Abstract
Mycoplasma pneumoniae accounts for 20 to 30% of all community-acquired pneumonia and has been associated with other airway pathologies, including asthma, and a range of extrapulmonary manifestations. Although the entire genomic sequence of M. pneumoniae has been completed, the functions of many of these genes in mycoplasma physiology are unknown. In this study, we focused on clpB, a well-known heat shock gene in other bacteria, to examine its role in mycoplasma growth. Transcriptional and translational analyses of heat shock in M. pneumoniae indicated that clpB is significantly upregulated, reinforcing its status as a critical responder to heat stress. Interestingly, M. pneumoniae ClpB does not use dual translational start points for ClpB synthesis, like other ClpB-characterized bacteria. Biochemical characterization of purified M. pneumoniae recombinant ClpB revealed casein- and lysine-independent ATPase activity and DnaK-DnaJ-GrpE-dependent chaperone activity. An M. pneumoniae mini-Tn4001-integrated, clpB-null mutant was impaired in its ability to replicate under permissive growth conditions, demonstrating the growth-promoting status of ClpB.
Mollicutes are unusual prokaryotes that lack bacterial cell walls and exhibit limited metabolic capabilities due to their streamlined genomes. Mycoplasma pneumoniae, with a genome of 816 kb, is confined to a parasitic life-style in humans (16). The importance of M. pneumoniae as a causative agent of acute and chronic respiratory diseases and extrapulmonary pathologies has been well documented by epidemiological studies in many settings. M. pneumoniae accounts for 20 to 30% of all community-acquired pneumonia and has been implicated in asthma and chronic obstructive pulmonary disease (3, 36). This mycoplasma utilizes several attachment mechanisms to bind to sialylated and sulfated receptors on human target cells and to abundant host proteins, like fibronectin and surfactant protein A (2, 7, 18, 19). An understanding of how M. pneumoniae responds to in vivo environmental signals and successfully establishes residence in the host has been elusive. This is compounded by the absence of well-established prokaryotic regulatory signaling, such as alternative sigma factors and two-component systems. For example, the presence of only one sigma factor (sigA, mpn352) in M. pneumoniae suggests that its response to external stimuli is not controlled by the level of expression of alternative sigma factors. However, the presence of the conserved inverted repeat sequence (CIRCE [controlling inverted repeat of chaperone expression]) immediately in front of four heat shock genes (dnaK, lonA, dnaJ, and clpB) and the identification of the CIRCE-associated repressor (hrcA, mpn124) provide evidence for negative regulation of gene expression in M. pneumoniae.
Recent transcriptional studies of heat shock responses in M. pneumoniae (39), Mycoplasma genitalium (29), and Mycoplasma hyopneumoniae (6) have provided additional insights concerning the upregulation of specific heat shock genes. For example, in M. pneumoniae, groE retained remnants of the CIRCE-like element with four mutations within both arms of the inverted repeat and a deletion within the 9-nucleotide spacer sequence. Among the three copies of dnaJ (mpn002, mpn021, and mpn119), only mpn021 possessed a CIRCE-like element with three mutations in both arms of the inverted repeats. The CIRCE element of dnaK had one mutation in one arm of the inverted repeat. Evidence that CIRCE might act as a negative cis element was obtained by the observation that mutations in one or both arms of specific inverted repeats resulted in elevated transcription of the downstream genes even at a normal growth temperature (35, 43). In contrast, the CIRCE elements of clpB and lonA were undisturbed, indicating that clpB and lonA are the only heat shock response genes that are fully under the control of the CIRCE element. Studies with M. genitalium and M. hyopneumoniae confirmed CIRCE-mediated interactions (6, 29). Interestingly, CIRCE is reported mostly in association with groE or dnaK operons in other bacteria (43), suggesting that different modes of stress-related gene regulation exist in M. pneumoniae. Therefore, the most-relevant regulatory transcriptional and translational distinctions between M. pneumoniae and other gram-positive bacteria are (i) the absence of alternative sigma factors, (ii) the presence of ClpB alone as representative of the Clp family, and (iii) the occurrence of CIRCE sequences upstream of clpB and their absence upstream of groESL. In both M. pneumoniae and M. genitalium transcriptional studies, the ClpB gene was highly upregulated during heat shock compared to all other heat shock genes.
ClpB belongs to the AAA protein (ATPases associated with a variety of cellular activities) superfamily, which includes one category of the Clp/Hsp100 proteins with its members ClpA, ClpX, and ClpY, which facilitate the activities of ClpP, and another category where ClpB does not interact with ClpP. Clp (caseinolytic protease) was first identified as a heat shock-inducible, ATP-dependent protease complex capable of hydrolyzing casein. In spite of the significance of Clp ATPases in stress responses, M. pneumoniae has conserved only one ClpB, and a single gene in eubacteria encodes two differently sized proteins (79 and 93 kDa) via dual translational initiation sites within the clpB transcript (34).
ClpB mediates the full recovery of aggregated proteins with the cooperation of the DnaK chaperone system (DnaK, DnaJ, and GrpE; termed KJE) (27, 28, 42). In some organisms, the functional interdependence of ClpB and KJE is reflected in their gene organization and/or coordinated expression. For example, in Mycoplasma capricolum clpB, dnaK, dnaJ, and grpE genes are transcribed as a single operon (9), whereas in M. pneumoniae and M. genitalium the unlinked clpB and dnaK genes belong to the same CIRCE regulon (10, 16).
Here, we describe specific attributes of M. pneumoniae ClpB and show that a ClpB-deficient mutant demonstrates markedly impaired cell growth.
MATERIALS AND METHODS
M. pneumoniae strains and growth conditions.
Wild-type M. pneumoniae clinical strain S1 and the S1ΔClpB mutant strain were grown in SP-4 broth medium at 37°C for 72 to 96 h in 150-cm2 tissue culture flasks as described earlier (7). For functional studies, M. pneumoniae S1 was inoculated into 300 ml SP-4 medium distributed equally into three 150-cm2 tissue flasks, incubated at 37°C for 72 h, and then harvested, concentrated, pooled, and added to three liters of fresh SP-4 broth. The latter was immediately transferred equally into 30 150-cm2 tissue culture flasks, which were divided into five groups of six flasks. One group was used for radiolabeling, and the remaining four groups were used for RNA isolation and immunoblotting.
Biosynthetic radiolabeling of M. pneumoniae proteins.
For radiolabeling, mycoplasmas were biosynthetically labeled with [35S]methionine-cysteine as follows: surface-attached M. pneumoniae cells grown to mid-log phase were washed three times with Dulbecco's modified Eagle's medium (DMEM) and placed at 37°C or 43°C. Surface-attached cells were immersed in 1/10 of their original volume in DMEM without cysteine or methionine but supplemented with 10% fetal bovine serum. Then, 100 μCi of [35S]methionine-cysteine was added, and mycoplasmas were incubated at 37°C or 43°C for 2 to 6 h, pelleted, and washed four times with phosphate-buffered saline (PBS). Cell aliquots were harvested for protein estimation and for scintillation counts. Based on protein concentration, the remaining cell pellets were resuspended individually in sample buffer A (4% sodium dodecyl sulfate [SDS], 125 mM Tris [pH 6.8], 10% β-mercaptoethanol, 10% glycerol, 0.2% bromophenol blue) and heated to 95°C for 5 min. Then, equal amounts of whole-cell lysates were analyzed on 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels and transferred to nitrocellulose membranes. Blots were then exposed to Kodak OMAT X-ray film for 1 to 2 weeks and autoradiograms analyzed. Quantitation of band intensities was performed by scanning autoradiograms and analyzing images with Kodak Image software. For immunoblot analysis, mycoplasma cells grown at 37°C or 43°C were washed two times, scraped into PBS, and characterized after protein quantification.
Bacterial strains, plasmids, and DNA manipulations.
