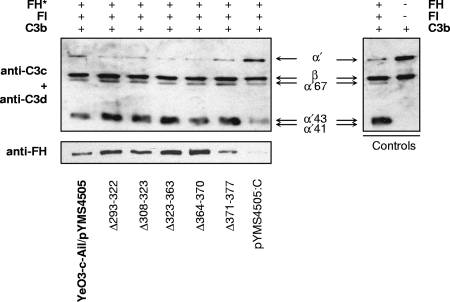
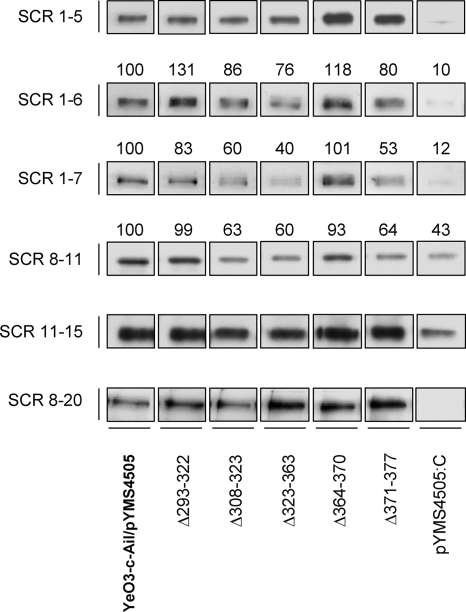
In order to understand the AP-sensitive phenotype of the YadA deletion mutants that bind serum FH, their ability to interact with purified FH or with truncated FH fragments was analyzed. All these YadA deletion mutants were introduced into YadA- and Ail-negative Y. enterocolitica strain YeO3-c-Ail to exclude the FH binding influence of Ail. Interestingly, the mutants bound purified FH at the wild-type YadA level or greater (Fig. 5). On the other hand, we could see some reduction in the binding of Δ308-323, Δ323-363, and Δ371-377 to FH recombinant fragments comprising SCRs 1 to 7 or 8 to 11 (Fig. 6). However, the importance of this reduction of binding remains elusive, as the purified FH bound to bacteria expressing these YadA mutants retained the cofactor activity for FI-mediated C3b cleavage (Fig. 5).
FIG. 5.
Cofactor activity of FH bound to bacteria expressing wild-type or mutated YadA. Bacteria were preincubated with FH (30-μg/ml final concentration) (left) and washed. Subsequently, the bacteria were exposed to factor I and C3b. The bacterial surface-bound FH is indicated by the asterisk, while in the control panel (right), reactions were carried out without bacteria. C3b and its cleavage products from the supernatants were detected using a mixture of rabbit anti-C3c and anti-C3d antisera. Inactivation of C3b can be seen by the reduced intensity of the C3b α′-chain and the appearance of α′-chain cleavage fragments of 67, 43, and 41 kDa. FH bound to the bacteria was detected from pellets by immunoblotting using preadsorbed goat anti-FH antiserum.
FIG. 6.
Binding of recombinant FH fragments to bacteria expressing wild-type or mutated YadA. Bacteria were incubated with truncated recombinant FH constructs representing SCRs 1 to 5, 1 to 6, 1 to 7, 8 to 11, 11 to 15, and 8 to 20. Following incubation, bacteria were washed, and whole-cell lysates were run on 12% SDS-PAGE gels and analyzed by immunoblotting using goat anti-FH and HRP-conjugated rabbit anti-goat antibodies for detection. The intensities of the bands for SCRs 1 to 6, 1 to 7, and 8 to 11 were measured. The intensity of the wild-type band was adjusted to 100, and those of the mutants are relative to that of the wild type. The relative intensity values are indicated above the bands.
The significant decrease in levels of FH binding by the YadA deletion mutants Δ247-262, Δ256-270, and Δ293-307 (P < 0.009 by ELISA) did not greatly affect CR. On the other hand, the decreased levels of FH binding of mutants Δ261-277 and Δ301-330 (P < 0.0027 by ELISA), although not as dramatic as that of the YadA-negative control pYMS4505:C, was reflected in decreased CR.
