Abstract
Escherichia coli O157:H7 is an important food-borne pathogen that specifically binds to the follicle-associated epithelium in the intestine, which rapidly brings this bacterial pathogen in contact with underlying human macrophages. Very little information is available about the interaction between E. coli O157:H7 and human macrophages. We evaluated the uptake and survival of strain EDL933 during infection of human macrophages. Surprisingly, EDL933 survived and multiplied in human macrophages at 24 h postinfection. The global gene expression profile of this pathogen during macrophage infection was determined. Inside human macrophages, upregulation of E. coli O157:H7 genes carried on O islands (such as pagC, the genes for both of the Shiga toxins, and the two iron transport system operons fit and chu) was observed. Genes involved in acid resistance and in the SOS response were upregulated. However, genes of the locus of enterocyte effacement or genes involved in peroxide resistance were not differentially expressed. Many genes with putative or unknown functions were upregulated inside human macrophages and may be newly discovered virulence factors. As the Shiga toxin genes were upregulated in macrophages, survival and cytotoxicity assays were performed with isogenic Shiga toxin mutants. The initial uptake of Shiga toxins mutants was higher than that of the wild type; however, the survival rates were significantly lower at 24 h postinfection. Thus, Shiga toxins are implicated in the interaction between E. coli O157:H7 and human macrophages. Understanding the molecular mechanisms used by E. coli to survive within macrophages may help in the identification of targets for new therapeutic agents.
Enterohemorragic Escherichia coli (EHEC) causes an acute gastroenteritis, bloody diarrhea, and hemorrhagic colitis. Severe complications develop in up to 10% of cases, including the hemolytic-uremic syndrome, which can be fatal (45, 56). EHEC strains share their genetic backbone with the laboratory strain K-12 and also contain 1.34 Mb of extra genetic material called O islands (OI) (46). In the EHEC strain EDL933 (O157:H7), 177 OI were identified; some of them harbor genes for important known virulence factors of EHEC, such as the locus of enterocyte effacement (LEE) and two different Shiga toxins, Stx1 and Stx2. Following the ingestion of contaminated food or water, EHEC enters the gastrointestinal tract, survives the acidic conditions of the stomach, and is released in the intestine. Adhesion of the bacteria to the intestinal wall is the first step in establishing infection. The LEE encodes a type III secretion system (T3SS) that injects effectors in host epithelial cells, creating an intimate binding, involving actin rearrangement and microvillus effacement, which results in the pathological lesions called attaching and effacing lesions (40, 52). In the large intestine, EHEC specifically adheres to the follicle-associated epithelium of Peyer's patches (47). Binding at the follicle-associated epithelium results in the rapid contact of E. coli O157:H7 with underlying human macrophages; however, there is very little information available about the interaction between EHEC and these cells. The interaction between EHEC and murine macrophages was investigated, and it was shown that phagocytosis of EHEC by murine macrophages caused actin rearrangements surrounding the phagosome; EHEC was rapidly killed, and high levels of Shiga toxins were produced (53). However, to our knowledge, the interaction between EHEC and human macrophages has not been investigated. This interaction could be important in the global pathogenesis of human EHEC infections.
One way to understand host-pathogen interactions is to dissect the molecular mechanisms by studying gene expression. There are very few studies on the transcriptional response of bacteria during infection, as there are many factors that make difficult bacterial RNA isolation in infected host cells or tissues, such as the small amount of bacterial RNA in vivo, the short half-life of bacterial mRNA, and the contamination of bacterial mRNA with rRNA and host RNA. Moreover, bacterial mRNA purification is complicated by the fact that there are very few bacterial transcripts that are polyadenylated. To overcome these problems, the selective capture of transcribed sequences (SCOTS) was developed to identify bacterial genes expressed during infection (20). This technique has been successfully used with many pathogens, such as Mycobacterium, Salmonella, Actinobacillus, Helicobacter, and Streptococcus (4, 8, 12, 15, 21, 27). SCOTS was recently used in combination with a DNA microarray in order to provide an effective way to elucidate the global bacterial expression profile, or transcriptome, from infected host cells (13). Until now, no study has investigated the transcriptome of E. coli O157:H7 in contact with human cells. In this study, we characterized the interactions between E. coli O157:H7 (EDL933) and human macrophages, the uptake and survival rates of the bacteria, and the cytotoxicity to macrophages. We have obtained the global bacterial gene expression profile during infection of human macrophages and evaluated the role of Shiga toxins in phagocytosis.
MATERIALS AND METHODS
Bacterial strains.
E. coli O157:H7 strain EDL933 (ATCC 700927) and its derivatives were grown routinely in Luria-Bertani (LB) medium at 37°C. Isogenic Stx mutants were described previously (19). The EDL933 green fluorescent protein (GFP)-positive strain was obtained by electroporation of a high-copy plasmid constitutively expressing GFP (pKen2) (7). When necessary, kanamycin or ampicillin was added to the medium at 50 μg/ml.
Cell culture and infection.
The human monocytic cell line THP-1 (ATCC TIB-202) was maintained and infected as described previously (8). A stock culture of these cells was maintained as monocyte-like, nonadherent cells at 37°C in an atmosphere containing 5% CO2. For macrophage infection, cells were seeded at 5 × 105 cells per well in 24-well tissue culture dishes and were differentiated by addition of 10−7 M phorbol 12-myristate 13-acetate for 48 h, and then bacteria were added to the cell monolayer at a multiplicity of infection of 10. The plate was centrifuged briefly to synchronize the infection and incubated for 20 min (0 h), and then the cells were washed and fresh medium containing 100 μg/ml of gentamicin was added to kill extracellular bacteria. After 2 h, the medium was discarded, the cells were washed, and the medium was changed to include 12 μg/ml of gentamicin. The infected monolayers were either lysed from the tissue culture dishes by addition of 0.1% sodium deoxycholate in phosphate-buffered saline or further incubated. The number of surviving bacteria was determined by bacterial plate counts (CFU). The level of phagocytosis, including associated bacteria, was calculated as a percentage of the initial inoculum. The survival level corresponds to the percentage of intracellular bacteria at a given time postinfection compared to the percentage at 0 h. Salmonella strain ISP1820 and nonpathogenic E. coli strain DH5α were used as positive and negative controls, respectively. For flow cytometry analysis, infections were performed as described above using a GFP-positive strain, except for the lysis step. The infected samples were gently scraped and fixed with 4% formaldehyde. The data from 20,000 acquired events on a BD FACSort were analyzed with the CellQuest program.