Escherichia coli Top10 (Invitrogen) and E. coli BL21(DE3) [F′ ompT hsdS (rB mB) gal dcm λ(DE3) pLysS] (Stratagene) were grown in Luria-Bertani (LB) broth and used to clone and express the mycoplasma clpB gene (mpn531). For DNA manipulations, the following vectors were used: pCR2.1 (Apr Kmr, TA cloning vector; Invitrogen) and pET19b (Apr, N-terminal His10 tag, expression vector; Novagen). M. pneumoniae chromosomal DNA was isolated using an Easy DNA isolation kit (Invitrogen). Plasmid DNA was purified using the QIAprep spin protocol according to the manufacturer's instructions (Qiagen). The M. pneumoniae clpB locus was amplified by PCR, using strain S1 chromosomal DNA as a template. PCR amplification and UGA corrections were performed as described earlier for CARDS TX (18). The specific clpB oligonucleotide primers were as follows: ClpB F1, 5′-CATATGGATTTTAGTTTTACACCAACACC-3′; ClpB R1, 5′-TTTCTCCTTTTTCCATTCGGCAGTTAGAG-3′; ClpB F2, 5′-CTCTAACTGCCGAATGGAAAAAGGAGAAA-3′; and ClpB R2, 5′-GGATCCTAAGATTTAACCTTATTGGCTAC-3′.
The resulting ca.-2.0-kb PCR product was cloned into pCR2.1, generating a plasmid designated pCR-ClpB, which was subsequently digested with NdeI and BamHI. Insert DNA was gel extracted and ligated into pET19b, which was digested with NdeI and BamHI to yield pET-ClpB. This plasmid was transformed into competent E. coli BL21(DE3) strains, and recombinant colonies were screened for resistance to ampicillin and expression of a protein product of the correct size. One recombinant clone was selected for further study. Verification of pET-ClpB was achieved by complete DNA sequencing (Department of Microbiology and Immunology Nucleic Acids Core Facility, The University of Texas Health Science Center at San Antonio). Sequences were analyzed using the BLAST program available in the NCBI database (www.ncbi.nlm.nih.gov).
Expression and purification of M. pneumoniae rClpB protein.
The pET-ClpB plasmid in E. coli BL21(DE3) cells was expressed as described earlier (18), and fusion proteins were purified by nickel affinity chromatography under native conditions (Qiagen). Recombinant ClpB (rClpB) was eluted with 2.5-ml aliquots of lysis buffer containing 250 mM imidazole (elution buffer). Each collected fraction was desalted in 50 mM Tris-HCl buffer (pH 8.0) and 5% glycerol (TG) by using PD-10 columns, and individual fractions were analyzed by SDS-PAGE. Fractions with the highest purity of rClpB were pooled and applied to 1 ml Mono Q HR anion-exchange columns (Amersham Bioscience) preequilibrated with TG plus 0.25 M NaCl. rClpB was eluted with a linear gradient of NaCl (0.25 to 1.0 M in TG buffer) (flow rate, 0.5 ml·min−1; fractions, 1.0 ml). Protein concentrations were monitored at an absorbance of 280 nm, and peak fractions were analyzed by SDS-PAGE. Fractions with the highest purity of rClpB were concentrated, and 100 μl of the concentrated sample was applied to a 25-ml gel filtration Superdex 200 HR 10/30 column (GE Healthcare) equilibrated with TG plus NaCl buffer (AKTA purifier) and eluted with the same buffer (flow rate, 0.5 ml·min−1; fractions, 0.5 ml). Protein concentrations were monitored at an absorbance of 280 nm. Samples obtained at all stages were analyzed by SDS-PAGE and silver staining. Fractions containing pure rClpB protein were dialyzed in storage buffer and placed at −80°C.
Generation of mouse anti-ClpB polyclonal antibodies.
Six-week-old BALB/c female mice (n = 5) were bled and screened by immunoblot analysis using rClpB to determine preexisting antibodies. As described earlier (18), mice were immunized with different concentrations of rClpB and later screened by immunoblot analysis using rClpB or whole M. pneumoniae cell lysates. All mice exhibited strong immune reactivity to rClpB and ClpB in whole-cell lysates. Sera were collected 2 weeks later, rescreened by immunoblot analysis and an enzyme-linked immunosorbent assay, and pooled.
RNA isolation.
RNase-free reagents and plasticware were used to isolate total RNA from M. pneumoniae clinical strain S1. Surface-adherent cells were washed twice with sterile PBS, and Tri Reagent (Sigma) was added to each flask to facilitate cell lysis. To eliminate minute DNA contamination in RNA samples, isolated total RNA was treated with DNase I (Gibco-BRL) prior to use in cDNA synthesis, reverse transcription-PCR (RT-PCR), or primer extension (PE).
PE.
For PE of clpB, oligonucleotide 5′-TGAGCACCTCATCGTTAATGCTA-3′ was end labeled with [γ-32P]ATP by using polynucleotide kinase (United States Biochemical) and annealed to 25 μg of total mycoplasma RNA. PE was performed as previously described (30). PE products were phenol-chloroform extracted, precipitated with ethanol, and dissolved in 10 μl of H2O. Two microliters of each sample was analyzed in 6% sequencing gels alongside sequencing reactions (double-stranded-DNA cycle sequencing system; Gibco-BRL) generated with the same oligonucleotides and appropriate recombinant plasmids. The sequencing template for identification of the clpB transcriptional start site was generated by PCR amplification of the corresponding region in S1 by using primers 5′-TAGCGCGTAAAGATGTACCGTT-3′ (532RTPCRP) and 5′-TGAGCACCTCATCGTTAATGCTA-3′ (reverse primer used for PE of clpB). The amplified ∼960-bp region was cloned in pCR2.1, and individual plasmids were purified from E. coli Top10 and sequenced.
RT-PCR analysis.
One microgram of DNase I-treated total RNA isolated from M. pneumoniae cells was reverse transcribed using gene-specific antisense primers (Table 1) and the SuperScript first-strand synthesis system (Gibco-BRL). PCR amplification was performed using Platinum Taq DNA polymerase (Invitrogen), and generated products were analyzed by electrophoresis on 1.2% agarose. To prove the absence of DNA in RNA preparations, a parallel reaction was run without reverse transcriptase. The specificity of all primers was demonstrated by PCR amplification of the predicted regions, using chromosomal DNA isolated from M. pneumoniae clinical strain S1. Regions amplified from the clinical strain S1 template were sequenced and compared with reference strain M129.
TABLE 1.
M. pneumoniae genes and their oligonucleotide primers used for transcriptional analysis by RT-PCR
| Gene | Primer namea | Primer sequence | Amplicon length (bp) |
|---|---|---|---|
| mpn529 | 529RTPCRP | ACAACATCGAGTAAGCCATTGTC | 317 |
| 529RTPCRM | TAGTCTGCGTATTTTCAGCGTACC | ||
| mpn530 | 530RTPCRP | GGAACCAAATAATTTAAAAGAAGAGCTAG | 404 |
| 530RTPCRM | TCGTTTAAGTAGTCAGCTATTTTTTCAG | ||
| clpB | 531RTPCRP1 | AATTTATGAACTTTCGTTGTCTGG | 426 |
| 531RTPCRM2 | CATTTCTACAGCAGCTACCAAGGC | ||
| 531RTPCRP2 | TTTACTAACTGAAGCAGTCCGACG | 544 | |
| 531RTPCRM2 | TCTCAACTATTACGTTGTAACTAACATC | ||
| licA | 532RTPCRP | TAGCGCGTAAAGATGTACCGTT | 557 |
| 532RTPCRM | CAGCAACACGTAAATTACCGTTTC | ||
| ackA | 533RTPCRP | GATTCGGTGATCGTAGATGCA | 558 |
| 533RTPCRM | AGCGAAAACATCACGCATATCC | ||
| mpn534 | 534RTPCRP | GAATACTTTTAAAGAAACCCTCTTTTCA | 408 |
| 534RTPCRM | ACCTTTCTAATTTGCTGTTTGG |
P, reverse primer; M, forward primer.