Collectively, these results showed that several deletions in the central part of the stalk and a deletion in the C-terminal part decreased CR, whereas deletions in the neck domain or in the N-terminal part of the stalk increased CR. Furthermore, the deletion mapping of YadA suggested that FH targets several regions of the YadA stalk and that the FH binding site on YadA is discontinuous and conformational. Surprisingly, FH binding by these YadA deletion derivatives did not always correlate with CR, suggesting that CR is a complex phenomenon. This is supported by our recent finding that YadA and Ail also bind C4BP, another complement regulator, from serum (21).
Serum resistance and FH binding phenotypes of the Ail mutants.
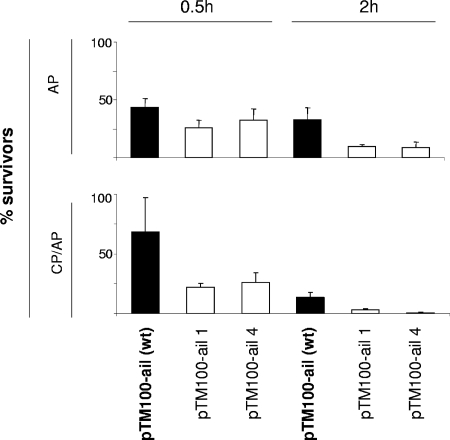
Similar to YadA, Ail was also shown to be a receptor for FH but solely in the absence of blocking O antigen and outer core (3). As lipopolysaccharide composition is known to be regulated by environmental cues, it is possible that Ail could be unveiled at some stages of infection. Therefore, to identify Ail regions responsible for FH binding, four point mutations were introduced into ail. The choice of the constructed point mutations was based on the mapping of the CR-associated residues of the Ail of Y. enterocolitica serotype O:8 (32). We engineered single-amino-acid substitutions in loop 2 (His65Ala expressed by pTM100-ail1) or in loop 3 (Ser100Asp expressed by pTM100-ail4) and substituted two amino acids critical for CR in loop 2 (Asp67Gly and Leu68Gly expressed by pTM100-ail2 and Asp67Ala and Leu68Arg expressed by pTM100-ail3). Unfortunately, the latter double-substitution ail mutants expressed very low levels of the mutated protein and were not studied further. Plasmid pTM100-ail, carrying wild-type ail, as well as plasmids pTM100-ail1 and pTM100-ail4, carrying mutated ail, were transferred into YeO3-c-Ail-OCR. YeO3-c-Ail-OCR lacks YadA, Ail, and, importantly, also the O antigen and outer core, which are known to block FH binding to Ail (3). YeO3-c-Ail-OCR expressing wild-type or mutated Ail from the plasmids were examined for resistance to CP/AP- and AP-mediated killing. YeO3-c-Ail-OCR carrying the vector alone was used as a negative control and was completely killed already at the early 0.5-h time point. Both mutants were more sensitive to NHS and EGTA-Mg-treated sera than the wild-type Ail-expressing strain (Fig. 7). The differences in CR between the mutants and the control strain expressing wild-type Ail were statistically significant (P < 0.0031), with the exception of YeO3-c-Ail-OCR/pTM100-ail4, for which the difference did not reach significance at 0.5 h of incubation in EGTA-Mg serum (P = 0.13). However, despite decreased serum resistance, the mutants bound amounts of FH similar to those of the wild type (data not shown), indicating that the introduced mutations did not affect the FH binding ability of Ail.
FIG. 7.
Effect of loop 2 and loop 3 substitutions of Ail on CR. Wild-type (wt) and mutated Ail proteins were expressed in strain YeO3-c-Ail-OCR. The survival percentages of the Ail mutants to CP/AP- and AP-mediated killing are presented for the 0.5-h and 2-h time points. The columns with standard deviation bars show the averages of data for three independent duplicate experiments. pTM100-ail1, His65Ala substitution in loop 2; pTM100-ail4, Ser100Asp substitution in loop 3.
DISCUSSION
YadA and Ail mediate the binding of FH to Y. enterocolitica (3). In this study, we further characterized regions on YadA and Ail involved in the serum resistance and interaction with FH.
