Abstract
The predominant organizational state of bacteria in nature is biofilms. Biofilms have been shown to increase bacterial resistance to a variety of stresses. We demonstrate for the first time that the anaerobic gram-positive pathogen Clostridium perfringens forms biofilms. At the same concentration of glucose in the medium, optimal biofilm formation depended on a functional CcpA protein. While the ratio of biofilm to planktonic growth was higher in the wild type than in a ccpA mutant strain in middle to late stages of biofilm development, the bacteria shifted from a predominantly biofilm state to planktonic growth as the concentration of glucose in the medium increased in a CcpA-independent manner. As is the case in some gram-negative bacteria, type IV pilus (TFP)-dependent gliding motility was necessary for efficient biofilm formation, as demonstrated by laser confocal and electron microscopy. However, TFP were not associated with the bacteria in the biofilm but with the extracellular matrix. Biofilms afforded C. perfringens protection from environmental stress, including exposure to atmospheric oxygen for 6 h and 24 h and to 10 mM H2O2 for 5 min. Biofilm cells also showed 5- to 15-fold-increased survival over planktonic cells after exposure to 20 μg/ml (27 times the MIC) of penicillin G for 6 h and 24 h, respectively. These results indicate C. perfringens biofilms play an important role in the persistence of the bacteria in response to environmental stress and that they may be a factor in diseases, such as antibiotic-associated diarrhea and gas gangrene, that are caused by C. perfringens.
Clostridium perfringens is a gram-positive, anaerobic pathogen that causes a number of important diseases in humans, including gas gangrene and intestinal diseases, as well as a variety of infections in animals (43). C. perfringens is also one of the most ubiquitous bacteria in natural environments and can be found anywhere mammals are present, including virtually all soils worldwide (26, 35, 47). In order to persist in the environment, C. perfringens, an obligate anaerobe, must have a means to deal with environmental stresses, such as oxygen, desiccation, and bactericidal compounds. Long-term resistance to these environmental stresses can be achieved by the formation of endospores. However, endospores exist in a dormant state, so they cannot adapt quickly to changes in the environment. Bacteria have evolved the ability to form biofilms, which provide some of the same protections afforded by endospores but can more quickly adapt to changes in the environment. Biofilms, the predominant state of bacteria in nature (13), are adherent bacterial populations that are encapsulated in a matrix composed of polysaccharides, nucleic acids, and proteins (7). Biofilms have been shown to provide increased resistance to oxidative stress and to antibiotics and other bactericidal agents (22).
In addition to environmental persistence, biofilms are also a factor in pathogenesis. The ability to form biofilms is a well-known trait of a number of pathogens, such as Vibrio cholerae (57), Escherichia coli (40), and Legionella pneumophila (42). Biofilms have been shown to be a major factor in the pathogenesis of Pseudomonas aeruginosa lung infections (12) and Staphylococcus epidermidis infections involving medical implants (19).
In the last several years, biofilms have been shown to comprise an important aspect of microbial persistence in the human colon (27, 28, 41). C. perfringens, which is a consistent but not major constituent of the microbiota in the human intestine (9), causes several intestinal diseases, including acute food-borne diarrhea, spontaneous non-food-borne diarrhea, and, most notably, antibiotic-associated diarrhea (AAD) (2, 10, 11). If the bacterium persists in the intestine in a biofilm or as endospores, the anticipated resistance to antimicrobials may explain how the bacterium can cause AAD after disruption of the normal microbiota by antibiotic treatment.
Regulation of biofilm production in gram-positive bacteria has been shown to be due to multiple factors, one of which is the availability of carbohydrates in the medium. A key regulator of the response to carbohydrate limitation is the catabolite control protein, CcpA (51). CcpA is a global transcriptional regulator that becomes active for DNA binding after formation of a complex with the Hpr protein phosphorylated at a serine residue (51). The phosphorylation of Hpr is controlled by the activity of a kinase that responds to the intracellular concentration of fructose 1-6-bisphosphate, which varies based on the rate of glucose transport into the cell (55). CcpA has been shown to be involved in biofilm formation in gram-positive bacteria, such as Bacillus subtilis and Staphylococcus aureus (45, 48). We have shown in previous work that a CcpA− strain of C. perfringens (strain SM120) is deficient in sporulation and has altered glucose-dependent regulation of several virulence factors, including capsule synthesis and enterotoxin production, as well as a defect in the initiation of gliding motility (52; J. J. Varga and S. B. Melville, unpublished results).
To our knowledge, C. perfringens has never been reported to form biofilms. Recently, we described type IV pilus (TFP)-mediated gliding motility in C. perfringens (53), which had not previously been clearly demonstrated in a gram-positive bacterium. TFP are thin filaments consisting of polymers of a single peptide subunit, PilA. The pilins are polymerized via a complex of proteins, including PilD (a signal peptidase that recognizes PilA), PilB (an extension motor), PilC (a membrane protein), and PilT (a retraction motor) (16, 30, 36). Since TFP are frequently involved in biofilm formation (23, 38, 50), we tested C. perfringens for the ability to form biofilms. The role of TFP in biofilm formation, the contribution of biofilms to stress resistance, and the roles of carbohydrates and CcpA-dependent catabolite repression in the regulation of biofilm formation were investigated.
MATERIALS AND METHODS
Growth of bacterial strains.
All bacterial strains used in this study are listed in Table 1. C. perfringens strains were routinely grown in PGY (per liter, 30 g proteose peptone no. 3, 20 g glucose, 10 g yeast extract, 1 g sodium thioglycolate) (31), with 15 g/liter of agar for solid medium. When appropriate, the medium was supplemented with 20 μg/ml of chloramphenicol or 30 μg/ml of erythromycin. The standard culture conditions were incubation in a Coy anaerobic chamber (Coy Laboratory Products, Grass Lake, MI) at 37°C.
TABLE 1.
Bacterial strains used in this study
| C. perfringens strain | Relevant characteristics | Source |
|---|---|---|
| ATCC 13124 | Type A strain, type strain, gangrene | ATCC |
| JGS 1495 | Type C strain | G. Songer |
| JGS 1721 | Type D strain | G. Songer |
| JGS 1987 | Type E strain | G. Songer |
| SM101 | Type A strain, food poisoning, transformable | 58 |
| SM120 | ccpA mutant derivative of SM101 | 52 |
| SM125 | pilC mutant derivative of strain 13, TFP−, nonmotile | 53 |
| SM126 | pilT mutant derivative of strain 13, TFP−, nonmotile | 53 |
| 13 | Type A strain, gangrene isolate, highly transformable | 29 |
Biofilm growth.