Cytotoxicity assay.
Human macrophages were seeded in 24-well plates and infected as described above. After 24 h of infection, the supernatant was collected and the amount of release of the cytoplasmic enzyme lactate dehydrogenase (LDH) was evaluated with the Cytotox96 kit (Promega) according to the manufacturer's instructions. The percentage of cytotoxicity was expressed as (experimental release − spontaneous release)/(maximum release − spontaneous release) × 100, in which the spontaneous release was the amount of LDH activity in the supernatant of uninfected cells and the maximum release was that when cells were lysed with the provided lysis buffer.
RNA extraction, SCOTS, and quantitative PCR (qPCR).
At each time point (2, 8, and 24 h postinfection), macrophage monolayers were washed, and infected macrophages from three different wells were combined and lysed in TRIzol (Invitrogen) for total RNA isolation according to the manufacturer's instructions. Similarly, total RNA was obtained from bacteria grown in the cell culture medium. Five micrograms of each RNA sample were treated with DNase and converted to cDNA using primer RB1-RNA, containing a defined 5′ end and a random nonamer in the 3′ end (9, 20) (Table 1). The second-strand cDNA was synthesized using the Klenow fragment of DNA polymerase I (NEB) as described by Froussard (17). As there are numerous limitations and difficulties in isolating bacterial RNA during infection to monitor gene expression (26), we used SCOTS. Each SCOTS sample represents the cDNAs captured from a mixture of three independent infected macrophage samples. Enrichment of EHEC cDNA from the mixture of host cDNA and bacterial rRNA genes by SCOTS was obtained as described previously (9, 13, 20).
TABLE 1.
Primers
| Technique and primer | Sequence |
|---|---|
| SCOTS | |
| RBI-RNA | CGGGATCCAGCTTCTCACGCANNNNNNNNN |
| RBI-PCR | CGGGATCCAGCTTCTCACGCA |
| qPCR | |
| gyrA-F | TAATGCTGTTCTCCGCCGAAGGTA |
| gyrA-R | TCGCCTAAACGAATACCGCGAACA |
| ler-F | TAATGTGCCTGATGATGGACTCGC |
| ler-R | ACAGTGCTTCTTTAAGCCAGCGTG |
| lexA-F | ATGACGAAGTTACCGTTAAGCGCC |
| lexA-R | TCAATGGTGAAGCTCTGCTGACGA |
| modE-F | AAACGCTGACAACCAATTACCGGG |
| modE-R | AAGTCGCTTCATTTACCGGCACTG |
| recA-F | GAATGCAACTGCCTGGCTGAAAGA |
| recA-R | ACAGAGAAATCCGGCGTTGAGTTC |
| stx1A-F | TAATGCAATTCTGGGAAGCGTGGC |
| stx1A-R | ATTGTGCGTAATCCCACGGACTCT |
| stx1B-F | ATCTTCAGTCTCTTCTTCTCAGTGCG |
| stx1B-R | CCCTCCATTATGACAGGCATTAGT |
| pagC-F | AGGCCCAGCTTATCGTCTGAATGA |
| pagC-R | ATCAGCACCCACACTGTTTCTCCA |
| soxR-F | AGAGTGGAAACAGCTTTCGTCCCA |
| soxR-R | AGCCACAACCAATACATCCGTCCA |
For qPCR analysis, RNA samples from three series of different infections (biological replicates) were extracted as described above, precipitated with 2.5 M LiCl (Ambion), washed with ice-cold 75% ethanol, and resuspended in diethyl pyrocarbonate-treated water. Rigorous DNase treatment was then performed to remove any trace of DNA (DNA-free kit; Ambion). cDNA was synthesized in triplicate using SuperScript II (Invitrogen) with random hexamers (Sigma), according to the manufacturer's instructions. For each sample, a reaction done without reverse transcriptase served as a no-template control. For each qPCR run, the calculated threshold cycle was normalized to the threshold cycle of an internal control gene amplified from the corresponding sample, and the fold change was calculated as previously described (13). The gene coding for the DNA gyrase subunit A (gyrA) was used as the reference gene for normalization. Primers are described in Table 1.
Microarray, labeling, hybridization, and data acquisition and analysis.
Corning Ultra-Gap II slides (Corning, Acton, MA) were spotted with the MWG E. coli O157:H7 array set (Ocimum) at the National Microbiology Laboratory (Winnipeg, Canada). The MWG array consists of 6,167 50-mer oligonucleotides covering the genomes of E. coli K-12 (MG1655) and E. coli O157:H7 strains Sakai (RIMD 0509952) and EDL933 (ATCC 700927), and 5,419 oligonucleotides correspond to the EDL933 genome, including 83 genes from the plasmid pO157. Four hybridization replicates were used for each time point (2 h, 8 h, and 24 h postinfection) in the microarray experiments, and the coefficient of correlation was calculated between each replicate. As the interaction between EHEC and macrophages was never previously reported, the experimental design was to sample a variety of conditions rather than to make repeat observations, an approach previously shown to be more valuable to get an overall picture (11). SCOTS cDNA or genomic DNA (400 ng) was labeled with the Bioprime DNA labeling system kit (Invitrogen) according to the manufacturer's instructions. SCOTS cDNA was labeled with Cy5, and genomic DNA was labeled with Cy3 (Amersham). The templates were purified, and 3 μg of each labeled SCOTS cDNA and genomic DNA sample were mixed in warmed digoxigenin hybridization buffer (Roche). The templates were denatured and added to the microarray, which had been prehybridized with a digoxigenin-5% bovine serum albumin buffer. The microarray was incubated at 42°C for 16 h and was then washed three time in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate for 5 min each at 37°C and one last time in 0.1× SSC. Data acquisition and normalization were performed as described elsewhere (13, 49). Bacterial genomic DNA was used as the reference channel on each slide to allow comparison of each time point and different samples (54). Statistical analysis was performed with TIGR Multiexperiment Viewer software by using the analysis of variance (ANOVA) function with default parameter (http://www.tm4.org). A P value of less than 0.0001 was considered statistically significant. Only the significant genes were considered for further studies, unless otherwise indicated. Moreover, for a gene to be considered upregulated or downregulated, the expression level of the gene needed to be two times superior or inferior, respectively, to the expression level in the control condition (bacteria grown in cell culture medium).
