Abstract
Previously, we identified a Plasmodium yoelii YM 140-kDa merozoite protein, designated PyP140, which formed a complex with apical membrane antigen 1 (AMA1). Furthermore, we produced a nonprotective monoclonal antibody (MAb), 48F8, that immunoprecipitated metabolically labeled PyP140 and localized the protein to the merozoite's apical end and, less frequently, to the merozoite surface, as observed by immunofluorescence assay (IFA). Here, using MAb 48F8, we have identified the pyp140 gene by screening a P. yoelii λ-Zap cDNA expression library. The pyp140 cDNA covers approximately 90% of the putative open reading frame (ORF) of PY02159 from the P. yoelii NL genome sequencing project. Analysis of the complete gene identified the presence of two introns. The ORF encodes a 102,407-Da protein with an amino-terminal signal sequence, a series of three unique types of repeats, and a cysteine-rich region. The binding site of MAb 48F8 was also identified. A BLAST search with the deduced amino acid sequence shows significant similarity with the Toxoplasma gondii RON4 protein and the Plasmodium falciparum RON4 protein, and the sequence is highly conserved in other Plasmodium species. We produced the cysteine-rich domain of PyP140/RON4 by using the Pichia pastoris expression system and characterized the recombinant protein biochemically and biophysically. BALB/c mice immunized with the protein formulated in oil-in-water adjuvants produced antibodies that recognize parasitized erythrocytes by IFA and native PyP140/RON4 by immunoblotting but failed to protect against a lethal P. yoelii YM infection. Our results show that PyP140/RON4 is located within the rhoptries or micronemes. It may associate in part with AMA1, but the conserved cysteine-rich domain does not appear to elicit inhibitory antibodies, a finding that is supported by the marked sequence conservation in this protein within Plasmodium spp., suggesting that it is not under immune pressure.
The leading blood-stage malaria vaccine candidate is apical membrane antigen 1 (AMA1). The protection observed in rodent (3, 20) and primate (7, 26) malaria challenge studies has led to AMA1's evaluation in multiple human clinical trials (8, 16, 24). AMA1 is a merozoite protein located within micronemes that is a target of antibodies that neutralize invasion of erythrocytes in vitro (13, 14, 18, 27). The specific function of AMA1 remains unclear. Rat monoclonal antibodies (MAbs) and their Fab fragments against native Plasmodium knowlesi AMA1 (PkAMA1; formerly called Pk66) inhibited merozoite invasion in vitro (6, 27), consistent with the idea that AMA1 is a parasite ligand involved in merozoite invasion of erythrocytes. Plasmodium yoelii AMA1 (PyAMA1) has been identified as an erythrocyte binding protein based on COS cell binding studies, and a PyAMA1-specific conformation-dependent MAb, 45B1, blocks this ligand-receptor interaction (9). PyAMA1 was also reported to form a complex with a 140-kDa protein (PyP140) located at the apical end of the merozoite. In addition, PyAMA1 and PyP140 were both identified on the surfaces of free merozoites, although the frequency of merozoites positive for surface PyP140 was markedly lower (20). The difference in the frequencies of merozoites with protein on the surface observed between PyP140 and PyAMA1 supported a differential release of these two proteins from the apical organelles (micronemes and/or rhoptries). Analysis of the crystal structure of Plasmodium falciparum AMA1 (PfAMA1) identified conserved PAN domain structures (23) and the presence of a conserved hydrophobic trough that is ringed by polymorphic residues (4), which together support a potential receptor-binding function during merozoite invasion.
An orthologue of AMA1 has been identified in the apicomplexan parasite Toxoplasma gondii (12). T. gondii AMA1 (TgAMA1) is a critical protein for host cell invasion (17) and is involved with the moving junction between parasite and host cell (2). Within the moving junction of the invasive tachyzoite, TgAMA1 is associated with rhoptry neck-associated proteins 2 and 4 (TgRON2 and TgRON4) (2). More recently, an orthologous protein to TgRON4 was identified in P. falciparum (PfRON4) by affinity purification from extracts of parasitized cells and by polypeptide analysis (1). No functional analysis of PfRON4 has been reported to date.
In this study, we report that the previously identified and characterized protein PyP140 (20) that associates with PyAMA1 is an orthologue of TgRON4 and PfRON4. Using a PyP140/RON4-specific MAb designated 48F8, we screened a P. yoelii λ-Zap cDNA expression library and identified and sequenced a clone which showed complete homology to part of the open reading frame of PY02159 from the P. yoelii NL genome sequencing project. We fully characterized the pyp140/ron4 gene and identified two introns. The primary deduced amino acid sequence comprises a signal peptide sequence, a region with a series of unique repetitive sequences, and a more-conserved high-complexity region defined by the presence of conserved cysteine residues. We also identified the likely epitope recognized by MAb 48F8 by using phage display. Given the interest in AMA1 as a vaccine and the fact that PyP140/RON4 associates with PyAMA1, we were interested in evaluating the vaccine potential of PyP140/RON4 in the lethal P. yoelii-rodent model. The cysteine-rich region, which presumably forms disulfide bridges, was produced using the methylotrophic yeast Pichia pastoris. Purified recombinant PpPyP140/RON4 was characterized biochemically and biophysically. PpPyP140/RON4 was formulated in oil-in-water adjuvants and used to immunize mice, which were subsequently challenged with P. yoelii YM parasites. We report here that immunization with PpPyP140/RON4 did not protect against a lethal challenge infection in vivo.