For the generation of biofilms, overnight PGY cultures of C. perfringens were washed in phosphate-buffered saline (PBS) and resuspended to an optical density at 600 nm (OD600) of 0.1 as measured with a Genesys 10 UV spectrophotometer. For visualization by light microscopy, 200-μl cultures were grown in Lab-Tek II Cell Culture-treated eight-chamber microscope slides (Nalgene Nunc International, Rochester, NY) using brain heart infusion (BHI) broth (Difco, Livonia, MI). For other applications, 400-μl biofilm cultures were grown in 24-well polystyrene tissue culture plates using T-Soy Broth (TSB) (Difco, Livonia, MI) supplemented with filter-sterilized carbohydrates. To allow biofilms to form and to prevent the evaporation of liquid, cultures were incubated anaerobically at 30°C for up to 5 days in a sealed container.
Determination of biomass distribution in biofilms.
The distribution between planktonic (free-floating) and sessile (biofilm) cells was determined using protocols derived from previously published methods (6, 17, 48). The supernatant from each well of a biofilm culture in a 24-well plate was transferred to three wells in a 96-well plate. The wells of the 24-well plate were then gently washed twice with PBS prior to incubation for 30 min at room temperature with 400 μl of filter-sterilized 1% crystal violet. Excess crystal violet was removed from the wells, followed by two additional PBS washes. The crystal violet was extracted by adding 400 μl methanol to each well and incubation for 30 min at room temperature, after which the liquid was transferred to three separate wells in a second 96-well plate. Background due to nonspecific staining of the 24-well plate by crystal violet was accounted for by subtracting the values obtained from wells that contained medium but were not inoculated with bacteria. The OD600 of the culture supernatants and the A570 of the methanol-extracted dye were then measured in a Spectrafluor Plus plate reader (Tecan, Salzburg, Austria). The value of the A570/OD600 ratio was used to represent the ratio of sessile biomass to planktonic cells (17).
Assessment of biofilm resistance to oxidative stress and antibiotics.
Biofilms of C. perfringens 13 were prepared as described above. TSB was supplemented with 10 mM lactose to optimize biofilm formation. After 3 days, the sessile and planktonic populations were quantitated from one set of tissue culture plates. Initial planktonic cell counts were determined by washing the pooled supernatants of three wells in PBS, diluting the samples, and plating them on PGY to determine the number of CFU. Biofilm cell populations were determined by adding PBS to the wells of the plate, detaching the cells by thorough scraping with a sterile pipette tip, pooling the contents of three wells, washing the cells in PBS, and plating them on PGY for determination of the number of CFU present in the biofilm.
To determine the resistance capabilities of planktonic and biofilm cells, the supernatants from biofilm cultures were washed and resuspended in PBS (for H2O2 and atmospheric-oxygen resistance) or TSB with lactose (for antibiotic resistance) and transferred to fresh tissue culture plates. The supernatants were replaced with PBS (for H2O2 and oxygen resistance) or TSB with lactose (for antibiotic resistance) in the original biofilm culture.
After the stress treatment, the samples were processed as described for the initial cell counts. Percent survival was calculated by dividing the posttreatment number of CFU by the initial number of CFU. All samples were pools of three wells, and the data are presented as the average of a minimum of two independent experiments, with error bars representing 1 standard deviation of the mean.
Visualization of biofilms.
To obtain images of biofilms formed by C. perfringens, the bacteria were cultured in eight-chamber Lab-Tek II glass slides. The cells were grown in BHI broth in each chamber for 24 h at 30°C under anaerobic conditions. The slides were then removed from the anaerobic chamber, and the wells were washed two times with PBS to remove unattached cells. PBS containing the fluorescent BacLight Live/Dead stain mixture (Molecular Probes, Carlsbad, CA), which contains the nucleic acid stains Syto 9 and propidium iodide, was added to the wells to allow visualization of individual bacteria by laser confocal microscopy. A Zeiss LSM 510 laser confocal microscope was used to collect three-dimensional images of the biofilms. An argon laser set at 488 nm was used to excite the Syto 9 dye, and a HeNe laser set at 543 nm was used to excite the propidium iodide stain. Confocal microscopy was performed on biofilms formed by C. perfringens strain 13 and derivatives lacking TFP, SM125 (pilC mutant), and SM126 (pilT mutant) (53). After 3 days of growth, biofilms contained in eight-chamber microscope slides were gently washed twice with PBS prior to being stained with BacLight Live/Dead stain (Molecular Probes) and visualized with a Zeiss LSM 510 laser scanning microscope. Enumeration of fluorescently labeled bacteria in biofilms was done using the CellC software package, which analyzes digital images to quantitate fluorescent particles (46).
To test for biofilm formation on animal tissues, SM101 cells were incubated with chopped-meat medium pellets for 3 days and stained with BacLight Live/Dead and 50 μg/ml calcofluor white dye, which is specific for polysaccharides (3). At that time, the meat particles were removed from the liquid suspension, and biofilms were observed on the meat particles using laser confocal microscopy with a UV laser at 364 nm to excite the calcofluor white dye, an argon laser set at 488 nm to excite the Syto 9 dye, and a HeNe laser set at 543 nm to excite the propidium iodide stain.
Field emission scanning electron microscopy (FE-SEM) was performed to visualize the surfaces of C. perfringens biofilms. C. perfringens strains were grown for 2 to 4 h in BHI broth, and a 100-μl droplet of the cell culture was added to a glass coverslip and allowed to incubate anaerobically at 37°C for 12 h to 24 h, depending on the experiment. Evaporation was prevented by sealing the coverslips in a plastic container with water reservoirs. The glass coverslips were then immersed in 2.5% glutaraldehyde to fix the cells, rinsed two times in distilled water, and allowed to dry. Samples were sputter coated under vacuum with a 5-nm-thick layer of gold using a Cressington 208 h rotary sputter coater. The coverslips and grids were then observed on a Leo 1550 field emission-scanning electron microscope with a beam acceleration of 1 kV.
