Abstract
Asymptomatic colonization of the nasopharynx by Streptococcus pneumoniae precedes pneumococcal disease, yet pneumococcal colonization factors remain poorly understood. Many bacterial infections involve biofilms which protect bacteria from host defenses and antibiotics. To gain insight into the genetics of biofilm formation by S. pneumoniae, we conducted an in vitro screen for biofilm-altered mutants with the serotype 4 clinical isolate TIGR4. In a first screen of 6,000 mariner transposon mutants, we repeatedly isolated biofilm-overproducing acapsular mutants, suggesting that the capsule was antagonistic to biofilm formation. Therefore, we screened 6,500 additional transposon mutants in an S. pneumoniae acapsular background. Following this approach, we isolated 69 insertions in 49 different genes. The collection of mutants includes genes encoding bona fide and putative choline binding proteins, adhesins, synthases of membrane and cell wall components, extracellular and cell wall proteases, efflux pumps, ABC and PTS transporters, and transcriptional regulators, as well as several conserved and novel hypothetical proteins. Interestingly, while four insertions mapped to rrgA, encoding a subunit of a recently described surface pilus, rrgB and rrgC (encoding the other two pilus subunits) mutants had no biofilm defects, implicating the RrgA adhesin but not the pilus structure per se in biofilm formation. To correlate our findings to the process of colonization, we transferred a set of 29 mutations into the wild-type encapsulated strain and then tested the fitness of the mutants in vivo. Strikingly, we found that 23 of these mutants were impaired for nasopharyngeal colonization, thus establishing a link between biofilm formation and colonization.
The life cycle of obligate commensal and pathogenic bacteria depends on efficient host colonization and transmission. It is increasingly being recognized that bacteria alternate between planktonic and sessile forms of growth, the latter in the form of surface-adherent biofilms—typically, complex microbial communities sometimes comprised of several species living in symbiotic relationships within a structured extracellular matrix of proteins, polysaccharides, and DNA. To form biofilms on a surface, bacteria rely on multiple genetic systems, including those involving attachment, intercellular interactions (e.g., quorum sensing), chemotaxis, carbon sensing, and stress responses (21, 23, 67, 87). Sessile bacteria differ strikingly physiologically and metabolically from their planktonic counterparts, and the establishment of bacterial biofilms on host tissues is thought to lead to expression of fitness determinants, protect against host defenses, and enhance resistance to antibiotics (29, 67). These observations and the fact that over half of all bacterial infections are believed to involve biofilms make the study of the role of biofilms in host-pathogen interactions an area of major scientific and clinical relevance (29, 69).
Streptococcus pneumoniae frequently colonizes the human oronasopharynx asymptomatically (9, 33, 84). This so-called carriage state not only allows for efficient transmission to new hosts but also precedes the onset of pneumococcal illnesses, such as pneumonia, septicemia, otitis media, and meningitis. Pneumococcal diseases constitute a major global health problem, being responsible for up to 1 million child deaths per year, 90% of which occur in developing countries (58). For these reasons, colonization represents the point of intervention most likely to be targeted by urgently needed protein-based non-serotype-specific vaccines (10); yet, the host-pathogen interactions underlying colonization remain poorly understood (9, 39, 44).
Bacteria use a variety of strategies to persist in their particular niche in the host (61). Among pathogens of the respiratory system, the ability to form biofilms seems critical for Pseudomonas aeruginosa and Staphylococcus aureus to cause chronic infections (32, 79, 91). Several oral streptococci, e.g., Streptococcus mutans, Streptococcus gordonii, and Streptococcus intermedius, persist in biofilms in the human oral cavity (22), where they are common colonizers of tooth surfaces and occasionally form cariogenic biofilms (15). Recently, Manetti and colleagues implicated biofilm formation by Streptococcus pyogenes in adherence to mucosal epithelial cells (55). Moreover, the ability to form biofilms has been proposed to play an important role in otitis media with effusion and recurrent otitis media, which commonly involve Haemophilus influenzae and S. pneumoniae (26, 28). Recently, it was demonstrated that S. pneumoniae can form biofilms in vitro (4, 26, 82) and that this mode of living is accompanied by the differential expression of about 30% of the S. pneumoniae proteome and the de novo synthesis of about 200 proteins (4). Moreover, Oggioni and colleagues (65) reported that the transcriptional profile of several known virulence-related genes in S. pneumoniae isolated from lungs and brains of infected mice is similar to that in biofilms formed in vitro, suggesting a possible biofilm-like state of S. pneumoniae in tissues. More recently, known surface molecules, including CbpA and PcpA, as well as murein hydrolases LytA, LytB, and LytC, were shown to contribute to S. pneumoniae biofilm formation on polystyrene surfaces (59).
After S. pneumoniae crosses the mucus layer of the oronasopharynx, it grows in intimate contact with the mucosal epithelium (7, 12, 56). We hypothesize that during colonization of this niche S. pneumoniae would benefit from growing as a biofilm and that an understanding of the genes involved in biofilm formation might give novel insights about colonization. In order to take a comprehensive approach to the identification of genes involved in biofilm formation by S. pneumoniae, we generated a large number of transposon insertion strains and screened them for biofilm formation in vitro. This led to the identification of 69 biofilm mutants. Mutations in genes encoding bona fide and putative adhesins and choline binding proteins, pili, synthases of membrane and cell wall components, extracellular and cell wall proteases, efflux pumps, ABC and PTS transporters, and transcriptional regulators, as well as several conserved proteins of unknown function, were isolated. When we systematically evaluated a subset of these mutants for their ability to colonize the mammalian host by using a mouse model of nasopharyngeal colonization, a striking correlation between biofilm defects in vitro and colonization in vivo was found.
MATERIALS AND METHODS
Bacterial growth conditions.
S. pneumoniae was routinely grown in Todd-Hewitt (BD) yeast extract (Fisher) (THY) broth supplemented with Oxyrase (5 μl/ml) by incubation at 37°C in a 5% CO2 incubator. The following antibiotic concentrations were used: chloramphenicol at 4 μg/ml, streptomycin (Sm) at 100 μg/ml, and spectinomycin (Spc) at 200 μg/ml. For biofilm growth, C+Y medium (59) and THY with or without 0.3% glucose were used. Growth on plates was done with tryptic soy agar (TSA) (B1676; Sigma)-5% sheep blood plates.
Bacterial strains, plasmids, and DNA manipulations.
Strains of serotypes 1, 2, 3, 4, 5, 6A, 8, 9V, 14, 15, 19F, and 23F were from our laboratory's strain collection. The transposon mutant library used for the first biofilm screen in vitro (encapsulated strain) consisted of a collection of circa 6,000 mariner transposon (Magellan 2, chloramphenicol-resistant) insertion mutants generated from strain AC353, described previously (37). Strain AC353 is an Sm-resistant encapsulated derivative of TIGR4, a serotype 4 clinical isolate (strain AC316). The second screen was carried out with an acapsular mutant from the AC353 strain background (gift of Ram Iyer). An additional Sm-resistant strain into which isolated transposon insertions were transferred was constructed in AC316 by incorporating a point mutation in residue 56 of the RpsL S12 protein (SP0271) (74). In vitro Magellan 5 transposon (also known as pR412 or pEMSPC) transposition reactions were carried out with purified MarC9 transposase essentially as described previously (37). DNA products obtained from transposition reactions were transformed into naturally competent acapsular S. pneumoniae (AC353), and transformants were selected in the presence of Spc after incubation in a CO2 incubator at 37°C overnight. All standard DNA manipulations were carried out according to established protocols.
Complementation of biofilm mutants.
All PCR amplifications were carried out using AC353 genomic DNA. For complementation studies, plasmid pLE1CATLE2 was constructed on the backbone of pAC1294, a shuttle vector that carries the Spc resistance cassette in place of the chloramphenicol (cat) cassette in PAC1000 (37). In pLE1CATLE2, LE1 and LE2 correspond to fragments of about 800 bp identical to regions upstream and downstream, respectively, of the lacE PTS operon (SP0474 to SP0478), which target the construct to this region of the chromosome (lacE is dispensable for normal growth of S. pneumoniae and has been used for mutant complementation in the past [43]). The CAT cassette (upstream promoter region and open reading frame [ORF] without a transcriptional terminator) is cloned downstream of LE1 and a multiple cloning site (XmaI, MluI, HindII, SpHI, and SacII) into which the gene of interest can be cloned for expression driven from the cat promoter. pLE1CATLE2 was constructed as follows. The lacE 3′ region (fragment LE2, 747 bp) was amplified using primers LE2F(H3SphISac2) and LE2R(XhoI). This amplicon was digested with HindIII and XhoI and cloned into pAC1294, which had previously been digested with the corresponding enzymes, thus resulting in pLE2AC1294. The lacE 5′ region (fragment LE1, 786 bp) was amplified with primers LE1F(AatII) and LE1R(cat); LE1R(cat) had a 10-bp overlap with the 5′ end of a cat cassette. A cat cassette fragment of 886 bp was amplified from pAC1000 using primers catF(LE1) and catR(XmaIMluIH3), where catF(LE1) had 10 bp of homology with the 3′ end of the LE1 fragment. The LE1 fragment and cat cassette fragments were joined using splicing by overlapping extension and primers LE1F(AatII) and catR(XmaIMluIH3), yielding a 1,660-bp product, which was digested with AatII and HindIII and cloned into pLE2AC1294, which had been digested with the same enzymes.