MATERIALS AND METHODS
Parasites.
The cloned rodent parasite P. yoelii yoelii YM (29) was obtained from David Walliker, University of Edinburgh, and grown in BALB/c mice. Slides were prepared with thin smears of blood parasitized with P. yoelii YM as described previously (19). For challenge infections, infected erythrocytes were collected into phosphate-buffered saline (PBS)-heparin.
Antibody.
The mouse MAb 48F8 has been described previously (20). It recognizes a 140-kDa P. yoelii apical merozoite protein identified as PyP140/RON4 and is not protective upon passive immunization.
Screening of λ-Zap cDNA library and phage display library and gene sequence analysis.
To identify the pyp140/ron4 gene, a λ-Zap cDNA library was screened as recommended by the manufacturer (Stratagene, La Jolla, CA). Following a third round of screening for binding to MAb 48F8, individual phage were isolated and both strands of the cDNA were sequenced using primers derived from the plasmid and internal sequence. To identify the epitope recognized by MAb 48F8, the linear seven-mer Ph.D.-7 phage display library (New England Biolabs, Beverly, Massachusetts) was screened as recommended by the manufacturer. Following a third round of amplification for binding to MAb 48F8, individual phage were isolated, and the peptide sequences they displayed were deduced following DNA sequencing. The 5′ region (covering the intron-exon boundaries) of the pyp140/ron4 gene was amplified (forward primer, ATGTCTGGCCTAAGGTTATTTTATG; and reverse primer, GGTTCGAAGGATTATGTTCTGG) from both cDNA and genomic DNA, prepared as described previously (22), and sequenced directly.
Recombinant production of PpPyP140/RON4.
Recombinant PpPyP140/RON4 was produced using the methylotrophic yeast Pichia pastoris (Invitrogen, Carlsbad, CA). A synthetic gene (pyp140/ron4; GenBank accession number EU348749) optimized for mammalian codon usage was used for expression. Five putative N-linked glycosylation sites were identified in the native deduced amino acid sequence, based on the consensus sequence NXS/T. These five sites were knocked out by replacing the N with positionally conserved amino acids identified in the orthologous protein P. falciparum chr11.genefinder_174r (PlasmoDB), which has been reported as PfRON4 (1). Referencing the full-length PyP140/RON4 protein, the specific amino acid substitutions within the recombinant protein were N714K, N760H, N782H, N789K, and N822H. The synthetic gene was cloned into the XhoI and XbaI sites of the Pichia expression vector pPICZαA following the manufacturer's guidelines, and then the sequence of one clone (pPICZαA/PyP140/RON4) was verified (data not shown). The plasmid was linearized with SacI and used to transform a modified P. pastoris GS115 Mut+ strain overexpressing P. pastoris protein disulfide isomerase, and recombinant protein was produced in 5-liter bioreactors similarly to the method described previously (28). The secreted protein was purified by immobilized metal affinity chromatography using Ni-Sepharose FF resin; captured protein was eluted with 300 mM imidazole in buffer. The elution pool was dialyzed, loaded onto a cation-exchange column (SP Sepharose FF; GE Healthcare) equilibrated with 20 mM citrate-phosphate buffer (pH 4.7), and eluted with a gradient from 0 to 1 M NaCl prepared in equilibration buffer. The eluted PpPyP140/RON4 protein was polished on a Superdex 75 gel permeation column equilibrated with PBS, pH 7.4. The recombinant protein was stored at −80°C.
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on 4 to 20% gradient Tris-glycine polyacrylamide gels as described by the manufacturer (Invitrogen). For analysis under reducing conditions, samples were mixed with 5% (vol/vol) β-mercaptoethanol before electrophoresis. For immunoblot analysis, native proteins that had been transferred electrophoretically onto nitrocellulose membranes and stored at −80°C were used, essentially as previously described (20).
Analytical reverse-phase high-performance liquid chromatography (HPLC), N-terminal sequencing, and liquid chromatography-mass spectrometry.
Purified PpPyP140/RON4 protein was acidified with 10% (vol/vol) trifluoroacetic acid (TFA; Fisher Scientific) to a final concentration of 0.1% TFA (vol/vol) and then microcentrifuged at 13,000 rpm for 2 min. Approximately 100 μg of PpPyP140/RON4 was analyzed on a Vydac (Hesperia, CA) C4 reverse-phase column (2.1-mm internal diameter [ID] × 250 mm). The initial mobile phase combined 99% mobile phase A (0.1% [wt/vol] TFA in water) and 1% mobile phase B (0.1% [wt/vol] TFA in acetonitrile). PpPyP140/RON4 was eluted with increasing concentrations of mobile phase B, with the flow rate maintained at 0.2 ml/min over 60 min. The primary peak was collected, vacuum dried in organic mode (Eppendorf Vacufuge), and submitted to the NIAID Research Technology Branch (RTB/NIAID/NIH, Rockville, MD) for mass spectrometry measurements and N-terminal sequencing.
Analytical SEC-MALS-HPLC and quasi-elastic light scattering.
