Abstract
Severe chlamydial disease typically occurs after previous infections and results from a hypersensitivity response that is also required for chlamydial elimination. Here, we quantitatively dissected the immune and disease responses to repeated Chlamydia pneumoniae lung infection by multivariate modeling with four dichotomous effects: mouse strain (A/J or C57BL/6), dietary protein content (14% protein and 0.3% l-cysteine-0.9% l-arginine, or 24% protein and 0.5% l-cysteine-2.0% l-arginine), dietary antioxidant content (90 IU α-tocopherol/kg body weight versus 450 IU α-tocopherol/kg and 0.1% g l-ascorbate), and time course (3 or 10 days postinfection). Following intranasal C. pneumoniae challenge, C57BL/6 mice on a low-protein/low-antioxidant diet, but not C57BL/6 mice on other diets or A/J mice, exhibited profoundly suppressed early lung inflammatory and pan-T-cell (CD3δ+) and helper T-cell (CD45) responses on day 3 but later strongly exacerbated disease on day 10. Contrast analyses characterized severe C. pneumoniae disease as being a delayed-type hypersensitivity (DTH) response with increased lung macrophage and Th1 cell marker transcripts, increased Th1:Th2 ratios, and Th1 cytokine-driven inflammation. Results from functional analyses by DTH, enzyme-linked immunospot, and immunohistofluorescence assays were consistent with the results obtained by transcript analysis. Thus, chlamydial disease after secondary infection is a temporal dysregulation of the T-cell response characterized by a profoundly delayed T-helper cell response that results in a failure to eliminate the pathogen and provokes later pathological Th1 inflammation. This delayed T-cell response is under host genetic control and nutritional influence. The mechanism that temporally and quantitatively regulates the host T-cell population is the critical determinant in chlamydial pathogenesis.
Chlamydia pneumoniae, an obligate intracellular bacterium, is almost exclusively transmitted by the respiratory route and occurs with high frequency in the human population (28, 59). These ubiquitous, mainly asymptomatic and chronic, infections have been associated with respiratory diseases such as bronchitis, pneumonia, and otitis (28). Furthermore, the impact of C. pneumoniae infections has also been extended to chronic inflammatory diseases of presumably noninfectious etiology. Under conditions such as atherosclerosis, coronary heart disease, Alzheimer's disease, metabolic syndrome, or insulin resistance, which are of major public health concern, C. pneumoniae is thought to act as an accelerator of disease progression (9, 15, 59). To advance prophylactic and therapeutic strategies, it is imperative to better understanding pathogenic versus protective host responses to C. pneumoniae infection and how these responses may be modulated by the nutritional status of the host.
The complex interaction between chlamydial replication and host response determines the outcome of the infection not by a single all-influencing factor but rather by a series of accumulated host- and pathogen-associated factors (47). The range of the primary host responses to chlamydial infection may vary from vigorous, with inflammatory exudates and overt clinical symptoms, to minimal, without symptoms (28). The initial events that elicit the host's innate immune response may control the type and degree of the repair response that ensues and ultimately influence the outcome of a chlamydial infection (47). The shared and most common disease manifestations for chlamydial infections are chronic granulomatous lesions of mononuclear cell aggregates and fibrosis (55). However, only a small subset of infected individuals develops these hallmark granulomatous lesions, even after repeated infections (2), implying that adaptive immune responses may be protective as well as immunopathological. Recurrent infection with C. trachomatis is associated with scarring sequelae such as trachoma (20). Repeated ocular challenge was required to produce the corneal pannus and conjunctival scarring characteristic of severe trachoma (20, 64). Similarly, children receiving semipurified C. trachomatis vaccines exhibited vaccine-induced delayed-type hypersensitivity (DTH) responses and severe disease with higher clinical scores (60).
In addition to repeated exposure, human studies indicated a major influence of host genetics on disease severity following chlamydial infection (41). That observation was confirmed and extended in animal experiments (4, 8, 38). Inbred A/J and B6 mice differ profoundly in their inflammatory responses and susceptibilities to pulmonary infection with C. pneumoniae in regard to the lung chlamydial loads and lung pathology (30). After intranasal inoculation with C. pneumoniae, A/J mice initially eliminate C. pneumoniae slowly and show a low inflammatory response but do not develop major lung lesions. In contrast, B6 mice initially eliminate C. pneumoniae efficiently and mount an excessive inflammatory response that results in substantial lung disease (30). Min-Oo et al. (38) mapped a recessive murine quantitative trait locus that accounts for ∼30% of the variance in C. pneumoniae lung loads to a region on chromosome 17 that overlaps with the major histocompatibility complex locus.
As is the case for many other intracellular pathogens, CD4+ lymphocytes play a key role in a protective host response to Chlamydia infection (40, 49). They restrict chlamydial replication via Th1-type effector cytokines, most prominently gamma interferon (IFN-γ), contributing to a DTH response (45, 52). Such protective DTH responses are characterized by tissue infiltration of CD4+ T cells and macrophages and the release of proinflammatory Th1 cytokines such as interleukin-1 (IL-1), IL-2, IL-12, IFN-γ, or tumor necrosis factor alpha (TNF-α).
Ultimately, Th1 cytokines exert their main proinflammatory effects by stimulating the production of free radical molecules such as reactive oxygen species (ROS) and reactive nitrogen oxide species (RNOS) (62). These central effectors of inflammation and pathogen elimination also play a pivotal role in cellular signaling and the regulation of the immune response to chlamydial infection (23, 48). The metabolism of RNOS and ROS is influenced by the dietary supply of precursors such as protein and amino acids (e.g., l-arginine [RNOS]) or of antioxidative vitamins (e.g., l-ascorbate or α-tocopherol [ROS]) that regulate the cellular redox status. Protein deficiency consistently interferes with resistance to infection and may induce impaired antibody formation, decreases in thymic function, splenic lymphocytes, and delayed cutaneous hypersensitivity (32). The amino acid l-arginine, a key substrate in protein/nitrogen metabolism, is a crucial component of the immune response as a substrate for both inducible nitric oxide synthase (iNOS) and arginase in the cytotoxic pathways of macrophages. l-Cysteine acts as an important structural and functional component of many proteins and enzymes and is a precursor to the antioxidant glutathione with antioxidant properties. The water-soluble vitamin l-ascorbate participates in the maintenance of redox status and enhances the activation and survival of immune cells (13, 16, 77). α-Tocopherol is a lipid-soluble antioxidant that stabilizes membrane redox status and prevents lipid oxidation (13, 77). Differences in macro- and micronutrient contents of diets may result in changed free radical production and profound variation in outcomes of infectious diseases in rodent models (10, 36).
Malnutrition combined with chronic infectious diseases still forms the major impediment for advances in public health (13, 16, 56). In developed countries, dietary imbalances represent the main nutritional stress (13), while in developing countries, chronic deficiencies in both macro- and micronutrients dominate (56). Individuals with diverse genetic makeups respond differently to malnutrition and infections, indicating a pivotal role of host genetics in the triad interaction of nutrition, infection, and immunity/inflammation (25).
In this investigation, we explored the multifaceted relationships among malnutrition, immunity/inflammation, host genetics, and C. pneumoniae infection. We addressed the mechanisms of immunoprotective versus immunopathological responses to C. pneumoniae in a mouse model of lung disease and used a nutrient-dependent perturbation approach to create differential outcomes in inflammation and immunity to C. pneumoniae on the divergent genetic backgrounds of immune A/J and B6 mice. These mice were primed with a low dose of C. pneumoniae and subsequently rechallenged with a high-dose intranasal inoculation. They received one of four isocaloric diets, which differed in protein/l-arginine/l-cysteine and l-ascorbate/α-tocopherol contents. Disease intensity and chlamydial lung loads were determined early after rechallenge, i.e., on day 3 and later on day 10 at peak disease. In addition, transcript levels for key markers of the immune and inflammatory response were measured. Distinct early response patterns correlated with protective and pathological outcomes of the infection, and functional analyses confirmed the causal role of an early pathological response in precipitating later disease.
MATERIALS AND METHODS
Animals and diets.
Mouse protocols followed NIH guidelines and were approved by the Auburn University Institutional Animal Care and Use Committee. Inbred A/J and B6 female mice at 5 weeks of age were obtained from Harlan Sprague Dawley and received a 19% protein-1.33% l-arginine standard rodent maintenance diet (Harlan Teklad). Starting 2 weeks before challenge infection until euthanasia, mice (n = 10/group) were fed one of four isocaloric diets with high or low contents of protein and of antioxidants (antioxidative vitamins C and E) (high protein and high antioxidant [HH], high protein and low antioxidant [HL], low protein and high antioxidant [LH], and low protein and low antioxidant [LL]) (Table 1), which were modeled after the AIN-93 series of rodent diets (51) and manufactured by Harlan Teklad.
TABLE 1.