Immunofluorescent staining of type IV pili also utilized 3-day-old biofilms formed in eight-chamber slides. Samples were fixed anaerobically for 15 min in 2.5% paraformaldehyde. The fixative was removed, and the samples were washed twice with 2% bovine serum albumin (BSA)/PBS. The samples were then incubated with a 1:100 dilution of rabbit polyclonal antibodies against the C. perfringens strain 13 pilin subunits PilA1 and PilA2 (53) for 1 h at 37°C; washed two times with BSA/PBS, followed by incubation with goat anti-rabbit antibodies (diluted 1:50 in BSA/PBS) conjugated to Alexa Fluor 588 (Molecular Probes); and incubated with the sample for 1 h at 37°C. After two washes with BSA/PBS to remove unbound antibodies, the samples were visualized on an Olympus IX81 upright fluorescence microscope at ×400 magnification, controlled by SlideBook software (Intelligent Imaging Innovations, Inc.) operating a Hamamatsu OHC1 charge-coupled device camera.
RESULTS
Visualization of C. perfringens biofilms.
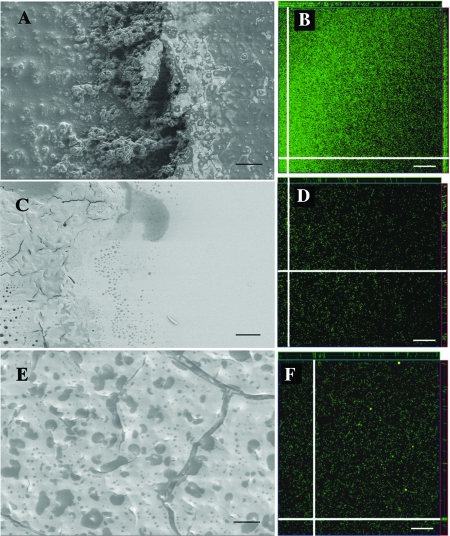
C. perfringens strain 13 cells were tested for biofilm formation after incubation in BHI broth on glass coverslips, examined for evidence of biofilm formation using FE-SEM, and incubated in eight-chamber microscope slides for laser confocal microscopy (Fig. 1A and B). In FE-SEM images, strain 13 appeared to form a flat biofilm. Underneath the surface of the biofilm, cells could be seen encased in a dense matrix material (Fig. 1A). Three-dimensional laser confocal microscopy images of fluorescently labeled bacteria revealed that the biofilms were densely packed with bacteria and 30 to 40 μm thick (see the z sections at the edges of Fig. 1B). Despite having been exposed for 2 to 3 h to aerobic conditions in the confocal microscope, the vast quantity of C. perfringens organisms in the biofilm appeared to be alive, as judged by green staining with the BacLight Live/Dead stain (i.e., live cells were green; dead cells would appear red) (Fig. 1B).
FIG. 1.
Appearances of biofilms formed by wild-type and mutant strains of C. perfringens on glass and plastic surfaces. The FE-SEM images show the relatively flat surfaces of biofilms formed by the wild-type (A), pilT mutant (C), and pilC mutant (E) strains. (A) The right side of the image shows the surface of the biofilm; in the center, the biofilm has been torn away, revealing a dense mixture of cells and matrix material. (C) The biofilm is present on the left side of the image, and the right side shows the glass surface used as a substrate for biofilm formation. (E) The entire surface shown is covered by a biofilm, with a crack in the surface visible on the right side. The material beneath the crack appears to be less thick and dense than the material under the surface of the biofilm formed by the wild-type strain shown in panel A. (B, D, and F) Laser confocal microscopy images of fluorescently labeled (Syto 9 and propidium iodide) wild-type (B), pilT mutant (D), and pilC mutant (F) bacteria. Representative images of quadruplicate samples are shown as single x-y sections. The white lines in each image indicate the locations of the z sections shown at the top or right edge of the corresponding figure. The thicknessesof the z sections shown at the edges correspond to the depths of the biofilms. Using compiled z sections, the thicknesses of the biofilms in panels B, D, and F were between 30 and 40 μm. Panels A, C, and E, bars = 10 μm; panels B, D, and F, bars = 20 μm.
In order to determine if C. perfringens can form a biofilm on a more natural surface than a tissue culture plate or glass slide, SM101 cells were incubated with chopped-meat medium pellets for 3 days and stained with the BacLight Live/Dead stain and calcofluor white dye. A representative image of a biofilm formed on a meat particle is shown in Fig. 2. The bacterial cells were present on the meat particle in significant numbers only in the region in which a biofilm was formed, identifiable as positive for calcofluor white staining, which represented an extended carbohydrate polysaccharide matrix. Meat particles that were not inoculated with bacteria did not show visible evidence of biofilms (data not shown).
FIG. 2.
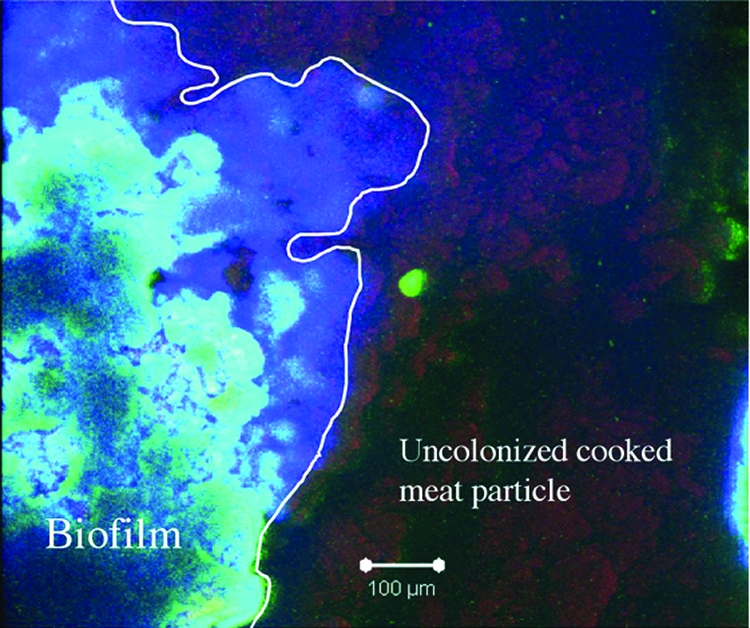
Laser confocal microscopy image of a C. perfringens biofilm formed on a cooked meat pellet. The sample was incubated with BacLight Live/Dead stain and calcofluor white, which binds to polysaccharides. The white line delineates the extent of the biofilm formed on the pellet. The colors visible in the image correspond to the following materials: red, meat pellet; blue, polysaccharides (calcofluor white stained); green, live bacteria.
TFP are required for efficient biofilm formation.