The purity, identity, and aggregation profile of purified PpPyP140/RON4 were analyzed using size-exclusion chromatography (SEC) with in-line multiangle light scattering (MALS), refractive index, and UV detection (25). A Waters (Milford, MA) model 2695 HPLC instrument, Waters model 2996 PDA detector, Wyatt (Santa Barbara, CA) Dawn EOS light-scattering detector, Wyatt quasi-elastic light-scattering detector, and Wyatt Optilab DSP refractive index detector were connected in series to acquire the data, and the Wyatt Astra V software suite was used for data processing. For protein separation, a Tosoh Bioscience (Montgomeryville, PA) TSK Gel G2000SWxl 7.8-mm-ID by 30-cm by 5-μm-particle-size analytical column was used with a TSK Gel Guard SWxl 6.0-mm-ID by 4-cm by 7-μm-particle-size guard column. The column was equilibrated in mobile phase (1.04 mM KH2PO4, 2.97 mM Na2HPO4, 308 mM NaCl, 0.02% azide, pH 7.4) for at least 60 min at 0.5 ml/min prior to sample injection. Prior to analysis, PpPyP140/RON4 was filtered through a 0.45-μm polyvinylidene difluoride centrifugal filtration device (Ultrafree-MC 0.45 μm PVDF; Durapore, Millipore, Bedford, MA) spun at 2,000 rpm for 2 min. Approximately 75 μg of sample was injected using an isocratic run at 0.50 ml/min in mobile phase for 60 min. For size comparisons, 10 μl of Bio-Rad (Hercules, CA) gel filtration standard HPLC standards was run, and a 25-μl injection of 5 mg/ml bovine serum albumin was run for EOS MALS detector normalization.
IFA.
For immunofluorescence assay (IFA), thin films of blood were prepared as previously described (21) and stored desiccated at −70°C. Thin films were made from blood with parasitemias ranging from 2 to 20% and were incubated with primary antibody at a 1/200 dilution for 60 min. The slides were washed and then incubated for 60 min with goat anti-mouse immunoglobulin G (IgG) conjugated to fluorescein (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD). Following further washes, the slides were examined by fluorescence microscopy.
ELISA.
Enzyme-linked immunosorbent assays (ELISAs) were performed using a standardized procedure as described by Miura et al. (18a). Briefly, ELISA plates were coated with 100 ng/well of PpPyP140/RON4 overnight, and after blocking of the plates, diluted test sera were added in triplicate and incubated for 2 h at room temperature. After being washed extensively, the plates were incubated with 0.1 μg/well of anti-mouse IgG(H+L) antibody (Kirkegaard & Perry). Bound antibodies were visualized by adding p-nitrophenyl phosphate (Sigma 104) substrate, and the absorbance at 405 nm was read using a SPECTRAmax 340PC microplate reader. Pooled mouse anti-PpPyP140/RON4-positive control sera served as a reference standard for the assay. Duplicates of serially diluted reference sera were included on each ELISA plate and were assigned ELISA unit values as the reciprocals of the dilutions giving an optical density at 405 nm of 1. The standard curve obtained with the reference sera was used to convert the absorbance values of individual test sera into antibody units (SOFTmax PRO v.3; Molecular Devices).
Immunization-challenge study.
Three different immunization-challenge studies were done with mice immunized with PpPyP140/RON4 protein in compliance with an NIH Animal Care and Use Committee-approved protocol (ASP MVDB118E). The first was done with two groups of 10 mice immunized using antigen formulated with Montanide ISA720 (Seppic) adjuvant; control mice received saline in the same adjuvant. Formulations were prepared by vortex mixing for 30 min. Mice were immunized three times subcutaneously at 3-week intervals with 25 μg antigen. Two additional experiments were performed following the schedule above, using groups of five BALB/c mice immunized three times with 25 μg PpPyP140/RON4 protein or groups of seven BALB/c mice immunized with 25 μg or 75 μg antigen emulsified in either complete Freund's adjuvant (first immunization) or incomplete Freund's adjuvant; control mice received saline in the same adjuvant. Two weeks after the last immunization, mice were inoculated intravenously or intraperitoneally with 10,000 P. yoelii YM-parasitized red blood cells. Resulting parasitemias were monitored beginning on day 3 postchallenge by determining the percentages of parasitized red blood cells in thin tail-blood smears stained with Giemsa stain. Mice were removed from the study and euthanized if the parasitemia exceeded 60% or they showed predetermined signs of morbidity. Such infections were classified as lethal.
Statistical analysis.
Parasitemias in immunized and control groups were compared on day 6 after parasite challenge infection by using Mann-Whitney analysis.
RESULTS
Isolation of the pyp140/ron4 gene and identification of a putative mimotope for the PyP140/RON4-specific MAb 48F8.
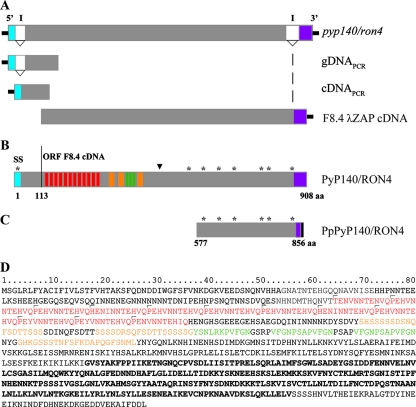
To identify the gene corresponding to the protein recognized by MAb 48F8, a λ-Zap cDNA expression library was screened by immunoblotting. One phage, designated F8.4, was identified as reacting with MAb 48F8. The cDNA was excised, and both DNA strands were sequenced. The open reading frame (ORF) corresponded to amino acids 113 to 908 (Fig. 1A and B) of PY02159 in the P. yoelii NL genome project database, identified previously as an orthologue of T. gondii RON4 (15). No nucleotide differences were identified between the P. yoelii NL and YM lines (data not shown). The 5′ end of the P. yoelii YM p140/ron4 gene was characterized by reverse transcription-PCR, using primers in the 5′ untranslated region and internal to the sequence within phage F8.4 and both genomic DNA and cDNA. Sequence analysis indicated that as predicted for the PY02159 gene, an intron was positioned between nucleotides 79 and 254 (data not shown). Based on the cDNA sequence and the ORF in phage clone F8.4, an additional intron was identified close to the 3′ end of the gene (nucleotide positions 2710 to 2946) (Fig. 1A and B).