Synthesis of radiolabeled cDNA.
Hybridization probes were also generated using the SuperScript first-strand synthesis system (Gibco-BRL). Twenty micrograms of total RNA plus a mixture containing 0.2 μM of each gene-specific antisense oligonucleotide (Table 2) was heated to 70°C for 10 min and cooled to room temperature. Reverse transcription was performed with both [α-32P]dCTP and [α-32P]TTP (NEN) replacing the corresponding nucleotides in the reaction buffer (29). After unincorporated nucleotides were removed by gel filtration using G-25 Sephadex column chromatography (Roche), an equal volume of formamide was added. Following denaturation (boiling for 5 min), individual probes were added to the hybridization buffer.
TABLE 2.
M. pneumoniae genes and their oligonucleotide primers used for RT-PCR quantification of transcripts by DNA macroarray analysisa
| Gene | Primer name | Primer sequence | Amplicon length (bp) | Ta (°C) |
|---|---|---|---|---|
| dnaJ1 | MPN002P | AGCGGAGTATGACGCTATGCTGCGC | 496 | 60 |
| MPN002M | TGTTGAGTTCGTACCACATTTGTCCAGGG | |||
| dnaJ2 | MPN021P | TTGCCCTATGATTTAGAGATTGCG | 477 | 55 |
| MPN021M | ATTGGCTACTACATTATGGTCAACC | |||
| dnaJ3 | MPN119P | AATAAGGCACCGGATGCGGC | 589 | 57 |
| MPN119M | TAACCGTACTCAGGATTACCAATCATTGC | |||
| grpE | MPN120P | TTACCGAAATTCTCAGTTCCATCCG | 553 | 55 |
| MPN120M | CGATACCTTCACCACCGTATTAGCCG | |||
| hrcA | MPN124P | ACCCGACAAGCCCAAATTCTCAAGG | 536 | 55 |
| MPN124M | GCGATCATTAAAGATCCTGACACATACC | |||
| tig | MPN331P | TTGATGCCATCTTCCAGCCG | 539 | 55 |
| MPN331M | TGATCTCTAACTGCTTAATTGCTTTGAGC | |||
| lon | MPN332P | AGCGTTAGATAAGTTGTTGGAACGG | 495 | 55 |
| MPN332M | TTTAATGTGCTCTGGATATGGG | |||
| sigA | MPN352P | AAGGAGAGTGATGTACCAAAGAAGCGTCG | 455 | 57 |
| MPN352M | GGAAGAATCGAGCGAACCTAGAAAGAAACG | |||
| nox | MPN394P | TTTGGGTTGTGGAATTGCCTTGG | 556 | 57 |
| MPN394M | GGTTAATGCCCGCTTTCTTCATGG | |||
| dnaK | MPN434P | CTACCGCCGCTGCATTGGCTTATGG | 446 | 60 |
| MPN434M | AAACATCCGAAATTGGGTTGCGAGTACG | |||
| clpB | MPN531P | AATTTATGAACTTTCGTTGTCTGG | 426 | 55 |
| MPN531M | CATTTCTACAGCAGCTACCAAGGC | |||
| groEL | MPN573P | GCTGTATCTACCAATGATATTGCTGG | 492 | 55 |
| MPN573M | TTTCCATGCTACCTTCTAGTAACGG | |||
| groES | MPN574P | AGTGCCCACAAGGGAACTGTCG | 103 | 55 |
| MPN574M | AAAGTAGATCGTGTCACCGATGCC | |||
| eno | MPN606P | AACTACGCGATGGTGATCCC | 553 | 55 |
| MPN606M | GCTGCCTTAATTGCTTCGACC | |||
| ftsH | MPN671P | GCCTACTACCAATTATTATCTTTGTCA | 523 | 55 |
| MPN671M | TTTAAGGTTTGTTCGACCACCG |
Reverse primers (M) were used to synthesize cDNA for hybridization. Ta, annealing temperature.
DNA macroarray construction, hybridization, and data analysis.
DNA regions of M. pneumoniae genes were amplified using specific primers (Table 2). All PCR products were purified by gel extraction, and 200 ng of each product was blotted in triplicate on ZETA probe membranes by using a Bio-Dot SF microfiltration apparatus as suggested by the manufacturer (Bio-Rad Laboratories). Each array contained two controls: one with no DNA, which served as the control for reagent contamination, and one with recA DNA, which served as the control for hybridization stringency. 32P-labeled cDNAs generated using total mRNA from heat-shocked and control M. pneumoniae genes served as probes. Blots were prehybridized and hybridized as suggested by the manufacturer (Bio-Rad Laboratories). Membranes were washed at room temperature for 15 min in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS, 30 min in 0.5× SSC-0.1% SDS, and 30 min in 0.1× SSC-0.1% SDS. After posthybridization washes, all membranes were sealed in plastic bags and placed in exposure cassettes with storage phosphor screens.
Individual screens were scanned using Typhoon 9400 (Molecular Dynamics), and resulting image files were assessed by determining pixel density for each band after background subtraction (ImageQuant 5.2; Molecular Dynamics). Numeric files were exported into a Microsoft Excel spreadsheet for analysis. To determine the transcript amounts for different experimental conditions, we expressed them as percentages of the nox or eno housekeeping gene. We further calculated average intensities for each gene under applied experimental conditions. To evaluate changes in specific transcripts, we determined the ratio of the corresponding average percent intensities of each gene-specific slot. These ratios represent relative transcript levels (n-fold) of each gene under the two experimental conditions. Triplicate values from three separate experiments were averaged, and comparative data between experiments differed by less than 10%.
ATPase activity of rClpB.
ClpB-ATPase activity was measured by estimating the concentration of inorganic phosphate produced from ATP by using the malachite green colorimetric assay (15, 21). Reactions were performed at 37°C for 15 min in assay buffer (100 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 5 mM ATP, 1 mM EDTA, 1 mM dithiothreitol) containing different concentrations of rClpB (0.1 μM to 10 μM). ATPase activities were also determined in the presence of 0.25 mg/ml α-casein (Sigma) or 0.04 mg/ml poly-l-lysine (Sigma).
Chaperone activity of ClpB.
ClpB chaperone refolding activity was measured as reported previously (22). In brief, firefly luciferase (Promega) was diluted 300-fold into PBS buffer containing 1 mg/ml bovine serum albumin and incubated at 45°C for 15 min. After heat treatment, luciferase samples were cooled to room temperature and diluted 20-fold into renaturation buffer (30 mM HEPES-KOH [pH 7.6], 120 mM KCl, 10 mM MgCl2, 5 mM ATP, 1 mM EDTA, 1 mM dithiothreitol, and 0.1 mg/ml bovine serum albumin). The reaction was initiated by addition of rClpB (0.1 to 10 μM), Mycoplasma genitalium rDnaK (1 to 5 μM) or rClpB (0.1 to 10 μM), and the KJE system (DnaK [0.5 to 5 μM], DnaJ [0.1 to 1 μM], and GrpE [0.05 to 0.5 μM]). DnaK, DnaJ, and GrpE were purchased from StressGen Biotechnologies (Victoria, BC, Canada). Refolded luciferase activities in refolding solution were determined using a Turner Biosystems luminometer.
Tn mutagenesis of M. pneumoniae.