YadA is the main FH receptor on the surface of Y. enterocolitica, and YadA-bound FH retains its biological function as an FI cofactor for C3b cleavage (3). To identify YadA regions involved in serum resistance and FH binding, short deletions were introduced into the neck and stalk, taking into consideration the proposed coiled-coil structure of the trimeric stalk (22). The YadA head domain was previously shown not to confer resistance against complement-mediated killing and was not mutagenized for this reason (43). None of the in-frame deletions within the stalk interfered with the surface exposure of the mutated YadA, except for a deletion of 22 residues overlapping the junction between the right-handed and left-handed parts of the stalk. YadA was not detected in this mutant, consistent with the work by Roggenkamp et al. demonstrating that this part of the stalk is indispensable for YadA translocation across the outer membrane (43). Additionally, all the stalk mutants bound collagen type I, indicating that the deletions did not affect the head and neck domains involved in collagen binding.
Functional analysis employing the YadA deletion mutants revealed that deletions in the N-terminal part of the stalk resulted in increased FH binding and CR. The reason for this is not clear. It is possible that in these deletions, FH gained better access to its binding site; however, since our stalk deletions were designed such that they left the collagen-binding head domain of YadA intact, the removal of the head was not the reason. A more likely possibility remains that the N-terminal part of the stalk contains the target site(s) for other serum proteins that could slow down or compete with FH binding. The deletion of these sites would then result in the facilitated access of FH to its binding site(s) on the YadA stalk. On the other hand, some deletions within the middle and C-terminal 15- and 7-mer regions of the stalk reduced the CR. In general, all serum-resistant YadA deletion mutants bound FH. The binding of FH to YadA seemed to depend on the recognition of several complementing and discontinuous structural YadA motifs rather than on one segment of the stalk, and the interaction seemed to be sensitive to both the conformation and the spacing of these motifs. This reasoning is based on the following observations.
Firstly, although most AP-sensitive mutants had not lost the FH binding activity, decreased FH binding was associated with decreased CR for some mutants. Since Δ261-277 disturbs both the pitch and the angle of the supercoil in the stalk, one could suggest that the reduction the FH binding of this mutant was due to a change in the protein conformation. Deletions Δ263-277 and Δ301-330, however, being in a pentadecad register, do not distort the supercoil. We can only speculate that the reduced ability to bind FH by these mutants is due to a change of a distance between the putative FH binding subsites on YadA. In addition, these three deletions were partially included in some other mutants (Fig. 1) whose ability to bind FH was not affected. Thus, the reduced FH binding to these three mutants was most probably not due to a deletion of the actual FH binding subsites of YadA.
Secondly, deletions Δ308-323, Δ323-363, and Δ371-377, which reduced CR but not FH binding, affected the MAb 2A9 epitope (see above), showing that the deletions introduced close to the C terminus could affect the N-terminal conformation of the YadA stalk. Even though FH bound strongly to these mutants, it was not protective. The cofactor assay, however, showed that FH bound to these mutants retained its cofactor activity (Fig. 5). Moreover, these mutants, similarly to wild-type YadA, bound FH fragments representing the SCRs of the entire polypeptide chain of FH (Fig. 6). We are forced to speculate that although FH bound to these mutants displays the cofactor activity, its ability to interfere with the formation of the alternative pathway convertases and/or to accelerate their decay must be affected. In addition, these mutants could be more susceptible to complement-mediated killing due to the defect(s) in the binding of other serum factors crucial for CR, e.g., C4bp. C4bp, the classical and lectin pathway regulator, has recently been shown to bind to YadA (21). The reduced binding of this regulator to bacteria could have an impact on CR, especially since C4bp is also able to stimulate the FI-mediated cleavage of C3b (6). Alternatively, the excessive binding of FH by the mutants could lead to the gradual depletion of FH from serum and, as a result, provoke an uncontrolled activation of the complement system.
Thirdly, although the YadA stalk has been shown to mediate serum resistance (43) and deletions generated in this study covered the whole stalk, none of these deletions, even those with broken periodicity (Fig. 2), totally abolished FH binding. This suggests that there is no single FH-binding site on the YadA stalk but instead that the binding is mediated by several conformational and discontinuous sites.
The notion of multiple FH binding sites on YadA may not be surprising given the very large size and extended nature of the FH protein. FH contains 20 short consensus repeats of different activities, and we have shown that SCRs throughout the length of FH are involved in the FH-Y. enterocolitica interaction (3). Thus, the abolishment of FH binding to one of the YadA sites involved in the interaction does not necessarily affect the other FH regions to find their complementary structural motifs elsewhere on YadA. We also note that the right-handed part of the YadA stalk is formed of 15-residue repeats and has a clear internal sequence symmetry so that a binding site could occur in similar forms several times along the stalk.