FIG. 1.
Molecular schematic of the PyP140/RON4 gene and deduced protein. (A) pyp140/ron4 gene schematic, including two introns (I) that have been confirmed by analysis of cDNA and genomic DNA products and a λ-Zap cDNA clone (F8.4). (B) ORF (908 residues) for the deduced full-length protein, including the signal sequence (SS; blue), the locations of the three types of repetitive regions (red, orange, and green), the positions of eight cysteine residues (*), and the location of the epitope recognized by MAb 48F8 (▾). Shown in purple is the ORF from exon 3. The solid line positioned at amino acid 113 identifies the first amino acid deduced from the ORF of the F8.4 phage clone. (C) Cysteine-rich region of PyP140/RON4 expressed in P. pastoris. The solid black region at the carboxyl end of PpPyP140/RON4 represents the His6 affinity tag. (D) Deduced amino acid sequence of Py140/RON4, with the different repeats shown in red, orange, and green and the region expressed in P. pastoris shown in bold.
pyp140/ron4 encodes a protein with a mass of 102,406.8 Da consisting of a putative signal peptide (residues 1 to 24), a region comprised of a unique set of three types of repetitive sequence, and a cysteine-rich region containing seven cysteine residues, which likely forms a highly structured domain (Fig. 1B and D). Analysis of the repetitive sequence using the rapid automatic detection and alignment of repeat (radar) algorithm (http://www.ebi.ac.uk/radar/) and manual alignment indicated the presence of a series of 12 11-amino-acid repeats with the consensus sequence P/H/TEH/N/YV/INNTEHV/IQ. This first region is followed by a series of three approximately 20-residue repeats and three approximately 11-residue repeats (Fig. 1D). These repetitive regions are unique to P140/RON4 proteins of the rodent malaria organisms (data not shown). Compared with PfRON4 and TgRON4, the greatest level of overall amino acid homology is within the cysteine-rich region. Of the cysteine residues, six of seven are positionally conserved in PfRON4, while five of seven are positionally conserved in TgRON4. The cysteine residues identified in P140/RON4 proteins of the rodent malaria parasites Plasmodium berghei and Plasmodium chabaudi are completely conserved, and there is also a marked conservation of the repeat regions (data not shown). No putative membrane-spanning domain was identified, suggesting that PyP140/RON4 is a soluble protein.
To try to identify the epitope recognized by MAb 48F8, a random linear seven-mer peptide phage display library was screened by panning, and phage binding to the antibody were isolated. The deduced seven-residue sequences derived from 10 selected phage clones were aligned with the PyP140/RON4 amino acid sequence. One unique clone yielded the best alignment, with four identical amino acids (bold), two conserved residues (underlined), and one neutral substitution (PyP140/RON4 sequence SFKDAPQ versus 48F8PEP12 sequence TFKDQPE). This potential PyP140/RON4 mimotope maps to residues 414 to 420, located between the amino-terminal repeat region and the carboxyl-terminal cysteine-rich region (Fig. 1B). Based on the homology, the result suggests that this sequence is likely to be the epitope recognized by MAb 48F8.
Expression, purification, and characterization of the cysteine-rich region of PyP140/RON4.
For many malarial antigens, domains that contain cysteine residues likely to form disulfide bridges and ordered three-dimensional structures are of interest for their biological function and as a target for antibodies, which may incriminate them as vaccine candidates. Therefore, we expressed the cysteine-rich region of PyP140/RON4 (Fig. 1D, amino acids 577 to 856 [in bold]), using the methylotrophic yeast P. pastoris, as a secreted protein with a carboxyl-terminal His6 tag. In order to eliminate the presence of N-linked mannosylation, which is commonly found in heterologous proteins expressed in P. pastoris (5, 10, 28), five putative N-linked sites were changed as described in Materials and Methods. The recombinant PpPyP140/RON4 protein was produced in 5-liter bioreactors and purified, again as described in Materials and Methods.