We and others have found Tn4001 to be useful in studies with M. pneumoniae and M. genitalium (14, 32). To avoid multiple insertions, we used transposon (Tn) pMT85-miniTn4001, which has a colE1 plasmid origin of replication and a selectable gentamicin/kanamycin marker within Tn4001 to allow for direct rescue of chromosomal sequences adjacent to the insertion site (41). pMT85-miniTn4001 was electroporated into competent mycoplasmas as reported previously (32). After growth in selective medium (two passages), the complete mixture of amplified Tn mutants was collected, and M. pneumoniae cell suspensions were passed through 25-gauge needles several times and filtered through membrane filter units with 0.45-μm pores (Millipore) to remove aggregates. Mycoplasma cells were diluted in SP-4 medium and plated on SP-4 agar plates containing 80 μg/ml gentamicin. Chromosomal DNAs from individual Tn-integrated colonies were HindIII digested, self-ligated, and transformed in E. coli Top10. Plasmid DNAs were isolated and subjected to DNA sequencing in order to identify the Tn integration site in the chromosomal DNA of M. pneumoniae. Sequencing across mini-Tn4001 Tn-plasmid junctions was facilitated by Tn4001 IS256 outer repeat sequences 5′-GATAAAGTCCGTATAATTGTGTAAAAG-3′ (forward) and 5′-CTTTTACACAATTATACGGACTTTATC-3′ (reverse), which anneal at the 5′ and 3′ ends of Tn4001, respectively. For each mini-Tn4001 insertion, a DNA sequence of 600 to 800 bp in each direction from the site of insertion was determined. To confirm the integration of Tn4001 within the clpB open reading frame, oligonucleotide primers clpB-F1 and clpB-R1 were used for PCR amplification, and chromosomal DNAs from S1 and the S1ΔClpB mutant were used as template DNAs.
Immunoblot analysis.
Whole-cell lysates were prepared from M. pneumoniae S1 and the S1ΔClpB mutant grown to mid-log phase in SP-4 medium at 37°C and 43°C as described above. Equal amounts of whole-cell lysates from each strain were separated on 4 to 12% Nu-PAGE gel (Invitrogen) and transferred to nitrocellulose membranes. Immunoblots were then performed with a 1:2,000 dilution of mouse polyclonal anti-ClpB antibody, polyclonal anti-M. genitalium DnaK antibody (W. Zhang and J. B. Baseman, unpublished results), rabbit polyclonal anti-LonA antibody (a gift from Richard Herrmann), and mouse monoclonal antibodies raised against P1 and the His tag (7). Sera from M. pneumoniae-infected BALB/c mice, uninfected control mice, M. pneumoniae-infected patients, and uninfected human controls were used to detect the immune response against ClpB.
RESULTS
Primary sequence and conserved domains of M. pneumoniae ClpB (MPN531).
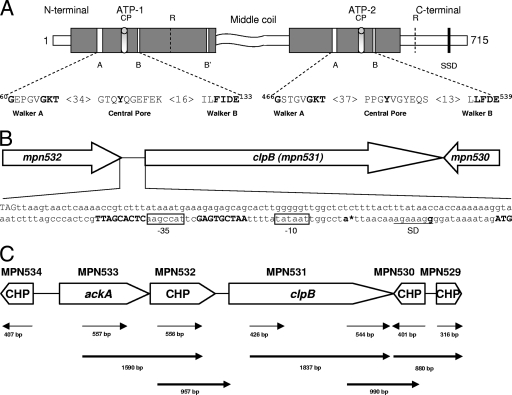
ClpB is the only member of the Clp family in M. pneumoniae, and we analyzed its primary sequence for the presence of specific motifs and essential amino acids involved in nucleotide and substrate binding as described for E. coli ClpB. An obvious difference is that M. pneumoniae ClpB comprises a 715-amino-acid sequence (Fig. 1A), compared to 857 amino acids for E. coli ClpB (16, 34). In E. coli, ClpB is synthesized in two forms, a 93-kDa (ClpBEC) and a 79-kDa (ClpB′EC) peptide (34), with ClpB′EC translation commencing at 149 codons downstream of the ClpBEC start site. Alignment of amino acid sequences showed that the translational start site of M. pneumoniae ClpB coincides with amino acid 143 of ClpBEC, which is near the start codon of E. coli ClpB′. As predicted by Scan Prosite, M. pneumoniae ClpB contains two well-conserved AAA domain regions (also designated ATP-1 and ATP-2), comprising 234 and 192 amino acids that share almost 80% similar and 50% identical amino acids compared to the most distantly related prokaryotes. Like E. coli, both ATP domains of M. pneumoniae ClpB possess a core region that forms the nucleotide binding pocket, containing Walker A and Walker B motifs (Fig. 1A) (37). Within the ATP-1 domain, 34 amino acid residues downstream of Walker A and 16 amino acids upstream of Walker B are separated by a central pore (CP) that contributes to protein substrate binding, with the essential tyrosine residues at position 105. A similar CP is observed within the ATP-2 domain at 37 residues downstream of Walker A and 13 residues upstream of Walker B (33). In addition, M. pneumoniae ClpB possesses a C-terminal region and a highly variable middle sequence as reported for other bacteria (11) but differs by exhibiting a shortened N terminus, as revealed by alignment with ClpBEC. The middle coiled-coil sequence is predicted to be involved in protein-protein interactions, and recent studies confirm its role in DnaK-dependent shuffling of aggregated proteins (13, 24). Besides sensing the nucleotide status of the core ATPase domain, the C-terminal domain of the ATP-2 domain has been proposed to mediate substrate interaction with the sensor and substrate discrimination domain (26). The essential Lys residues of the Walker A motif (K-68 and K-472), the Glu residues of the Walker B motif (E-133 and E-539), and the Arg residue of the GAR motif (666GAR668 in ClpB) are conserved in M. pneumoniae ClpB (Fig. 1A). However, the conserved Ser residue at amino acid 407, a putative phosphorylation site for casein kinase II with the consensus pattern SKPE in ClpBEC, is not detected in M. pneumoniae ClpB.
FIG. 1.
M. pneumoniae ClpB organization and transcription. (A) Proposed domain organization of M. pneumoniae ClpB and conserved motifs involved in ATP binding and ATP hydrolysis. M. pneumoniae ClpB retains only a shortened N-terminal domain, two nucleotide binding domains (ATP-1, positions 115 to 265; and ATP-2, positions 392 to 583) which are separated by the middle region, and a C-terminal region. Both nucleotide binding domains contain the Walker A (60GX4GKT69 and 466GX4GKT473) and Walker B (130Hy2DE133 and 536Hy2DE539) motifs, where Xn represents n amino acids of any kind and Hyn represents n hydrophobic amino acids. ATP-1 has one additional nucleotide binding motif, termed Walker B′ (249Hy2DE252). CPs (positions 102 to 111 and 511 to 519) contribute to protein substrate binding. Arg (R) residues (positions 187 and 609) are proposed to serve as Arg fingers, contacting ATP bound to an adjacent subunit. The sensor and substrate domain (SSD) (666GAR668) potentially senses the nucleotide status of ATP-2. The middle coiled-coil region is predicted to be involved in protein-protein interactions. (B) Promoter region of clpB. The bold ATG (far right) indicates the putative translational start site of clpB (mpn531). Inverted repeats on each side of the 9-nucleotide spacer sequence of the CIRCE element are shown in bold. The promoter region contains a Pribnow box (−10 site), which is an exact match for the consensus sequence (TATAAT) for the housekeeping sigma factor (σ70). A putative −35 site (boxed) was also identified upstream of the Pribnow box. The stop codon of mpn532 (TAG, far left) is 175 nucleotides upstream of the start codon of clpB. An asterisk designates the transcriptional start site (bold letter) as determined by PE. A Shine-Dalgarno (SD) sequence (underlined) is located downstream of the putative transcriptional start site. (C) Transcriptional analysis of genes surrounding clpB of M. pneumoniae. RT-PCR was performed on total RNA isolated from M. pneumoniae cells as described in Materials and Methods. A schematic of clpB and its surrounding genes is presented. Conserved hypothetical proteins (CHP) are indicated. Thin arrow lines represent regions that were amplified to confirm transcription of individual genes. Bold arrow lines represent RT-PCR products to indicate cotranscription of corresponding genes. Numbers represent the sizes of RT-PCR-amplified products.