For complementation of 9F2 (SP0199), rrgA (60F5, SP0462), 3F2 (SP1537), and 22A (SP2192) mutants, the corresponding ORFs were amplified by PCR using primer pairs C9H2F/C9H2R, C60F5F/C60F5R, C3F2F/C3F2R, and C22A4F/C22AR, respectively. The amplicons were digested with XmaI/SphI and cloned into a similarly digested pLE1CATLE2. Each mutant was transformed with the corresponding complementation construct, and transformants were selected by plating on plates containing chloramphenicol. Four to eight clones were grown and tested for their ability to form biofilms compared with the abilities of the wild-type and mutant parental strains. Primers used are listed in Table S1 in the supplemental material.
For complementation of srtA, plasmid pLE1SPCLE2, in which the SPC cassette drives the expression of the downstream gene, was constructed as described above but using the backbone of pAC1000 (37), which carries a chloramphenical resistance cassette. The wild-type copy of srtA was amplified using primers CsrtAF1 and CsrtAR1.
In vitro screening for biofilm formation.
In each round of screening, 320 Spc-resistant clones obtained after mariner in vitro transposition and transformation were picked and inoculated individually into wells of 96-well polystyrene plates (Costar 3596; Corning, Inc., NY) containing 200 μl of THY, 0.3% glucose, Sm, and Oxyrase (Oxyrase, Inc.). After 4 h of growth, each plate was replica plated onto a square TSA-5% sheep blood agar plate. The acapsular wild-type parental strain was spotted on each plate as a control, and the plate was incubated for ∼18 h in a CO2 incubator at 37°C. Each plate was then replica plated (in triplicate) onto 96-well plates containing 200 μl of THY, 0.3% glucose, Sm, and Oxyrase per well, and these were incubated in a CO2 incubator at 37°C and evaluated for biofilm formation 14 to 16 h later.
For quantitation of CFU in biofilms, bacteria were grown in 12-well polystyrene plates (Costar 3513; Corning, Inc., NY) as described above for 96-well plates. At the desired time, the supernatants were removed by pipetting and the attached bacteria were washed five times by gentle squirting of sterile phosphate-buffered saline (PBS). One milliliter of sterile PBS was added per well, and the attached bacteria were lifted using a cell lifter (Costar 3008; Corning, Inc., NY) and then collected by pipetting, followed by vigorous vortexing for 30 s, serial dilution, and plating for CFU.
Direct sequencing of transposon/genomic junctions.
In order to identify the location of the mariner transposon insertions, highly concentrated genomic DNA was prepared from the mutants harboring the transposon insertions by following the DNA easy kit (Qiagen) protocol, with slight modifications. Bacterial cultures (5 ml) were grown to saturation (6 to 8 h), pelleted by centrifugation at 4,000 × g for 10 min, and resuspended in the kit's tissue lysis buffer. From this point on, the protocol was followed according to the manufacturer's recommendations except that the samples were bead beaten in a BeatBeater (BioSpec Products, Inc.) with 0.1-mm zirconia beads for 2 min prior to passage through the kit's columns. Genomic DNA at a concentration of ∼500 ng/μl was sent for sequencing along with primer Mag2F3 (5′-GGAATCATTTGAAGGTTGGTA-3′), which reads out from the mariner transposon (Magellan 2 or Magellan 5). Direct sequence reads of ∼500 to 800 bp were obtained by the Tufts University Core Sequencing Facility. The location of the transposon insertions as well as the direction relative to the closest ORF was determined by comparing the obtained sequences with the TIGR4 genome sequence using BLAST. The predicted protein sequence of each disrupted ORF was used to search the nonredundant NCBI protein database by PSI-BLAST for homology/function prediction and was analyzed by SMART BLAST (51).
Animal infections.
In vitro competitions were carried out for each mutant strain as follows. Mutant and wild-type parental strains were grown separately to early log phase and then diluted 200-fold into prewarmed THY, 0.3% glucose, Sm, and Oxyrase and grown to late log phase (∼5 h), serially diluted, and differentially plated on TSA-5% blood agar plates containing either Sm or Spc to determine the numbers of mutant and wild-type parental strains at the end of the competition. Competitive index (CI) values were calculated by dividing the ratio of mutant to wild-type bacteria (Spcr/Smr) recovered at 5 h by the input ratio. The in vitro CI values were used in statistical comparisons against the in vivo CI values obtained in the colonization competition experiments.
In all animal infections, 6- to 10-week-old female C57BL/6 mice (Taconic Labs) were used. Mice were housed according to Tufts University Department of Lab Animal Medicine guidelines and given access to food and water ad libitum. Mutants tested in competition experiments for colonization were first backcrossed into a wild-type Sm-resistant strain. For each competition, mutant and parental strains were grown overnight on TSA-5% blood agar plates, inoculated into THY broth, and allowed to reach mid-log phase (∼1 × 108 CFU/ml) before 1-ml aliquots were centrifuged (2,500 × g, 5 min), washed once in prewarmed PBS, pH 7.4 (Gibco), and resuspended in 100 μl of prewarmed PBS. Mutant and wild-type bacteria were mixed 1:1, and 10 μl of the mixture (∼1 × 107 CFU) was inoculated into the nares of mice that had been lightly anesthetized by isoflurane/oxygen inhalation (5 μl per nostril; five mice per mutant tested). Mice were sacrificed by CO2 asphyxiation 5 days later. Bacteria colonizing the nasopharynx were recovered by washing the nasopharynx through the trachea with 1 ml of sterile PBS and plating serial dilutions onto TSA-5% blood agar plates basically as described previously (37). The ratio of the numbers of mutant to wild-type bacteria after colonization from each animal (in vivo CI) was determined as for the in vitro CI.
Statistics.
Nonpaired Student t tests and nonparametrical Mann-Whitney tests were carried out using GraphPad Prism Graph (GraphPad Software, Inc.).
RESULTS
In vitro biofilm model used for the screen.
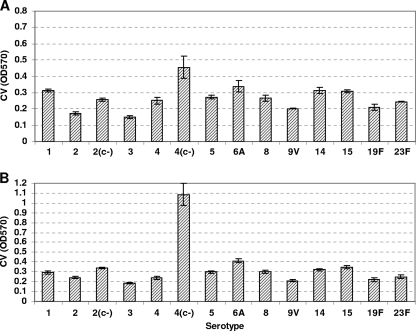
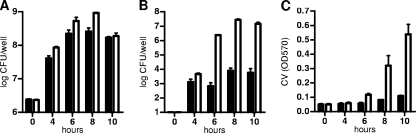
In order to set up the conditions to carry out a genetic screen for the identification of genes contributing to biofilm formation by S. pneumoniae, we began by assaying the ability of S. pneumoniae to form biofilms on microtiter polystyrene plates, as described originally for Pseudomonas fluorescens (68) and more recently for S. pneumoniae (59, 65). In preliminary experiments, we observed biofilm formation in either defined C+Y medium (59) or THY medium (4, 65) by using crystal violet (CV) staining from 8 to 16 h postinoculation of 5 × 105 CFU/ml in 96-well plates (not shown). We next evaluated an array of S. pneumoniae serotypes for biofilm formation (Fig. 1) and found that strains from different serotypes varied in their levels of biofilm formation; furthermore, we observed that acapsular strains made more biofilm than encapsulated strains (see serotypes 2 and 4 in Fig. 1), a difference that became accentuated when the medium was supplemented with 0.3% glucose, especially for serotype 4 strain TIGR4 (Fig. 1A versus B). We next looked at the kinetics of S. pneumoniae TIGR4 sessile growth in more detail by determining the number of CFU attached to the bottom of polystyrene wells over time and by staining with CV. In 12-well plates seeded with 5 × 105 CFU/ml, bacteria began to attach at around 4 h postinoculation, accumulation continued over the next few hours, and the maximum number of attached cells was seen at ∼8 to 10 h (Fig. 2B). This was in contrast to planktonic growth, where the number of bacteria reached a maximum by 6 h (Fig. 2A). CV staining began to be detected at 6 h, increased over the next 4 h, and then dropped slightly between 12 and 16 h (∼2-fold drop); when the assays were done with 96-well plates, the CV staining remained constant from 12 to 16 h. We have observed that the washing of the attached biofilm in the larger wells of the 12-well plates tends to be more disruptive than in the smaller wells of the 96-well plate. In contrast, the CFU did not change significantly between 8 and 10 h (Fig. 2C) but then dropped rapidly: 3.3 × 105 CFU/ml at 12 h, 333 CFU/ml at 14 h, and undetectable at 16 h (limit of detection, 20 CFU/ml). After 18 h, a sharp decline in the overall attached material was seen (not shown). At this point, we do not know whether the bacteria in the biofilm at 14 and 16 h are viable but unable to form CFU on a plate or are no longer viable, but similar in vitro studies by Moscoso and colleagues (59) indicate that, although prolonged incubation results in increased numbers of dead cells, a considerable proportion of the bacteria in the biofilm are still viable. As reported previously (59), we observed that the biofilm was severely impaired by treatment with proteinase K before or after biofilm formation (no effect on planktonic growth), although no effect of DNase I treatment was detected (not shown).
FIG. 1.