YadA deletion mutants proved to be useful for the epitope mapping of three MAbs specific for YadA (3G12-15, 2A9, and 2G12). Previous attempts to map the epitopes of these MAbs using overlapping 16-mer YadA peptides were not successful (10). The deletion mutants generated in this study allowed us to localize the epitopes within the N terminus of the stalk. This suggested that the epitopes are conformational, as several overlapping deletions destroyed the epitopes, and the MAb 2A9 epitope seemed to be affected by deletions several nanometers apart. This long-distance effect was shown by a reduced signal in the dot blotting of the C-terminal deletions Δ308-323, Δ323-363, and Δ371-377 (Fig. 1). In the case of Δ371-377, this could be due to problems of protein folding and/or trimerization, also reflected as a lower level of collagen binding (Fig. 1). Our modeling analyses suggested that deletions Δ308-323 and Δ323-363, however, would affect the periodicity of the stalk and thus the supercoil structure. Interestingly, MAb 2A9 binding was much less influenced by two other deletions, Δ247-262 and Δ261-277, which also perturb the supercoil and are located even closer to the actual MAb A9 epitope than Δ308-323 and Δ323-363. The reason for this observation is unclear to us at present.
The conformational and discontinuous nature of FH binding was reported previously for OspE and BBA68 of Borrelia burgdorferi and FhbA of Borrelia hermsii (19, 28-30). All three proteins were predicted to form coiled coils, and these structural motifs were shown to be involved in forming the binding site for FH. The distortion of one of the three widely spaced coiled coils of OspE resulted in the attenuation or complete abolishment of FH binding (29, 30), while the destabilization of the BBA68 coiled coils resulted in a reduced FH binding capacity (28). Interestingly, FhbA shares some similarity with YadA, as a deletion of the N-terminal coiled-coil segment of FhbA did not affect FH binding. The FhbA region responsible for the interaction with FH was mapped to the C-terminal coiled coils and the loop that they flank (19). Although the loop might be a contact point for FH, the proper presentation of this region by the flanking coiled coils seems to be critical for the interaction to occur (19). The binding of FH to streptococcal M protein was also shown to involve coiled-coil conformation (13); hence, several FH binding proteins appear to present the conformational and discontinuous epitope for FH.
Ail acts as a receptor for FH on Y. enterocolitica provided that it is well surface exposed. Moreover, FH bound to Ail showed cofactor activity for FI (3). An attempt to locate Ail regions involved in FH binding was undertaken in this work. Amino acids shown to be crucial for serotype O:8 Ail-mediated serum resistance (32) were substituted in loop 2 or loop 3 of serotype O:3 Ail, and the mutants showed decreased CR in both NHS and EGTA-Mg serum. Despite this, their FH binding was not affected. This suggests (i) that these Ail mutants would bind FH in a way that prevents the biological activity of FH, resulting in reduced serum resistance, or, more likely, (ii) that these Ail mutants fail to interact with another serum factor(s) important for CR of Y. enterocolitica. In support of the latter hypothesis, we have evidence that, similar to YadA, Ail is also able to bind C4bp (21). However, a more comprehensive set of Ail mutants is needed to elucidate the structural requirements of Ail-mediated FH and C4bp binding and the relative importance of these binding activities to Ail-mediated CR. On the other hand, the biological significance of FH binding by Ail, a factor promoting Yersinia entry into tissue culture cells in vitro (34), could also be related to bacterial adherence to the host cells as FH binds to glycosaminoglycans and sialic acids.
In conclusion, we have demonstrated that FH binding seems to involve multiple higher-order structural motifs on the YadA stalk. As a rule, YadA deletion mutants that bound FH survived well in serum. The FH binding-deficient deletion mutants identified in this study can be used to study the biological significance of this interaction in vivo. On the other hand, our results demonstrated that the two serum-sensitive Ail mutants were not affected in their abilities to bind FH. Thus, it appears that FH binding might not be the main mechanism of Ail-mediated CR.
Acknowledgments
This work has been supported by the Academy of Finland (projects 114075, 201358, and 104361 to M.S.) and the European Union Network of Excellence EuroPathoGenomics (contract LSHB-CT-2005-512061). M.B.-S. has been supported by the University of Helsinki Graduate School in Biotechnology and Molecular Biology.