Microarray data accession number.
The microarray data have been deposited in NCBI's Gene Expression Omnibus (GEO accession number GSE9671) (http://www.ncbi.nlm.nih.gov/projects/geo/).
RESULTS
Uptake, survival, and multiplication of E. coli O157:H7 within human macrophages.
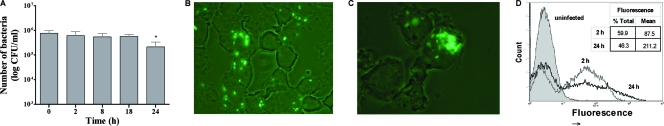
The level of phagocytosis and survival of E. coli O157:H7 strain EDL933 in contact with human macrophages was determined (Fig. 1A). Twelve percent of the initial bacterial inoculum was associated with the macrophages after 20 min of interaction (0 h) (data not shown). The number of internalized bacteria remained relatively constant during the infection, at approximately 5.5 × 105 CFU/ml, and the level of survival decreased at 24 h postinfection but still remained high, at about 1.9 × 105 CFU/ml (Fig. 1A). EDL933 constitutively expressing GFP was used in order to visualize bacteria within macrophages by fluorescence microscopy. Initially, many macrophages were infected with a single bacterium or a few bacteria (Fig. 1B). However, at 24 h postinfection, only a few macrophages were infected, suggesting that many of the macrophages managed to clear the bacteria, but the infected macrophages contained many bacterial clusters, suggesting that the bacteria had replicated (Fig. 1C). This observation was quantified by cytometry in order to estimate the percentage of macrophages infected with GFP-expressing bacteria, as well as the mean level of fluorescence. A higher number of fluorescent macrophages (60%) was observed at the beginning of the infection than at 24 h postinfection (46%) (Fig. 1D). However, the mean fluorescence was twice as high at 24 h postinfection (Fig. 1D). This corroborates the fluorescence microscopy data and indicates that later in the infection, fewer macrophages were infected but infected macrophages contained more bacteria.
FIG. 1.
Infection of THP-1 macrophages with EDL933. (A) Quantification of intracellular bacteria at different times postinfection, where 0 h corresponds to 20 min of phagocytosis. (B and C) Fluorescence microscopy of infected macrophages, with EDL933-GFP and human macrophages at 2 h (B) or 24 h (C) postinfection (magnification, ×1,000). (D) Cytometry analysis of EDL933-GFP in contact with human macrophages at 2 h (gray) and 24 h (black) postinfection. All assays were conducted in duplicate and repeated independently at least three times. Results are expressed as means ± standard deviations for the replicate experiments, and the Student t test was used for statistical analysis of the data.
Transcriptome of EHEC within macrophages.
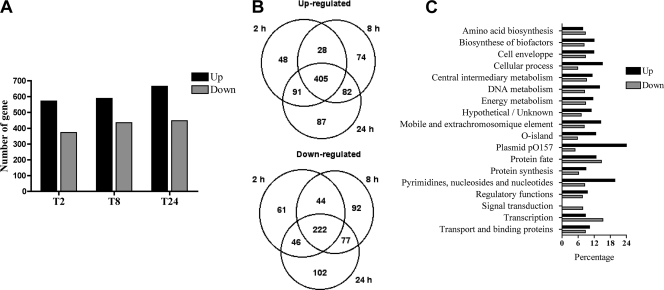
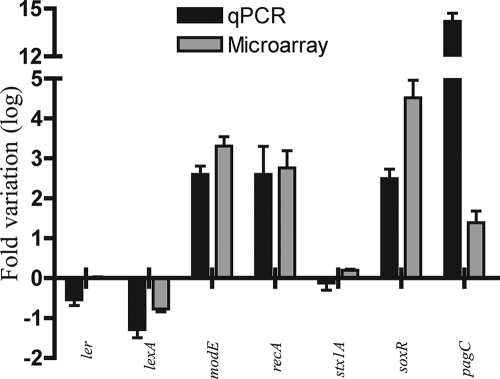
To determine the global bacterial gene expression profile of E. coli O157:H7 in human macrophages, strain EDL933 was used to infect macrophages. Three intracellular postinfection time points were chosen, i.e., 2 h, 8 h, and 24 h. Bacteria grown in RPMI, the culture medium for human macrophages, were used as an in vitro control. In order to obtain a complex mixture of bacterial transcripts, three rounds of SCOTS were performed on each of these samples, as described previously (9, 20). The bacterial cDNAs were hybridized to the E. coli microarray, which includes the entire genome of EDL933. The percentage of detected genes was approximately 78% for each condition tested, confirming that SCOTS allowed the recovery of a great variety of transcripts. The intracellular expression profiles were analyzed by comparison with the expression profile of bacteria grown in RPMI. A total of 1,221 genes of EDL933 were significantly differentially expressed within macrophages (ANOVA, P < 0.0001). For each intracellular time point, approximately 600 genes were upregulated (11%) and 400 genes downregulated (7%) (Fig. 2A). Interestingly, most of the differentially expressed genes showed a similar expression profile at all intracellular time points, as more than 60% (405) of upregulated genes and more than 50% (222) of downregulated were conserved at each of the intracellular time points (Fig. 2B). Similar detection profiles for many of the transcripts at three different time points validated the use of SCOTS cDNA to reveal the EHEC transcriptome from infected host cells. Because the transcription profile observed was comparable for numerous genes throughout the infection course and the largest number of genes were expressed at 24 h postinfection, our analysis focused mainly on this time point. E. coli O157:H7 differentially expressed genes were classified according to their orthologous groups (COG) in the TIGR annotation (www.cmr.tigr.org) (Fig. 2C). The profile of highly upregulated or downregulated genes was not found to be specific to an orthologous group. The microarray data were confirmed by qPCR, and a correlation of 0.9 was obtained between the expression ratios determined by microarrays versus qPCR (Fig. 3). Two different technologies applied to different biological samples were consistent, supporting the validity of SCOTS-microarray analysis as an effective way to study host-pathogen interaction.
FIG. 2.
Global analysis of differentially expressed genes. (A) Number of upregulated or downregulated genes at each intracellular time point of macrophage infection. (B) Venn diagram of the number of up- or downregulated genes at each intracellular time point. (C) Distribution of E. coli genes in functional categories (COG). Percentages of upregulated and downregulated genes for each functional class at 24 h postinfection are shown. Data are the means from four replicates.