We have previously described derivatives of strain 13 in which pilC (strain SM125) and pilT (strain SM126) mutants were shown to lack both gliding motility and TFP on the surfaces of the bacteria (53). Strains SM125 and SM126 were examined for biofilm formation using electron and laser confocal microscopes, as described above for strain 13. The surfaces of biofilms made by strains SM125 (Fig. 1C) and SM126 (Fig. 1E) appeared in the FE-SEM to be flat, similar in appearance to those of the wild-type strain (Fig. 1B). However, the amounts of cellular and matrix material beneath the surfaces of the biofilms, visible at the edges and in cracks in the surfaces of the biofilms, appeared to be significantly less than that seen with the wild-type strain (compare Fig. 1A to C and E). This observation was confirmed by examining laser confocal microscopy images, which showed that the pilC and pilT mutants had five and six times, respectively, fewer bacteria present in the biofilms they made than in those produced by the wild-type strain (compare the fluorescent staining in Fig. 1B to D and F). However, the pilC and pilT mutants still produced biofilms that were 30 to 40 μm thick, as judged by examination of the z sections in Fig. 1D and F. We interpret the FE-SEM images and laser confocal images to indicate that the pilC and pilT mutants produce biofilms that, when hydrated (as seen in the laser confocal microscopy experiments), are the same thickness as those made by the wild-type strain, but the biofilms contain significantly fewer bacteria and less intercellular matrix material than those of the wild-type strain.
Pilin subunits are present but do not colocalize with C. perfringens in biofilms.
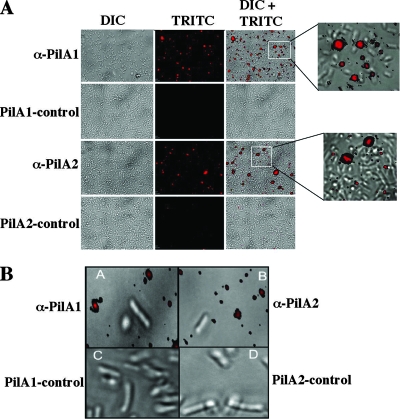
Since TFP were needed for efficient biofilm formation, we performed immunofluorescent staining of biofilms to determine if the two pilin proteins shown to be made by C. perfringens, PilA1 and PilA2, were present. Biofilms were incubated with polyclonal antibodies to C. perfringens strain 13 PilA1 and PilA2 pilin subunits (53). Figure 3 shows representative images displaying the results of the immunofluorescent staining. Significant levels of fluorescence were seen using both anti-PilA1 and -PilA2 antibodies, while little or no fluorescence was visible using the prebleed serum controls. Higher-magnification images of regions with high cell density (Fig. 3A, insets) showed that pilin proteins were present, but the bacteria were too densely packed to determine if the pili were cell associated. Therefore, we examined images from lower-cell-density areas of the biofilm and observed that the majority of PilA1- and PilA2-dependent fluorescence did not colocalize with the bacteria (Fig. 3B), suggesting pili had been shed from the bacterial surfaces inside the biofilm.
FIG. 3.
Localization of PilA in C. perfringens biofilms. (A) Three-day-old C. perfringens strain 13 biofilms were incubated with antibodies to either the C. perfringens PilA1 or PilA2 protein and then stained with Alexa Fluor 594-conjugated goat anti-rabbit antibodies. Images labeled as PilA1-control and PilA2-control were biofilms incubated with prebleed sera for PilA1 and PilA2, respectively. (B) High-magnification images showing differential interference contrast (DIC) and tetramethyl rhodamine isothiocyanate (TRITC) staining in biofilms. Antibodies directed against PilA proteins did not colocalize with bacteria.
All sequenced strains of C. perfringens can form biofilms.
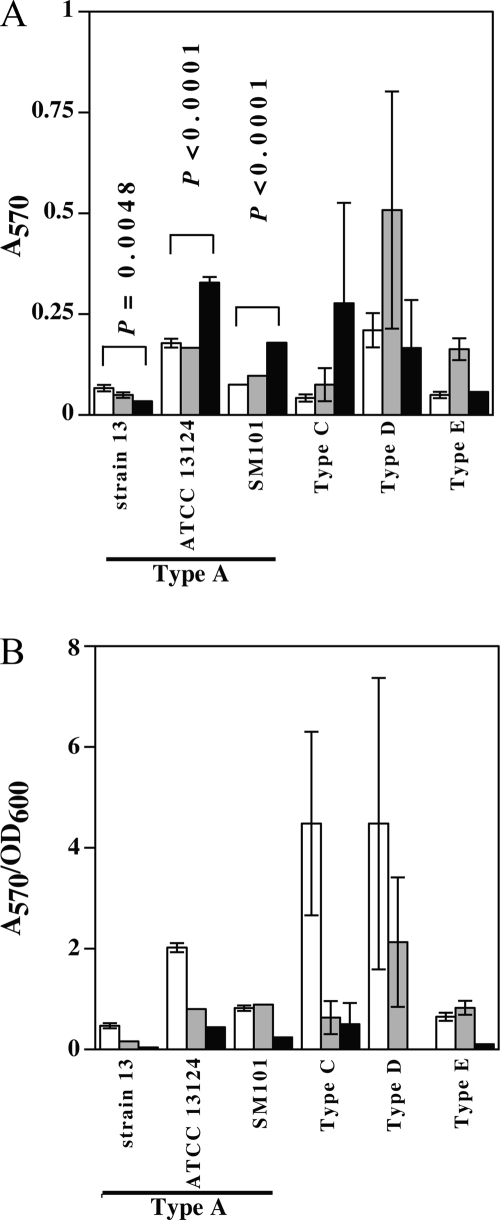
C. perfringens strains of biotypes A (ATCC 13124, 13, and SM101), C, D, and E were assayed for the ability to form biofilms in various concentrations of glucose. Sessile biomass (biofilm cells) was measured by staining the biofilm with crystal violet, extracting the stain with methanol, and measuring the A570 of the methanol extract. All tested strains demonstrated the ability to form biofilms after 3 days in the absence of glucose and in the presence of 10 or 100 mM glucose (Fig. 4A). The strains varied in the ability to form biofilms in the presence of increased glucose, with strains ATCC 13124 and SM101 showing significantly higher levels of biofilm formation in the presence of the highest level (100 mM) of glucose than in the absence of glucose. Strain 13 showed the opposite effect, with significantly less biofilm made in the presence of 100 mM glucose than in the absence of glucose (Fig. 4A), while the other three strains did not show a statistically significant difference under these conditions (Fig. 4A).
FIG. 4.