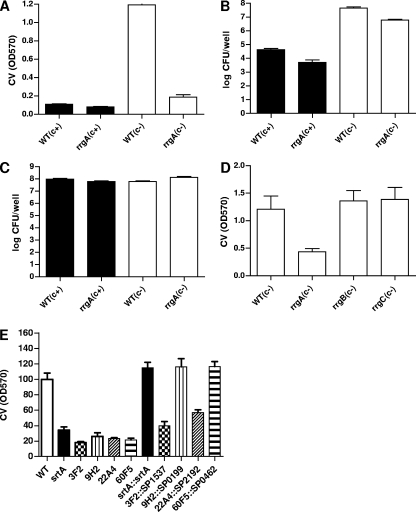
Comparison of biofilm formation by several S. pneumoniae serotypes on polystyrene plates. Laboratory and clinical isolates of the indicated serotypes were inoculated in 96-well microtiter plates and allowed to statically form a biofilm. Bacteria were inoculated in THY (A) or THY with 0.3% glucose (B) and incubated for 14 h, at which time supernatants containing planktonic cells were discarded and attached biofilms were washed three times with PBS, stained with 0.05% CV for 30 min, dissolved in 95% ethanol, and quantified by measuring the absorbance at 570 nm (OD570). c−, acapsular. Strains were inoculated in quadruplicate. Results are representative of three experiments. Error bars show standard deviations.
FIG. 2.
Dynamics of biofilm formation by S. pneumoniae TIGR4. Wild-type (filled bars) and acapsular mutant (open bars) bacteria were grown planktonically (A) and as biofilm (B). The corresponding CFU were determined at the indicated time points. (C) The total biofilm attached (adhered bacteria plus extracellular material) was estimated by staining with CV and measuring the absorbance at 570 nm (OD570). Bacteria were grown in 12-well polystyrene plates in THY plus 0.3% glucose in a 5% CO2 incubator at 37°C. Results are representative of three experiments. Error bars show standard deviations.
Isolation of biofilm mutants in an S. pneumoniae TIGR4 background.
Having established conditions that were amenable to a biofilm mutant screen in vitro, we set out to screen a collection of transposon insertion S. pneumoniae mutants previously described (37) for attachment to polystyrene in 96-well plates. Similar screens have been conducted successfully with P. fluorescens (68), Vibrio cholerae (83), S. gordonii (52), and Klebsiella pneumoniae (49). In an initial screen, 6,000 individual mutants that had been kept at −80°C in 96-well plates were replica plated onto fresh TSA-5% sheep blood agar plates and then inoculated in triplicate into 96-well plates containing THY plus 0.3% glucose and incubated in a 5% CO2 incubator at 37°C for 14 to 16 h. Wells were then washed three times with PBS and stained with CV for 30 min, and the attached material was solubilized with 95% ethanol for 2 h prior to determination of the absorbance at 570 nm. The results from triplicate wells were averaged, and mutants which were 30% different from the wild type were analyzed further.
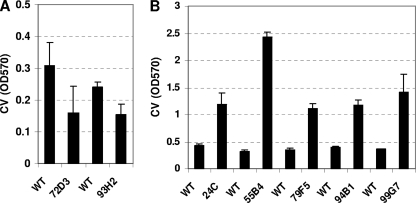
Commonly, transposon insertion sites from desired mutants are identified by restriction digest and subcloning of genomic DNA or by PCR using a transposon-specific randomized primer, followed by sequencing. Both methods are time-consuming and labor-intensive. Instead, we used a modified protocol for S. pneumoniae DNA preparation to obtain high concentrations of genomic DNA (see Materials and Methods) and determined the locations of the insertions by direct sequencing of the transposon-genome junctions in the isolated genomic DNA from each mutant using a primer that read out of the transposon. As shown in Fig. 3, seven mutants were isolated in this initial screen. Two of these mutants were poor biofilm formers and had independent insertions in lytC (SP1593) (Fig. 3A), a gene encoding one of the three murein hydrolases in S. pneumoniae (60) recently implicated together with lytA and lytB in biofilm formation by unencapsulated S. pneumoniae (59). The other five mutants were hyperbiofilm formers, and their transposon insertions were all mapped to different locations within cps4E (SP0350) (Fig. 3B), a gene in the capsule locus that encodes a glycosyl-phosphotransferase that transfers glucose-1-phosphate units to the growing undecaprenyl phosphate glycolipid (17); incidentally, cps4E is one of the few genes in the serotype 4 capsule locus that can be disrupted without compromising bacterial viability (37, 90), which might explain why it was the only gene hit in the 15-kb capsule locus in our screen. All of the cps4E mutants isolated were acapsular, as determined by the Quellung reaction using an antibody against the serotype 4 capsule polysaccharide (not shown).
FIG. 3.
Biofilm mutants isolated in a screen of transposon insertion mutants in an S. pneumoniae TIGR4 background. A collection of 6,000 mariner transposon insertion mutants was screened for biofilm formation. (A) Mutants 72D3 and 93H2 were poor biofilm formers and had insertions in lytC (SP1573). (B) Mutants 24C, 55B4, 79F5, 94B1, and 99G7 were all hyperbiofilm formers and had insertions mapping to cps4E (SP0350) of the capsule locus. The total biofilm attached was estimated by staining with CV and measuring the absorbance at 570 nm (OD570). Bacteria were grown in 96-well polystyrene plates in THY plus 0.3% glucose in a 5% CO2 incubator at 37°C. Results are representative of three experiments. Error bars show standard deviations. WT, wild type.
Influence of the capsular polysaccharide on biofilm formation.
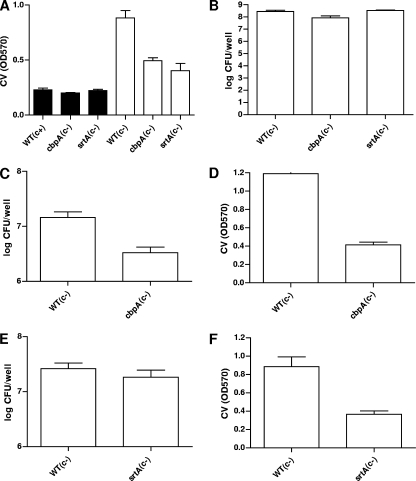
Initially, we had surmised that the polysaccharide capsule of S. pneumoniae would contribute to biofilm formation, as has been reported for the hyaluronic acid capsule of S. pyogenes (19). Although Oggioni et al. (65) reported no role for the capsule in biofilm formation, in agreement with Moscoso et al. (59) we found that the capsule reduced the level of biofilm formation in S. pneumoniae serotypes 2 and 4 (Fig. 1). This observation, together with the fact that screening of 6,000 mutants resulted in the isolation of insertions in only two genes (lytC and cps4E), an unexpectedly low number, and that mutations in cps4E resulted in increased biofilm formation, led us to hypothesize that the presence of the capsule and the consequent reduction in biofilm formation could have masked differences in biofilm phenotypes in our screen. To evaluate this possibility, we compared the in vitro biofilm-forming abilities of a cbpA mutant and a srtA mutant to that of the wild type in both encapsulated and acapsular backgrounds (Fig. 4). CbpA is a well-described choline binding surface protein known to contribute to S. pneumoniae adherence to epithelial cells and colonization in the infant rat model (73) that was recently shown to contribute to biofilm formation in S. pneumoniae (59). srtA encodes a housekeeping sortase which mediates the covalent surface attachment of up to 23 proteins containing an LPXTG anchoring motif in S. pneumoniae (81). S. pneumoniae srtA mutants are defective in attachment to epithelial cells and in the mouse and chinchilla models of colonization (18, 46, 71). Recently, an S. gordonii srtA mutant was reported to be defective in biofilm formation (64). Thus, we expected S. pneumoniae cbpA and srtA mutants to have biofilm defects in our polystyrene in vitro assay, making them good candidates to determine whether the capsule could mask biofilm phenotypes under our assay conditions. As can be seen in Fig. 4, cpbA and srtA mutants in the encapsulated background showed levels of biofilm formation comparable to that of the parental wild-type strain. However, when the same mutations were transferred to a capsule-deficient background, both mutations resulted in reduced biofilms (Fig. 4A, D, and F). Both cbpA and srtA mutants grew similarly to the wild type regardless of the presence (not shown) or absence (Fig. 4B) of capsule during planktonic conditions. These observations lend support to the notion that the capsule might have hindered our ability to identify biofilm mutants in our initial screen.
FIG. 4.
Biofilm formation by mutants in the presence or absence of capsule. Biofilm formation by isogenic cbpA and srtA mutants in the parental encapsulated (c+, filled bars) or acapsular (c−, open bars) strain background during sessile (A and C to F) or planktonic (B) growth. Bacteria were inoculated and incubated for 14 h (A) or 10 h (B to F), at which times supernatants were removed and attached biofilms were washed with PBS and quantified by staining with CV (A, D, and F) or by plating for CFU (C and E). Planktonic cells in the supernantants were enumerated by plating for CFU (B). Bacteria were grown in 12-well polystyrene plates in THY plus 0.3% glucose in a 5% CO2 incubator at 37°C. Results are representative of three experiments. Error bars show standard deviations. WT, wild type; OD570, absorbance at 570 nm.
Identification of biofilm mutants in an acapsular TIGR4 strain background.
We thus decided to carry out a second in vitro biofilm screen with an acapsular TIGR4 strain. To this end, a total of 6,500 mutants were generated by in vitro transposition using the mariner-derived minitransposon Magellan 5 (37), as described previously (3, 48). Following transformation of the Magellan 5 mutagenized genomic DNA into an acapsular S. pneumoniae TIGR4 strain, groups of transformants were tested for biofilm formation as described above. Mutants that had no apparent planktonic growth defects, as determined by optical density after 14 h of static growth in a 5% CO2 incubator at 37°C, but that differed from the wild type by at least 30% in biofilm formation by the CV assay were grown individually and stored for further study. A total of 80 mutants were isolated, and after retesting, 11 were discarded as false positives. The sequences of the transposon-genome junctions for the remaining 69 mutants were determined as described above and were used to find the exact location and direction of the transposon insertions by comparison against the TIGR4 genome sequence. Table 1 lists these 69 mutants, which have insertions in 42 different genes and eight promoters, along with the position of the mariner transposon insertion, the mean values of biofilm formation percentages relative to that of the wild type, and the functional information for each gene based on the TIGR4 genome annotation and BLAST analysis (PSI-BLAST) of each predicted protein.