FIG. 3.
Real-time qPCR and microarray results for a set of seven representative genes at 24 h postinfection compared to RPMI. Data are the means and standard deviations for RNA extracted from three different infections. See text for details.
Differential expression of OI.
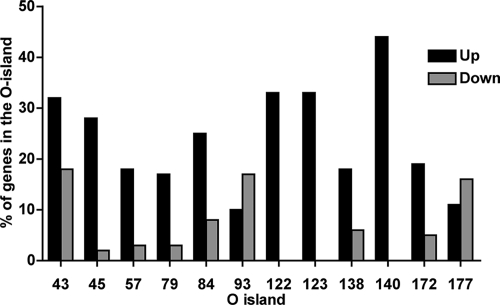
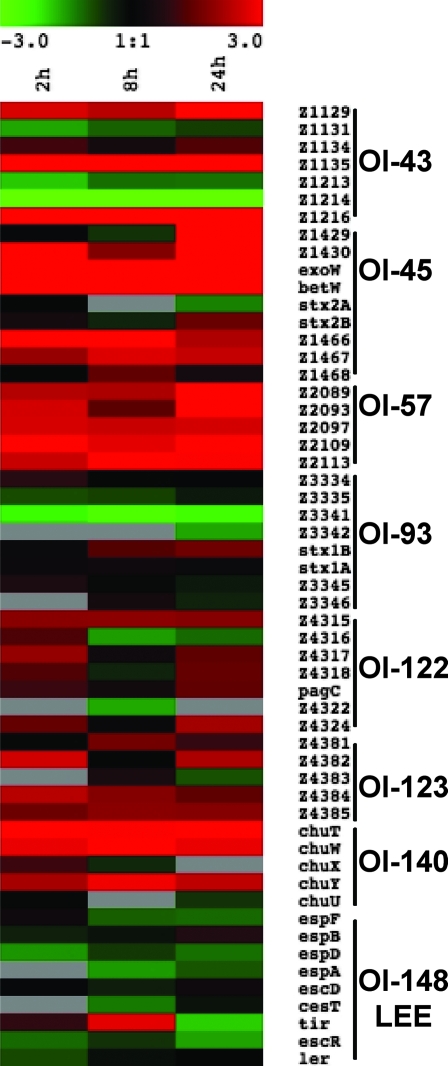
OI represent about 20% of the EDL933 genome, and 13% of the genes within OI were upregulated, whereas 6% were downregulated, during infection. The OI that harbor at least six genes and contain at least 15% of differentially expressed genes were selected (Fig. 4 and 5). OI-148, carrying the LEE, did not meet these criteria, as only 4 genes were downregulated (sepZ, espD, tir, and Z5094) and 2 genes were upregulated (Z5095 and Z5088), of a total of 48 genes. In most of the selected OI, genes were upregulated (Fig. 4). OI-122, OI-123, and OI-140 contain many genes that were upregulated (Fig. 4 and 5). Among upregulated genes in OI-122, the virulence gene pagC was upregulated at all intracellular time points. The upregulated genes of OI-123 include the putative iron transport operon fit (44), and the upregulated genes of OI-140 include the iron uptake operon chu (39). In OI-93, harboring genes of phage CP-933V as well as Stx1, many genes were downregulated, with the exception of stx1B, which was upregulated. In OI-45, encoding genes of phage BP-933W and Stx2, genes were mostly upregulated, including stx2B. In OI-43, encoding the tellurite resistance- and adherence-conferring island (55), genes for tellurite resistance (terA and terC) were upregulated. Most of the other islands with differentially expressed genes harbor prophage genes or genes encoding hypothetical proteins with unknown functions.
FIG. 4.
Percentage of upregulated and downregulated genes for selected OI (OI containing more than six genes and showing a minimum of 15% of differentially expressed genes) at 24 h postinfection. Data are the means from four technical replicates.
FIG. 5.
Heat map of expression levels of predicted open reading frame selected from specific OI over time during macrophage infection. Expression values were determined from the ANOVA and are represented colorimetrically, with red representing upregulation (ratio of 3) and green representing downregulation (ratio of −3) on a log2 scale. Data are the means from four technical replicates.
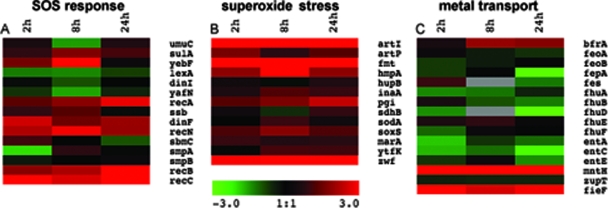
Genes involved in stress responses and metal ion availability.
As the macrophage environment is hostile, expression of genes involved in the bacterial response to DNA damage (SOS response), to superoxide, and to peroxide and in acid resistance were analyzed. Genes related to DNA damage, such as recA, recB, recC, recN, ruvC, dinF, dinD, and sulA (14), as well as genes involved in superoxide stress, such as soxRS, hmpA, and zwf (48), were upregulated (Fig. 6). The peroxide resistance genes oxyR, katP, katE, and katG were not differentially expressed. The acid regulator genes yhiF and gadE were upregulated, as were many acid resistance genes such as hdeA, hdeD, gadA, adiY, asr, and adiC. Metal ion-related genes for transport and utilization are important, as metal ions are scarce in the host. The transcription of ion uptake systems mntH, chu (OI-140), fit (OI-123), bfrA, and fieF was upregulated in macrophages, whereas transcription of feo, zupT, ent, or fhu, involved in metal ion transport, was not upregulated (Fig. 6).
FIG. 6.
Heat map of gene expression from the SOS response, oxidative stress, and metal transport gene classes. Data are the means from four technical replicates.