Analysis of the clpB promoter region of M. pneumoniae clinical strain S1 revealed the presence of conserved CIRCE, suggesting that clpB is transcribed from its own promoter. Promoter mapping of S1 revealed a single transcriptional start site 17 nucleotides downstream of the CIRCE element (Fig. 1B), whereas clpB transcriptional studies with the M. pneumoniae M129 type strain revealed the presence of an additional transcriptional start site (38). clpB is preceded by genes encoding an acetate kinase (ackA, mpn533) and a hypothetical protein (mpn532) (Fig. 1B and C). Downstream of clpB are genes encoding a conserved hypothetical protein (mpn530) and a histone-like protein (mpn529). Genes mpn529 to mpn533 are transcribed in the same orientation, except for mpn530, which is transcribed on the complementary strand. To analyze the organization and expression of these genes in strain S1, we performed RT-PCR analysis using primers based on the available M. pneumoniae M129 genomic sequences (Table 1). For all reactions, genomic DNA isolated from strain S1 was used to ensure that all targeted regions were present and could be amplified with the designed primers. Evidence indicated that all genes (mpn529 to mpn534) are transcribed in the predicted orientations. Further, bicistronic transcriptional studies showed that ackA and mpn532 (Fig. 1C) were cotranscribed as predicted from sequence analysis (short intergenic region: 7 nucleotides). Even in the presence of the CIRCE element, RT-PCR demonstrated cotranscription of clpB with mpn532. Additionally, mpn530 was found to be transcribed in both orientations, and a possible cotranscription of clpB and mpn529 was demonstrated.
Cloning, expression, and purification of the M. pneumoniae ClpB protein.
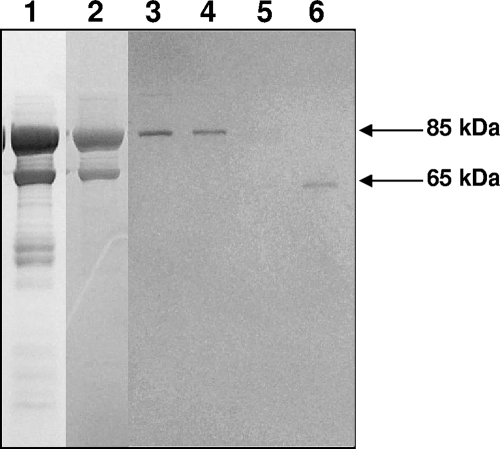
To obtain sufficient amounts of M. pneumoniae ClpB for functional characterization and generation of antiserum, we utilized the His tag expression system. ClpB featured a single UGA-encoded tryptophan at amino acid position 322, which corresponds to the TGA codon at nucleotide positions 964 to 966. The UGA was corrected, permitting clpB to be expressed as a 715-amino-acid recombinant protein fused at the NH2-terminal end with the His10 tag. The purified rClpB protein preparation was analyzed on SDS-PAGE gel, revealing a predicted 85-kDa protein; however, a 65-kDa protein and some low-molecular-weight peptides were also observed (Fig. 2, lane 1). The 85- and 65-kDa proteins were further separated by Mono Q anion exchange (Fig. 2, lane 2) and Superdex 200 gel filtration columns (Fig. 2, lanes 3 to 6) and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis and immunoblotting. MALDI-TOF analysis and immunoblots with anti-mouse rClpB and His-tagged antibodies confirmed that the 85- and 65-kDa proteins were indeed ClpB, and both employed similar translational start points (data not shown). However, MALDI-TOF analysis did not identify the C-terminal region (amino acid residues 600 to 715) in the 65-kDa ClpB peptide.
FIG. 2.
Purification of ClpB of M. pneumoniae. rClpB protein was purified through different columns as described in Materials and Methods, separated on SDS-PAGE gel, and Coomassie blue stained. Protein fractions were from a Ni-nitrilotriacetic acid column (lane 1), a Mono Q anion exchange column (lane 2), and a Superdex 200 gel filtration column eluted using a linear NaCl gradient (lanes 3 to 6).
Transcriptional and translational analysis of the M. pneumoniae ClpB gene at an elevated temperature.
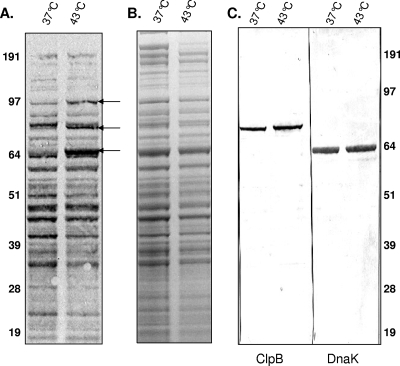
In order to further clarify ClpB properties, we assessed the effects of a temperature shift from 37°C to 43°C on the expression of ClpB at the transcriptional and translational levels and compared these effects with those for other heat shock and specific housekeeping genes (Table 2). Using the clpB probe, we detected clpB-specific mRNA in cells grown at 37°C, with a very strong signal (∼22-fold increase) in cells grown for 2 h at 43°C; lonA transcription was equally increased (Table 3). Transcription of other CIRCE-regulated genes, such as dnaK and dnaJ2, increased much less dramatically (Table 3). Except for dnaJ1 (∼7-fold increase), all other non-CIRCE regulated hsp genes (groEL, groES, and hrcA) demonstrated various responses, ranging from three- to sixfold, under the same conditions (six-, three-, and threefold increases, respectively) (Table 3). Parallel analysis of [35S]methionine-cysteine biosynthetically labeled proteins showed enhanced syntheses of three heat shock proteins (Fig. 3A). Densitometer analysis revealed 3.3-, 4.5-, and 2.1-fold increases in the syntheses of the 65-, 82-, and 94-kDa proteins, respectively. However, these proteins were not visible in parallel Coomassie blue-stained gels (Fig. 3B).
TABLE 3.
Transcriptional response of M. pneumoniae during heat shocka
| Gene | Ratio (fold) |
|---|---|
| dnaJ1 | 6.83 |
| dnaJ2 | 1.68 |
| dnaJ3 | 2.21 |
| grpE | 2.11 |
| hrcA | 2.82 |
| tig | 1.12 |
| lon | 21.71 |
| sigA | 2.39 |
| nox | 1.00 |
| dnaK | 5.96 |
| clpB | 22.28 |
| groEL | 5.54 |
| groES | 2.82 |
| eno | 1.44 |
| ftsH | 1.97 |
M. pneumoniae S1 cells grown in DMEM were placed at 43°C for 2 h. The housekeeping gene nox was used as a normalizer. Ratios represent changes in transcript amounts (at 43°C/37°C). Intensities were determined by measuring spots in individual circles. Each value represents the average for three separate experiments. Similar transcriptional responses were observed when eno was used as a normalizer.
FIG. 3.
Translational response of M. pneumoniae during heat shock. [35S]methionine-cysteine was added to M. pneumoniae S1 cells grown in DMEM at 37°C and 43°C, and levels of radiolabeled proteins were analyzed after heat shock as described in Materials and Methods. M. pneumoniae proteins were separated on 4 to 12% Nu-PAGE gel and transferred to nitrocellulose membranes and exposed to X-ray film (A), Coomassie blue stained (B), and transferred to nitrocellulose membranes and immunoblotted using anti-ClpB and anti-DnaK antisera (C). Arrows indicate the upregulated heat shock proteins.
We used hsp-specific antibodies (see Materials and Methods) to further identify the upregulated M. pneumoniae protein patterns. The abundant 82-kDa M. pneumoniae heat shock protein was immunoreactive with antibodies raised against purified rClpB (Fig. 3C). The prominent 65-kDa M. pneumoniae protein was immunoreactive with antibodies against DnaK protein (Fig. 3C). The 94-kDa M. pneumoniae protein was also identified by immunoblot analysis using LonA protease-specific antibodies (data not shown).