TABLE 1.
Magellan 5 tranposon insertion biofilm mutants
| Mutanta | Geneb | Functional annotationb | Transposon positionc | % Biofilm formationd |
|---|---|---|---|---|
| 30B10 (2) | SP0013, or ftsH | Putative single-chain AAA protease | 50 | 34 ± 8 |
| 46E5 (2) | SP0060 | Putative glycosyl hydrolase | 1727 | 244 ± 18 |
| 63A5 | SP0137 | Putative ABC transporter, ATP binding | 1108 | 54 ± 6 |
| 12G2 | SP0140, or ugd | UDP-glucose 6-dehydrogenase, authentic frameshift | 648 | 192 ± 26 |
| 9H2 (5) | SP0199, or cls | Putative cardiolipin synthetase | 20 | 50 ± 10 |
| 76G6 | SP0200, or ccs4 | Putative competence-induced protein | 620 | 60 ± 5 |
| 70E2 (2) | SP0391, or cbpF | Choline binding protein F | 729 | 36 ± 6 |
| 12D4 | SP0393 | Putative IS3-SPN1 hypothetical protein, point mutation | 1068 | 52 ± 31 |
| 70B2 (1) | SP0461 | rlrA islet transcriptional regulator | 693 | 55 ± 10 |
| 60F5 (6) | SP0462, or rrgA | Pilus | 996 | 45 ± 8 |
| 40F1 | SP0615, or fibA | Putative beta-lactam resistance factor | 938 | 47 ± 9 |
| 72F3 (2) | SP0619 | Conserved hypothetical protein | 400 | 38 ± 11 |
| 8A10 | SP0646 | Putative PTS system, IIB component | 137 | 337 ± 94 |
| 36C1 | SP0648, or bgaA | Putative beta-galactosidase | 36 (5′) | 309 ± 143 |
| 67C10 | SP0693 | Putative hypothetical protein | 64 | 36 ± 7 |
| 5F10 | SP0694 | Putative conserved-domain protein | 46 (5′) | 33 ± 5 |
| 35C8 (3) | SP0695 | Putative HesA/MoeB/ThiF family protein | 5 (5′) | 42 ± 5 |
| 75F5 | SP0696 | Putative hypothetical protein | 149 | 44 ± 11 |
| 61H6 | SP0698 | Hypothetical protein | 647 | 37 ± 12 |
| 1E9 | SP0746 | ATP-dependent Clp protease, proteolytic subunit (ClpP) | 58 (5′) | 48 ± 9 |
| 70E9 (3) | SP0785 | Conserved hypothetical protein | 217 | 53 ± 17 |
| 69B2 | SP0799, or ciaH | Sensor histidine kinase | 388 | 62 ± 11 |
| 62A2 | SP0801 | Conserved hypothetical protein | 19 | 56 ± 7 |
| 56G6 | SP0975 | Putative exoribonuclease VacB/Rnb family | 498 | 60 ± 12 |
| 29D8 | SP0979, or pepF | Putative oligoendopeptidase F | 1540 | 53 ± 13 |
| 4E1 | SP1154 | ZmpA, immunoglobulin A1 protease | 5580 | 165 ± 38 |
| 67F4 | SP1182, or lacR2 | Putative lactose PTS repressor | 279 | 333 ± 83 |
| 69H1 | SP1370, or aroK | Putative shikimate kinase | 105 | 45 ± 6 |
| 61A1 | SP1390, or murB | UDP-N-acetylmuramate dehydrogenase | 63 (5′) | 46 ± 15 |
| 30E4 | SP1492 | Putative cell wall surface anchor family protein | 16 (5′) | 49 ± 15 |
| 37C3 (2) | SP1527, or aliB | Putative oligopeptide ABC transporter | 751 | 60 ± 23 |
| 20H6 | SP1531, or murE | UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate ligase | 45 (5′) | 55 ± 9 |
| 3F2 | SP1537 | General stress protein 13 | 238 | 41 ± 12 |
| 6H6 | SP1538 | Protein/peptidyl-prolyl cis-trans isomerase, Cof family | 783 | 39 ± 7 |
| 32F5 | SP1570, or clpX | ATP-binding subunit of Clp protease | 144 | 54 ± 18 |
| 27B9 | SP1586 | RNA helicase | 653 | 47 ± 5 |
| 61D9 | SP1664, or ylmF | Conserved protein of unknown function | 339 | 38 ± 4 |
| 61H10 | SP1682 | Putative sugar ABC transporter, permease | 869 | 58 ± 13 |
| 6A3 (2) | SP1687, or nanB | Neuraminidase B | 748 | 68 ± 15 |
| 69D7 | SP1731 | Conserved hypothetical protein | 55 | 42 ± 9 |
| 29D5 | SP1745 | Isochorismate protein | 15 | 67 ± 33 |
| 1D4 | SP1739 | Putative KH domain protein | 878 | 51 ± 15 |
| 1C7 (2) | SP1772 | Cell wall surface anchor family protein | 300 (5′) | 157 ± 48 |
| 35C7 | SP1903 | Conserved hypothetical protein | 219 | 56 ± 15 |
| 9F3 | SP1939 | Putative MATE efflux family protein DinF | 1245 | 52 ± 16 |
| 12H7 | SP2131 | Putative transcriptional regulator of the BglG family | 657 | 203 ± 43 |
| 27C9 | SP2190, or cbpA | Choline binding protein A | 267 | 47 ± 4 |
| 22A4 | SP2192 | Sensor histidine kinase | 246 | 44 ± 10 |
| 75H4 | SP2194, or clpC | ATP-binding subunit of Clp protease | 1793 | 49 ± 13 |
Numbers in parentheses next to the mutant names refer to the number of transposon insertions isolated in that gene; the name of only one of the mutants is given.
Gene names and assigned functions are from the TIGR4 genome annotation at the Comprehensive Microbial Resource site of the JCVI (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi).
The transposon insertion position represents the number of nucleotides within the corresponding ORF or 5′ of the corresponding ORF (for insertions in promoters).
Percentages of biofilm formation for each mutant relative to that of the wild type represent the means (±standard deviations) from four to five independent experiments.
Biofilm bacteria differ from their planktonic counterparts in many respects and rely on multiple genetic systems to accomplish the physical and physiological adaptations required for sessile growth (21, 29, 67). As anticipated, the collection of mutants obtained contained genes belonging to multiple genetic systems (Table 1). One prominent class of insertions comprised genes that are known to or are predicted to encode cell surface proteins, such as the cell wall choline binding proteins. Mutants 26H4 and 27C9 had insertions in cbpA (SP2190), which was recently shown to contribute to biofilm growth (Fig. 4) (59), while 31G5 and 70E2 had insertions in cbpF (SP0391), an abundant protein attached noncovalently to the bacterial cell wall that contributes to colonization of the rat nasopharynx (31). 1C7 and 73C8 had insertions 5′ of SP1772, which encodes a putative LPTXG cell wall protein, potentially affecting its expression. Mutant 30E4 had an insertion in SP1492, predicted to encode another cell wall surface protein. 6A3 and 45A8 had insertions in nanB (SP1687), encoding a cell surface neuraminidase, one of several glycosidases which is thought to modify host glycoconjugates (such as sialic acids) affecting both attachment and availability of carbon sources in vivo and whose function seems to overlap partially with that of neuraminidase A (14, 54, 92); parenthetically, both nanA and nanB are transcriptionally induced during biofilm growth (65). Another group of mutants had insertions in genes involved in the biosynthesis and degradation of peptidoglycan, including 40F1, which had an insertion in fibA (SP0615), predicted to encode a beta-lactam resistance factor, and 20H6 and 61A1, which had insertions affecting murE (SP1531) and murB (SP1390), respectively, both predicted to encode proteins catalyzing intracellular steps of peptidoglycan modification. Another group of mutants (9H2 and two others [not shown]) had insertions in SP0199, which encodes a putative cardiolipin synthase. An increase in cardiolipin content is an adaptation to stationary phase in Escherichia coli and might also contribute to survival in high osmolality (72). Several of the mutants isolated had mutations in genes encoding putative membrane proteins involved in transport of peptides and sugars. These included 4B9 and 37C3, which had insertions in aliB (SP1527), encoding a putative oligopeptide transporter previously shown to contribute to nasopharyngeal colonization (45); 61H10 (SP1682), encoding a putative sugar ABC transporter/permease; and 63A5 (SP0137), encoding the ATP-binding unit of another predicted ABC transporter. Mutants 9F3 and 62G9 had insertions affecting the function of a putative MATE efflux family protein; efflux pumps have been suggested to contribute to antibiotic resistance in biofilms (24). Another class of mutants comprised genes related to stress responses and protein folding and stabilization, including ATP-dependent proteases of the Clp family. 1E9 had an insertion in the promoter of clpP (SP746), which encodes the proteolytic subunit of an ATP-dependent Clp protease, while 32F5 and 75H4 had insertions potentially affecting clpX (SP1569) and clpC (SP1294), respectively, both of which encode ATP-binding subunits of the ATP-dependent Clp protease. Clp proteases have been studied in the context of S. pneumoniae resistance to stress and virulence, and their contribution seems to be serotype dependent. ClpC is required for the release of autolysin A and pneumolysin in serotype 2 (strain D39), while ClpP is required for thermotolerance and virulence in invasive disease in both serotype 2 and serotype 4 (41). Interestingly, Clp proteases have recently been shown to contribute to biofilm formation and intracellular invasion by Porphyromonas gingivalis (16). Mutant 3F2 had an insertion in SP1537, which encodes a putative general stress protein, while 6H6 (SP1538) was found to encode a Cof family protein/peptidyl-prolyl cis-trans isomerase. Mutant 29D8 had an insertion in SP0979, which encodes one of two putative oligoendopeptidases F found in the S. pneumoniae genome. Mutations in signal transduction systems included an insertion in SP2192 (22A4) and another in ciaH (SP0799), both of which encode the sensor histidine kinase partner of two different two-component systems (70); ciaH's partner, ciaR, was recently shown to be upregulated during biofilm growth (65). Two insertions in transcriptional regulators were also isolated: 12H7 had an insertion in SP2131, predicted to encode a transcriptional regulator of the BglG family and likely in an operon with a PTS transport system, while 67F4 had an insertion in lacR2 (SP1182), a putative transcriptional repressor. Of note, these two mutants were hyperbiofilm formers. Two mutants isolated, 69H1 (aroK, SP1370) and 29D5 (SP1745), had insertions in genes predicted to encode components of the shikimate pathway for synthesis of isochorismate, a precursor of aromatic amino acids and secondary metabolites. In E. coli, the two-component Cpx system modulates expression of shikimate pathway genes in response to a variety of stresses, including membrane protein damage, starvation, and high osmolarity (25), and has been postulated to be involved in biofilm growth (27). Finally, a number of insertions isolated were in genes predicted to encode hypothetical and conserved proteins of unknown function (Table 1).