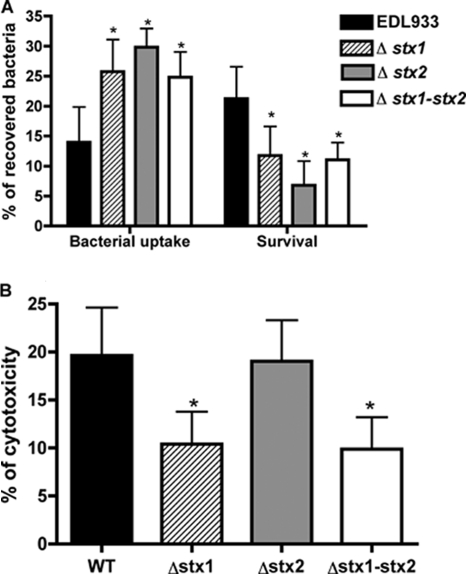
Role of Shiga toxins in E. coli O157:H7 survival in human macrophages.
As the Shiga toxins are very important virulence factors in EHEC and as its gene transcription was modulated during infection of macrophages, we investigated the impact of Shiga toxins on the phagocytosis, survival, and cytotoxicity level of E. coli O157:H7 in human macrophages. The absence of either or both of the Shiga toxins significantly increased the level of phagocytosis compared to the wild-type level (Fig. 7A). The survival rate of each stx mutant significantly decreased at 24 h postinfection compared to that of the wild type and was significantly more affected by the absence of Stx2 than by that of Stx1 (Fig. 7A). As the macrophages’ viability could be affected by toxins, the cytotoxicity level of infected macrophages was evaluated. A twofold decrease of cytotoxicity with the stx1 mutant or the stx1 stx2 mutant was observed, whereas the cytotoxicity of the stx2 mutant was similar to that of the wild-type strain (Fig. 7B). Shiga toxins appear to play an important role during interaction with macrophages: they reduced bacterial uptake by macrophages and promoted bacterial survival.
FIG. 7.
Role of Stx during macrophages infection. (A) Bacterial uptake (percent compared to inoculum) and survival (percent compared to 0 h) of the stx mutants. (B) Cytotoxicity of strain EDL933 O157:H7 and isogenic mutants measured by LDH release after 24 h of infection. The figures show the means ± standard deviations from at least three independent experiments. An asterisk indicates a significant difference compared to EDL933 (P < 0.05).
DISCUSSION
EHEC of serotype O157:H7 interacts with intestinal cells and causes microvillus effacement, which results in mild to bloody diarrhea. A strong inflammatory response is observed during EHEC infection, and cytokines produced by infected macrophages were previously shown to contribute to the severe inflammation associated with hemolytic-uremic syndrome (58). Thus, the interaction between EHEC and human macrophages could be important in the global pathogenesis of EHEC during human infections, but this interaction is very poorly characterized. In this study, we have examined the phagocytosis and survival rate of E. coli O157:H7 and of its isogenic Shiga toxins mutants during infection of human macrophages, as well as the global bacterial gene expression profile. We observed that E. coli O157:H7 can survive in human macrophages and was even able to replicate in this milieu. Effectively, even if many macrophages manage to clear the bacteria, some infected macrophages contain higher numbers of bacteria after 24 h of infection than at earlier time points, suggesting replication of the bacteria (Fig. 1). There was a 20% mortality of macrophage cells following 24 h of infection (Fig. 7), which may be due to lysis of cells that were killed by replicating bacteria or may be due to toxin release. Although not reported for E. coli O157:H7, it was previously shown that some extracellular bacterial pathogens are able to survive within professional phagocytes, such as uropathogenic E. coli (42), group A Streptococcus (37, 59, 60), Staphylococcus aureus (22), and Enterococcus faecalis (3, 18). However, only E. faecalis has been shown to multiply in macrophages at 24 h postinfection (18). We have shown that EHEC O157:H7 can resist macrophage killing and can also multiply, although the total number of viable bacteria decreased throughout the course of infection (Fig. 1). There may be two distinct populations of bacteria inside the macrophages, one that is able to survive and multiply and another that is killed. This hypothesis of a “double population” has been proposed for other pathogens infecting macrophages, such as E. faecalis, Salmonella, and Listeria monocytogenes (1, 10, 18). Our microarray analysis cannot distinguish between these populations. Yet, EHEC has not been detected in blood or organs, and intestinal infection does not appear to lead to a systemic infection; however, prolonged survival in macrophages may be important in the global course of infection and may significantly influence the intestinal inflammatory response.
During infection of human macrophages, E. coli O157:H7 faces a hostile environment and adapts by modifying its global transcriptome in order to survive and multiply. There was 22% of the genome of EDL933 that was differentially regulated intracellularly. Interestingly, up to one-third of the upregulated genes (210) encode proteins with unknown functions, and these could potentially be new virulence genes. OI are regions of the EDL933 genome that are present in E. coli O157 but not in E. coli K-12, and some OI are known to encode virulence determinants. Genes in some OI were upregulated in macrophages. OI-122 has been shown to be present in Stx-producing E. coli strains associated with epidemic and/or serious disease, which suggests that genes of this region could be important for virulence (28). One of the OI-122 genes, pagC, was constitutively upregulated. A pagC mutant of Citrobacter rodentium, which causes attaching and effacing lesions in mice, had a significantly lower competitive index, indicating that this gene is important for the establishment of in vivo infection (61). In Salmonella, PagC is important for bacterial survival in macrophages (38) and was upregulated in macrophages (13), and PagC antibodies were detected in human blood (24). Thus, pagC might also be involved in survival of E. coli O157:H7 in human macrophages.
The iron uptake system operons fit and chu, harbored by OI-123 and OI-140, respectively, were upregulated. The fit (“ferrous or ferric iron transport”) system was previously found to be expressed during mouse infection with a human strain of E. coli O25:H causing septicemia (30). Chu is an iron uptake system based on heme and is sufficient for bacterial survival when the heme is the only available iron source (39, 62). In uropathogenic E. coli, the transcription of chuW, a member of the Chu system, was found to be upregulated in the urine of mice and, moreover, antibodies against the hemin receptor, ChuA, were detected in infected mice (23). In avian-pathogenic E. coli, a chuA mutant did not survive in a chicken infection model (32). These iron uptake genes are therefore important during in vivo infections and may also confer a survival advantage to E. coli O157:H7 during macrophage infection. Other metal ion uptake systems, such as mntH, bfrA, fieF, feo, zupT, ent, and fhu, are located outside OI. MntH is a metal ion transporter involved in the uptake of Mn2+, specifically in acidic conditions, and to a lesser extent of Fe2+ and was upregulated, as were bfrA and fieF. BfrA is transcribed under conditions of iron abundance, and FieF is believed to pump iron out from the cytoplasm in the extracellular environment. In human THP-1 macrophages, E. coli O157:H7 does not seem to face iron-limiting conditions but may use heme or other metal ion sources. Similarly, transcriptional profiles of intracellular pathogens such as Salmonella and Mycobacterium in THP-1 macrophages did not correspond with a predicted low-iron environment either (13, 16).