Compositions of four isocaloric custom rodent diets
| Dieta | Amt of component (g/kg)
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Casein | L-Cysteine | L-Arginine- HCl | Cornstarch | Maltodextrin | Sucrose | Soybean oil | Cellulose (fiber) | Mineral mix, AIN- 93G-MX | Calcium phosphate monobasic | Calcium carbonate | Vitamin mix, AIN- 93-VX | Choline bitartrate | TBHQ (antioxidant) | Vitamin E acetate (500 U/g) | Stay-C 35b | Food color (soluble) | |
| HH | 275.9 | 4.14 | 9.37 | 303.09 | 132 | 100 | 70 | 50 | 35 | 2.31 | 12 | 2.5 | 0.01 | 0.72 | 2.86 | 0.1 | |
| HL | 275.9 | 4.14 | 9.37 | 306.67 | 132 | 100 | 70 | 50 | 35 | 2.31 | 12 | 2.5 | 0.01 | 0.1 | |||
| LH | 160.92 | 2.41 | 2.46 | 426.88 | 132 | 100 | 70 | 50 | 35 | 2.02 | 0.12 | 12 | 2.5 | 0.01 | 0.72 | 2.86 | 0.1 |
| LL | 160.92 | 2.41 | 2.46 | 430.46 | 132 | 100 | 70 | 50 | 35 | 2.02 | 0.12 | 12 | 2.5 | 0.01 | 0.1 | ||
HH, 24% protein, 0.5% l-cysteine, 2.0% l-arginine, 450 IU α-tocopherol/kg, and 0.1% l-ascorbate; HL, 24% protein, 0.5% l-cysteine, 2.0% l-arginine, and 90 IU α-tocopherol/kg; LH, 14% protein, 0.3% l-cysteine, 0.9% l-arginine, 450 IU α-tocopherol/kg, and 0.1% l-ascorbate; LL, 14% protein, 0.3% l-cysteine, 0.9% l-arginine, and 90 IU α-tocopherol/kg.
Stable form of vitamin C, mainly monophosphate.
C. pneumoniae lung challenge infection.
C. pneumoniae strain CDC/CWL-029 (ATCC VR-1310) was grown, purified, and quantified as described previously (31). Six-week-old mice received a low-dose intranasal inoculation of 5 × 106 viable C. pneumoniae elementary bodies in 30 μl sucrose-phosphate-glutamate buffer. Four weeks later, the mice were challenged with 1 × 108 C. pneumoniae bacteria and sacrificed by CO2 inhalation 3 or 10 days later. Lungs were weighed, snap-frozen in liquid nitrogen, and stored at −80°C. The percent lung weight increase was calculated based on naïve lung weights of 138.4 mg for adult A/J mice and 133.0 mg for adult B6 mice (23, 24).
Nucleic acid extraction.
Mouse lungs were homogenized in guanidinium isothiocyanate Triton X-100-based RNA/DNA stabilization reagent in disposable tissue grinders (Fisher Scientific) to create a 10% (wt/vol) tissue suspension, and total nucleic acids were extracted by glass fiber matrix binding and elution (13). Poly(A)+ RNA was extracted after DNase treatment by use of oligo(dT)20-coated silica beads (Kisker GbR) (73).
Analysis of lung nucleic acids by real-time PCR.
The copy number of C. pneumoniae genomes per lung was determined by Chlamydia genus-specific 23S rRNA real-time fluorescence resonance energy transfer (FRET) PCR (14). Copy numbers of murine genomes per lung, as a marker of the total number of cells, were determined by real-time FRET PCR of an intron sequence of the single-copy porphobilinogen deaminase (PBGD) gene. The parameters for the PBGD intron PCR were 40 for 0 s at 95°C, 8 s at 58°C followed by fluorescence acquisition, and 8 s at 72°C. Concentrations of 24 transcripts (Table 2) were determined by one-step duplex FRET reverse transcription (RT)-PCR (72). Hypoxanthine guanine phosphoribosyltransferase mRNA was the internal reference for the Arg1 transcript, and PBGD mRNA was the internal reference for all other targets. Transcript numbers are expressed as copies per 1,000 reference transcripts. To prevent amplification from genomic DNA, PCRs were designed with primers and probes that bridged exons (see Table S1 in the supplemental material). For each target, duplex PCRs with different concentrations of the analyte and reference standards were used to obtain an equation that corrected sample PCR raw data according to the analyte/reference ratio of each sample (72).
TABLE 2.
Transcripts of cellular markers and inflammatory regulator genes
| Transcript | Marker fora: | Reference(s) |
|---|---|---|
| PBGD intron (no RT-PCR) | Total cells (single-copy gene; 3rd enzyme of the heme biosynthetic pathway) | 3 |
| Lactoferrin | Neutrophils (neutrophil granule antimicrobial protein) | 58 |
| F4/80 | Macrophages (macrophage surface antigen) | 35 |
| NKp46 | Natural killer cells (NK cell natural cytotoxicity-triggering receptor) | 46 |
| CD3δ | T cells (T cell specific, expressed T-cell receptor component) | 68 |
| Tim3 | T-helper 1 cells (CD4+ Th1 cell surface protein) | 39 |
| GATA3 | T-helper 2 cells (CD4+ Th2 cell-specific transcription factor) | 81 |
| CD45RB | Naïve T cells (surface protein expressed mainly on naïve T cells) | 22 |
| CD45RO | Memory T cells (surface protein expressed mainly on memory T cells) | 22, 54 |
| Perforin1 | Cytotoxic T cells (CD8+ cytotoxic T lymphocyte and NK cell cytolytic granule protein) | 66 |
| CD19 | B cells (B-cell surface protein) | 82 |
| Arg1 | Arginase 1 (cytosolic enzyme, liver isoform; regulation of arginine metabolism, NO production, and inflammation) | 63 |
| Arg2 | Arginase 2 (mitochondrial enzyme, extrahepatic isoform; regulation of arginine metabolism, NO production, and inflammation) | 57 |
| NOS2 | NOS2 (macrophage-produced nitric oxide synthase 2) | 34 |
| Cybb | Oxidative burst oxidase (cytochrome b-245, beta polypeptide [formerly gp91phox subunit of the phagocyte oxidative burst NADPH oxidase]) | 6 |
| Ptgs2 | Cyclooxygenase 2 (prostaglandin-endoperoxide synthase 2 [COX-2]) | 42 |
| IL-6 | IL-6 (inflammatory cytokine) | 70 |
| TNF-α | TNF-α (inflammatory cytokine) | 18 |
| IFN-γ | IFN-γ (main Th1 cytokine) | 19 |
| Cxcl2 | MIP-2 (neutrophil-recruiting chemokine, C-X-C motif ligand 2; homolog to human IL-8) | 65 |
| IL-4 | IL-4 (main Th2 cytokine) | 29 |
| IL-10 | IL-10 (Th2 cytokine) | 27 |
| Ccl2 | MCP-1 (C-C motif ligand 2) | 7 |
| CRP | CRP (inflammatory marker) | 43 |
| Serpine1 | Plasminogen activator inhibitor 1 (serine proteinase inhibitor E1, intravascular inhibition of fibrinolysis, modulator of tissue repair) | 78 |
MIP-2, macrophage inflammatory protein 2; MCP-1, monocyte chemotactic protein 1; CRP, C-reactive protein.
Immunofluorescence.
Mice were sacrificed, 2 ml Neg-50 (Richard-Allan Scientific) was infused into the lung through an incision in the trachea, and the lung was snap-frozen and stored at −80°C. Lung sections were cut to 5 μm, mounted onto Superfrost/Plus microscope slides (Fisher Scientific), fixed for 5 min with ice-cold acetone, air dried, and blocked with antibody dilution buffer (10% donkey serum-5% bovine serum albumin in phosphate-buffered saline [PBS]). Sections were stained with monoclonal rabbit anti-CD3δ (1:100 for 1 h) (clone SP7; Neomarkers), followed by Alexa Fluor 598 donkey anti-rabbit immunoglobulin G conjugate (1:400 for 1 h) (Invitrogen) and fluorescein isothiocyanate-conjugated anti-Chlamydia lipopolysaccharide monoclonal antibody (MAb) (1:10 for 0.5 h) (Fitzgerald). Nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) in mounting medium (Invitrogen). Immunofluorescence was examined with a Nikon Eclipse E800M microscope, and all images were captured with a digital Spot RT camera (Diagnostic Instruments). Microscope and camera settings were identical within experiments, and representative images are shown.
DTH and enzyme-linked immunospot (ELISPOT) assays.
Mice were intranasally challenged with C. pneumoniae (n = 10 mice/group) and were injected on day 2 postinfection (p.i.) into the footpads with 1.25 μg C. pneumoniae elementary body lysate in 20 μl PBS in one paw and PBS in the contralateral paw (23). Twenty-four hours after injection, footpad thickness was measured with a spring dial thickness gauge (Swiss Precision Instruments) to assess the DTH response as thickness increase (mm × 10−2) of antigen-injected over PBS-injected control footpads.