The mutants listed in Table 1 were isolated by screening for differences in CV staining at 14 to 16 h postinoculation. We next evaluated a number of the mutants by CV staining and plating for CFU at an early time point (8 h). We generally found a good agreement between the level of biofilm detected by CV staining and CFU (not shown). However, we also observed that several mutants, including 6A3, 9F3, 37C3, 61D9, 61H6, 61H10, 69B2, and 69H1, had no defects at this early point in biofilm formation. Thus, these genes are apparently dispensable for initial attachment but are required for maintenance of the biofilm structure after prolonged incubation.
Identification of pilus protein RrgA as an adhesin involved in biofilm formation.
The rlrA islet, initially identified in a signature-tagged mutagenesis screen and reported to be required for lung and nasopharyngeal colonization (8, 37), encodes a pilus composed of a major structural subunit encoded by rrgB and two additional accessory subunits encoded by rrgA and rrgC (8, 50). We isolated seven mutants representing six unique insertions in this islet. Mutants 12A8, 32C8, 44B9, and 60F5 had insertions in rrgA, 70B2 had an insertion in rlrA, and 23G8 and 39A4 had identical insertions 194 bp 5′ of the transcriptional start of rrgA. The last two identical insertions likely affect transcription of rrgA and/or the divergently transcribed rlrA, the positive transcriptional regulator of the pilus-encoding genes (38). Intriguingly, no insertions in rrgB (the main pilus subunit), rrgC, or any of three putative sortases were obtained, prompting us to test the idea that the RrgA subunit but not the pilus structure per se may play a role in biofilm formation. We first confirmed the difference in biofilm formation between the rrgA mutant and the wild type by CV staining and CFU determination. As expected, a difference was discernible only when acapsular strains were compared, and this difference was most noticeable by CV staining (Fig. 5). In contrast, no differences in biofilm were found between the rrgB or rrgC mutant and the wild type in the encapsulated background (not shown) or the acapsular background (Fig. 5D). Thus, it seems that surface expression of RrgA either within the context of a pilus or on the surface itself in the absence of the pilus backbone (8) is sufficient to affect biofilm formation. Interestingly, it has recently been reported that RrgA functions as an adhesin during S. pneumoniae interaction with epithelial cells ex vivo independently of RrgB (62). In vivo, RrgA is likely to bind a host molecule(s), the nature of which remains to be defined. However, it is also possible that RrgA acts as a self-recognition adhesin, similarly to antigen 43 in E. coli (75). In the pilus, RrgA is found in clusters along the main shaft (50), and this positioning could facilitate interaction between pili from the same bacterium or adjacent bacteria. In the absence of the pilus, RrgA could mediate self-recognition because it is still found on the surface (8), possibly explaining why an RrgB mutant, which lacks the main pilus subunit, still forms wild-type biofilm levels. Similarly to our observations regarding RrgA and capsule expression in S. pneumoniae, in E. coli, the capsule shields the function of the self-recognition adhesin antigen 43, an adhesin that contributes to self-aggregation and biofilm formation in this gram-negative organism (75).
FIG. 5.
Biofilm formation by pilus-deficient S. pneumoniae. (A to D) rrgA, rrgB, and rrgC mutants were compared to wild-type (WT) isogenic encapsulated (c+, filled bars) or acapsular (c−, open bars) bacteria for their ability to grow planktonically or as biofilms. Bacteria were inoculated and incubated for 10 h in a 5% CO2 incubator at 37°C, at which time supernatants were removed and attached biofilms were washed with PBS and quantified by staining with CV (A and D) or by plating for CFU (B). Planktonic cells in the supernantants were enumerated by plating for CFU (C). (E) Complementation of srtA, 3F2, 9H2, 22A4, and 60F5 biofilm mutants. The indicated mutants were transformed with the corresponding complementing plasmid carrying the wild-type gene under the control of the upstream CAT (3F2, 9H2, 22A4, and 60F5) or SPC (srtA) cassette. Results are representative of three experiments. Error bars show standard deviations. OD570, absorbance at 570 nm.
Complementation of biofilm mutants.
The possibility that some of the phenotypes observed are due not to the transposon insertion(s) identified but instead to some unlinked unidentified secondary mutation exists. Ruling out this possibility awaits construction of individual deletion mutants and/or their complementation. We have begun such studies by engineering a construct in which the gene of interest with its ribosomal binding site is cloned downstream of a chloramphenicol (or Spc, for complementation of the srtA mutant) resistance marker, which has its own promoter and drives expression of the antibiotic resistance gene and the gene downstream of it. The construct is integrated into the chromosome (see Materials and Methods). Using this strategy, we were able to complement the 9H2, 22A4 (partially), 60F5, and srtA mutants but failed to complement the 3F2 mutant (Fig. 5E).
Analysis of biofilm mutants in the mouse model of nasopharyngeal colonization.
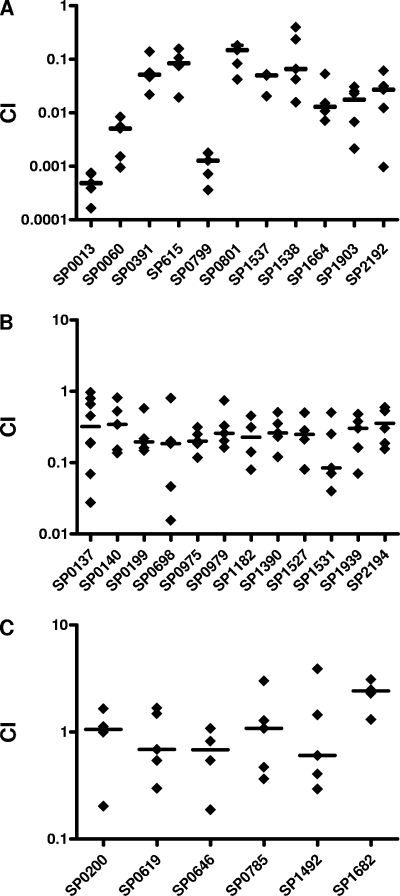
Our working hypothesis was that, during colonization, S. pneumoniae exists primarily in a biofilm-like state and that genes important for biofilm formation would play roles in colonization. In support of this idea, several of the mutants isolated in our biofilm screen (Table 1) had insertions in genes previously implicated in colonization, e.g., aliB, cbpA, rlrA, rrgA, and ciaH (39, 44). To examine this hypothesis further, we tested a subset of the biofilm mutants listed in Table 1 (only mutants with in vitro CI values between 0.5 and 1.5 were considered) for their ability to colonize the mammalian nasopharynx in competition with the wild-type strain by using the mouse model of colonization (89). All of the mutants isolated in this screen were in the acapsular background, and it is well established that S. pneumoniae requires at least some level of capsule expression for colonization (53). Therefore, before testing the biofilm mutants in vivo, we backcrossed the mutations into an encapsulated strain. As expected, when these biofilm mutants in the encapsulated background were tested for biofilm formation in vitro, they were not significantly different from the wild type (not shown). Nonetheless, when tested for colonization of the nasopharynx, a total of 23 out of 29 mutants tested were defective in colonization (Fig. 6); 11 had mean CI values of ≤0.1 and were classified as highly deficient for colonization (Fig. 6A), 12 had mean CI values between 0.1 and 0.3 and were considered colonization deficient, and 6 had mean CI values of ≥0.4 and were considered fit. For each mutant, the in vivo CI values were compared to the CI values obtained during planktonic growth competitions, and the differences were analyzed for statistical significance using a nonparametrical Mann-Whitney test. All differences were significant (P < 0.05) except in the case of mutant 63A5, which had an apparent bimodal distribution of CI values (Fig. 6B).