ATPase activities of ClpB.
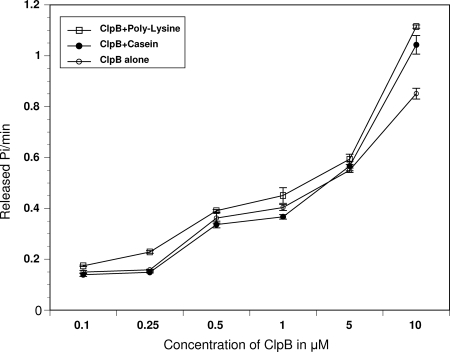
Amino acid sequence analysis predicted the presence of two well-conserved ATP domains in M. pneumoniae ClpB (Fig. 1A). To corroborate their role in ATPase activities, we analyzed rClpB in the absence or presence of the artificial substrates α-casein and poly-lysine, which are known to stimulate hydrolysis of ATP by ClpB (22, 40). M. pneumoniae rClpB exhibited ATPase activity over a range of concentrations. However, at low concentrations, there were small differences in ATPase activity in the absence and presence of casein or lysine, while at a 10 μM concentration of rClpB, we observed a 20% increase in ATPase activity in the presence of casein or lysine (Fig. 4). However, these increases were modest compared to the 5- to 20-fold stimulations observed with ClpBEC under similar conditions (22, 40).
FIG. 4.
ATPase activities of ClpB. ATPase activities of ClpB were measured at different ClpB protein concentrations as described in Materials and Methods in the absence or presence of α-casein (0.25 mg/ml) or poly-l-lysine (0.04 mg/ml).
Chaperone activities of ClpB.
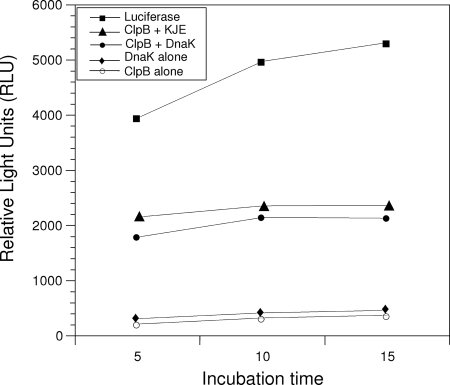
Since ClpB has the biologically important capacity to rescue proteins from an aggregated state by mediating the disaggregation of stress-damaged proteins, we tested the chaperone activity of ClpB in vitro by following ClpB/DnaK-dependent reactivation of heat-aggregated firefly luciferase (Fig. 5). rClpB exhibited significant disaggregation activity in the presence of KJE as shown by the reactivated luciferase activity. Surprisingly, ClpB also exhibited a significant disaggregation activity in the presence of mycoplasma rDnaK alone. However, mycoplasma ClpB or DnaK alone did not enhance the refolding of luciferase under similar experimental conditions (Fig. 5). KJE alone gave the same results as DnaK alone.
FIG. 5.
Chaperone activities of ClpB. The activities of firefly luciferase during its chaperone-assisted reactivation were monitored using luminescence (see Materials and Methods). Activities of rClpB alone; rClpB with DnaK; rClpB with DnaK, DnaJ, and GrpE (KJE); DnaK alone; and unheated luciferase alone are compared. The protein concentrations used were 33 nM luciferase, 2.0 μM ClpB, 2.0 μM M. genitalium rDnaK, 1.0 μM E. coli DnaK, 1.0 μM DnaJ, and 1.0 μM GrpE.
ClpB mutation impairs cell growth and alters morphology.
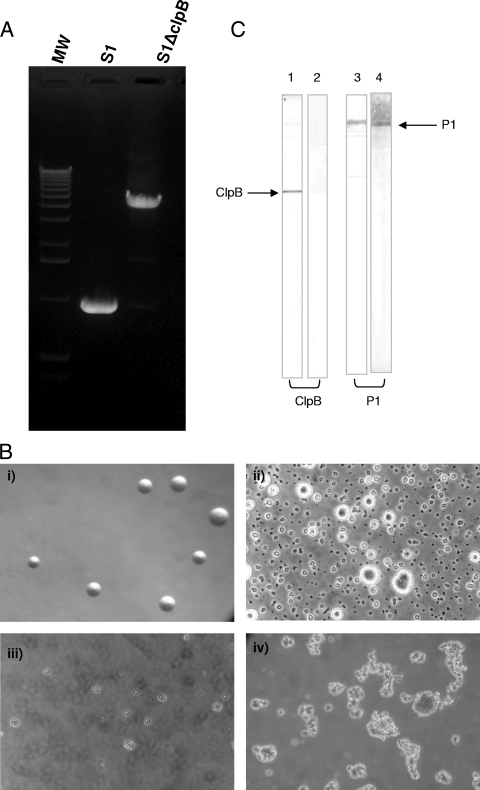
To explore the role of ClpB in M. pneumoniae, we performed Tn mutagenesis. Chromosomal DNAs from individual mutants were isolated and plasmids generated. Sequencing of one of the plasmids revealed that the Tn was integrated in direct orientation within the coding region of clpB (76 nucleotides downstream of the start codon). By using primers specific for clpB, we reconfirmed mini-Tn4001 integration (Fig. 6A) within the mutant S1ΔClpB. The ClpB mutant colony sizes were similar to those of the wild type on SP-4 agar plates, but in liquid culture, the ClpB mutant grew slowly and displayed obvious differences in cell morphology over time (Fig. 6B). Interestingly, subsequent passage of the S1ΔClpB mutant in broth resulted in no growth. However, whole-cell lysates of the first passage of liquid-grown cultures were analyzed for the presence of ClpB by immunoblotting with mouse polyclonal antibodies raised against rClpB. As expected, the ClpB mutant lacked ClpB protein whereas whole-cell lysates of the wild-type S1 strain recognized a single band at 82 kDa. For comparative purposes, we analyzed the ClpB mutant and the wild-type S1 strain for the synthesis of adhesin protein P1 (MPN141) and detected P1 in both by immunoblotting (Fig. 6C).
FIG. 6.
Construction of ClpB-null mutant M. pneumoniae S1ΔClpB. (A) PCR amplification of clpB in ClpB mutant and wild-type M. pneumoniae S1. After sequence confirmation of integration of mini-Tn4001 in clpB, the integrated region was amplified by region-specific mpn531 primers. Due to integration of Tn, an extra DNA fragment, 3 kb larger than that in wild-type M. pneumoniae S1, was amplified in mutant S1ΔClpB. MW, molecular weight. (B) Morphology of ClpB mutant of M. pneumoniae S1 strain. (i) ClpB mutant on SP-4 agar plate (similar to wild-type result [not shown]). (ii to iv) Morphologies of wild-type and ClpB mutant cells grown in SP-4 broth: (ii) M. pneumoniae S1 after 48 h (showing well-established adhering cells/colonies), (iii) ClpB mutant after 1 week (showing mostly floating cells/colonies), and (iv) ClpB mutant after 3 weeks (showing large, floating clumps). (C) ClpB expression in M. pneumoniae. Total cell lysates from wild-type M. pneumoniae strain S1 and the ClpB mutant (S1ΔClpB) were separated on SDS-PAGE gel, transferred to a nitrocellulose membrane, and treated with anti-ClpB and anti-P1 adhesin antibodies. Lanes: 1, S1 anti-ClpB antibodies; 2, S1ΔClpB anti-ClpB antibodies; 3, S1 anti-P1 antibodies; 4, S1ΔClpB anti-P1 antibodies.
Immune response against ClpB during M. pneumoniae infection.
Analysis of M. pneumoniae-infected mice showed a strong immune response against rClpB at 35 days postinfection that was undetected in control mice. Similarly, patients infected with M. pneumoniae demonstrated a strong immune response against rClpB in contrast to uninfected controls.