We are grateful to Nathalie Friberg and Hanna Tossavainen for the purification of the truncated FH fragments and Heli Mönttinen and Laura Kalin for excellent technical assistance.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 2 September 2008.
REFERENCES
- 1.Areschoug, T., M. Stalhammar-Carlemalm, I. Karlsson, and G. Lindahl. 2002. Streptococcal beta protein has separate binding sites for human factor H and IgA-Fc. J. Biol. Chem. 27712642-12648. [DOI] [PubMed] [Google Scholar]
- 2.Balligand, G., Y. Laroche, and G. Cornelis. 1985. Genetic analysis of a virulence plasmid from a serogroup 9 Yersinia enterocolitica strain: role of outer membrane protein P1 in resistance to human serum and autoagglutination. Infect. Immun. 48782-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biedzka-Sarek, M., H. Jarva, H. Hyytiäinen, S. Meri, and M. Skurnik. 2008. Characterization of complement factor H binding to Yersinia enterocolitica serotype O:3. Infect. Immun. 764100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biedzka-Sarek, M., R. Venho, and M. Skurnik. 2005. Role of YadA, Ail, and lipopolysaccharide in serum resistance of Yersinia enterocolitica serotype O:3. Infect. Immun. 732232-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bliska, J. B., and S. Falkow. 1992. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc. Natl. Acad. Sci. USA 893561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blom, A. M., L. Kask, and B. Dahlbäck. 2003. CCP1-4 of the C4b-binding protein alpha-chain are required for factor I mediated cleavage of complement factor C3b. Mol. Immunol. 39547-556. [DOI] [PubMed] [Google Scholar]
- 7.Byrappa, S., D. K. Gavin, and K. C. Gupta. 1995. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 5404-407. [DOI] [PubMed] [Google Scholar]
- 8.Caswell, C. C., R. Han, K. M. Hovis, P. Ciborowski, D. R. Keene, R. T. Marconi, and S. Lukomski. 2008. The Scl1 protein of M6-type group A Streptococcus binds the human complement regulatory protein, factor H, and inhibits the alternative pathway of complement. Mol. Microbiol. 67584-596. [DOI] [PubMed] [Google Scholar]
- 9.China, B., M. P. Sory, B. T. N’Guyen, M. De Bruyere, and G. R. Cornelis. 1993. Role of the YadA protein in prevention of opsonization of Yersinia enterocolitica by C3b molecules. Infect. Immun. 613129-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Tahir, Y., P. Kuusela, and M. Skurnik. 2000. Functional mapping of the Yersinia enterocolitica adhesin YadA. Identification of eight NSVAIG-S motifs on the amino-terminal half of the protein involved in collagen binding. Mol. Microbiol. 37192-206. [DOI] [PubMed] [Google Scholar]
- 11.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291209-218. [DOI] [PubMed] [Google Scholar]
- 12.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 761648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischetti, V. A., R. D. Horstmann, and V. Pancholi. 1995. Location of the complement factor H binding site on streptococcal M6 protein. Infect. Immun. 63149-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gemski, P., J. R. Lazere, and T. Casey. 1980. Plasmid associated with pathogenicity and calcium dependency of Yersinia enterocolitica. Infect. Immun. 27682-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heesemann, J., C. Keller, R. Morawa, N. Schmidt, H. J. Siemens, and R. Laufs. 1983. Plasmids of human strains of Yersinia enterocolitica: molecular relatedness and possible importance for pathogenesis. J. Infect. Dis. 147107-115. [DOI] [PubMed] [Google Scholar]
- 16.Hellwage, J., T. Meri, T. Heikkilä, A. Alitalo, J. Panelius, P. Lahdenne, I. J. Seppälä, and S. Meri. 2001. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 2768427-8435. [DOI] [PubMed] [Google Scholar]