FIG. 6.
Biofilm mutants are severely compromised in their ability to colonize the mouse nasopharynx. A set of 29 biofilm mutants isolated in vitro were tested for their ability to colonize the mouse nasopharynx. These fell into three categories of colonization fitness: (A) highly deficient, (B) deficient, or (C) competent. Mutant and wild-type bacteria were grown to mid-log phase, washed in PBS, mixed 1:1, and then inoculated into the nares of C57BL/6 mice (1 × 107 CFU/mouse, 5 μl/nostril). Mice were sacrificed 5 days later, and bacterial counts in their nasopharyngeal lavage fluids were determined by plating for CFU. CI values were calculated by dividing the numbers of CFU for the mutant by those for the wild type. The median for each group is shown by the horizontal bar. In vivo CI values were significantly different from in vitro CI values (P ≤ 0.005).
Mutations leading to biofilm defects in a serotype 2 background.
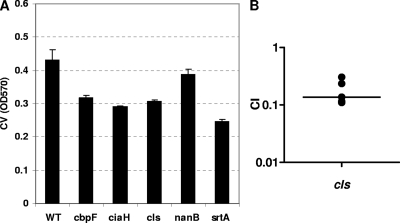
Natural immunity to S. pneumoniae seems to be non-serotype specific, as suggested by the fact that children become less susceptible to colonization by all serotypes as they age (44). Thus, identifying proteins important for colonization might contribute to the development of effective non-serotype-specific protein-based vaccines (66, 88). Therefore, we sought to extend the observations made for the serotype 4 TIGR4 strain to a different serotype. We transferred a subset of mutations from strain TIGR4 into strain Rx1, an acapsular serotype 2 strain, and compared the insertion mutants to the parental strain for biofilm formation. In all cases, the mutants had lower levels of biofilm than did the parental strain (Fig. 7A), albeit the differences were not as marked as those seen between mutants with mutations in the same genes and the wild type in the TIGR4 strain background. One insertion, that in cls (SP0199), which encodes a putative cardiolipin synthase and which was repeatedly isolated in the screen with the acapsular TIGR4 strain, was also transferred into an encapsulated serotype 2 strain (D39). Similarly to TIGR4 (Fig. 6B), the ability of a cardiolipin synthase mutant in serotype 2 to colonize the mouse nasopharynx was compromised (Fig. 7B).
FIG. 7.
Biofilm mutants in an S. pneumoniae serotype 2 strain background. (A) The mutations in several biofilm mutants isolated from the serotype 4 (TIGR4 strain) screen were transferred into an acapsular serotype 2 strain (Rx1), and the resulting mutants were tested for their ability to form a biofilm in vitro on polystyrene plates. The experiment was done three times. Error bars show standard deviations. WT, wild type; OD570, absorbance at 570 nm. (B) The cls (SP0199) mutant was also generated from an encapsulated serotype 2 strain (D39) and tested for its ability to colonize the mouse nasopharynx. Mutant and wild-type bacteria were grown to mid-log phase, washed in PBS, mixed 1:1, and then inoculated into the nares of C57BL/6 mice (1 × 107 CFU/mouse, 5 μl/nostril). Mice were sacrificed 5 days later, and bacterial counts in their nasopharyngeal lavage fluids were determined by plating for CFU. CI values were calculated by dividing the numbers of CFU for the mutant by those for the wild type. The median is shown by the horizontal bar. The experiment was done once.
DISCUSSION
Genetic and molecular studies of S. pneumoniae in vitro have utilized primarily axenic planktonic cultures. Accumulating evidence suggests that the ability to form biofilms is important for host infection by several clinically relevant bacteria, including E. coli (6), P. aeruginosa (30, 91), Bordetella species (42, 78), H. influenzae (34, 40), several Streptococcus species (22), and possibly S. pneumoniae (65). We hypothesized that S. pneumoniae adopted a biofilm-like mode of growth as a strategy for persistence during colonization. If sessile growth indeed reflected conditions during colonization more accurately than planktonic growth, some of the genes required for biofilm formation in vitro could also play a role in colonization in vivo. To address this question in a straightforward and unbiased manner, we screened two transposon libraries for defects in biofilm formation in vitro. This led to the identification of a set of 69 mutants involved in S. pneumoniae biofilm formation in vitro. Remarkably, a large percentage of the implicated genes were also shown to represent novel nasopharyngeal colonization factors.
The structure and physiology of bacterial biofilms vary widely in different species and environments. Typically, biofilms have extracellular matrices consisting of polysaccharides, proteins, membrane vesicles, and DNA (11, 76). Many different serotypes of S. pneumoniae are capable of producing biofilms in vitro in both flow cell and static culture conditions (13, 26, 82), but little is known about their composition or structure. Moscoso et al. (59) reported that S. pneumoniae biofilms are susceptible to proteinase K and DNase I treatment, and we obtained similar results with proteinase K treatment, indicating that proteins are important components of S. pneumoniae biofilms. However, in contrast to the results of Moscoso and colleagues (59), we found that DNase treatment did not significantly affect biofilm formation in vitro. There are currently a few variations of S. pneumoniae in vitro biofilm models. At present, it is not known which of these models will turn out to be the most useful to study pneumococcal factors related to colonization and virulence. The lack of DNase sensitivity observed in our study suggests that DNA is not likely to be an essential constituent of the biofilms formed under our experimental conditions. Thus, a limitation of our screen is that it did not allow the identification of factors that contribute to this aspect of biofilm formation in vitro. Interestingly, biofilm formation by S. pneumoniae serotype 3 in vitro results in the enrichment of acapsular phase variants (5, 82), which express less than 10% of wild-type capsule levels (57). Thus, the capsule polysaccharide may not constitute an important component of the S. pneumoniae biofilm. Indeed, our findings reported here suggest that biofilm formation is enhanced by the absence of the capsule.
In the present study, we began investigating the molecular mechanisms required for biofilm formation in vitro by conducting a genetic screen for biofilm mutants in an encapsulated serotype 4 TIGR4 strain background. Unexpectedly, despite near-saturating mutagenesis, we obtained transposon insertions in only two genes: lytC and cps4E. In similar screens with other microorganisms, a much larger number of genes was implicated in biofilm formation (49, 52, 68). This discrepancy, together with the observation that the cps4E mutants, all of which were acapsular, made more biofilm than the wild type, led us to suggest that the capsule might have prevented detection of mutants with biofilm defects. Indeed, the finding that we could uncover biofilm defects for only cbpA and srtA mutants in the acapsular background seemed to confirm this suspicion. One explanation for this could be that the low levels of biofilm made in the encapsulated background simply made it difficult to detect relatively small differences in biofilm formation between the mutants and the wild type. Alternatively, capsule expression could be epistatic to genes involved in biofilm formation, a distinct possibility that merits further investigation. The regulation of capsule expression in S. pneumoniae is poorly understood, but it seems to be affected by the environment (35, 47, 77, 86) and controlled at both the transcriptional and posttranscriptional levels (57, 63, 90).
We thus decided to perform a second screen, this time using an acapsular serotype 4 TIGR4 strain. This new screen resulted in the isolation of 69 biofilm-altered mutants with insertions in genes belonging to multiple genetic systems. Although this screen allowed the identification of a number of candidate genes implicated in biofilm formation, given that the mutants have not been reconstructed and that several of them represent a single transposon insertion, we cannot rule out the possibility that secondary unlinked mutations are responsible for the observed phenotypes. However, the fact that we successfully complemented three out of four biofilm mutants suggests that at least some of the phenotypes can be attributed to the transposon insertions isolated in the screen.
In support of a plausible link between biofilm formation and colonization, several of the mutants isolated had insertions in genes already known to play roles in colonization, including aliB (45), cbpA (73), and rlrA or rrgA (37, 62). Since all of the mutants isolated in the second screen were in the acapsular background and the capsule is required for colonization (53), we backcrossed all of the mutations into a wild-type encapsulated strain before testing them for colonization. Although the capsule, S. pneumoniae's quintessential virulence factor, has been studied extensively during invasive disease (2, 36), little is known about its role during colonization. Recent work by Nelson and colleagues (63) suggests that one of its functions is to help S. pneumoniae evade entrapment by the mucus covering the mucosa, allowing bacteria to reach the underlying epithelial layer, where it is thought stable colonization can take place. Interestingly, it seems that once the bacteria cross the mucus layer, the capsule contributes less to the ability of S. pneumoniae to persist. Moreover, clinical isolates from the nasopharynx often express low levels of capsule compared to those of wild-type laboratory strains (53), and isolates from otitis media patients have even lower levels of capsule expression (57). In addition, it has long been known that acapsular strains of S. pneumoniae or low-capsule-expressing variants adhere to epithelial layers much more efficiently than wild-type isogenic strains (77) and that colonization of the nasopharynx favors S. pneumoniae transparent variants which express lower capsule levels than their opaque counterparts (47, 85). Recent direct observation of S. pneumoniae in contact with epithelial monolayers ex vivo and in vivo indicates that S. pneumoniae that comes in contact with host cells and/or is internalized by them expresses no capsule or very low levels of capsule (35). Interestingly, downmodulation of encapsulation in order to facilitate colonization has been proposed for H. influenzae type b, another frequent cocolonizer of the nasopharynx (80). Reflecting on these data, we reasoned that even though transferring the biofilm mutations into the wild-type encapsulated background would likely result in no detectable biofilm defects in vitro (as indeed proved to be the case for all mutants tested), downregulation of the capsule after the bacteria crossed the mucus layer and began colonizing the nasopharyngeal epithelial layer would allow expressivity of the biofilm phenotypes. Indeed, of the 29 mutants we selected for testing in vivo, 80% were found to be defective in the mouse model of colonization.