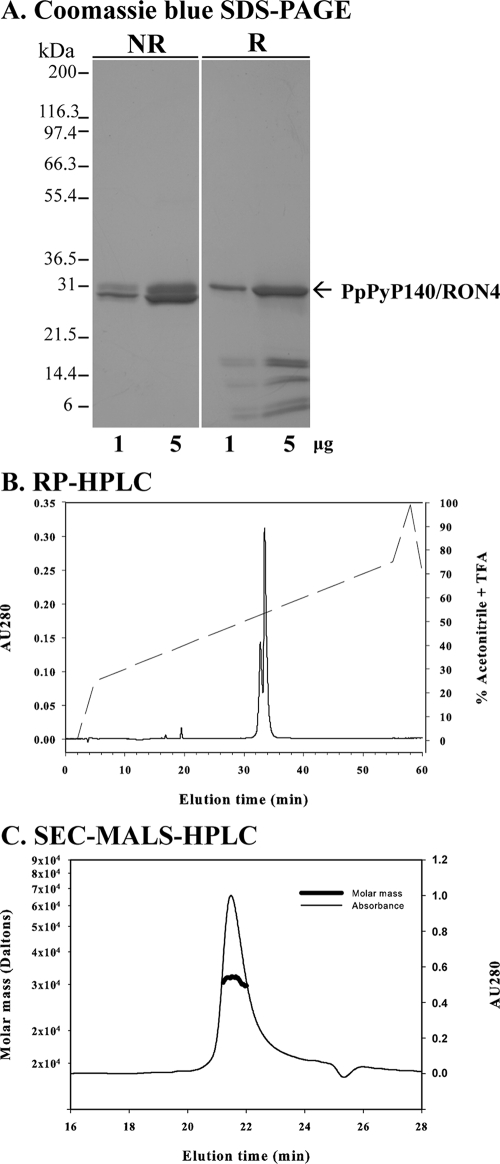
The purified PpPyP140/RON4 protein was biochemically and biophysically characterized. Analysis by Coomassie blue-stained SDS-PAGE showed that the purified PpPyP140/RON4 protein migrated as expected, at around 31 kDa, under reducing conditions (Fig. 2A). A proportion of the recombinant protein was observed to be nicked and visible as several bands under reducing conditions, which has been observed for other malarial proteins expressed in P. pastoris (11). Analysis by reverse-phase HPLC identified two forms of PpPyP140/RON4 protein, representing ∼70% and 30% of the protein (Fig. 2B). It is likely that the smaller peak represents the upper band(s) observed by SDS-PAGE under nonreducing conditions (Fig. 2A). Analysis of the amino terminus by Edman degradation identified three sequences: a portion of the purified protein had an intact amino terminus (GVSYAKFPPIIK), and there was a mixture of two amino-terminal truncations, identified as AKFPPIIK and KFPPIIK. Mass spectroscopy also identified the three major molecular forms observed by amino-terminal sequencing. The observed masses of the oxidized forms were 32,383.6 Da, 31,977.3 Da, and 31,907.1 Da (data not shown), in agreement with the theoretical masses, 32,379.9 Da, 31,973.6 Da, and 31,902.5 Da, respectively, which indicate that the recombinant protein formed disulfide bridges and contained no additional posttranslational modifications. Finally, we analyzed PpPyP140/RON4 in solution by SEC-MALS-HPLC. The purified recombinant protein migrated as a monomeric tightly folded globular protein (hydrodynamic radius, 2.2 nm) in PBS (Fig. 2C).
FIG. 2.
Biochemical and biophysical analysis of purified PyP140/RON4. (A) Purified recombinant PyP140/RON4 protein analyzed by Coomassie blue-stained SDS-PAGE under nonreducing (NR) and reducing (R) conditions. (B and C) Results obtained by reverse-phase HPLC (RP-HPLC) (B) and analytical SEC-MALS-HPLC (thick line indicates molar mass) (C). AU, absorbance units.
Immunization with PpPyP140/RON4, antibody specificity, and outcome of parasite challenge.
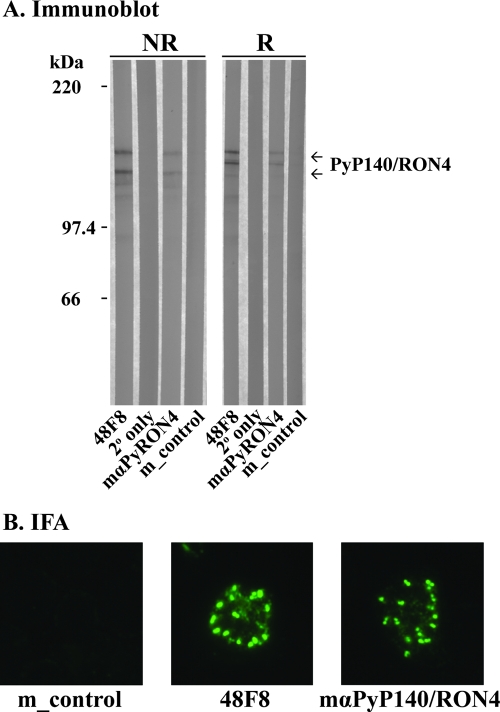
Groups of 10 or 5 BALB/c mice were immunized with PpPyP140/RON4 protein or PBS formulated in either Montanide ISA 720 or Freund's adjuvant. The PBS group was used as a negative control. ELISA titers of the individual mice from the Montanide ISA 720 group on day 42 ranged from 30,491 to 194,656 units (average ± standard deviation, 122,531 ± 59,435 units). The antibodies from mice immunized with PpPyP140/RON4, using either adjuvant, recognized native PyP140/RON4 protein in immunoblots (Fig. 3A) and parasitized erythrocytes by IFA (Fig. 3B). MAb 48F8 was used as a positive control for each test (Fig. 3A and B). Two forms of native PyP140/RON4 were identified by immunoblotting, as previously observed (20). The antibodies located PyP140/RON4 at the apical end of the merozoite, yielding a double-dot punctate pattern typical of a rhoptry protein. After challenge with P. yoelii YM-infected parasites, the antibody titer was measured again, and parasitemias were scored on consecutive days.
FIG. 3.