Genes involved in nucleotide synthesis and in DNA synthesis and repair were upregulated, which could correspond to the multiplication of the bacteria or their encounter with different stressors. During macrophage infection, expression of genes of the SOS response, such as recA, recB, recC, recN, sulA, ruvC, dinD, and dinF, was upregulated, whereas the expression of lexA, the repressor of the SOS system, was downregulated. Activation of the SOS response will lead to induction of phage lytic cycles, including that of Stx prophages, which will lead in turn to an increase in production of Stx, corresponding with the phenotypes of stx mutants observed during macrophage infection and with stx upregulation detected by microarray analysis. We identified many upregulated genes that belong to acid resistance systems. The two extreme acid resistance systems, as well as a group of genes called the “acid fitness island,” were activated during macrophage infection. Furthermore, it was also observed that some of these acid resistance genes were specifically upregulated in human macrophages by Shigella dysenteriae (35). E. coli O157:H7 may face unsuitable acidic conditions within human macrophages, particularly late in infection. Interestingly, no peroxide stress response was noted, as the transcription of critical genes such as oxyR, katP, katE, and katG was not upregulated.
In other pathogens, T3SSs are implicated in bacterial survival in macrophages, such as the T3SS located on SPI-2 in Salmonella (43) or on the plasmid in Shigella (2). In enteropathogenic E. coli, the T3SS encoded by the LEE was shown to inhibit phagocytosis by murine macrophages and to inhibit uptake by M cells (36), which can be a way of escaping underlying macrophages. Our results show that the majority of LEE genes were not differentially regulated during macrophage infection. Tissue culture media may be an inappropriate control in this situation for gene expression comparison, as LEE genes are expressed in Dulbecco modified Eagle medium (29, 31). LEE genes may be already expressed in RPMI, our control, explaining the absence of differential expression. Moreover, two genes, yhiF and gadE (yhiE), which are known repressors of the LEE (57), were upregulated, suggesting that the LEE may not be involved in EHEC survival in human macrophages, but further studies are needed to confirm this observation.
Shiga toxins are important virulence factors of EHEC that recognize the Gb3 receptor on host cells. It was previously shown that differentiated THP-1 human macrophages express Gb3 (50). At this stage of the study, we cannot say if the Gb3 status of the macrophages is important for the observed effect of Shiga toxin during interaction with macrophages. However, the Shiga toxins are also known to affect epithelial cells that do not express Gb3 (51). In this study, we notice that the transcription of stx1B and stx2B was induced in human macrophages compared to growth in RPMI. A concomitant upregulation of genes encoding the Stx subunit A, stx1A and stx2A, was not observed. Differential regulation of Shiga toxin subunit genes was previously observed during growth in vitro. Effectively, in a study that investigated global gene expression during growth transitions of E. coli O157:H7, the transcript levels of stx2A decreased 1.5-fold from late exponential to stationary phase, while levels of stx2B remained higher in stationary phase than in exponential phase (5). In contrast, when global expression of EDL933 in response to norfloxacin was obtained, the transcript levels of stx2A were at least twice as high as those of stx2B (25).
The involvement of Shiga toxins in phagocytosis and survival level in THP-1 human macrophages was examined. We have shown that there is a correlation between the level of phagocytosis of EDL933 by macrophages and the presence of Shiga toxins. The Stx mutants, lacking one or both toxins, were phagocytosed to a greater extent than the wild-type strain. The Shiga toxins were also involved in bacterial survival in macrophages, as Stx mutants showed a lower survival rate than the wild type. The stx2 mutant has a significantly lower survival rate than the stx1 mutant or the double stx1 stx2 mutant. Interestingly, both the stx1 and stx1 stx2 mutants were less cytotoxic to the macrophages than the stx2 mutant, whose cytotoxicity was similar to that of the wild-type strain. Thus, the presence of both toxins contributed to bacterial survival in macrophages, and Stx1 is more cytotoxic than Stx2 for macrophages. Differential activities of the two Shiga toxins have been previously shown on cultured human endothelial cells (34) and epithelial intestinal cells (41). The mechanism by which these toxins influence the uptake and survival of E. coli O157:H7 and its cytotoxicity on human macrophages is unknown, and more research will be needed to answer this question.
EHEC preferably binds to Peyer's patches (47), where it encounters subadjacent macrophages. EHEC infections are known to induce a strong inflammatory response. The presence of EHEC and Shiga toxins in the gut recruits and activates cells of the immune system. The activated cells produce proinflammatory cytokines that intensify the inflammatory response and lead to the disruption of the intestinal barrier, permitting transport of the Stx toxins into the circulatory system (6). Furthermore, these cytokines have also been shown to increase Gb3 expression on endothelial cells, allowing more binding of Stx and thus further exacerbating the disease process (33, 58). Thus, the extended survival of EHEC in underlying macrophages could have many functions. First, EHEC survival may enhance inflammatory responses responsible for intestinal epithelial barrier disruption, which could lead to an increase of Shiga toxins entering the bloodstream. Second, as Shiga toxins are expressed and participate in bacterial survival in macrophages, the production of Shiga toxins within macrophages may be used to increase the toxin concentration in the bloodstream, as it was shown that bacteria residing in macrophages release these toxins (53). Understanding the interactions between host and pathogen is of fundamental importance in developing vaccines and other new therapeutic agents, particularly in the case of E. coli O157:H7, as no effective treatment is presently available for humans infected with this bacterium.
Acknowledgments
This paper is dedicated to the memory of our colleague Roland Brousseau.
This work was supported by a Natural Sciences and Engineering Research Council (NSERC) Strategic Grant (to J.H.), Discovery Grant 251114 (to F.D.), and the Commission Permanente de Coopération Franco-Québecoise (61-116). S.P.F. was supported by an NSERC scholarship, and K.P. was supported by a scholarship from Fonds de la Recherche en Santé du Québec (FRSQ).
We thank Serge Sénéchal for technical assistance with flow cytometry experiments and C. M. Dozois and M. Mourez for critical reading of the manuscript.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 25 August 2008.