Following determinations of DTH levels, mice were sacrificed. Lungs were perfused with PBS, and mononuclear cells were obtained by cutting the lungs into small pieces and digesting them with collagenase type IV (Sigma-Aldrich), followed by filtration through a nylon strainer. The resulting cells were centrifuged over a Ficoll gradient for an ELISPOT assay (69). Mononuclear cells of each mouse were loaded in triplicates onto ELISPOT plates (Millipore) coated with anti-mouse IFN-γ MAb (BD Biosciences) and incubated overnight at 37°C. After extensive washing, biotinylated IFN-γ MAb (BD Biosciences) was added to the wells and incubated overnight at 4°C. Peroxidase-labeled goat anti-biotin (Vector Laboratories) was added to each well for 1 h at room temperature. The spots were developed for 15 to 30 min with the substrate 3-amino-9-ethylcarbazole and counted with a stereomicroscope.
Statistical analysis.
All statistical analyses were performed with the Statistica 7.0 software package (StatSoft, Inc.). C. pneumoniae genomes were log10 transformed, and transcript numbers were log2 transformed. Dietary protein and antioxidant effects and genetic effects on 26 outcome parameters were visualized in plots of the log2-transformed ratios of the mean values at the dichotomous-effect levels (67). Normal distribution of data was confirmed by Shapiro-Wilk's W test, and homogeneity of variances was confirmed by Levene's test. Data were analyzed by mean plots ± 95% confidence intervals (CI) in one-way or factorial analysis of variance. Comparisons of means under the assumption of no a priori hypothesis were performed by two-tailed Tukey honest significant difference (HSD) test. Differences at P values of ≤0.05 in the Tukey HSD test were considered to be significant.
RESULTS
The balanced multivariate design quantitatively dissected the outcome of C. pneumoniae lung infection in immune mice for four bimodal effects (24 = 16 groups, 10 mice/group). These effects were (i) mouse strain (A/J or B6), (ii) time after inoculation (3 or 10 days p.i.), (iii) dietary protein content (low, 14% protein, 0.3% l-cysteine, and 0.9% l-arginine; high, 24% protein, 0.5% l-cysteine, and 2.0% l-arginine) (Table 1), and (iv) dietary antioxidant content (low, 90 IU α-tocopherol/kg body weight; high, 0.1% g l-ascorbate and 450 IU α-tocopherol/kg) (Table 1). Two weeks after low-dose priming, the mice received one of four isocaloric diets: HH, HL, LH, and LL. The protein and antioxidant concentrations were close to the lower and upper limits but within the ranges recommended for rodent diets. After another 2 weeks, the mice received an intranasal high-dose C. pneumoniae challenge and were sacrificed 3 or 10 days p.i. These time points served to evaluate the early inflammatory response and approximately the peak of disease that typically occurs in week 2 p.i. (30, 80). Parameters measured were lung weight increase as disease intensity, total C. pneumoniae lung burden, and lung transcript profiles of cellular and inflammatory markers (Table 2 and see Table S1 in the supplemental material). The deduced protective versus pathological mechanisms were functionally confirmed and extended by DTH measurements, lung ELISPOT assay, and lung immunofluorescent histology.
B6 mice on low dietary protein/antioxidants develop severe disease and do not eliminate C. pneumoniae.
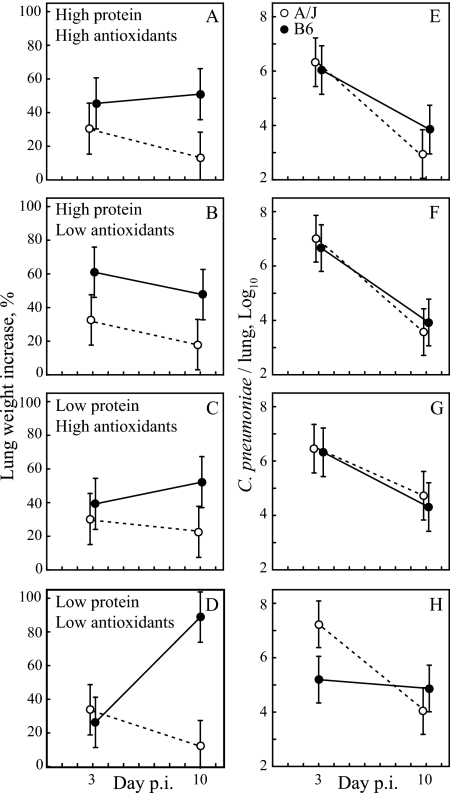
Figure 1 shows the time course of disease intensity and C. pneumoniae lung burdens for two mouse strains and four diets. Previous studies established that the degrees of interstitial cellular infiltration and cytokine production are proportional to the lung weight increase, which provides an accurate measurement of disease intensity (23, 24, 30). On HH, HL, and LH diets, B6 mice exhibited generally more severe disease than did A/J mice (Fig. 1A to C). Both mouse strains had similar early chlamydial lung loads of 106 to 107 organisms per lung (Fig. 1E to G) and reduced these loads by 100- to 1,000-fold between day 3 and day 10. The peak disease phenotype on day 10 conforms to data from previous reports of C. pneumoniae lung disease in murine models and is histopathologically characterized by extensive interstitial infiltrates of macrophages, lymphocytes, and neutrophils (24, 26, 80).
FIG. 1.
B6 mice, but not A/J mice, on an LL diet exhibit exacerbated late disease on day 10. Lung weight increases (over unchallenged mice) as a measure of disease (A to D) and lung C. pneumoniae loads (E to H) are shown for each of the four diets, two mouse strains, and two sacrificing time points. Immune B6 mice on the LL diet had significantly higher lung weight increases on day 10 p.i. than on day 3 (89% versus 26%, respectively; P = 0.0009 by Tukey HSD test) (D), while the lung weight increase of all other groups did not differ significantly between days 3 and 10 (A to D). C. pneumoniae lung loads of B6 mice on an LL diet on day 3 were significantly lower than those of all combined other groups (105.2 versus 106.6, respectively; P = 0.006) but significantly higher on day 10 (104.9 versus 103.9, respectively; P = 0.042). C. pneumoniae loads did not differ between days 3 and 10 for B6 mice on the LL diet (H) but were significantly lower (P ≤ 0.02) on day 10 for all other groups (E to H). Open circles, A/J mice; filled circles, B6 mice. Error bar, ±95% CI (n = 10 mice/group).
On the LL diet, A/J mice show disease and pathogen loads similar to those for both mouse strains on the three other diets (Fig. 1D and H). In stark contrast, B6 mice on the LL diet (B6-LL mice) showed minimal disease on day 3 but profoundly and significantly exacerbated peak disease on day 10 (Fig. 1D). Interestingly, B6-LL mice had a significantly lower C. pneumoniae lung load on day 3 than did other mice but a significantly higher pathogen load on day 10 than did the other groups combined (Fig. 1D). Overall, B6-LL mice progressed from minimal disease on day 3 to maximal disease on day 10, while they did not eliminate C. pneumoniae during that time. In contrast, B6 mice on any other diet or A/J mice showed no significant change in disease between days 3 and 10 but showed significant reductions of the chlamydial lung burden during that time. These results confirm the resistance of A/J mice to C. pneumoniae-induced disease independent of dietary protein and antioxidant contents. Histopathologically, A/J mice exhibited prominent peribronchiolar lymphocytic cuffs but virtually no interstitial pneumonitis. The data also identify low-protein and low-antioxidant dietary conditions under which the disease-prone B6 mice develop significantly more severe disease than they do under conditions of high-protein or high-antioxidant nutrition.
Low dietary protein exacerbates day 10 inflammation and disease in B6 mice.
The phenotype of B6 mice on the LL diet allowed us to identify the unique transcriptional attributes that separate severe disease in B6 mice from low-level disease in B6 or A/J mice. Figure 2 displays the relative impact of dietary protein, antioxidants, and host genetics on day 10 levels of marker transcripts (Table 2). (Original absolute data are shown in Table S2 in the supplemental material.) The difference is expressed as the log2 ratio of each B6 transcript (low protein or low antioxidants) divided by the respective A/J transcript (low protein or low antioxidants). Thus, a value of 1 for a parameter indicates a twofold increase, and a value of −1 indicates a twofold decrease to 50%. Transcripts of IL-4 and C-reactive protein were not detected in any specimen and were therefore not included in analyses.
FIG. 2.
Low dietary protein increases day 10 disease and inflammation in B6 mice. Day 10 lung parameters are contrasted between high-disease (B6-LL) and low-disease outcomes for the main effects dietary protein (A), dietary antioxidants (B), and host genetics (mouse strain) (C). The ratios are log2 transformed, and black bars represent significant differences, while gray bars indicate nonsignificance (n = 10 mice/group; P < 0.05 by Tukey HSD test). Protein reduction (LL versus HL diet) resulted in pronounced disease (lung weight [LW] increase) in B6 mice and prominently increased inflammation-associated transcripts such as IFN-γ, IL-10, IL-6, and TNF-α but essentially not cellular marker transcripts (A). Antioxidant reduction (LL versus LH diet) increased disease in B6 mice, with marginal transcript changes (B). The effect of host genetics showed the most profound differential of disease on the LL diet, with almost eightfold-increased levels of disease and 16-fold-increased levels of IFN-γ transcripts in B6 over A/J mice (C). Similar to the protein effect, the enhanced disease in B6 mice was accompanied by increased inflammatory transcripts but also cellular marker transcripts for a DTH response (F4/80, Tim3, and Tim3/GATA3). CPN, C. pneumoniae.