Evaluation of PpPyP140/RON4-specific mouse antiserum. (A) Immunoblot. Pooled anti-PpPyP140/RON4 specific mouse serum (mαPyRON4) was blotted against purified native PyP140/RON4 protein under reducing (R) and nonreducing (NR) conditions. MAb 48F8 was included as a positive control, and appropriate negative controls are shown (secondary antibody alone [2° only] or pooled sera from mice immunized with PBS alone [m_control]). (B) IFA of PyP140/RON4 location in mature schizont-infected erythrocytes stained with MAb 48F8, antisera from mice immunized with PpPyP140/RON4, or control mouse serum (m_control).
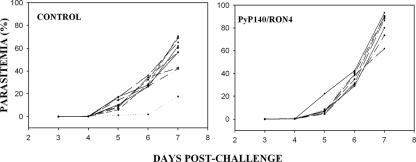
To determine whether immunization with PyP140/RON4 could elicit immune responses which could protect mice against an otherwise lethal challenge infection, mice immunized three times with antigen formulated with Montanide ISA720 were infected with P. yoelii YM-parasitized red cells; mice given saline in the same adjuvant served as controls. After challenge, parasitemias were scored on consecutive days, beginning on day 3. The results are shown in Fig. 4. All of the control mice but one were sacrificed on day 7, with parasitemias of >60% or signs of severe morbidity (Fig. 4A). Similarly, all of the test mice were euthanized on day 7, so there was no apparent protection conferred by the immunization regimen. To confirm and extend these results, two additional immunization-challenge studies were performed with test and control mice immunized using Freund's adjuvant with 25 μg or a dose escalation from 25 to 75 μg. Similarly, all mice either died or were euthanized on or before day 8, so there was no apparent protection as a result of PyP140/RON4 immunization (data not shown).
FIG. 4.
Course of P. yoelii YM infection in individual BALB/c mice within groups of 9 or 10 mice immunized with either PpPyP140/RON4 or PBS (control) formulated in Montanide ISA720. Following challenge with parasites, the parasitemia was monitored by microscopy each day for 7 days.
DISCUSSION
Our earlier studies to characterize the role of PyAMA1 demonstrated that it formed a protein complex with a 140-kDa apical protein, identified as PyP140 (20). We reported that PyP140 was localized to the apical end of the merozoite, likely within the rhoptries or micronemes, and that PyP140 could be distributed to the merozoite surface independent of AMA1 (20). Regarding P. falciparum, a protein called PfRON4 was more recently identified as forming a complex with PfAMA1 (1). PfRON4 was identified by similarity with a protein in another apicomplexan parasite, T. gondii, which contains proteins orthologous to PfAMA1 and PfRON4. Both of these apical proteins in T. gondii play a significant role in tachyzoite invasion of cells; in particular, TgRON4 is localized to the junctional ring of the invading parasite (2).
We report here the identification of the gene coding for PyP140 and its further characterization. The PyP140 gene corresponds to the gene designated PY02159 in the P. yoelii NL genomic sequencing project. By comparison of genomic DNA and cDNA, we identified two introns and no nucleotide polymorphisms in the gene between the two parasite lines. Most importantly, the protein is orthologous to PfRON4 and TgRON4 and therefore is designated PyP140/RON4. The deduced amino acid sequence encodes a protein of 102,407 Da, which is smaller than the 140-kDa mass previously determined by migration on SDS-PAGE under reducing conditions (20). It is likely that the difference is due to an anomalous migration on SDS-PAGE, perhaps due to the repetitive amino acid sequence, which has been observed for other malarial proteins, in particular PfRON4 (1).
Comparison of the PyP140/RON4 amino acid sequence with others in the databases identified a unique set of repeats which was shared by other rodent (P. berghei and P. chabaudi) malaria parasites. The function of these repeats is unknown. Based on their overall conservation, it appears unlikely that they are low-complexity regions present to “mask” or bias the immune response against a more structured carboxyl-terminal domain of PyP140/RON4. With respect to the 12 11-amino-acid repeats, the presence of proline and histidine suggests that these repeats may be a structural element, although computational modeling failed to identify any known structure (D. Hurt and D. L. Narum, unpublished results). The region of RON4 proteins that shares the greatest degree of similarity between P. yoelii, P. falciparum, and T. gondii is toward the carboxyl end of the proteins, which contain seven, six, and five cysteines, respectively. Five cysteines are conserved within all three proteins, while six of seven are conserved between P. yoelii and P. falciparum RON4 proteins. Other structural amino acids, such as proline and tryptophan, are also conserved.
PyP140/RON4 has a signal peptide and is therefore probably targeted to the secretory pathway and the lumen of the rhoptries, but in the absence of a putative transmembrane region PyP140/RON4 is likely to be a soluble protein. PyP140/RON4 appears to be present in both free and bound forms, and its association with AMA1 is therefore likely to be mediated by interaction with the ectodomain of AMA1. The structural basis of this interaction is unclear, but it is unlikely to include sequences that include the epitope of the AMA1-specific MAb 45B1 (19), since this antibody precipitates the PyAMA1/RON4 complex (20). The epitope for MAb 48F8 has been identified in this study in a region between the N-terminal repetitive sequence and the C-terminal cysteine-rich domain of PyP140/RON4; this region could be important in the interaction between PyP140/RON4 and AMA1 because MAb 48F8 appears to recognize free PyP140/RON4 and does not appear to precipitate the complex (20). Once AMA1 is bound, the epitope for MAb 48F8 may be occluded.