Biofilm formation by multiple C. perfringens toxinotypes. (A) Biofilm formation in 24-well plates, measured as described in Materials and Methods. (B) Biofilm (A570)/planktonic-growth (OD600) ratio. White bars, 0 mM glucose; gray bars, 10 mM glucose; black bars, 100 mM glucose. In panel A, the P values shown represent a statistical comparison, using Student's t test, of biofilm formation between 0 mM and 100 mM glucose concentrations. For types C, D, and E, no statistically significant difference was found at these glucose concentrations. The error bars represent standard deviations.
Microscopic observations of wells containing biofilms indicated that planktonic cells were also present in various amounts (data not shown). To quantitate how the ratio of cells shifted from biofilms to planktonic cells in response to different levels of glucose, the ratio of biofilm to planktonic cells was measured (see Materials and Methods). Glucose repressed biofilm formation relative to planktonic cells in all six strains (Fig. 4B). The highest ratios of biofilm cells to planktonic cells were seen in the absence of glucose with strains ATCC 13124 and the type C and type D strains (Fig. 4B). Given that more biofilm material was produced by strains ATCC 13124 and SM101 in response to higher glucose levels (Fig. 4A), it appears that both biofilm and planktonic cells increased with increasing glucose but that planktonic cells increased to a greater extent, which would account for the A570/OD600 ratios for these strains seen in Fig. 4B. For strain 13, in which biofilm formation was repressed by glucose (Fig. 4A), the number of planktonic cells increased in high levels of glucose, leading to the more dramatic shift in this ratio seen with this strain (Fig. 4B).
Roles of glucose and ccpA in limiting biofilm formation.
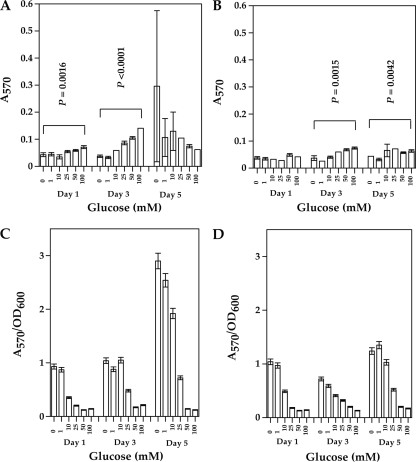
In B. subtilis, a mutation in ccpA was shown to alleviate glucose repression of biofilm formation (48). Since the results of experiments shown in Fig. 4 suggested that glucose has the ability to stimulate biofilm formation in some strains but also to lower the amounts of biofilm relative to planktonic cells, we investigated the role of CcpA in catabolite control of biofilm formation in C. perfringens strain SM101 and a previously described ccpA mutant derivative of C. perfringens SM101, strain SM120 (52). SM101 and its ccpA mutant derivative were used in these experiments because the role of CcpA in catabolite regulation of sporulation, toxin production, and capsule synthesis in this strain had been previously determined (52). C. perfringens biofilm formation was determined by testing the effects of adding increasing concentrations of glucose and measuring biofilm formation after 1-, 3-, and 5-day periods (Fig. 5). At the higher concentrations of glucose (25, 50, and 100 mM), the wild-type strain formed more biofilm mass than the ccpA mutant strain, except at day 5 (Fig. 5A and B), indicating that CcpA is important for maximal biofilm formation. A statistically significant difference in biofilm formation at 0 and 100 mM glucose was seen in the wild-type strain at days 1 and 3 and in the ccpA mutant strain at days 3 and 5 (Fig. 5A and B). After 5 days at 0 mM glucose, formation of biofilms was highly variable in the wild-type strain, with some wells containing very small amounts of biofilm material, which is indicated by the large error bars for the standard deviation value. We believe this represents the dissolution of previously formed biofilms in some of the wells. The ratio of biofilm to planktonic growth was higher in the wild type than the ccpA mutant strain at days 3 and 5 at lower glucose concentrations (0 to 25 mM); however, the ratio decreased with increasing concentrations of glucose in both strains (Fig. 5C and D).
FIG. 5.
Effects of glucose concentrations on biofilm formation in the wild type and a ccpA mutant strain of C. perfringens. (A and B) Biofilm formation by strains SM101 (wild type) (A) and SM120 (ccpA mutant) (B). (C and D) Ratio of biofilm/planktonic growth by strains SM101 (wild type) (C) and SM120 (ccpA mutant) (D). In panels A and B, the P values shown represent a statistical comparison, using Student's t test, of biofilm formation between 0 mM and 100 mM glucose concentrations. No statistically significant difference was found at these glucose concentrations for the wild-type strain at day 5 and the ccpA mutant strain at day 1. The error bars represent standard deviations.
C. perfringens biofilms protect cells from environmental stresses.
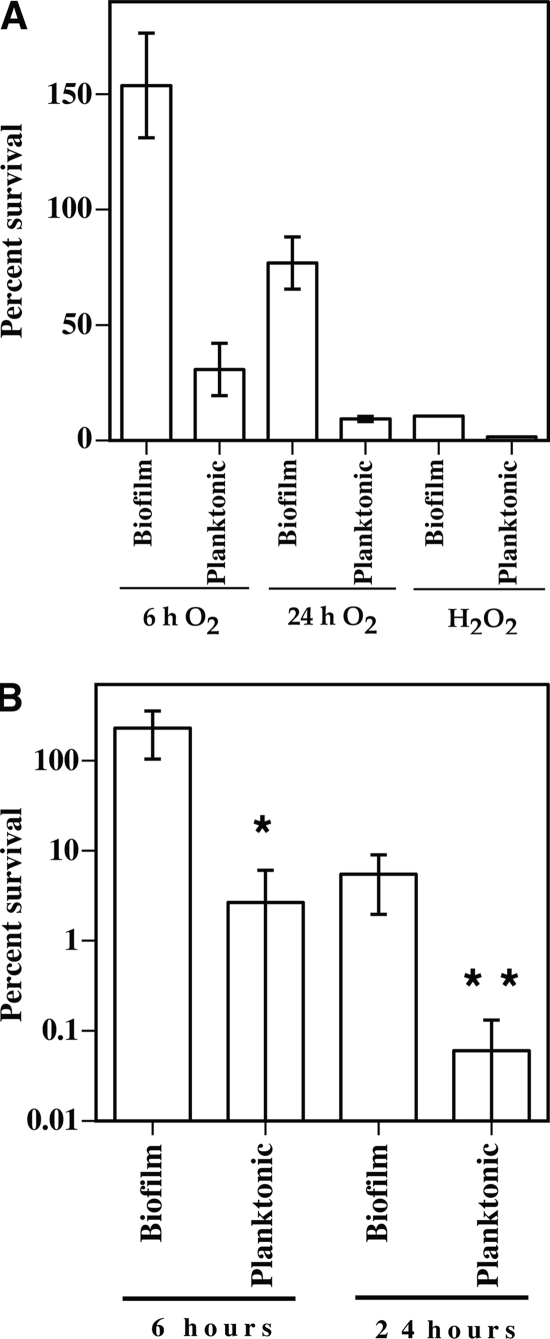
The abilities of C. perfringens biofilms and planktonic cells to survive oxidative stress in the form of atmospheric oxygen and hydrogen peroxide were assayed. Cells of C. perfringens strain 13 were subjected to atmospheric oxygen for 6 h and 24 h or to 10 mM H2O2 for 5 min (15). Strain 13 was used for these experiments because it is a gangrene-causing strain and antibiotics are used in the treatment of gangrene but not acute food poisoning caused by strains such as SM101. C. perfringens is known to be aerotolerant (8), and our results indicated that the survival rates of planktonic cells in atmospheric oxygen were 30.6% and 9.2% for 6 h and 24 h, respectively (Fig. 6A). However, biofilm cells increased in number to 153% by 6 h, and after 24 h, biofilm survival was 76.6% (Fig. 6A), five and eight times the number seen in planktonic cells. To measure protection against a more stringent oxidative stress, cells were exposed to H2O2, in which biofilm cells had a sevenfold-higher rate of survival than planktonic cells, 10.5% to 1.5% (Fig. 6A).