The striking observation is that dietary protein and host genetics (Fig. 2A and C), but not dietary antioxidants (Fig. 2B), profoundly affect the day 10 disease outcome. Protein reduction exacerbates disease from a 52% lung weight increase in B6-HL mice to 89% in B6-LL mice (Fig. 1B and D and see Table S2 in the supplemental material). The exacerbated disease in B6-LL mice is accompanied by strongly elevated global inflammatory effector and regulator transcripts (Ptgs2 Th1-associated IFN-γ and TNF-α and Th2-associated IL-6, IL-10, Arg1, and Arg2) but essentially unchanged cellular marker transcripts and total lung cells (Fig. 2A). These data indicate that enhanced lung cytokine production, but not an increased lung immune cell population, characterizes the disease induced in B6 mice by relative protein deficiency.
Host genetics exert an even greater influence on disease outcome, as evident in the 89% lung weight increase in B6 mice compared to the 12% increase in A/J mice on the same LL diet (Fig. 1D and see Table S2 in the supplemental material). This exacerbation is accompanied by an elevation not only of inflammatory transcripts (Arg1, Arg2, IL-6, and IFN-γ) but also of cellular marker transcripts (F4/80, Tim3, and Tim3/GATA3) (Fig. 2C). These data indicate that the severe disease of B6 mice in comparison to the completely disease-protected A/J mice is characterized by increased lung infiltration with macrophages and Th1 lymphocytes and by increased levels of inflammatory cytokine production in the lung. The B6-LL transcript profile encompasses the full spectrum of Th1 inflammation and is consistent with the DTH nature of C. pneumoniae lung disease.
Lower day 10 disease outcomes were observed for all other diets containing low protein but high antioxidants or high protein and either low or high antioxidants. Protein reduction on the background of high levels of dietary antioxidants also resulted in only B6 mice in a significant increase of day 10 inflammation but not of disease (see Fig. S1A1 to S1A3 and Table S2 in the supplemental material). Antioxidant reduction decreases the already low level of disease in A/J mice by almost twofold (22.6% versus 12.4% LW increase) and significantly modifies several leukocyte- and inflammation-associated transcripts (see Fig. S1B1 to S1B3 and Table S2 in the supplemental material). On all diets, B6 mice showed more severe disease than did A/J mice and, in general, elevated cellular marker transcripts (see Fig. S1C1 to S1C3 and Table S2 in the supplemental material) except for reduced pan-T-cell (CD3δ) transcripts with high dietary protein. Other changes in day 10 transcripts were marginal.
Overall, the LL diet reveals the complete range of C. pneumoniae disease outcomes in dependence on the host genetic background. Protein deficiency precipitates maximum disease in the susceptible B6 mice and maximum protection from disease in the resistant A/J mice in a genetically restricted manner. The disease phenotype is characterized by profoundly increased inflammation. These conditions therefore define both the dietary and genetic control of maximum amplitude between disease and protection (B6-LL mice versus B6-HL mice and B6-LL mice versus A/J-LL mice) and can be explored for the analysis of early events that precipitate later disease.
Delayed early T-cell and inflammatory responses prime immune B6 mice for severe disease.
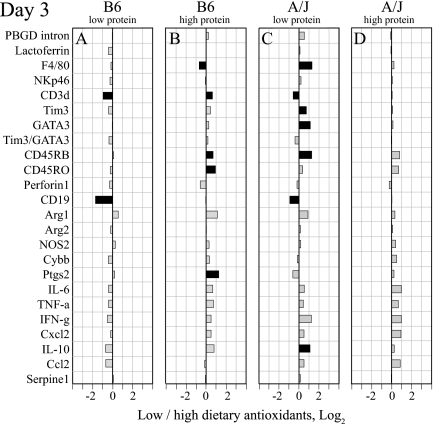
To develop an understanding of the mechanisms that result in pathological versus protective responses to C. pneumoniae infection, we contrasted day 3 lung transcripts by dietary protein levels (Fig. 3), dietary antioxidant levels (Fig. 4), and mouse strains (Fig. 5). (The original absolute data are shown in Table S2 in the supplemental material.) The contrast approach allowed us to identify unique transcript changes in B6-LL mice that associate with the dietary or genetic control of chlamydial disease.
FIG. 3.
Low dietary protein reduces day 3 T-cell counts and inflammation in B6 mice. The ratio of parameter levels between low- and high-protein diets is shown for B6 and A/J mice at low (A and C) and high (B and D) dietary antioxidant levels. Black bars indicate significant differences (n = 10 mice/group; P < 0.05 by Tukey HSD test). Protein reduction consistently decreased inflammation (Tim3/GATA3, Arg1, NOS2, and Ptgs2) in B6 mice (A and B). This decrease was more pronounced at low antioxidant levels with lower cell numbers (PBGD intron), pan-T cells (CD3δ), Th1 cells (Tim3), and naïve and memory T cells (CD45RB and CD45RO), and lower inflammatory cytokines (IL-6, TNF-α, IFN-γ, CxCl2, and IL-10) (A). However, protein reduction at both antioxidant levels in A/J mice increased day 3 inflammation (Tim3/GATA3 and Ptgs2) and T-cell populations (CD3δ and perforin 1) but decreased anti-inflammatory Th2 cell (GATA3) and naïve T-cell (CD45RB) populations (C and D).
FIG. 4.
Low levels of dietary antioxidants marginally influence day 3 lung transcripts. The ratio of parameter levels between low- and high-antioxidant diets is shown for B6 and A/J mice at low (A and C) and high dietary protein levels (B and D). Black bars indicate significant differences (n = 10 mice/group; P < 0.05 by Tukey HSD test). Overall, antioxidant reduction did not exert consistent effects across mouse strains and dietary protein levels. Most significantly, antioxidant reduction at low dietary protein concentrations reduced pan-T (CD3δ) and B (CD19) lymphocytes in B6 as well as A/J mice (A and C) and increased levels of macrophages (F4/80), Th1 and Th2 cells (Tim3 and GATA3), naïve T cells (CD45RB), and IL-10 in A/J mice (C). At high dietary protein concentrations, an antioxidant reduction increased levels of T-cell and inflammation transcripts (CD3δ, CD45RB, CD45RO, and Ptgs2) and decreased levels of macrophages (F4/80) in B6 mice (B).
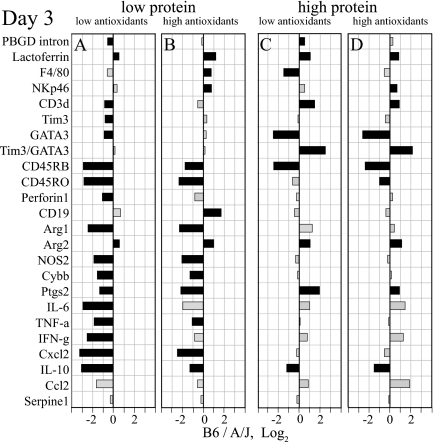
FIG. 5.
Host genetic background determines profound and diet-dependent differences in early response to C. pneumoniae. The ratio of parameter levels between B6 and A/J mice is shown for all diets (A, LL; B, LH; C, HL; D, HH). Black bars indicate significant differences (n = 10 mice/group; P < 0.05 by Tukey HSD test). The dominant protein modulation of the response to C. pneumoniae is evident in the conserved patterns with low dietary protein (A and B) and the different conserved patterns with high dietary protein (C and D) irrespective of dietary antioxidant levels. Compared to A/J mice, B6 mice showed increased levels of neutrophils (lactoferrin) and arginase 2 (Arg2) but reduced levels of naïve and memory T cells (CD45RB and CD45RO) and Th2 anti-inflammatory markers (GATA3 and IL-10) virtually across all diets. Low dietary protein, particularly when combined with low levels of antioxidants (A), induced in B6 mice profoundly lowers cellular infiltration (PBGD intron, CD3δ, Tim3, GATA3, CD45RB, CD45RO, and perforin 1) and inflammation (Arg1, NOS2, Cybb, Ptgs2, IL-6, TNF-α, IFN-γ, Cxcl2, and IL-10).
Protein is the predominant dietary modulator of the day 3 transcript profile compared to dietary antioxidants, and this modulation has more pronounced effects in B6 mice than in A/J mice (Fig. 3 and 4). In particular, B6 mice on the LL diet compared to the HL diet (Fig. 3A) exhibited profound and consistent decreases in transcript levels related to the lung cellular infiltrate (PBGD intron, CD3δ, Tim3, Tim3/GATA3, CD45RB, and CD45RO), inflammatory effectors (Arg1, NOS2, Cybb, and Ptgs2), and global cytokine responses (IL-6, TNF-α, IFN-γ, CxCl2, and IL-10). A reduction in dietary protein in B6 mice at high antioxidant levels results in similar but much less pronounced changes (Fig. 3B). In contrast, the protein reduction in A/J mice changed few transcripts, and these changes were more pronounced in a high dietary antioxidant background (Fig. 3C and D) and suggested stronger Th1 inflammation (decreased GATA3 and increased Tim3/GATA3, Cybb, and Ptgs2 levels).