It was of interest to examine the potential of PyP140/RON4 as a vaccine candidate. AMA1 is an important blood-stage malaria vaccine target, and we showed previously that AMA1 purified from the parasite was capable of inducing a host-protective response (20). However, such material would include PyP140/RON4-AMA1 complexes in addition to free AMA1. Although the PyP140/RON4-specific MAb 48F8 was not protective on passive immunization (20), the possibility remained that immunity to PyP140/RON4 was an important component in the protection induced by immunization with the complex. To address this issue, we expressed the conserved cysteine-rich region of PyP140/RON4 in yeast cells, purified the protein, and used it in vaccination and challenge experiments.
The purified recombinant protein delivered in two different strong adjuvant systems was immunogenic and induced high-titer antibodies that reacted with the native Py140/RON4 protein by Western blotting and localized to the apical end of merozoites by IFAs. Despite this good response, immunization was unable to protect groups of mice against challenge infection. It is possible that other parts of the molecule, such as the N-terminal repetitive sequences, may be the target of protective immunity, but this was not tested here.
Further studies are required to understand the functional significance of the binding of PyP140/RON4 to AMA1 and the role of this protein in the invasion of red blood cells by the malarial merozoite.
Acknowledgments
We express our gratitude to Louis Miller for insightful discussions regarding PyP140, to Kim Lee Sim for support of this project while D.L.N. was employed by EntreMed, Inc., and to Michael Fay for assistance with the statistical analysis of parasitemias. We also thank Tin Luu (EntreMed, Inc.) for performing the phage display assays, Kelly Magee and Angela Lunger for assistance with the mouse immunization-challenge study, and Ababacar Diouf for performing the ELISAs.
This work was supported in part by the Intramural Research Program of the NIH and by the Medical Research Council (United Kingdom).
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Alexander, D. L., S. Arastu-Kapur, J. F. Dubremetz, and J. C. Boothroyd. 2006. Plasmodium falciparum AMA1 binds a rhoptry neck protein homologous to TgRON4, a component of the moving junction in Toxoplasma gondii. Eukaryot. Cell 51169-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, D. L., J. Mital, G. E. Ward, P. Bradley, and J. C. Boothroyd. 2005. Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 1e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16240-247. [DOI] [PubMed] [Google Scholar]
- 4.Bai, T., M. Becker, A. Gupta, P. Strike, V. J. Murphy, R. F. Anders, and A. H. Batchelor. 2005. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc. Natl. Acad. Sci. USA 10212736-12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly, R., and M. T. Hearn. 2005. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J. Mol. Recognit. 18119-138. [DOI] [PubMed] [Google Scholar]
- 6.Deans, J. A., T. Alderson, A. W. Thomas, G. H. Mitchell, E. S. Lennox, and S. Cohen. 1982. Rat monoclonal antibodies which inhibit the in vitro multiplication of Plasmodium knowlesi. Clin. Exp. Immunol. 49297-309. [PMC free article] [PubMed] [Google Scholar]
- 7.Deans, J. A., A. M. Knight, W. C. Jean, A. P. Waters, S. Cohen, and G. H. Mitchell. 1988. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol. 10535-552. [DOI] [PubMed] [Google Scholar]
- 8.Dicko, A., D. J. Diemert, I. Sagara, M. Sogoba, M. B. Niambele, M. H. Assadou, O. Guindo, B. Kamate, M. Baby, M. Sissoko, E. M. Malkin, M. P. Fay, M. A. Thera, K. Miura, A. Dolo, D. A. Diallo, G. E. Mullen, C. A. Long, A. Saul, O. Doumbo, and L. H. Miller. 2007. Impact of a Plasmodium falciparum AMA1 vaccine on antibody responses in adult Malians. PLoS ONE 2e1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser, T. S., S. H. Kappe, D. L. Narum, K. M. VanBuskirk, and J. H. Adams. 2001. Erythrocyte-binding activity of Plasmodium yoelii apical membrane antigen-1 expressed on the surface of transfected COS-7 cells. Mol. Biochem. Parasitol. 11749-59. [DOI] [PubMed] [Google Scholar]
- 10.Gemmill, T. R., and R. B. Trimble. 1999. Overview of N- and O-linked oligosaccharide structures found in various yeast species. Biochim. Biophys. Acta 1426227-237. [DOI] [PubMed] [Google Scholar]
- 11.Giersing, B., K. Miura, R. Shimp, J. Wang, H. Zhou, A. Orcutt, A. Stowers, A. Saul, L. H. Miller, C. Long, and S. Singh. 2005. Posttranslational modification of recombinant Plasmodium falciparum apical membrane antigen 1: impact on functional immune responses to a malaria vaccine candidate. Infect. Immun. 733963-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hehl, A. B., C. Lekutis, M. E. Grigg, P. J. Bradley, J. F. Dubremetz, E. Ortega-Barria, and J. C. Boothroyd. 2000. Toxoplasma gondii homologue of Plasmodium apical membrane antigen 1 is involved in invasion of host cells. Infect. Immun. 687078-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kocken, C. H., D. L. Narum, A. Massougbodji, B. Ayivi, M. A. Dubbeld, A. van der Wel, D. J. Conway, A. Sanni, and A. W. Thomas. 2000. Molecular characterisation of Plasmodium reichenowi apical membrane antigen-1 (AMA-1), comparison with P. falciparum AMA-1, and antibody-mediated inhibition of red cell invasion. Mol. Biochem. Parasitol. 109147-156. [DOI] [PubMed] [Google Scholar]