Preliminary assays indicate that the rrgA mutant is only barely defective for adherence to D562 and A549 epithelial cell lines in the encapsulated background but markedly defective in the acapsular background (not shown). Similarly, cbpA and srtA mutants show deficiencies in adherence to epithelial cell lines only in the acapsular background (46, 71, 73). Interestingly, for S. pyogenes, pili could be shown to mediate adherence to host cells only in primary epithelial cells but not in HEp-2 or A549 cell lines, highlighting important limitations of ex vivo studies using immortalized cell lines (1). In future studies, we plan to evaluate the adherence properties of some of the biofilm mutants to human cells by using primary nasopharyngeal cells.
Obligate pathogens have to cause disease in their hosts in order to be transmitted successfully to a new host. Thus, in addition to fitness factors, i.e., those required for the pathogen's survival and growth in vivo, obligate pathogens have virulence factors, i.e., disease-causing factors. In these pathogens, both classes of factors are under selective pressure. In contrast, commensal organisms, which do not harm their hosts as part of their life cycles, lack virulence factors per se. S. pneumoniae is a commensal that often colonizes the oronasopharynx of a large percentage of healthy individuals asymptomatically, only occasionally causing disease. Traditionally, when searching for novel drug and vaccine targets in microorganisms, researchers have focused on virulence factors (37, 44). However, some of S. pneumoniae's so-called virulence factors whose maintenance is not under strong selective pressure, i.e., those factors that are not required for colonization, can be lost or rapidly modified by S. pneumoniae, a bacterium with remarkable genetic plasticity (20). Thus, in the case of a commensal colonizer such as S. pneumoniae, the focus should be on factors that are first and foremost required for fitness during colonization, the stage during which the bacteria experience selective pressure.
Supplementary Material
Acknowledgments
We acknowledge Mario Cepeda for technical assistance as well as the Camilli lab for helpful suggestions and discussions.
E.J.M.-E. acknowledges the financial support of the Irvington Institute Fellowship Program of the Cancer Research Institute. A.C. is an investigator of the Howard Hughes Medical Institute.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 15 September 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Abbot, E. L., W. D. Smith, G. P. Siou, C. Chiriboga, R. J. Smith, J. A. Wilson, B. H. Hirst, and M. A. Kehoe. 2007. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell. Microbiol. 91822-1833. [DOI] [PubMed] [Google Scholar]
- 2.Abeyta, M., G. G. Hardy, and J. Yother. 2003. Genetic alteration of capsule type but not PspA type affects accessibility of surface-bound complement and surface antigens of Streptococcus pneumoniae. Infect. Immun. 71218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akerley, B. J., E. J. Rubin, A. Camilli, D. J. Lampe, H. M. Robertson, and J. J. Mekalanos. 1998. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. USA 958927-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allegrucci, M., F. Z. Hu, K. Shen, J. Hayes, G. D. Ehrlich, J. C. Post, and K. Sauer. 2006. Phenotypic characterization of Streptococcus pneumoniae biofilm development. J. Bacteriol. 1882325-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allegrucci, M., and K. Sauer. 2007. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J. Bacteriol. 1892030-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301105-107. [DOI] [PubMed] [Google Scholar]
- 7.Austrian, R. 1986. Some aspects of the pneumococcal carrier state. J. Antimicrob. Chemother. 18(Suppl. A)35-45. [DOI] [PubMed] [Google Scholar]
- 8.Barocchi, M. A., J. Ries, X. Zogaj, C. Hemsley, B. Albiger, A. Kanth, S. Dahlberg, J. Fernebro, M. Moschioni, V. Masignani, K. Hultenby, A. R. Taddei, K. Beiter, F. Wartha, A. von Euler, A. Covacci, D. W. Holden, S. Normark, R. Rappuoli, and B. Henriques-Normark. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. USA 1032857-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bogaert, D., R. de Groot, and P. Hermans. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4144-154. [DOI] [PubMed] [Google Scholar]
- 10.Bogaert, D., P. W. M. Hermans, P. V. Adrian, H. C. Rumke, and R. de Groot. 2004. Pneumococcal vaccines: an update on current strategies. Vaccine 222209-2220. [DOI] [PubMed] [Google Scholar]
- 11.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 1320-26. [DOI] [PubMed] [Google Scholar]
- 12.Briles, D. E., L. Novak, M. Hotomi, F. W. van Ginkel, and J. King. 2005. Nasal colonization with Streptococcus pneumoniae includes subpopulations of surface and invasive pneumococci. Infect. Immun. 736945-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Budhani, R. K., and J. K. Struthers. 1997. The use of Sorbarod biofilms to study the antimicrobial susceptibility of a strain of Streptococcus pneumoniae. J. Antimicrob. Chemother. 40601-602. [DOI] [PubMed] [Google Scholar]
- 14.Burnaugh, A. M., L. J. Frantz, and S. J. King. 2008. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burne, R. A. 1998. Oral streptococci…products of their environment. J. Dent. Res. 77445-452. [DOI] [PubMed] [Google Scholar]
- 16.Capestany, C. A., G. D. Tribble, K. Maeda, D. R. Demuth, and R. J. Lamont. 2008. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J. Bacteriol. 1901436-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cartee, R. T., W. T. Forsee, M. H. Bender, K. D. Ambrose, and J. Yother. 2005. CpsE from type 2 Streptococcus pneumoniae catalyzes the reversible addition of glucose-1-phosphate to a polyprenyl phosphate acceptor, initiating type 2 capsule repeat unit formation J. Bacteriol. 1877425-7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, S., G. K. Paterson, H. H. Tong, T. J. Mitchell, and T. F. DeMaria. 2005. Sortase A contributes to pneumococcal nasopharyngeal colonization in the chinchilla model. FEMS Microbiol. Lett. 253151-154. [DOI] [PubMed] [Google Scholar]
- 19.Cho, K. H., and M. G. Caparon. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 571545-1556. [DOI] [PubMed] [Google Scholar]
- 20.Claverys, J. P., M. Prudhomme, I. Mortier-Barriere, and B. Martin. 2000. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35251-259. [DOI] [PubMed] [Google Scholar]
- 21.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 2841318-1322. [DOI] [PubMed] [Google Scholar]
- 22.Cvitkovitch, D. G., Y. H. Li, and R. P. Ellen. 2003. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Investig. 1121626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280295-298. [DOI] [PubMed] [Google Scholar]
- 24.De Kievit, T. R., M. D. Parkins, R. J. Gillis, R. Srikumar, H. Ceri, K. Poole, B. H. Iglewski, and D. G. Storey. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 451761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 27726652-26661. [DOI] [PubMed] [Google Scholar]
- 26.Donlan, R. M., J. A. Piede, C. D. Heyes, L. Sanii, R. Murga, P. Edmonds, I. El-Sayed, and M. A. El-Sayed. 2004. Model system for growing and quantifying Streptococcus pneumoniae biofilms in situ and in real time. Appl. Environ. Microbiol. 704980-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorel, C., P. Lejeune, and A. Rodriguez. 2006. The Cpx system of Escherichia coli, a strategic signaling pathway for confronting adverse conditions and for settling biofilm communities? Res. Microbiol. 157306-314. [DOI] [PubMed] [Google Scholar]
- 28.Ehrlich, G. D., R. Veeh, X. Wang, J. W. Costerton, J. D. Hayes, F. Z. Hu, B. J. Daigle, M. D. Ehrlich, and J. C. Post. 2002. Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 2871710-1715. [DOI] [PubMed] [Google Scholar]
- 29.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 1334-40. [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Medina, R., W. M. Dunne, P. K. Singh, and S. L. Brody. 2005. Pseudomonas aeruginosa acquires biofilm-like properties within airway epithelial cells. Infect. Immun. 738298-8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gosink, K. K., E. R. Mann, C. Guglielmo, E. I. Tuomanen, and H. R. Masure. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect. Immun. 685690-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 431367-1378. [DOI] [PubMed] [Google Scholar]
- 33.Gray, B. M., G. M. Converse III, and H. C. Dillon, Jr. 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J. Infect. Dis. 142923-933. [DOI] [PubMed] [Google Scholar]
- 34.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. A. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammerschmidt, S., S. Wolff, A. Hocke, S. Rosseau, E. Muller, and M. Rohde. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect. Immun. 734653-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy, G. G., A. D. Magee, C. L. Ventura, M. J. Caimano, and J. Yother. 2001. Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae. Infect. Immun. 692309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 451389-1406. [PMC free article] [PubMed] [Google Scholar]
- 38.Hava, D. L., C. J. Hemsley, and A. Camilli. 2003. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 185413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hava, D. L., J. LeMieux, and A. Camilli. 2003. From nose to lung: the regulation behind Streptococcus pneumoniae virulence factors. Mol. Microbiol. 501103-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong, W., K. Mason, J. Jurcisek, L. Novotny, L. O. Bakaletz, and W. E. Swords. 2007. Phosphorylcholine decreases early inflammation and promotes the establishment of stable biofilm communities of nontypeable Haemophilus influenzae strain 86-028NP in a chinchilla model of otitis media. Infect. Immun. 75958-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibrahim, Y. M., A. R. Kerr, N. A. Silva, and T. J. Mitchell. 2005. Contribution of the ATP-dependent protease ClpCP to the autolysis and virulence of Streptococcus pneumoniae. Infect. Immun. 73730-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Irie, Y., A. Preston, and M. H. Yuk. 2006. Expression of the primary carbohydrate component of the Bordetella bronchiseptica biofilm matrix is dependent on growth phase but independent of Bvg regulation. J. Bacteriol. 1886680-6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer, R., N. S. Baliga, and A. Camilli. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 1878340-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kadioglu, A., J. N. Weiser, J. C. Paton, and P. W. Andrew. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat. Rev. Microbiol. 6288-301. [DOI] [PubMed] [Google Scholar]