- 17.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 195989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horstmann, R. D., H. J. Sievertsen, J. Knobloch, and V. A. Fischetti. 1988. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc. Natl. Acad. Sci. USA 851657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hovis, K. M., J. P. Jones, T. Sadlon, G. Raval, D. L. Gordon, and R. T. Marconi. 2006. Molecular analyses of the interaction of Borrelia hermsii FhbA with the complement regulatory proteins factor H and factor H-like protein 1. Infect. Immun. 742007-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kask, L., B. O. Villoutreix, M. Steen, B. Ramesh, B. Dahlback, and A. M. Blom. 2004. Structural stability and heat-induced conformational change of two complement inhibitors: C4b-binding protein and factor H. Protein Sci. 131356-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirjavainen, V., H. Jarva, M. Biedzka-Sarek, A. M. Blom, M. Skurnik, and S. Meri. 2008. Yersinia enterocolitica serum resistance proteins YadA and Ail bind the complement regulator C4b-binding protein. PLoS Pathog. 4e1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koretke, K. K., P. Szczesny, M. Grüber, and A. N. Lupas. 2006. Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J. Struct. Biol. 155154-161. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn, S., C. Skerka, and P. F. Zipfel. 1995. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H1. J. Immunology 1555663-5670. [PubMed] [Google Scholar]
- 24.Lambert de Rouvroit, C., C. Sluiters, and G. R. Cornelis. 1992. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 6395-409. [PubMed] [Google Scholar]
- 25.Lupas, A. N., and M. Grüber. 2005. The structure of alpha-helical coiled coils. Adv. Protein Chem. 7037-78. [DOI] [PubMed] [Google Scholar]
- 26.Martinez, R. J. 1983. Plasmid-mediated and temperature-regulated surface properties of Yersinia enterocolitica. Infect. Immun. 41921-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez, R. J. 1989. Thermoregulation-dependent expression of Yersinia enterocolitica protein P1 imparts serum resistance to Escherichia coli K-12. J. Bacteriol. 1713732-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDowell, J. V., M. E. Harlin, E. A. Rogers, and R. T. Marconi. 2005. Putative coiled-coil structural elements of the BBA68 protein of Lyme disease spirochetes are required for formation of its factor H binding site. J. Bacteriol. 1871317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDowell, J. V., J. Wolfgang, L. Senty, C. M. Sundy, M. J. Noto, and R. T. Marconi. 2004. Demonstration of the involvement of outer surface protein E coiled coil structural domains and higher order structural elements in the binding of infection-induced antibody and the complement-regulatory protein, factor H. J. Immunol. 1737471-7480. [DOI] [PubMed] [Google Scholar]
- 30.Metts, M. S., J. V. McDowell, M. Theisen, P. R. Hansen, and R. T. Marconi. 2003. Analysis of the OspE determinants involved in binding of factor H and OspE-targeting antibodies elicited during Borrelia burgdorferi infection in mice. Infect. Immun. 713587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michiels, T., and G. R. Cornelis. 1991. Secretion of hybrid proteins by the Yersinia Yop export system. J. Bacteriol. 1731677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, V. L., K. B. Beer, G. Heusipp, B. M. Young, and M. R. Wachtel. 2001. Identification of regions of Ail required for the invasion and serum resistance phenotypes. Mol. Microbiol. 411053-1062. [DOI] [PubMed] [Google Scholar]
- 33.Miller, V. L., J. B. Bliska, and S. Falkow. 1990. Nucleotide sequence of the Yersinia enterocolitica ail gene and characterization of the Ail protein product. J. Bacteriol. 1721062-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, V. L., and S. Falkow. 1988. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect. Immun. 561242-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ngampasutadol, J., S. Ram, S. Gulati, S. Agarwal, C. Li, A. Visintin, B. Monks, G. Madico, and P. A. Rice. 2008. Human factor H interacts selectively with Neisseria gonorrhoeae and results in species-specific complement evasion. J. Immunol. 1803426-3435. [DOI] [PubMed] [Google Scholar]
- 36.Nummelin, H., M. C. Merckel, J. C. Leo, H. Lankinen, M. Skurnik, and A. Goldman. 2004. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel β-roll. EMBO J. 23701-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Offer, G., M. R. Hicks, and D. N. Woolfson. 2002. Generalized Crick equations for modeling noncanonical coiled coils. J. Struct. Biol. 13741-53. [DOI] [PubMed] [Google Scholar]