Similar to day 10, dietary antioxidants by themselves do not exert a major influence on day 3 transcripts (Fig. 4). The main influence is a small but significant reduction of T-and B-lymphocyte transcripts (CD3δ and CD19) in both mouse strains on the LL diet compared to the LH diet (Fig. 4A and C). To assess the genetic control of the C. pneumoniae disease outcome, we compared day 3 lung transcripts of B6 and A/J mice on each of the four diets. Profound strain differences are evident with distinct patterns for low or high dietary protein content irrespective of dietary antioxidant content (Fig. 5). On the LL diet, B6 mice show reduced levels of transcripts for cell and inflammatory markers compared to those of A/J mice (PBGD intron, CD3δ, Tim3, GATA3, CD45RB, CD45RO, perforin 1, Arg1, NOS2, Cybb, Ptgs2, IL-6, TNF-α, IFN-γ, CxCl2, and IL-10) (Fig. 5A).
This pattern of differences with low dietary protein in day 3 lung transcripts between disease-prone B6 mice and disease-resistant A/J mice, particularly with low dietary antioxidants (Fig. 5A), strikingly resembles the differences between B6 mice on the disease-precipitating LL diet and the disease-protective HL diet (Fig. 3A). Thus, a shared early transcriptional response is associated with later C. pneumoniae disease precipitated either by dietary protein deficiency or by host genetic regulation. This result suggests that similarly regulated transcripts determine protection from disease in B6 mice on a high-protein diet and in A/J mice on any diet, i.e., that the dietary and genetic control of pathological responses to C. pneumoniae infection functions by the same mechanism. Among all day 3 transcripts that are reduced under conditions that lead to severe disease, the pattern of up-to-fourfold-reduced pan-T-cell (CD3δ) and 1.7-fold-reduced Th1 cell (Tim3) transcripts is present only in B6-LL mice (Fig. 3A and 5A) but not in B6 and A/J mice with less disease (Fig. 3B to D and 5B to D). Thus, the delayed early T-cell response is the unique feature that determines the severe disease of immune B6 mice.
B6 mice at low dietary protein levels have a reduced early DTH response to C. pneumoniae and low lung CD3δ+ and IFN-γ+ lymphocyte numbers.
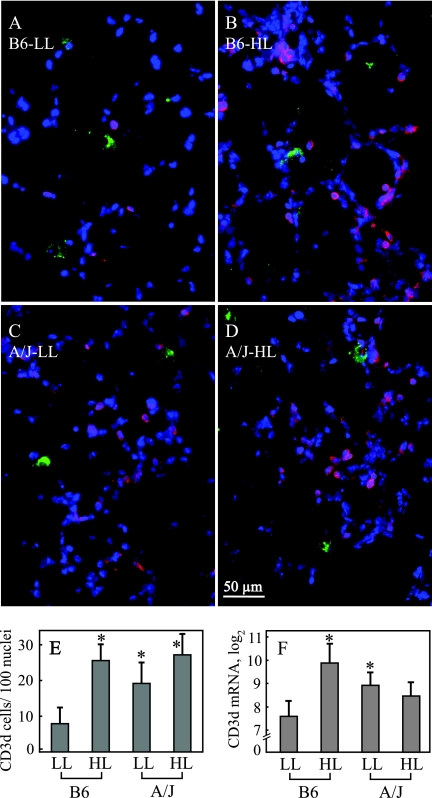
To ascertain the results obtained by analysis of early lung transcripts and to further scrutinize the distinctly different attributes of B6-LL mice, we performed functional tests for the early T-cell response by immunohistofluorescence, DTH, and ELISPOT assays. B6-LL mice on day 3 p.i. have decreased early lung CD3δ+ cells as determined by immunohistology (Fig. 6A to E), confirming reduced levels of CD3δ transcripts (Fig. 6F). Lungs of B6-LL mice show 7.91 CD3δ+ cells/100 nuclei, a significantly lower number than the 25.59 CD3δ+ cells/100 nuclei of B6-HL mice, the 19.18 CD3δ+ cells/100 nuclei in A/J-LL mice, and the 27.19 CD3δ+ cells/100 nuclei in A/J-HL mice (Fig. 6A to E).
FIG. 6.
Low dietary protein/antioxidants decrease day 3 lung CD3δ+ T-cell numbers in B6 mice. Shown is immunofluorescent staining for Chlamydia spp. (green), CD3δ (red), and cellular nuclei (DAPI) (blue) in lung sections of B6 and A/J mice on LL or HL diets sacrificed on day 3 p.i. (A to D). For each mouse (n = 5), the number of DAPI- and CD3δ-stained cells was counted in two microscopic fields of two lung sections showing similar chlamydial staining. B6 mice on the LL diet (A) exhibited significantly lower numbers of CD3δ+ cells than other groups (B to E). Determination of CD3δ transcripts (per 1,000 copies of PBGD mRNA) by duplex real-time PCR confirmed the reduced CD3δ+ cell numbers (F) (n = 10). Asterisks indicate significant differences compared to B6-LL mice (P < 0.05 by Tukey HSD test). Error bar, ±95% CI.
To evaluate the functional competence of the C. pneumoniae-specific T-cell response on day 3, we determined the DTH response to the intracutaneous injection of C. pneumoniae antigen. B6-LL mice show a minimal DTH response with an increase in paw thickness of 4.5 × 10−2 mm, significantly lower than the increase of 21.0 × 10−2 mm for B6-HL mice and the increase of 21.9 × 10−2 mm for A/J-LL mice. To assess parameters essential for DTH responsiveness, i.e., numbers and IFN-γ production of lymphocytes, we performed ELISPOT assays of infected lungs. B6-LL mice sacrificed on day 3 p.i. had a total of 7.34 × 106 lung lymphocytes, significantly lower than the 1.69 × 107 lymphocytes for B6-HL mice (Fig. 7). IFN-γ-producing cells are significantly lower in B6-LL mice than in B6-HL mice (2.70 cells per 106 lung lymphocytes versus 20.2 cells per 106 lung lymphocytes, respectively) or in A/J-LL mice (2.70 cells per 106 lung lymphocytes versus 16.99 cells per 106 lung lymphocytes, respectively). Collectively, B6 mice on the LL diet exhibit a decreased DTH response coupled with lower numbers of total lung lymphocytes and CD3δ+ and IFN-γ+ cells.
FIG. 7.
Low dietary protein and antioxidants reduce day 3 lung lymphocytes, IFN-γ-producing cells, and DTH responses to C. pneumoniae in B6 mice. B6 mice on LL or HL diets and A/J mice on LL diet were rechallenged with C. pneumoniae and injected into the paws with C. pneumoniae antigen 2 days later. After 24 h, the DTH response was determined as an increase in paw thickness (mm−2) over PBS-injected paws. Lung cells from sacrificed mice were obtained by collagenase digestion for the quantification of lung lymphocytes (×200,000), and IFN-γ-producing cells (per 106 lung lymphocytes) were quantified by ELISPOT assay. Asterisks indicate significant differences between B6-LL and B6-HL or A/J-LL mice (P < 0.05 by Tukey HSD test). Error bar, ±95% CI (n = 10 mice/group).
DISCUSSION
The efficient elimination of chlamydial infection requires IFN-γ-producing Th1 helper lymphocytes (45, 50, 52, 61, 71), and the ablation of Th1 cells or effector functions results in increased chlamydial disease (12, 33, 74). The clinical correlate to protective Th1 cells is a vigorous DTH reaction to chlamydial antigens (11, 17, 53, 75). However, a pronounced hypersensitivity response after repeated infections also drives the progression of the pathological lesion in chlamydial diseases such as trachoma (60, 64). This conundrum, i.e., the same type of immune reactivity to chlamydiae appears to be protective as well as pathological, has perplexed Chlamydia research for decades. Investigators have addressed this conundrum using genetically modified mice lacking essential genes of the agonistic Th1 or antagonistic Th2 response pathways (12, 79). Not surprisingly, mice with deletions of Th1 pathway genes such as IFN-γ, IFN-γR, or IL-12 develop exacerbated chlamydial disease due to the reduced clearance of the bacteria (12, 33, 52). Unexpectedly, however, the deletion of Th2 pathway genes such as IL-10 also exacerbates disease due to excessive inflammation despite an increased chlamydial clearance by a Th1-biased immune response (44). Thus, neither Th1- nor Th2-polarized immunity to chlamydiae, as created in the genetically modified mouse models, avoids disease in response to chlamydial infection.
In this investigation, we created a spectrum from protective through pathological outcomes of repeated C. pneumoniae infection in a design that modeled the disease in an outbred population. The use of the genetically intact A/J and B6 mouse strains, but not of genetically modified strains, created non-Th-polarized host-dependent responses that were perturbed by differentials in dietary protein and antioxidants. The temporal resolution of day 3 and day 10 outcomes by transcript analysis allowed strictly quantitative reconstruction of early response patterns that are associated with later disease or protection from disease. A specific strength is the statistical power of the balanced multivariate design coupled to highly accurate one-step duplex real-time RT-PCR, which simultaneously determines analyte and reference transcripts (72). By targeting fewer transcripts (24 transcripts) than typical microarray experiments (thousands of transcripts), but those critical transcripts at the appropriate group size (n = 10/group), our design incurs little penalty for multiple comparisons and minimizes statistical type I errors (67). The discrete, bimodal-effect layout intrinsically incorporates a reversal of outcomes and provides stronger evidence for causality than continuous effects that are typically analyzed in associative epidemiological studies of outbred populations.