FIG. 6.
Biofilms provide protection against oxidative and antibiotic stresses. (A) C. perfringens 3-day-old cultures were separated into biofilm and planktonic fractions and then exposed to various oxidative stresses, as indicated. (B) C. perfringens 3-day-old cultures were separated into biofilm and planktonic fractions and then exposed to 20 μg/ml penicillin G (27 times the MIC [37]) for the times indicated. In panel A, the P values of differences in survival of biofilm versus planktonic cells were each <0.002, by Student's t test, under the conditions described for each experiment. In panel B, differences in survival of biofilm and planktonic cells at each time were compared using Student's t test. *, P = 0.0156; **, P = 0.0254. The error bars represent standard deviations.
C. perfringens is extremely sensitive to penicillin G, a bactericidal antibiotic that inhibits cell wall synthesis. The MIC for strain 13, used in this study, is 0.75 μg/ml (37). Exposure to high levels of penicillin (20 μg/ml) for 6 h failed to kill C. perfringens cells in a biofilm (i.e., 229% survival), while identical exposure of planktonic cells to the antibiotic resulted in a survival rate of 15%, a 15-fold difference (Fig. 6B). After 24 h of exposure to penicillin, the survival of C. perfringens in a biofilm dropped to 4% while planktonic survival decreased to 0.8%, a fivefold difference (Fig. 6B).
DISCUSSION
In this report, we have established for the first time that C. perfringens is capable of forming biofilms. The conditions we used to induce biofilm formation were static cultures grown anaerobically for up to 5 days. Under these conditions, C. perfringens formed a flat biofilm 30 to 40 μm thick that had bacteria encased in a dense intercellular matrix (Fig. 1A). Strains that lacked motility due to mutations in genes encoding TFP-associated functions showed significantly reduced levels of bacteria and intercellular matrix material in the biofilms, a result seen in many gram-negative bacteria but not previously reported in a gram-positive bacterium. The TFP-dependent gliding motility exhibited by C. perfringens involves the extension of filaments of bacteria that are aligned end to end on a semisolid (e.g., agar) or solid surface (53). pilT and pilC mutants are unable to carry out this movement and do not produce pili on their surfaces (53). Since the pili appear to be a component of the biofilm matrix themselves (Fig. 3), the lack of these two functions may be the reason for reduced cell numbers and biofilm matrix density seen with these mutant strains.
We have presented evidence that the extracellular matrix is composed of at least two components, carbohydrates and type IV pilin proteins (Fig. 2 and 3). Based on studies describing the biofilm matrix material produced by other gram-positive bacteria, it is likely that additional components are present, including nucleic acids and proteins other than pilins (24, 34).
We examined the effects of changing glucose levels on biofilm formation in C. perfringens (Fig. 4 and 5). The levels of glucose used in this study (0 to 100 mM) span the range found in the human body, where plasma has 5.07 mM and skeletal muscle has 4.41 mM glucose (44).
Published reports on the role of catabolite repression in biofilm formation show that there is no unifying theme for the carbohydrate-based regulation of biofilm formation. In Streptococcus gordonii, carbohydrates induce biofilm formation (18); in Streptococcus mutans, the phosphotransferase system component EIIABman activates biofilm formation in the presence of glucose (1); and in B. subtilis, CcpA represses biofilm formation under high glucose levels, but some glucose is required for biofilm formation (48). In S. aureus, biofilm formation under static and flow conditions was lost when a mutation was introduced into the ccpA gene (45). Our results with C. perfringens and CcpA differ somewhat from those seen with these gram-positive bacteria. The wild-type strain (SM101) produced more biofilm material than the ccpA mutant strain (SM120) at days 3 and 5, and both strains produced increasing amounts of biofilm material with increasing glucose concentrations at day 3 (Fig. 5A and B). CcpA also played a role in regulating the ratio of biofilm to planktonic cells, since the wild-type strain had a higher ratio than did the ccpA mutant strain at lower glucose concentrations, but both strains suppressed biofilm formation in favor of planktonic growth as the concentration of glucose in the medium increased (Fig. 5C and D), suggesting this regulation is independent of CcpA. The highest ratio of biofilm to planktonic cells was seen in the absence of added glucose (Fig. 5C and D), which suggests biofilm formation is induced in the absence of exogenous carbohydrates, which corresponds to its potential role as a mechanism for cells to survive a starvation stress response without having to make an endospore.
While tolerant of the oxygen in air, C. perfringens has been previously reported to be sensitive to H2O2 (8), which was corroborated by the low survival rate of C. perfringens SM101 in H2O2 (Fig. 6). However, there was a marked increase in survival after H2O2 exposure: 10.5% in biofilms compared with 1.5% in planktonic cultures, which demonstrates one of the protective effects conferred on cells in the biofilm.