- 38.Pangburn, M. K., R. D. Schreiber, and H. J. Müller-Eberhard. 1977. Human complement C3b inactivator: isolation, characterization, and demonstration of an absolute requirement for the serum protein β1H for cleavage of C3b and C4b in solution. J. Exp. Med. 146257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry, R. D., and R. R. Brubaker. 1983. Vwa+ phenotype of Yersinia enterocolitica. Infect. Immun. 40166-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilz, D., T. Vocke, J. Heesemann, and V. Brade. 1992. Mechanism of YadA-mediated serum resistance of Yersinia enterocolitica serotype O3. Infect. Immun. 60189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Portnoy, D. A., and R. J. Martinez. 1985. Role of plasmid in the pathogenicity of Yersinia species. Curr. Top. Microbiol. Immunol. 11829-51. [DOI] [PubMed] [Google Scholar]
- 42.Ram, S., D. P. McQuillen, S. Gulati, C. Elkins, M. K. Pangburn, and P. A. Rice. 1998. Binding of complement factor H to loop 5 of porin protein 1A: a molecular mechanism of serum resistance of nonsialylated Neisseria gonorrhoeae. J. Exp. Med. 188671-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 1853735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roggenkamp, A., K. Rückdeschel, L. Leitritz, R. Schmitt, and J. Heesemann. 1996. Deletion of amino acids 29 to 81 in adhesion protein YadA of Yersinia enterocolitica serotype O:8 results in selective abrogation of adherence to neutrophils. Infect. Immun. 642506-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossmann, E., P. Kraiczy, P. Herzberger, C. Skerka, M. Kirschfink, M. M. Simon, P. F. Zipfel, and R. Wallich. 2007. Dual binding specificity of a Borrelia hermsii-associated complement regulator-acquiring surface protein for factor H and plasminogen discloses a putative virulence factor of relapsing fever spirochetes. J. Immunol. 1787292-7301. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt, C. Q., A. P. Herbert, H. G. Hocking, D. Uhrin, and P. N. Barlow. 2008. Translational mini-review series on complement factor H: structural and functional correlations for factor H. Clin. Exp. Immunol. 15114-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skurnik, M. 1984. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Appl. Bacteriol. 56355-363. [DOI] [PubMed] [Google Scholar]
- 48.Skurnik, M., I. Bölin, H. Heikkinen, S. Piha, and H. Wolf-Watz. 1984. Virulence plasmid-associated autoagglutination in Yersinia spp. J. Bacteriol. 1581033-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skurnik, M., Y. El Tahir, M. Saarinen, S. Jalkanen, and P. Toivanen. 1994. YadA mediates specific binding of enteropathogenic Yersinia enterocolitica to human intestinal submucosa. Infect. Immun. 621252-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skurnik, M., and P. Toivanen. 1992. LcrF is the temperature-regulated activator of the yadA gene of Yersinia enterocolitica and Yersinia pseudotuberculosis. J. Bacteriol. 1742047-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoorvogel, J., M. J. van Bussel, J. Tommassen, and J. A. van de Klundert. 1991. Molecular characterization of an Enterobacter cloacae outer membrane protein (OmpX). J. Bacteriol. 173156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tertti, R., E. Eerola, O.-P. Lehtonen, T. Ståhlberg, M. Viander, and A. Toivanen. 1987. Virulence-plasmid is associated with the inhibition of opsonization in Yersinia enterocolitica and Yersinia pseudotuberculosis. Clin. Exp. Immunol. 68266-274. [PMC free article] [PubMed] [Google Scholar]
- 53.Vesikari, T., T. Nurmi, M. Mäki, M. Skurnik, C. Sundqvist, K. Granfors, and P. Grönroos. 1981. Plasmids in Yersinia enterocolitica serotypes O:3 and O:9: correlation with epithelial cell adherence in vitro. Infect. Immun. 33870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogt, J., and G. E. Schulz. 1999. The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence. Structure 71301-1309. [DOI] [PubMed] [Google Scholar]
- 55.Whaley, K., and S. Ruddy. 1976. Modulation of the alternative complement pathways by beta 1 H globulin. J. Exp. Med. 1441147-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 57.Zink, D. L., J. C. Feeley, J. G. Wells, C. Vanderzant, J. C. Vickery, W. D. Roofs, and G. A. O'Donovan. 1980. Plasmid-mediated tissue invasiveness in Yersinia enterocolitica. Nature 283224-226. [DOI] [PubMed] [Google Scholar]