Our results reaffirm the hypersensitivity nature of chlamydial disease. B6-LL mice with the most severe lung weight increase on day 10 p.i. show the hallmark of exaggerated Th1 inflammation with 10- to 40-fold-higher IFN-γ transcripts than protected B6-HL mice or A/J mice on any diet (Fig. 2 and see Table S2 in the supplemental material). The dietary protein-controlled C. pneumoniae disease (B6-LL versus B6-HL mice) reflects elevated cytokine production by hyperactivated cells since the lung cell populations in B6-LL mice were largely unchanged (Fig. 2A). The more severe disease exacerbation under genetic control (B6-LL versus A/J-LL mice) is driven by enlarged lung macrophage and Th1 lymphocyte populations and inflammatory cytokine overproduction (Fig. 2C). The cellular infiltrate under genetic disease control as well as the Th1-cytokine-driven inflammation under both dietary protein and genetic control encompass all classical characteristics of a DTH response (75, 76). This result confirms that, in fact, chlamydial disease is caused by the DTH reaction, which is nevertheless also required for the elimination of chlamydiae (40, 49).
The clear delineation of day 10 disease outcomes enabled us to identify the early response patterns that determined later disease. Specifically, contrast analyses (Fig. 3 to 5) allowed us to pinpoint day 3 transcript patterns that uniquely differentiate B6-LL mice from B6-HL and A/J-LL mice. B6-LL mice show highly reduced levels of T-cell (pan-T, Th1, naïve, and memory Th cells) and inflammatory transcripts compared to those for B6-HL and A/J-LL mice (Fig. 3A and 5A). Thus, B6 mice on the LL diet that precipitates severe day 10 disease show a day 3 transcript profile of a profoundly lower Th1 cellular immune response. The comprehensive quantitative comparison of the full interaction effects of dietary and genetic effectors on early transcripts revealed that reduced transcript levels of the pan-T-cell marker CD3δ and Th1 cell marker Tim3 uniquely differentiate B6 on the LL diet from B6 mice on any other diet and from all A/J mice (see Table S2 in the supplemental material). Related, but not absolutely unique as a high disease differential, is the strong reduction of naïve and memory T-cell marker transcripts (CD45RO) in B6-LL mice (Fig. 3A and B and 5). Thus, the delayed day 3 T-cell response is uniquely and significantly associated with late severe disease in immune B6 mice fed an LL diet. B6-LL mice also exhibit a much lower day 3 DTH response to C. pneumoniae as well as decreased numbers of lung lymphocytes and CD3δ+- and IFN-γ-secreting lung lymphocytes compared to B6-HL and A/J-LL mice (Fig. 6 and 7), confirming the concept of a delayed early T-cell response based on transcript analysis. This delay in the recruitment/expansion of a robust lung T-cell population in response to the C. pneumoniae stimulus results in significantly slowed elimination of the organisms (Fig. 1H).
Interestingly, B6-LL mice on the low-protein diet exhibited highly significantly reduced day 3 chlamydial loads that were approximately 100-fold lower than those of B6 mice on other diets or of A/J mice (Fig. 1E to H) and resembled those of naïve inoculated mice of any strain. We speculate that the high early cytokine stimulus in immune mice triggers metabolic changes in C. pneumoniae-infected cells that initially enhance chlamydial replication prior to IFN-γ-mediated tryptophan depletion and growth retardation (37). A corollary of this effect may be an observation from a study in which the therapeutic vaccination of Chlamydophila abortus-infected dairy cows resulted in highly significantly increased chlamydial shedding for approximately 1 week (5). These cows were vaccinated with either an inactivated Chlamydia, a mock, or another unrelated vaccine, and we assume that the adjuvant, the common factor among these vaccines, triggered a response similar to the one that we saw in this mouse model.
On day 10 under dietary protein control within the B6 mouse strain, the slow T-cell expansion and high chlamydial loads vastly exacerbate late inflammation with increases in both Th1 and Th2 cytokine transcripts. Under genetic control in stark contrast between B6-LL and A/J-LL mice, the pathogenetic principle of chlamydial disease comes into even clearer view on day 10, with the full phenotype of a DTH response including increased macrophage levels, Th1 cell levels, Th1:Th2 polarity, and exclusively Th1 cytokine-driven inflammation in B6-LL mice. Thus, chlamydial disease is characterized by a delayed immune response with a profound reduction of the early T-cell population that results in a failure to eliminate the pathogen and provokes later pathological Th1 inflammation.
Our results invite speculation as to the molecular control mechanisms of chlamydial disease. Of note are (i) the low early baseline of T cells (CD45) in B6 mice on all diets compared to A/J mice, (ii) the A/J phenotype with a four- to eightfold-higher early naïve lung T-cell population (CD45RB) than the B6 phenotype that is completely refractory to the delayed T-cell response associated with protein deficiency, and (iii) the low early T-cell levels under conditions of low-protein and low-antioxidant nutrition in B6 mice. These observations suggest that the control of T-cell homeostasis, with a B6:A/J differential, is central to chlamydial pathogenesis. The apoptotic death of T cells, of both thymocytes and peripheral lymphocytes, is one of the essential regulators of T-cell population dynamics (1). In fact, A/J background mice show significantly lower dexamethasone-induced thymocyte apoptosis and cyclophosphamide-induced peripheral lymphocyte apoptosis than B6 background mice (21). It will be of interest to determine if quantitative trait loci for this trait that map to chromosomes 1 and 6 (21) are also associated with differential susceptibility to chlamydial disease.
Of major medical interest is our observation that even minor short-term protein deficiency may cause severe T-cell immunosuppression in a susceptible host under simultaneous relative antioxidant deficiency. Our study extends the exacerbation of an infectious disease such as tuberculosis and immunosuppression under severe protein malnutrition (e.g., 2% dietary protein) (10) to much more prevalent, relative protein deficiency (14% dietary protein) at which the dietary protein concentration is low but within accepted limits. Also, profound host genetic differences in susceptibility to chlamydial disease triggered by protein-dependent reduced T-cell immunity do exist. Combined with frequent reinfection due to high population density, protein malnutrition and host genetic susceptibility may well be pivotal factors that determine the prevalence of chlamydial disease in the developing world as well as the developed world.
Supplementary Material
Acknowledgments
This investigation was supported by NIH grant AI47202 (to B.K.) and by an Auburn University graduate research fellowship (to T.K.).