Biofilm formation may play a role in diseases caused by C. perfringens. C. perfringens is known to cause a form of non-food-borne enteritis associated with antibiotic use (i.e., AAD) (4, 5, 56). Biofilms are well known for their ability to confer antibiotic resistance on the constituent bacteria through a variety of mechanisms (14, 39), and in our work, biofilms appear to contribute to the resistance of C. perfringens to penicillin G. Studies by other research groups have shown that biofilms constitute a major form of existence of the microbiota in the intestine (27, 28, 41). Together, these results led us to hypothesize that biofilm formation by C. perfringens in the small intestine can contribute to AAD by aiding in bacterial persistence through antibiotic treatment. However, as the C. perfringens enterotoxin is the causative effect of the symptoms of AAD (33) and C. perfringens enterotoxin is expressed only by sporulating cells (21), for biofilms to play a role in AAD, the cells must either be sporulating in the biofilm, as has been observed in B. subtilis (20, 25) and Bacillus cereus (54), or at the cessation of antibiotic treatment, they must escape the biofilm and begin sporulating.
C. perfringens type C, D, and E strains are known to colonize the guts of mammals (47) and to cause economically important enteric infections in animals, such as cattle, pigs, and sheep (reviewed in reference 47). We found that representatives of type C, D, and E strains were capable of biofilm formation (Fig. 4). Therefore, biofilm formation may represent a mechanism for colonization and persistence in the intestinal tracts of these animals until a change occurs in the condition of the animal that results in the onset of disease.
The role that biofilms play in gas gangrene (myonecrosis) is more difficult to ascertain. Gas gangrene is a quickly spreading infection involving large numbers of rapidly growing bacteria (49), characteristics which, at first glance, are not consistent with biofilm formation. However, when incubated with cooked meat pellets, C. perfringens readily formed a biofilm on the surfaces of the pellets (Fig. 2). This result may indicate that C. perfringens can in fact form biofilms in vivo in a soft tissue infection after a certain time has elapsed, and this biofilm would also be resistant to antibiotics (Fig. 6). These findings underscore the necessity for complete removal by debridement or amputation of any tissue that is potentially infected with C. perfringens.
The ability of C. perfringens to form biofilms results in a multifaceted response by the bacterium to environmental stresses, including carbohydrate limitation. The bacterium can also respond to carbohydrate limitation by producing an endospore or initiating TFP-dependent gliding motility (32, 52, 53). All three responses include a different carbohydrate-responsive profile, where some functions are CcpA dependent and others are CcpA independent. When exposed to carbohydrate limitation, the bacterium must decide which developmental pathway is most likely to enhance its survival. It is likely that a complex regulatory network that senses factors other than the carbohydrate level is required for C. perfringens to manage these three distinct stress responses and to utilize the most advantageous response for the particular environment that it encounters.
Acknowledgments
We thank Kristi DeCourcy for assistance with confocal microscopy and Steve McCartney for help with the FE-SEM. We also thank Wesley Black for discussions concerning TFP.
This work was supported by grants 2000-02621 and 2003-35201-13580 from NRICGP/USDA awarded to S.B.M.
Editor: A. Camilli
Footnotes
Published ahead of print on 2 September 2008.
REFERENCES
- 1.Abranches, J., M. M. Candella, Z. T. Wen, H. V. Baker, and R. A. Burne. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 1883748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asha, N. J., D. Tompkins, and M. H. Wilcox. 2006. Comparative analysis of prevalence, risk factors, and molecular epidemiology of antibiotic-associated diarrhea due to Clostridium difficile, Clostridium perfringens, and Staphylococcus aureus. J. Clin. Microbiol. 442785-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, W. P., and Z. Yang. 2004. Myxococcus xanthus chemotaxis homologs DifD and DifG negatively regulate fibril polysaccharide production. J. Bacteriol. 1861001-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borriello, S. P., H. E. Larson, A. R. Welch, F. Barclay, M. F. Stringer, and B. A. Bartholomew. 1984. Enterotoxigenic Clostridium perfringens: a possible cause of antibiotic-associated diarrhoea. Lancet i305-307. [DOI] [PubMed] [Google Scholar]
- 5.Borriello, S. P., and R. K. Williams. 1985. Treatment of Clostridium perfringens enterotoxin-associated diarrhoea with metronidazole. J. Infect. 1065-67. [DOI] [PubMed] [Google Scholar]
- 6.Branda, S. S., F. Chu, D. B. Kearns, R. Losick, and R. Kolter. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 591229-1238. [DOI] [PubMed] [Google Scholar]
- 7.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 1320-26. [DOI] [PubMed] [Google Scholar]
- 8.Briolat, V., and G. Reysset. 2002. Identification of the Clostridium perfringens genes involved in the adaptive response to oxidative stress. J. Bacteriol. 1842333-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carman, R. J., S. Sayeed, J. Li, C. W. Genheimer, M. F. Hiltonsmith, T. D. Wilkins, and B. A. McClane. 2008. Clostridium perfringens toxin genotypes in the feces of healthy North Americans. Anaerobe 14102-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carney, T., J. D. Perry, M. Ford, S. Majumdar, and F. K. Gould. 2002. Evidence for antibiotic induced Clostridium perfringens diarrhoea. J. Clin. Pathol. 55240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collie, R. E., J. F. Kokai-Kun, and B. A. McClane. 1998. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foodborne human gastrointestinal diseases. Anaerobe 469-79. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W. 2001. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 950-52. [DOI] [PubMed] [Google Scholar]
- 13.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, D. 2003. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discov. 2114-122. [DOI] [PubMed] [Google Scholar]
- 15.Elkins, J. G., D. J. Hassett, P. S. Stewart, H. P. Schweizer, and T. R. McDermott. 1999. Protective role of catalase in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide. Appl. Environ. Microbiol. 654594-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forest, K. T. 2005. Structure and assembly of type IV pilins. In G. Waksman, M. Caparon, and S. Hultgren (ed.), Structural biology of bacterial pathogenesis. ASM Press, Washington, DC.