- 14.Kocken, C. H., A. M. van der Wel, M. A. Dubbeld, D. L. Narum, F. M. van de Rijke, G. J. van Gemert, X. van der Linde, L. H. Bannister, C. Janse, A. P. Waters, and A. W. Thomas. 1998. Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. J. Biol. Chem. 27315119-15124. [DOI] [PubMed] [Google Scholar]
- 15.Lebrun, M., A. Michelin, H. El Hajj, J. Poncet, P. J. Bradley, H. Vial, and J. F. Dubremetz. 2005. The rhoptry neck protein RON4 re-localizes at the moving junction during Toxoplasma gondii invasion. Cell. Microbiol. 71823-1833. [DOI] [PubMed] [Google Scholar]
- 16.Malkin, E. M., D. J. Diemert, J. H. McArthur, J. R. Perreault, A. P. Miles, B. K. Giersing, G. E. Mullen, A. Orcutt, O. Muratova, M. Awkal, H. Zhou, J. Wang, A. Stowers, C. A. Long, S. Mahanty, L. H. Miller, A. Saul, and A. P. Durbin. 2005. Phase 1 clinical trial of apical membrane antigen 1: an asexual blood-stage vaccine for Plasmodium falciparum malaria. Infect. Immun. 733677-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mital, J., M. Meissner, D. Soldati, and G. E. Ward. 2005. Conditional expression of Toxoplasma gondii apical membrane antigen-1 (TgAMA1) demonstrates that TgAMA1 plays a critical role in host cell invasion. Mol. Biol. Cell 164341-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell, G. H., A. W. Thomas, G. Margos, A. R. Dluzewski, and L. H. Bannister. 2004. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect. Immun. 72154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Miura, K., A. C. Orcutt, O. V. Muratova, L. H. Miller, A. Saul, and C. A. Long. 2008. Development and characterization of a standardized ELISA including a reference serum on each plate to detect antibodies by experimental malaria vaccines. Vaccine 26193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Narum, D. L., S. A. Ogun, A. H. Batchelor, and A. A. Holder. 2006. Passive immunization with a multicomponent vaccine against conserved domains of apical membrane antigen 1 and 235-kilodalton rhoptry proteins protects mice against Plasmodium yoelii blood-stage challenge infection. Infect. Immun. 745529-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narum, D. L., S. A. Ogun, A. W. Thomas, and A. A. Holder. 2000. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect. Immun. 682899-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narum, D. L., and A. W. Thomas. 1994. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 6759-68. [DOI] [PubMed] [Google Scholar]
- 22.Ogun, S. A., S. A. Howell, H. M. Taylor, and A. A. Holder. 2006. A member of the py235 gene family of Plasmodium yoelii encodes an erythrocyte binding protein recognised by a protective monoclonal antibody. Mol. Biochem. Parasitol. 147140-143. [DOI] [PubMed] [Google Scholar]
- 23.Pizarro, J. C., B. Vulliez-Le Normand, M. L. Chesne-Seck, C. R. Collins, C. Withers-Martinez, F. Hackett, M. J. Blackman, B. W. Faber, E. J. Remarque, C. H. Kocken, A. W. Thomas, and G. A. Bentley. 2005. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science 308408-411. [DOI] [PubMed] [Google Scholar]
- 24.Saul, A., G. Lawrence, A. Allworth, S. Elliott, K. Anderson, C. Rzepczyk, L. B. Martin, D. Taylor, D. P. Eisen, D. O. Irving, D. Pye, P. E. Crewther, A. N. Hodder, V. J. Murphy, and R. F. Anders. 2005. A human phase 1 vaccine clinical trial of the Plasmodium falciparum malaria vaccine candidate apical membrane antigen 1 in Montanide ISA720 adjuvant. Vaccine 233076-3083. [DOI] [PubMed] [Google Scholar]
- 25.Shimp, R. L., Jr., L. B. Martin, Y. Zhang, B. S. Henderson, P. Duggan, N. J. MacDonald, J. Lebowitz, A. Saul, and D. L. Narum. 2006. Production and characterization of clinical grade Escherichia coli derived Plasmodium falciparum 42 kDa merozoite surface protein 1 (MSP1(42)) in the absence of an affinity tag. Protein Expr. Purif. 5058-67. [DOI] [PubMed] [Google Scholar]
- 26.Stowers, A. W., M. C. Kennedy, B. P. Keegan, A. Saul, C. A. Long, and L. H. Miller. 2002. Vaccination of monkeys with recombinant Plasmodium falciparum apical membrane antigen 1 confers protection against blood-stage malaria. Infect. Immun. 706961-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas, A. W., J. A. Deans, G. H. Mitchell, T. Alderson, and S. Cohen. 1984. The Fab fragments of monoclonal IgG to a merozoite surface antigen inhibit Plasmodium knowlesi invasion of erythrocytes. Mol. Biochem. Parasitol. 13187-199. [DOI] [PubMed] [Google Scholar]
- 28.Tsai, C. W., P. F. Duggan, R. L. Shimp, Jr., L. H. Miller, and D. L. Narum. 2006. Overproduction of Pichia pastoris or Plasmodium falciparum protein disulfide isomerase affects expression, folding and O-linked glycosylation of a malaria vaccine candidate expressed in P. pastoris. J. Biotechnol. 121458-470. [DOI] [PubMed] [Google Scholar]
- 29.Yoeli, M., B. Hargreaves, R. Carter, and D. Walliker. 1975. Sudden increase in virulence in a strain of Plasmodium berghei yoelii. Ann. Trop. Med. Parasitol. 69173-178. [DOI] [PubMed] [Google Scholar]