We thank Dongya Gao and Sudhir Ahluwalia for excellent technical assistance and Lei Wang for fruitful discussions.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 25 August 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Baumann, S., A. Krueger, S. Kirchhoff, and P. H. Krammer. 2002. Regulation of T cell apoptosis during the immune response. Curr. Mol. Med. 2257-272. [DOI] [PubMed] [Google Scholar]
- 2.Beatty, W. L., G. I. Byrne, and R. P. Morrison. 1994. Repeated and persistent infection with Chlamydia and the development of chronic inflammation and disease. Trends Microbiol. 294-98. [DOI] [PubMed] [Google Scholar]
- 3.Beaumont, C., C. Porcher, C. Picat, Y. Nordmann, and B. Grandchamp. 1989. The mouse porphobilinogen deaminase gene. Structural organization, sequence, and transcriptional analysis. J. Biol. Chem. 26414829-14834. [PubMed] [Google Scholar]
- 4.Bernstein-Hanley, I., Z. R. Balsara, W. Ulmer, J. Coers, M. N. Starnbach, and W. F. Dietrich. 2006. Genetic analysis of susceptibility to Chlamydia trachomatis in mouse. Genes Immun. 7122-129. [DOI] [PubMed] [Google Scholar]
- 5.Biesenkamp-Uhe, C., Y. Li, H. R. Hehnen, K. Sachse, and B. Kaltenboeck. 2007. Therapeutic Chlamydophila abortus and C. pecorum vaccination transiently reduces bovine mastitis associated with Chlamydophila infection. Infect. Immun. 75870-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorgvinsdottir, H., L. Zhen, and M. C. Dinauer. 1996. Cloning of murine gp91phox cDNA and functional expression in a human X-linked chronic granulomatous disease cell line. Blood 872005-2010. [PubMed] [Google Scholar]
- 7.Boring, L., J. Gosling, F. S. Monteclaro, A. J. Lusis, C. L. Tsou, and I. F. Charo. 1996. Molecular cloning and functional expression of murine JE (monocyte chemoattractant protein 1) and murine macrophage inflammatory protein 1 alpha receptors: evidence for two closely linked C-C chemokine receptors on chromosome 9. J. Biol. Chem. 2717551-7558. [DOI] [PubMed] [Google Scholar]
- 8.Buzoni-Gatel, D., F. Bernard, M. Pla, A. Rodolakis, and F. Lantier. 1994. Role of H-2 and non-H-2-related genes in mouse susceptibility to Chlamydia psittaci. Microb. Pathog. 16229-233. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, L. A., and C. C. Kuo. 2004. Chlamydia pneumoniae—an infectious risk factor for atherosclerosis? Nat. Rev. Microbiol. 223-32. [DOI] [PubMed] [Google Scholar]
- 10.Chan, J., Y. Tian, K. E. Tanaka, M. S. Tsang, K. Yu, P. Salgame, D. Carroll, Y. Kress, R. Teitelbaum, and B. R. Bloom. 1996. Effects of protein calorie malnutrition on tuberculosis in mice. Proc. Natl. Acad. Sci. USA 9314857-14861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cher, D. J., and T. R. Mosmann. 1987. Two types of murine helper T cell clone. II. Delayed-type hypersensitivity is mediated by TH1 clones. J. Immunol. 1383688-3694. [PubMed] [Google Scholar]
- 12.Cotter, T. W., K. H. Ramsey, G. S. Miranpuri, C. E. Poulsen, and G. I. Byrne. 1997. Dissemination of Chlamydia trachomatis chronic genital tract infection in gamma interferon gene knockout mice. Infect. Immun. 652145-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cunningham-Rundles, S., D. F. McNeeley, and A. Moon. 2005. Mechanisms of nutrient modulation of the immune response. J. Allergy Clin. Immunol. 1151119-1128. [DOI] [PubMed] [Google Scholar]
- 14.DeGraves, F. J., D. Gao, and B. Kaltenboeck. 2003. High-sensitivity quantitative PCR platform. BioTechniques 34106-115. [DOI] [PubMed] [Google Scholar]
- 15.Fernández-Real, J. M., A. López-Bermejo, J. Vendrell, M. J. Ferri, M. Recasens, and W. Ricart. 2006. Burden of infection and insulin resistance in healthy middle-aged men. Diabetes Care 291058-1064. [DOI] [PubMed] [Google Scholar]
- 16.Field, C. J., I. R. Johnson, and P. D. Schley. 2002. Nutrients and their role in host resistance to infection. J. Leukoc. Biol. 7116-32. [PubMed] [Google Scholar]
- 17.Fong, T. A., and T. R. Mosmann. 1989. The role of IFN-gamma in delayed-type hypersensitivity mediated by Th1 clones. J. Immunol. 1432887-2893. [PubMed] [Google Scholar]
- 18.Fransen, L., R. Muller, A. Marmenout, J. Tavernier, J. Van der Heyden, E. Kawashima, A. Chollet, R. Tizard, H. Van Heuverswyn, and A. Van Vliet. 1985. Molecular cloning of mouse tumor necrosis factor cDNA and its eukaryotic expression. Nucleic Acids Res. 134417-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray, P. W., and D. V. Goeddel. 1983. Cloning and expression of murine immune interferon cDNA. Proc. Natl. Acad. Sci. USA 805842-5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grayston, J. T., S. P. Wang, L. J. Yeh, and C. C. Kuo. 1985. Importance of reinfection in the pathogenesis of trachoma. Rev. Infect. Dis. 7717-725. [DOI] [PubMed] [Google Scholar]
- 21.Guler, M. L., D. L. Ligons, Y. Wang, M. Bianco, K. W. Broman, and N. R. Rose. 2005. Two autoimmune diabetes loci influencing T cell apoptosis control susceptibility to experimental autoimmune myocarditis. J. Immunol. 1742167-2173. [DOI] [PubMed] [Google Scholar]
- 22.Hermiston, M. L., Z. Xu, and A. Weiss. 2003. CD45: a critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol. 21107-137. [DOI] [PubMed] [Google Scholar]
- 23.Huang, J., F. J. DeGraves, S. D. Lenz, D. Gao, P. Feng, D. Li, T. Schlapp, and B. Kaltenboeck. 2002. The quantity of nitric oxide released by macrophages regulates Chlamydia-induced disease. Proc. Natl. Acad. Sci. USA 993914-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, J., M.-D. Wang, S. D. Lenz, D. Gao, and B. Kaltenboeck. 1999. Interleukin-12 administered during Chlamydia psittaci lung infection in mice confers immediate and long-term protection and reduces MIP-2 level and neutrophil infiltration in lung tissue. J. Immunol. 1622217-2226. [PubMed] [Google Scholar]
- 25.Kaput, J. 2004. Diet-disease gene interactions. Nutrition 2026-31. [DOI] [PubMed] [Google Scholar]
- 26.Kaukoranta-Tolvanen, S. S., A. L. Laurila, P. Saikku, M. Leinonen, L. Liesirova, and K. Laitinen. 1993. Experimental infection of Chlamydia pneumoniae in mice. Microb. Pathog. 15293-302. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J. M., C. I. Brannan, N. G. Copeland, N. A. Jenkins, T. A. Khan, and K. W. Moore. 1992. Structure of the mouse IL-10 gene and chromosomal localization of the mouse and human genes. J. Immunol. 1483618-3623. [PubMed] [Google Scholar]
- 28.Kuo, C. C., L. A. Jackson, L. A. Campbell, and J. T. Grayston. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, F., T. Yokota, T. Otsuka, P. Meyerson, D. Villaret, R. Coffman, T. Mosmann, D. Rennick, N. Roehm, C. Smith, A. Zlotnik, and K.-I. Arai. 1986. Isolation and characterization of a mouse interleukin cDNA clone that expresses B-cell stimulatory factor 1 activities and T-cell and mast-cell-stimulating activities. Proc. Natl. Acad. Sci. USA 832061-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, D., A. Borovkov, A. Vaglenov, C. Wang, T. Kim, D. Gao, K. F. Sykes, and B. Kaltenboeck. 2006. Mouse model of respiratory Chlamydia pneumoniae infection for a genomic screen of subunit vaccine candidates. Vaccine 242917-2927. [DOI] [PubMed] [Google Scholar]
- 31.Li, D., A. Vaglenov, T. Kim, C. Wang, D. Gao, and B. Kaltenboeck. 2005. High-yield culture and purification of Chlamydiaceae bacteria. J. Microbiol. Methods 6117-24. [DOI] [PubMed] [Google Scholar]
- 32.Li, P., Y. L. Yin, D. Li, S. W. Kim, and G. Wu. 2007. Amino acids and immune function. Br. J. Nutr. 98237-252. [DOI] [PubMed] [Google Scholar]
- 33.Lu, H., X. Yang, K. Takeda, D. Zhang, Y. Fan, M. Luo, C. Shen, S. Wang, S. Akira, and R. C. Brunham. 2000. Chlamydia trachomatis mouse pneumonitis lung infection in IL-18 and IL-12 knockout mice: IL-12 is dominant over IL-18 for protective immunity. Mol. Med. 6604-612. [PMC free article] [PubMed] [Google Scholar]
- 34.Lyons, C. R., G. J. Orloff, and J. M. Cunningham. 1992. Molecular cloning and functional expression of an inducible nitric oxide synthase from a murine macrophage cell line. J. Biol. Chem. 2676370-6374. [PubMed] [Google Scholar]
- 35.McKnight, A. J., A. J. Macfarlane, P. Dri, L. Turley, A. C. Willis, and S. Gordon. 1996. Molecular cloning of F4/80, a murine macrophage-restricted cell surface glycoprotein with homology to the G-protein-linked transmembrane 7 hormone receptor family. J. Biol. Chem. 271486-489. [DOI] [PubMed] [Google Scholar]
- 36.McMurray, D. N., and R. A. Bartow. 1992. Immunosuppression and alteration of resistance to pulmonary tuberculosis in guinea pigs by protein undernutrition. J. Nutr. 122(Suppl. 3)738-743. [DOI] [PubMed] [Google Scholar]
- 37.Mehta, S. J., R. D. Miller, J. A. Ramirez, and J. T. Summersgill. 1998. Inhibition of Chlamydia pneumoniae replication in HEp-2 cells by interferon-gamma: role of tryptophan catabolism. J. Infect. Dis. 1771326-1331. [DOI] [PubMed] [Google Scholar]
- 38.Min-Oo, G., L. Lindqvist, C. Wang, P. Fortin, Y. Li, B. Kaltenboeck, and P. Gros. 2008. Genetic control of susceptibility to pulmonary infection with Chlamydia pneumoniae in the mouse. Genes Immun. 9383-388. [DOI] [PubMed] [Google Scholar]
- 39.Monney, L., C. A. Sabatos, J. L. Gaglia, A. Ryu, H. Waldner, T. Chernova, S. Manning, E. A. Greenfield, A. J. Coyle, R. A. Sobel, G. J. Freeman, and V. K. Kuchroo. 2002. Th-1 specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 415536-540. [DOI] [PubMed] [Google Scholar]
- 40.Morrison, R. P., K. Feilzer, and D. B. Tumas. 1995. Gene knockout mice establish a primary protective role for major histocompatibility complex class II-restricted responses in Chlamydia trachomatis genital tract infection. Infect. Immun. 634661-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Natividad, A., N. Hanchard, M. J. Holland, O. S. Mahdi, M. Diakite, K. Rockett, O. Jallow, H. M. Joof, D. P. Kwiatkowski, D. C. Mabey, and R. L. Bailey. 2007. Genetic variation at the TNF locus and the risk of severe sequelae of ocular Chlamydia trachomatis infection in Gambians. Genes Immun. 8288-295. [DOI] [PubMed] [Google Scholar]
- 42.O'Banion, M. K., V. D. Winn, and D. A. Young. 1992. cDNA cloning and functional activity of a glucocorticoid-regulated inflammatory cyclooxygenase. Proc. Natl. Acad. Sci. USA 894888-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohnishi, S., S. Maeda, S. Nishiguchi, T. Arao, and K. Shimada. 1988. Structure of the mouse C-reactive protein gene. Biochem. Biophys. Res. Commun. 156814-822. [DOI] [PubMed] [Google Scholar]
- 44.Penttilä, T., A. Haveri, A. Tammiruusu, J. M. Vuola, R. Lahesma, and M. Puolakkainen. 2006. Enhanced clearance but severe inflammation during pulmonary Chlamydia pneumoniae infection in IL-10 knockout mice, p. 535-538. In M. Chernesky, H. Caldwell, G. Christiansen, I. N. Clarke, B. Kaltenboeck, C. Knirsch, C.-C. Kuo, J. Mahony, R. G. Rank, P. Saikku, J. Schachter, W. E. Stamm, R. S. Stephens, J. T. Summersgill, P. Timms, and P. B. Wyrick (ed.), Chlamydial infections. Proceedings of the Eleventh International Symposium on Human Chlamydial Infections, San Francisco, CA.