REFERENCES
- 1.Abshire, K. Z., and F. C. Neidhardt. 1993. Growth rate paradox of Salmonella typhimurium within host macrophages. J. Bacteriol. 1753744-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, G. P., A. E. Hromockyj, C. Coker, and A. T. Maurelli. 1991. Two novel virulence loci, mxiA and mxiB, in Shigella flexneri 2a facilitate excretion of invasion plasmid antigens. Infect. Immun. 591997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldassarri, L., L. Bertuccini, R. Creti, P. Filippini, M. G. Ammendolia, S. Koch, J. Huebner, and G. Orefici. 2005. Glycosaminoglycans mediate invasion and survival of Enterococcus faecalis into macrophages. J. Infect. Dis. 1911253-1262. [DOI] [PubMed] [Google Scholar]
- 4.Baltes, N., F. F. Buettner, and G. F. Gerlach. 2007. Selective capture of transcribed sequences (SCOTS) of Actinobacillus pleuropneumoniae in the chronic stage of disease reveals an HlyX-regulated autotransporter protein. Vet. Microbiol. 123110-121. [DOI] [PubMed] [Google Scholar]
- 5.Bergholz, T. M., L. M. Wick, W. Qi, J. T. Riordan, L. M. Ouellette, and T. S. Whittam. 2007. Global transcriptional response of Escherichia coli O157:H7 to growth transitions in glucose minimal medium. BMC Microbiol. 797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brigotti, M., A. Caprioli, A. E. Tozzi, P. L. Tazzari, F. Ricci, R. Conte, D. Carnicelli, M. A. Procaccino, F. Minelli, A. V. Ferretti, F. Paglialonga, A. Edefonti, and G. Rizzoni. 2006. Shiga toxins present in the gut and in the polymorphonuclear leukocytes circulating in the blood of children with hemolytic-uremic syndrome. J. Clin. Microbiol. 44313-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 17333-38. [DOI] [PubMed] [Google Scholar]
- 8.Daigle, F., J. E. Graham, and R. Curtiss III. 2001. Identification of Salmonella typhi genes expressed within macrophages by selective capture of transcribed sequences (SCOTS). Mol. Microbiol. 411211-1222. [DOI] [PubMed] [Google Scholar]
- 9.Daigle, F., J. Y. Hou, and J. E. Clark-Curtiss. 2002. Microbial gene expression elucidated by selective capture of transcribed sequences (SCOTS). Methods Enzymol. 358108-122. [DOI] [PubMed] [Google Scholar]
- 10.de Chastellier, C., and P. Berche. 1994. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun. 62543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 9514863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faucher, S. P., R. Curtiss III, and F. Daigle. 2005. Selective capture of Salmonella enterica serovar Typhi genes expressed in macrophages that are absent from the Salmonella enterica serovar Typhimurium genome. Infect. Immun. 735217-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faucher, S. P., S. Porwollik, C. M. Dozois, M. McClelland, and F. Daigle. 2006. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. USA 1031906-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez De Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 351560-1572. [DOI] [PubMed] [Google Scholar]
- 15.Fittipaldi, N., M. Gottschalk, G. Vanier, F. Daigle, and J. Harel. 2007. Use of selective capture of transcribed sequences to identify genes preferentially expressed by Streptococcus suis upon interaction with porcine brain microvascular endothelial cells. Appl. Environ. Microbiol. 734359-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontan, P., V. Aris, S. Ghanny, P. Soteropoulos, and I. Smith. 2008. Global transcriptional profile of Mycobacterium tuberculosis during THP-1 human macrophage infection. Infect. Immun. 76717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froussard, P. 1992. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 202900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gentry-Weeks, C. R., R. Karkhoff-Schweizer, A. Pikis, M. Estay, and J. M. Keith. 1999. Survival of Enterococcus faecalis in mouse peritoneal macrophages. Infect. Immun. 672160-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gobert, A. P., M. Vareille, A. L. Glasser, T. Hindre, T. de Sablet, and C. Martin. 2007. Shiga toxin produced by enterohemorrhagic Escherichia coli inhibits PI3K/NF-kappaB signaling pathway in globotriaosylceramide-3-negative human intestinal epithelial cells. J. Immunol. 1788168-8174. [DOI] [PubMed] [Google Scholar]
- 20.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 9611554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham, J. E., R. M. Peek, Jr., U. Krishna, and T. L. Cover. 2002. Global analysis of Helicobacter pylori gene expression in human gastric mucosa. Gastroenterology 1231637-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gresham, H. D., J. H. Lowrance, T. E. Caver, B. S. Wilson, A. L. Cheung, and F. P. Lindberg. 2000. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J. Immunol. 1643713-3722. [DOI] [PubMed] [Google Scholar]
- 23.Hagan, E. C., and H. L. Mobley. 2007. Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect. Immun. 753941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris, J. B., A. Baresch-Bernal, S. M. Rollins, A. Alam, R. C. LaRocque, M. Bikowski, A. F. Peppercorn, M. Handfield, J. D. Hillman, F. Qadri, S. B. Calderwood, E. Hohmann, R. F. Breiman, W. A. Brooks, and E. T. Ryan. 2006. Identification of in vivo-induced bacterial protein antigens during human infection with Salmonella enterica serovar Typhi. Infect. Immun. 745161-5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herold, S., J. Siebert, A. Huber, and H. Schmidt. 2005. Global expression of prophage genes in Escherichia coli O157:H7 strain EDL933 in response to norfloxacin. Antimicrob. Agents Chemother. 49931-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinton, J. C., I. Hautefort, S. Eriksson, A. Thompson, and M. Rhen. 2004. Benefits and pitfalls of using microarrays to monitor bacterial gene expression during infection. Curr. Opin. Microbiol. 7277-282. [DOI] [PubMed] [Google Scholar]