- 17.Genevaux, P., S. Muller, and P. Bauda. 1996. A rapid screening procedure to identify mini-Tn10 insertion mutants of Escherichia coli K-12 with altered adhesion properties. FEMS Microbiol. Lett. 14227-30. [DOI] [PubMed] [Google Scholar]
- 18.Gilmore, K. S., P. Srinivas, D. R. Akins, K. L. Hatter, and M. S. Gilmore. 2003. Growth, development, and gene expression in a persistent Streptococcus gordonii biofilm. Infect. Immun. 714759-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ha, K. Y., Y. G. Chung, and S. J. Ryoo. 2005. Adherence and biofilm formation of Staphylococcus epidermidis and Mycobacterium tuberculosis on various spinal implants. Spine 3038-43. [DOI] [PubMed] [Google Scholar]
- 20.Hamon, M. A., and B. A. Lazazzera. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 421199-1209. [DOI] [PubMed] [Google Scholar]
- 21.Hauschild, A. H., L. Niilo, and W. J. Dorward. 1970. Enteropathogenic factors of food-poisoning Clostridium perfringens type A. Can. J. Microbiol. 16331-338. [DOI] [PubMed] [Google Scholar]
- 22.Jefferson, K. K. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236163-173. [DOI] [PubMed] [Google Scholar]
- 23.Kang, Y., H. Liu, S. Genin, M. A. Schell, and T. P. Denny. 2002. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46427-437. [DOI] [PubMed] [Google Scholar]
- 24.Latasa, C., C. Solano, J. R. Penades, and I. Lasa. 2006. Biofilm-associated proteins. C. R. Biol. 329849-857. [DOI] [PubMed] [Google Scholar]
- 25.Lindsay, D., V. S. Brozel, and A. von Holy. 2005. Spore formation in Bacillus subtilis biofilms. J. Food Prot. 68860-865. [DOI] [PubMed] [Google Scholar]
- 26.Lisle, J. T., J. J. Smith, D. D. Edwards, and G. A. McFeters. 2004. Occurrence of microbial indicators and Clostridium perfringens in wastewater, water column samples, sediments, drinking water, and Weddell seal feces collected at McMurdo Station, Antarctica. Appl. Environ. Microbiol. 707269-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macfarlane, G. T., and S. Macfarlane. 2000. Growth of mucin degrading bacteria in biofilms. Methods Mol. Biol. 125439-452. [DOI] [PubMed] [Google Scholar]
- 28.Macfarlane, S., and J. F. Dillon. 2007. Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 1021187-1196. [DOI] [PubMed] [Google Scholar]
- 29.Mahony, D. E. 1977. Stable L-forms of Clostridium perfringens: growth, toxin production, and pathogenicity. Infect. Immun. 1519-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56289-314. [DOI] [PubMed] [Google Scholar]
- 31.Melville, S. B., R. Labbe, and A. L. Sonenshein. 1994. Expression from the Clostridium perfringens cpe promoter in C. perfringens and Bacillus subtilis. Infect. Immun. 625550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendez, M., I. H. Huang, K. Ohtani, R. Grau, T. Shimizu, and M. R. Sarker. 2008. Carbon catabolite repression of type IV pilus-dependent gliding motility in the anaerobic pathogen Clostridium perfringens. J. Bacteriol. 19048-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Modi, N., and M. H. Wilcox. 2001. Evidence for antibiotic induced Clostridium perfringens diarrhoea. J. Clin. Pathol. 54748-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohamed, J. A., and D. B. Huang. 2007. Biofilm formation by enterococci. J. Med. Microbiol. 561581-1588. [DOI] [PubMed] [Google Scholar]
- 35.Narayan, K. G. 1982. Food borne infection with Clostridium perfringens type A. Int. J. Zoonoses 912-32. [PubMed] [Google Scholar]
- 36.Nudleman, E., and D. Kaiser. 2004. Pulling together with type IV pili. J. Mol. Microbiol. Biotechnol. 752-62. [DOI] [PubMed] [Google Scholar]
- 37.O'Brien, D. K., and S. B. Melville. 2000. The anaerobic pathogen Clostridium perfringens can escape the phagosome of macrophages under aerobic conditions. Cell. Microbiol. 2505-519. [DOI] [PubMed] [Google Scholar]
- 38.Paranjpye, R. N., and M. S. Strom. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 731411-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel, R. 2005. Biofilms and antimicrobial resistance. Clin. Orthop. Relat Res. 43741-47. [DOI] [PubMed] [Google Scholar]
- 40.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 591354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Randal Bollinger, R., A. S. Barbas, E. L. Bush, S. S. Lin, and W. Parker. 2007. Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J. Theor. Biol. 249826-831. [DOI] [PubMed] [Google Scholar]
- 42.Rogers, J., and C. W. Keevil. 1992. Immunogold and fluorescein immunolabelling of Legionella pneumophila within an aquatic biofilm visualized by using episcopic differential interference contrast microscopy. Appl. Environ. Microbiol. 582326-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rood, J. I., and S. T. Cole. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55621-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosdahl, H., K. Hamrin, U. Ungerstedt, and J. Henriksson. 1998. Metabolite levels in human skeletal muscle and adipose tissue studied with microdialysis at low perfusion flow. Am. J. Physiol. 274E936—E945. [DOI] [PubMed] [Google Scholar]
- 45.Seidl, K., C. Goerke, C. Wolz, D. Mack, B. Berger-Bachi, and M. Bischoff. 2008. Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 762044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selinummi, J., J. Seppala, O. Yli-Harja, and J. A. Puhakka. 2005. Software for quantification of labeled bacteria from digital microscope images by automated image analysis. BioTechniques 39859-863. [DOI] [PubMed] [Google Scholar]
- 47.Songer, J. G. 1997. Clostridial diseases of animals, p. 153-182. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathogenesis. Academic Press, Inc., San Diego, CA.
- 48.Stanley, N. R., R. A. Britton, A. D. Grossman, and B. A. Lazazzera. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 1851951-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stevens, D. L. 1997. Necrotizing clostridial soft tissue infections, p. 141-152. In J. I. Rood, B. A. McClane, J. G. Songer, and R. W. Titball (ed.), The clostridia: molecular biology and pathognesis. Academic Press, San Diego, CA.
- 50.Thormann, K. M., R. M. Saville, S. Shukla, D. A. Pelletier, and A. M. Spormann. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 1868096-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Titgemeyer, F., and W. Hillen. 2002. Global control of sugar metabolism: a gram-positive solution. Antonie van Leeuwenhoek 8259-71. [PubMed] [Google Scholar]
- 52.Varga, J., V. L. Stirewalt, and S. B. Melville. 2004. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 1865221-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varga, J. J., V. Nguyen, K. O'Brien, D., K. Rodgers, R. A. Walker, and S. B. Melville. 2006. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Mol. Microbiol. 62680-694. [DOI] [PubMed] [Google Scholar]
- 54.Vilain, S., Y. Luo, M. B. Hildreth, and V. S. Brozel. 2006. Analysis of the life cycle of the soil saprophyte Bacillus cereus in liquid soil extract and in soil. Appl. Environ. Microbiol. 724970-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warner, J. B., and J. S. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilcox, M. H., and E. T. Smyth. 1998. Incidence and impact of Clostridium difficile infection in the UK, 1993-1996. J. Hosp. Infect. 39181-187. [DOI] [PubMed] [Google Scholar]
- 57.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 964028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]