DISCUSSION
The heat shock response in mycoplasmas illustrates diversity in prokaryotic gene regulation, highlighted in M. pneumoniae by the presence of the CIRCE element and the absence of alternative sigma factors. Since relatively little is known about the mechanisms underlying stress response in Mollicutes, we initiated studies to characterize transcriptional and translational regulation in M. pneumoniae, focusing on the heat shock protein ClpB. We chose ClpB because of its fundamental importance in regulation and the fact that it is the only representative of the Clp AAA protein superfamily in M. pneumoniae (Fig. 1A). Further, the shortened M. pneumoniae ClpB variable N-terminal region differs from those of other bacterial ClpB proteins (8, 12, 17, 34).
Unlike M. pneumoniae reference strain M129, which uses two transcriptional start sites to express the full-length ClpB protein (38), clinical strain S1 used only one transcriptional start site, indicating the possibility of expression differences among clinical isolates (Fig. 1B). Nucleotide sequence analysis of the S1-ClpB open reading frame and its upstream promoter did not reveal any sequence variation from M129. Thus, the factors that determine the transcriptional start site of this gene remain unidentified.
We observed cotranscription of four genes of disparate functions (mpn533, mpn532, mpn530, and mpn529), along with clpB (Fig. 1C). Surprisingly, we detected transcription of clpB with the antisense strand of mpn530 (Fig. 1C). In addition, RT-PCR analysis showed a relatively small amount of transcription from the antisense strands between mpn529-530 and mpn531-530, suggesting that these genes are transcribed in single long polycistronic mRNAs. Cotranscription of functionally unrelated genes, along with antisense strands of downstream genes, has been reported for M. genitalium and M. pneumoniae, possibly due to the lack of transcriptional terminators (5).
The M. pneumoniae rClpB protein is expressed in two forms (85-kDa and 65-kDa proteins) in E. coli (Fig. 2). Monoclonal His-tagged antibodies recognized the NH2-terminal regions in both rClpB forms, indicating that the NH2 terminus is not processed in E. coli. However, MALDI-TOF analysis revealed C-terminal deletion of M. pneumoniae rClpB, possibly by E. coli proteases.
In M. pneumoniae, the conserved CIRCE element was observed only with heat-shock genes dnaJ2 (mpn021), lonA (mpn332), dnaK (mpn434), and clpB (mpn531) and not with groESL (mpn573 and mpn574). Even though a remnant of the CIRCE element has been observed upstream of groESL, it did not favor the upregulation of these products during heat shock (Fig. 3A). The absence of CIRCE-controlled GroE raises the question as to how the HrcA (MPN124) repressor is transiently inactivated to induce clpB, dnaJ, dnaK, and lonA transcription in M. pneumoniae. For example, earlier studies with Bacillus and Bradyrhizobium species suggest that the GroE-CIRCE-HrcA complex plays an important role in monitoring protein folding (23, 25). On the other hand, a nonpolar ΔdnaK mutant of Lactococcus lactis exhibited elevated levels of hsp, raising the possibility that DnaK-DnaJ chaperones could be involved in the functionality of the HrcA repressor (20). In M. pneumoniae, only the CIRCE-controlled heat shock proteins ClpB, DnaK, and LonA exhibited upregulation during heat shock (Fig. 3A). Although several other heat shock genes were increased at transcriptional levels upon heat shock (Table 3), the encoded proteins were not detected during pulse-chase experiments (Fig. 3A). Therefore, in M. pneumoniae, it appears that DnaK and ClpB chaperones assist in protein folding and renaturation and that the affinities of these chaperone systems for the respective HrcA proteins may determine their relative levels of expression and functional effectiveness in M. pneumoniae. This possibility is supported by in vitro chaperone activities of DnaK and ClpB on the refolding of luciferase (Fig. 5). The increased heat shock-driven synthesis of the CIRCE-controlled LonA protease, along with ClpB and DnaK, suggests that degradation of dissociated proteins is also an important function of these specific heat shock proteins (Fig. 3A). In addition to translational upregulation during heat shock, the display of a single ClpB band in immunoblots of whole-cell lysates of wild-type M. pneumoniae confirmed that M. pneumoniae ClpB was translationally processed in only one form (Fig. 3C and 6C), possibly due to the absence of the extended variable N-terminal region observed in bacterial ClpBs (8, 12, 17, 34).
We also observed that heat shock genes not under the control of the CIRCE element were induced transcriptionally (Table 3). However, we did not detect increases in protein levels for these upregulated genes (Fig. 3A), suggesting that a complex control network of interactions exists between specific heat shock genes and chaperones that regulate stress response.
Casein stimulates the ATPase activity of ClpB by binding to the N domain, which is abbreviated in M. pneumoniae. Even at high concentrations of ClpB, casein and lysine had limited effects on ATPase activity (Fig. 4), which is consistent with the role that the full-length N terminus of ClpB plays in other bacteria (1, 4), in contrast to the role of the shortened ClpB protein of M. pneumoniae. However, the chaperone function of M. pneumoniae ClpB appears intact, as it restores luciferase activity in the presence of KJE, which agrees with the results for ClpBΔN of Thermus thermophilus (4) and contrasts with the result for ClpBΔN of E. coli (1).
Although our ability to obtain a clpB-null mutant suggests that ClpB is not essential for the initial growth of M. pneumoniae in SP-4 agar and broth media, we observed that, upon clone selection, mutant cells lacking ClpB were fragile and aggregated during next-passage growth in SP-4 liquid culture. Like M. pneumoniae, cyanobacterium mutants lacking ClpB differ in growth from wild-type cells (31). Therefore, the absence of ClpB alone in M. pneumoniae triggers impaired growth, with subsequent failure to passage sequentially, suggesting that yet-uncharacterized critical regulatory circuits are perturbed and metabolic pathways altered by the absence of ClpB. This is consistent with the lack in M. pneumoniae of other Clp-related proteins that have diverse functions often associated with stress adaptation in bacteria. Based on our data, ClpB appears to be a key chaperone involved in resolubilization of protein aggregates in M. pneumoniae. It is possible that the slow and eventual aborted growth of the ClpB mutant may arise from its incompetence to adequately compensate for the loss of ClpB and to refold proteins for normal growth-related functions. Interestingly, ClpB elicits immune responsiveness in experimentally infected mice and M. pneumoniae-infected patients, indicating that it is both expressed and immunogenic during infection. Clearly, understanding how each heat shock protein is regulated under a range of stress conditions will contribute to an appreciation of mycoplasma survival and pathogenicity during acute and chronic stages of disease.
Acknowledgments
We thank Marianna Cagle for generating mouse polyclonal antibodies; Richard Herrmann for providing helpful comments, the pMT85 plasmid, and anti-LonA antibodies; and William Haldenwang for manuscript review and constructive suggestions. We thank Rose Garza for assistance with finalizing the manuscript.
This work was supported by National Institutes of Health grants AI045737 and AI070412 and The Kleberg Foundation.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Editor: A. Camilli
Footnotes
Published ahead of print on 8 September 2008.