- 45.Kerr, A. R., P. V. Adrian, S. Estevao, R. de Groot, G. Alloing, J. P. Claverys, T. J. Mitchell, and P. W. Hermans. 2004. The Ami-AliA/AliB permease of Streptococcus pneumoniae is involved in nasopharyngeal colonization but not in invasive disease. Infect. Immun. 723902-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kharat, A. S., and A. Tomasz. 2003. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect. Immun. 712758-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim, J. O., and J. N. Weiser. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J. Infect. Dis. 177368-377. [DOI] [PubMed] [Google Scholar]
- 48.Lampe, D. J., M. E. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 155470-5479. [PMC free article] [PubMed] [Google Scholar]
- 49.Lavender, H. F., J. R. Jagnow, and S. Clegg. 2004. Biofilm formation in vitro and virulence in vivo of mutants of Klebsiella pneumoniae. Infect. Immun. 724888-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LeMieux, J., D. L. Hava, A. Basset, and A. Camilli. 2006. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect. Immun. 742453-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 1821374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Magee, A. D., and J. Yother. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect. Immun. 693755-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manco, S., F. Hernon, H. Yesilkaya, J. C. Paton, P. W. Andrew, and A. Kadioglu. 2006. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect. Immun. 744014-4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manetti, A. G., C. Zingaretti, F. Falugi, S. Capo, M. Bombaci, F. Bagnoli, G. Gambellini, G. Bensi, M. Mora, A. M. Edwards, J. M. Musser, E. A. Graviss, J. L. Telford, G. Grandi, and I. Margarit. 2007. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol. Microbiol. 64968-983. [DOI] [PubMed] [Google Scholar]
- 56.McCool, T. L., T. R. Cate, G. Moy, and J. N. Weiser. 2002. The immune response to pneumococcal proteins during experimental human carriage. J. Exp. Med. 195359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McEllistrem, M. C., J. V. Ransford, and S. A. Khan. 2007. Characterization of in vitro biofilm-associated pneumococcal phase variants of a clinically relevant serotype 3 clone. J. Clin. Microbiol. 4597-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell, T. J. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat. Rev. Microbiol. 1219-230. [DOI] [PubMed] [Google Scholar]
- 59.Moscoso, M., E. Garcia, and R. Lopez. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 1887785-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moscoso, M., E. Lopez, E. Garcia, and R. Lopez. 2005. Implications of physiological studies based on genomic sequences: Streptococcus pneumoniae TIGR4 synthesizes a functional LytC lysozyme. J. Bacteriol. 1876238-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Muñoz-Elías, E. J., and J. D. McKinney. 2002. Bacterial persistence: strategies for survival, p. 331-355. In S. H. E. Kaufmann, A. Sher, and R. Ahmed (ed.), Immunology of infectious diseases. ASM Press, Washington, DC.
- 62.Nelson, A. L., J. Ries, F. Bagnoli, S. Dahlberg, S. Falker, S. Rounioja, J. Tschop, E. Morfeldt, I. Ferlenghi, M. Hilleringmann, D. W. Holden, R. Rappuoli, S. Normark, M. A. Barocchi, and B. Henriques-Normark. 2007. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol. Microbiol. 66329-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson, A. L., A. M. Roche, J. M. Gould, K. Chim, A. J. Ratner, and J. N. Weiser. 2007. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect. Immun. 7583-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nobbs, A. H., R. M. Vajna, J. R. Johnson, Y. Zhang, S. L. Erlandsen, M. W. Oli, J. Kreth, L. J. Brady, and M. C. Herzberg. 2007. Consequences of a sortase A mutation in Streptococcus gordonii. Microbiology 1534088-4097. [DOI] [PubMed] [Google Scholar]
- 65.Oggioni, M. R., C. Trappetti, A. Kadioglu, M. Cassone, F. Iannelli, S. Ricci, P. W. Andrew, and G. Pozzi. 2006. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol. Microbiol. 611196-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ogunniyi, A. D., M. Grabowicz, D. E. Briles, J. Cook, and J. C. Paton. 2007. Development of a vaccine against invasive pneumococcal disease based on combinations of virulence proteins of Streptococcus pneumoniae. Infect. Immun. 75350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 5449-79. [DOI] [PubMed] [Google Scholar]
- 68.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28449-461. [DOI] [PubMed] [Google Scholar]
- 69.Parsek, M. R., and E. P. Greenberg. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 1327-33. [DOI] [PubMed] [Google Scholar]
- 70.Paterson, G. K., C. E. Blue, and T. J. Mitchell. 2006. Role of two-component systems in the virulence of Streptococcus pneumoniae. J. Med. Microbiol. 55355-363. [DOI] [PubMed] [Google Scholar]
- 71.Paterson, G. K., and T. J. Mitchell. 2006. The role of Streptococcus pneumoniae sortase A in colonisation and pathogenesis. Microbes Infect. 8145-153. [DOI] [PubMed] [Google Scholar]
- 72.Romantsov, T., S. Helbig, D. E. Culham, C. Gill, L. Stalker, and J. M. Wood. 2007. Cardiolipin promotes polar localization of osmosensory transporter ProP in Escherichia coli. Mol. Microbiol. 641455-1465. [DOI] [PubMed] [Google Scholar]
- 73.Rosenow, C., P. Ryan, J. N. Weiser, S. Johnson, P. Fontan, A. Ortqvist, and H. R. Masure. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol. Microbiol. 25819-829. [DOI] [PubMed] [Google Scholar]
- 74.Salles, C., L. Creancier, J. P. Claverys, and V. Mejean. 1992. The high level streptomycin resistance gene from Streptococcus pneumoniae is a homologue of the ribosomal protein S12 gene from Escherichia coli. Nucleic Acids Res. 206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schembri, M. A., D. Dalsgaard, and P. Klemm. 2004. Capsule shields the function of short bacterial adhesins. J. Bacteriol. 1861249-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schooling, S. R., and T. J. Beveridge. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 1885945-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Selinger, D. S., and W. P. Reed. 1979. Pneumococcal adherence to human epithelial cells. Infect. Immun. 23545-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sloan, G. P., C. F. Love, N. Sukumar, M. Mishra, and R. Deora. 2007. The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J. Bacteriol. 1898270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soong, G., A. Muir, M. I. Gomez, J. Waks, B. Reddy, P. Planet, P. K. Singh, Y. Kaneko, M. C. Wolfgang, Y. S. Hsiao, L. Tong, and A. Prince. 2006. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J. Clin. Investig. 1162297-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.St. Geme, J. W., III, and D. Cutter. 1996. Influence of pili, fibrils, and capsule on in vitro adherence by Haemophilus influenzae type b. Mol. Microbiol. 2121-31. [DOI] [PubMed] [Google Scholar]
- 81.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293498-506. [DOI] [PubMed] [Google Scholar]
- 82.Waite, R. D., J. K. Struthers, and C. G. Dowson. 2001. Spontaneous sequence duplication within an open reading frame of the pneumococcal type 3 capsule locus causes high-frequency phase variation. Mol. Microbiol. 421223-1232. [DOI] [PubMed] [Google Scholar]
- 83.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 1822675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Webster, L. T., and T. P. Hughes. 1931. The epidemiology of pneumococcus infection. J. Exp. Med. 53535-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weiser, J. N., R. Austrian, P. K. Sreenivasan, and H. R. Masure. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect. Immun. 622582-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weiser, J. N., D. Bae, H. Epino, S. B. Gordon, M. Kapoor, L. A. Zenewicz, and M. Shchepetov. 2001. Changes in availability of oxygen accentuate differences in capsular polysaccharide expression by phenotypic variants and clinical isolates of Streptococcus pneumoniae. Infect. Immun. 695430-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413860-864. [DOI] [PubMed] [Google Scholar]
- 88.Wu, H. Y., M. H. Nahm, Y. Guo, M. W. Russell, and D. E. Briles. 1997. Intranasal immunization of mice with PspA (pneumococcal surface protein A) can prevent intranasal carriage, pulmonary infection, and sepsis with Streptococcus pneumoniae. J. Infect. Dis. 175839-846. [DOI] [PubMed] [Google Scholar]
- 89.Wu, H. Y., A. Virolainen, B. Mathews, J. King, M. W. Russell, and D. E. Briles. 1997. Establishment of a Streptococcus pneumoniae nasopharyngeal colonization model in adult mice. Microb. Pathog. 23127-137. [DOI] [PubMed] [Google Scholar]
- 90.Xayarath, B., and J. Yother. 2007. Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J. Bacteriol. 1893369-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang, L., J. A. J. Haagensen, L. Jelsbak, H. K. Johansen, C. Sternberg, N. Hoiby, and S. Molin. 2008. In situ growth rates and biofilm development of Pseudomonas aeruginosa populations in chronic lung infections. J. Bacteriol. 1902767-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yesilkaya, H., S. Manco, A. Kadioglu, V. S. Terra, and P. W. Andrew. 2008. The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol. Lett. 278231-235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.