- 27.Hou, J. Y., J. E. Graham, and J. E. Clark-Curtiss. 2002. Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences. Infect. Immun. 703714-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karmali, M. A., M. Mascarenhas, S. Shen, K. Ziebell, S. Johnson, R. Reid-Smith, J. Isaac-Renton, C. Clark, K. Rahn, and J. B. Kaper. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 414930-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kenny, B., A. Abe, M. Stein, and B. B. Finlay. 1997. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect. Immun. 652606-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan, M. A., and R. E. Isaacson. 2002. Identification of Escherichia coli genes that are specifically expressed in a murine model of septicemic infection. Infect. Immun. 703404-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leverton, L. Q., and J. B. Kaper. 2005. Temporal expression of enteropathogenic Escherichia coli virulence genes in an in vitro model of infection. Infect. Immun. 731034-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, G., C. Laturnus, C. Ewers, and L. H. Wieler. 2005. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect. Immun. 732818-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louise, C. B., and T. G. Obrig. 1991. Shiga toxin-associated hemolytic-uremic syndrome: combined cytotoxic effects of Shiga toxin, interleukin-1 beta, and tumor necrosis factor alpha on human vascular endothelial cells in vitro. Infect. Immun. 594173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louise, C. B., and T. G. Obrig. 1995. Specific interaction of Escherichia coli O157:H7-derived Shiga-like toxin II with human renal endothelial cells. J. Infect. Dis. 1721397-1401. [DOI] [PubMed] [Google Scholar]
- 35.Lucchini, S., H. Liu, Q. Jin, J. C. Hinton, and J. Yu. 2005. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect. Immun. 7388-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Argudo, I., C. Sands, and M. A. Jepson. 2007. Translocation of enteropathogenic Escherichia coli across an in vitro M cell model is regulated by its type III secretion system. Cell. Microbiol. 91538-1546. [DOI] [PubMed] [Google Scholar]
- 37.Medina, E., O. Goldmann, A. W. Toppel, and G. S. Chhatwal. 2003. Survival of Streptococcus pyogenes within host phagocytic cells: a pathogenic mechanism for persistence and systemic invasion. J. Infect. Dis. 187597-603. [DOI] [PubMed] [Google Scholar]
- 38.Miller, V. L., K. B. Beer, W. P. Loomis, J. A. Olson, and S. I. Miller. 1992. An unusual pagC::TnphoA mutation leads to an invasion- and virulence-defective phenotype in salmonellae. Infect. Immun. 603763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mills, M., and S. M. Payne. 1995. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J. Bacteriol. 1773004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 411340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukhopadhyay, P., M. Zheng, L. A. Bedzyk, R. A. LaRossa, and G. Storz. 2004. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. USA 101745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nazareth, H., S. A. Genagon, and T. A. Russo. 2007. Extraintestinal pathogenic Escherichia coli survives within neutrophils. Infect. Immun. 752776-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ochman, H., F. C. Soncini, F. Solomon, and E. A. Groisman. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. USA 937800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouyang, Z., and R. Isaacson. 2006. Identification and characterization of a novel ABC iron transport system, Fit, in Escherichia coli. Infect. Immun. 746949-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga toxin-producing Escherichia coli infections. Clin. Microbiol. Rev. 11450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409529-533. [DOI] [PubMed] [Google Scholar]
- 47.Phillips, A. D., and G. Frankel. 2000. Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J. Infect. Dis. 1811496-1500. [DOI] [PubMed] [Google Scholar]
- 48.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 1833890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porwollik, S., J. Frye, L. D. Florea, F. Blackmer, and M. McClelland. 2003. A non-redundant microarray of genes for two related bacteria. Nucleic Acids Res. 311869-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramegowda, B., and V. L. Tesh. 1996. Differentiation-associated toxin receptor modulation, cytokine production, and sensitivity to Shiga-like toxins in human monocytes and monocytic cell lines. Infect. Immun. 641173-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuller, S., G. Frankel, and A. D. Phillips. 2004. Interaction of Shiga toxin from Escherichia coli with human intestinal epithelial cell lines and explants: Stx2 induces epithelial damage in organ culture. Cell. Microbiol. 6289-301. [DOI] [PubMed] [Google Scholar]
- 52.Sherman, P., R. Soni, and M. Karmali. 1988. Attaching and effacing adherence of Vero cytotoxin-producing Escherichia coli to rabbit intestinal epithelium in vivo. Infect. Immun. 56756-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimada, O., H. Ishikawa, H. Tosaka-Shimada, and S. Atsumi. 1999. Exocytotic secretion of toxins from macrophages infected with Escherichia coli O157. Cell Struct. Function 24247-253. [DOI] [PubMed] [Google Scholar]
- 54.Talaat, A. M., S. T. Howard, W. t. Hale, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 681400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarr, P. I., C. A. Gordon, and W. L. Chandler. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 3651073-1086. [DOI] [PubMed] [Google Scholar]
- 57.Tatsuno, I., K. Nagano, K. Taguchi, L. Rong, H. Mori, and C. Sasakawa. 2003. Increased adherence to Caco-2 cells caused by disruption of the yhiE and yhiF genes in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 712598-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van de Kar, N. C., L. A. Monnens, M. A. Karmali, and V. W. van Hinsbergh. 1992. Tumor necrosis factor and interleukin-1 induce expression of the verocytotoxin receptor globotriaosylceramide on human endothelial cells: implications for the pathogenesis of the hemolytic uremic syndrome. Blood 802755-2764. [PubMed] [Google Scholar]
- 59.Voyich, J. M., K. R. Braughton, D. E. Sturdevant, C. Vuong, S. D. Kobayashi, S. F. Porcella, M. Otto, J. M. Musser, and F. R. DeLeo. 2004. Engagement of the pathogen survival response used by group A Streptococcus to avert destruction by innate host defense. J. Immunol. 1731194-1201. [DOI] [PubMed] [Google Scholar]
- 60.Voyich, J. M., D. E. Sturdevant, K. R. Braughton, S. D. Kobayashi, B. Lei, K. Virtaneva, D. W. Dorward, J. M. Musser, and F. R. DeLeo. 2003. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc. Natl. Acad. Sci. USA 1001996-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wickham, M. E., C. Lupp, M. Mascarenhas, A. Vazquez, B. K. Coombes, N. F. Brown, B. A. Coburn, W. Deng, J. L. Puente, M. A. Karmali, and B. B. Finlay. 2006. Bacterial genetic determinants of non-O157 STEC outbreaks and hemolytic-uremic syndrome after infection. J. Infect. Dis. 194819-827. [DOI] [PubMed] [Google Scholar]
- 62.Wyckoff, E. E., D. Duncan, A. G. Torres, M. Mills, K. Maase, and S. M. Payne. 1998. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 281139-1152. [DOI] [PubMed] [Google Scholar]