REFERENCES
- 1.Barnett, M. E., A. Zolkiewska, and M. Zolkiewski. 2000. Structure and activity of ClpB from Escherichia coli. Role of the amino- and carboxyl-terminal domains. J. Biol. Chem. 27537565-37571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baseman, J. B. 1993. The cytadhesins of Mycoplasma pneumoniae and M. genitalium. Subcell. Biochem. 20243-259. [DOI] [PubMed] [Google Scholar]
- 3.Baseman, J. B., and J. G. Tully. 1997. Mycoplasmas: sophisticated, reemerging, and burdened by their notoriety. Emerg. Infect. Dis. 321-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beinker, P., S. Schlee, Y. Groemping, R. Seidel, and J. Reinstein. 2002. The N terminus of ClpB from Thermus thermophilus is not essential for the chaperone activity. J. Biol. Chem. 27747160-47166. [DOI] [PubMed] [Google Scholar]
- 5.Benders, G. A., B. C. Powell, and C. A. Hutchison III. 2005. Transcriptional analysis of the conserved ftsZ gene cluster in Mycoplasma genitalium and Mycoplasma pneumoniae. J. Bacteriol. 1874542-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, L. J., W. H. Chen, F. C. Minion, and D. Shiuan. 2008. Mycoplasmas regulate the expression of heat-shock protein genes through CIRCE-HrcA interactions. Biochem. Biophys. Res. Commun. 367213-218. [DOI] [PubMed] [Google Scholar]
- 7.Dallo, S. F., T. R. Kannan, M. W. Blaylock, and J. B. Baseman. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 461041-1051. [DOI] [PubMed] [Google Scholar]
- 8.Eriksson, M. J., and A. K. Clarke. 1996. The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 1784839-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falah, M., and R. S. Gupta. 1997. Phylogenetic analysis of mycoplasmas based on Hsp70 sequences: cloning of the dnaK (hsp70) gene region of Mycoplasma capricolum. Int. J. Syst. Bacteriol. 4738-45. [DOI] [PubMed] [Google Scholar]
- 10.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270397-403. [DOI] [PubMed] [Google Scholar]
- 11.Gottesman, S., C. Squires, E. Pichersky, M. Carrington, M. Hobbs, J. S. Mattick, B. Dalrymple, H. Kuramitsu, T. Shiroza, T. Foster, et al. 1990. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc. Natl. Acad. Sci. USA 873513-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandvalet, C., V. de Crecy-Lagard, and P. Mazodier. 1999. The ClpB ATPase of Streptomyces albus G belongs to the HspR heat shock regulon. Mol. Microbiol. 31521-532. [DOI] [PubMed] [Google Scholar]
- 13.Haslberger, T., J. Weibezahn, R. Zahn, S. Lee, F. T. Tsai, B. Bukau, and A. Mogk. 2007. M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol. Cell 25247-260. [DOI] [PubMed] [Google Scholar]
- 14.Hedreyda, C. T., K. K. Lee, and D. C. Krause. 1993. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid 30170-175. [DOI] [PubMed] [Google Scholar]
- 15.Hess, H. H., and J. E. Derr. 1975. Assay of inorganic and organic phosphorus in the 0.1-5 nanomole range. Anal. Biochem. 63607-613. [DOI] [PubMed] [Google Scholar]
- 16.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 244420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingmer, H., F. K. Vogensen, K. Hammer, and M. Kilstrup. 1999. Disruption and analysis of the clpB, clpC, and clpE genes in Lactococcus lactis: ClpE, a new Clp family in gram-positive bacteria. J. Bacteriol. 1812075-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kannan, T. R., and J. B. Baseman. 2006. ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. Proc. Natl. Acad. Sci. USA 1036724-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kannan, T. R., D. Provenzano, J. R. Wright, and J. B. Baseman. 2005. Identification and characterization of human surfactant protein A binding protein of Mycoplasma pneumoniae. Infect. Immun. 732828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koch, B., M. Kilstrup, F. K. Vogensen, and K. Hammer. 1998. Induced levels of heat shock proteins in a dnaK mutant of Lactococcus lactis. J. Bacteriol. 1803873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanzetta, P. A., L. J. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 10095-97. [DOI] [PubMed] [Google Scholar]
- 22.Liu, Z., V. Tek, V. Akoev, and M. Zolkiewski. 2002. Conserved amino acid residues within the amino-terminal domain of ClpB are essential for the chaperone activity. J. Mol. Biol. 321111-120. [DOI] [PubMed] [Google Scholar]
- 23.Minder, A. C., H. M. Fischer, H. Hennecke, and F. Narberhaus. 2000. Role of HrcA and CIRCE in the heat shock regulatory network of Bradyrhizobium japonicum. J. Bacteriol. 18214-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogk, A., T. Haslberger, P. Tessarz, and B. Bukau. 2008. Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem. Soc. Trans. 36120-125. [DOI] [PubMed] [Google Scholar]
- 25.Mogk, A., G. Homuth, C. Scholz, L. Kim, F. X. Schmid, and W. Schumann. 1997. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 164579-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mogk, A., C. Schlieker, C. Strub, W. Rist, J. Weibezahn, and B. Bukau. 2003. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J. Biol. Chem. 27817615-17624. [DOI] [PubMed] [Google Scholar]
- 27.Mogk, A., T. Tomoyasu, P. Goloubinoff, S. Rudiger, D. Roder, H. Langen, and B. Bukau. 1999. Identification of thermolabile Escherichia coli proteins: prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 186934-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motohashi, K., Y. Watanabe, M. Yohda, and M. Yoshida. 1999. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc. Natl. Acad. Sci. USA 967184-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musatovova, O., S. Dhandayuthapani, and J. B. Baseman. 2006. Transcriptional heat shock response in the smallest known self-replicating cell, Mycoplasma genitalium. J. Bacteriol. 1882845-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musatovova, O., S. Dhandayuthapani, and J. B. Baseman. 2003. Transcriptional starts for cytadherence-related operons of Mycoplasma genitalium. FEMS Microbiol. Lett. 22973-81. [DOI] [PubMed] [Google Scholar]
- 31.Porankiewicz, J., and A. K. Clarke. 1997. Induction of the heat shock protein ClpB affects cold acclimation in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 1795111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy, S. P., W. G. Rasmussen, and J. B. Baseman. 1996. Isolation and characterization of transposon Tn4001-generated, cytadherence-deficient transformants of Mycoplasma pneumoniae and Mycoplasma genitalium. FEMS Immunol. Med. Microbiol. 15199-211. [DOI] [PubMed] [Google Scholar]
- 33.Schlieker, C., J. Weibezahn, H. Patzelt, P. Tessarz, C. Strub, K. Zeth, A. Erbse, J. Schneider-Mergener, J. W. Chin, P. G. Schultz, B. Bukau, and A. Mogk. 2004. Substrate recognition by the AAA+ chaperone ClpB. Nat. Struct. Mol. Biol. 11607-615. [DOI] [PubMed] [Google Scholar]
- 34.Squires, C. L., S. Pedersen, B. M. Ross, and C. Squires. 1991. ClpB is the Escherichia coli heat shock protein F84.1. J. Bacteriol. 1734254-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Asseldonk, M., A. Simons, H. Visser, W. M. de Vos, and G. Simons. 1993. Cloning, nucleotide sequence, and regulatory analysis of the Lactococcus lactis dnaJ gene. J. Bacteriol. 1751637-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waites, K. B., and D. F. Talkington. 2004. Mycoplasma pneumoniae and its role as a human pathogen. Clin. Microbiol. Rev. 17697-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiner, J., III, R. Herrmann, and G. F. Browning. 2000. Transcription in Mycoplasma pneumoniae. Nucleic Acids Res. 284488-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiner, J., III, C. U. Zimmerman, H. W. Gohlmann, and R. Herrmann. 2003. Transcription profiles of the bacterium Mycoplasma pneumoniae grown at different temperatures. Nucleic Acids Res. 316306-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woo, K. M., K. I. Kim, A. L. Goldberg, D. B. Ha, and C. H. Chung. 1992. The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J. Biol. Chem. 26720429-20434. [PubMed] [Google Scholar]
- 41.Zimmerman, C. U., and R. Herrmann. 2005. Synthesis of a small, cysteine-rich, 29 amino acids long peptide in Mycoplasma pneumoniae. FEMS Microbiol. Lett. 253315-321. [DOI] [PubMed] [Google Scholar]
- 42.Zolkiewski, M. 1999. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J. Biol. Chem. 27428083-28086. [DOI] [PubMed] [Google Scholar]
- 43.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 1761359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]