- 45.Perry, L. L., K. Feilzer, and H. D. Caldwell. 1997. Immunity to Chlamydia trachomatis is mediated by T helper 1 cells through IFN-gamma-dependent and -independent pathways. J. Immunol. 1583344-3352. [PubMed] [Google Scholar]
- 46.Pessino, A., S. Sivori, C. Bottino, A. Malaspina, L. Morelli, L. Moretta, R. Biassoni, and A. Moretta. 1998. Molecular cloning of NKp46: a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J. Exp. Med. 188953-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsey, K. H. 2006. Alternative mechanisms of pathogenesis, p. 435-473. In P. M. Bavoil and B. Wyrick (ed.), Chlamydia genomics and pathogenesis. Horizon Bioscience Press, Norfolk, United Kingdom.
- 48.Ramsey, K. H., I. M. Sigar, S. V. Rana, J. Gupta, S. M. Holland, and G. I. Byrne. 2001. Role for inducible nitric oxide synthase in protection from chronic Chlamydia trachomatis urogenital disease in mice and its regulation by oxygen free radicals. Infect. Immun. 697374-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rank, R. G. 2006. The role of the CD4 T cell in the host response to Chlamydia, p. 365-379. In P. M. Bavoil and B. Wyrick (ed.), Chlamydia genomics and pathogenesis. Horizon Bioscience Press, Norfolk, United Kingdom.
- 50.Rank, R. G., L. S. Soderberg, M. M. Sanders, and B. E. Batteiger. 1989. Role of cell-mediated immunity in the resolution of secondary chlamydial genital infection in guinea pigs infected with the agent of guinea pig inclusion conjunctivitis. Infect. Immun. 57706-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reeves, P. G., F. H. Nielsen, and G. C. Fahey, Jr. 1993. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1231939-1951. [DOI] [PubMed] [Google Scholar]
- 52.Rottenberg, M. E., A. C. G. Rothfuchs, D. Gigliotti, M. Ceausu, C. Une, V. Levitsky, and H. Wigzell. 2000. Regulation and role of IFN-gamma in the innate resistance to infection with Chlamydia pneumoniae. J. Immunol. 1644812-4818. [DOI] [PubMed] [Google Scholar]
- 53.Sacks, D. L., W. J. Todd, and A. B. Macdonald. 1978. Cell-mediated immune responses in owl monkeys (Aotus trivirgatus) with trachoma to soluble antigens of Chlamydia trachomatis. Clin. Exp. Immunol. 3357-64. [PMC free article] [PubMed] [Google Scholar]
- 54.Saga, Y., J. S. Lee, C. Sariya, and E. A. Boyse. 1990. Regulation of alternative splicing in the generation of isoforms of the mouse Ly-5 (CD45) glycoprotein. Proc. Natl. Acad. Sci. USA 873728-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schachter, J. 1999. Infection and disease epidemiology, p. 139-170. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC.
- 56.Scrimshaw, N. S. 2003. Historical concepts of interactions, synergism and antagonism between nutrition and infection. J. Nutr. 133316S-321S. [DOI] [PubMed] [Google Scholar]
- 57.Shi, O., D. Kepka-Lenhart, S. M. Morris, and W. E. O'Brien. 1998. Structure of the murine arginase II gene. Mammal Genome 9822-824. [DOI] [PubMed] [Google Scholar]
- 58.Shirsat, N. V., S. Bittenbender, B. L. Kreider, and G. Rovera. 1992. Structure of the murine lactotransferrin gene is similar to the structure of the other transferrin-encoding genes and shares a putative regulatory region with the murine myeloperoxidase gene. Gene 110229-234. [DOI] [PubMed] [Google Scholar]
- 59.Smieja, M., J. Mahony, A. Petrich, J. Boman, and M. Chernesky. 2002. Association of circulating Chlamydia pneumoniae DNA with cardiovascular disease: a systematic review. BMC Infect. Dis. 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sowa, S., J. Sowa, L. H. Collier, and W. A. Blyth. 1969. Trachoma vaccine field trials in The Gambia. J. Hyg. (London) 167699-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su, H., and H. D. Caldwell. 1995. CD4+ T cells play a significant role in adoptive immunity to Chlamydia trachomatis infection of the mouse genital tract. Infect. Immun. 633302-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Swindle, E. J., and D. D. Metcalfe. 2007. The role of reactive oxygen species and nitric oxide in mast cell-dependent inflammatory processes. Immunol. Rev. 217186-205. [DOI] [PubMed] [Google Scholar]
- 63.Takiguchi, M., Y. Haraguchi, and M. Mori. 1988. Human liver-type arginase gene: structure of the gene and analysis of the promoter region. Nucleic Acids Res. 268789-8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor, H. R., S. L. Johnson, R. A. Prendergast, J. Schachter, C. R. Dawson, and A. M. Silverstein. 1982. An animal model of trachoma. II. The importance of repeated infection. Investig. Ophthalmol. Vis. Sci. 23507-515. [PubMed] [Google Scholar]
- 65.Tekamp-Olson, P., C. Gallegos, D. Bauer, J. McClain, B. Sherry, M. Fabre, S. van Deventer, and A. Cerami. 1990. Cloning and characterization of cDNAs for murine macrophage inflammatory protein 2 and its human homologues. J. Exp. Med. 172911-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trapani, J. A., B. S. Kwon, C. A. Kozak, C. Chintamaneni, J. D-E. Young, and B. Dupont. 1990. Genomic organization of the mouse pore-forming protein (perforin) gene and localization to chromosome 10, similarities to and differences from C9. J. Exp. Med. 171545-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Belle, G. 2002. Statistical rules of thumb. John-Wiley & Sons, Inc., New York, NY.
- 68.van den Elsen, P., K. Georgopoulos, B.-A. Shepley, S. Orkin, and C. Terhorst. 1986. Exon/intron organization of the genes coding for the δ chains of the human and murine T-cell receptor/T3 complex. Proc. Natl. Acad. Sci. USA 832944-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Ginkel, F. W., J. R. McGhee, C. Liu, J. W. Simecka, M. Yamamoto, R. A. Frizzell, E. J. Sorscher, H. Kiyono, and D. W. Pascual. 1997. Adenoviral gene delivery elicits distinct pulmonary-associated T helper cell responses to the vector and to its transgene. J. Immunol. 159685-693. [PubMed] [Google Scholar]
- 70.Van Snick, J. S., J. P. Cayphas, J. C. Szikora, J. C. Renauld, E. Van Roost, T. Boon, and R. J. Simpson. 1988. cDNA cloning of murine interleukin-HP1: homology with human interleukin 6. Eur. J. Immunol. 183618-3623. [DOI] [PubMed] [Google Scholar]
- 71.Vuola, J. M., V. Puurula, M. Anttila, P. H. Mäkelä, and N. Rautonen. 2000. Acquired immunity to Chlamydia pneumoniae is dependent on gamma interferon in two mouse strains that initially differ in this respect after primary challenge. Infect. Immun. 68960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, C., D. Gao, A. Vaglenov, and B. Kaltenboeck. 2004. One-step real-time duplex reverse transcription PCRs simultaneously quantify analyte and reference gene mRNAs. BioTechniques 36508-519. [DOI] [PubMed] [Google Scholar]
- 73.Wang, C., T. Kim, D. Gao, A. Vaglenov, and B. Kaltenboeck. 2007. Rapid high-yield mRNA extraction for reverse-transcription PCR. J. Biochem. Biophys. Methods 70507-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, S., Y. Fan, R. C. Brunham, and X. Yang. 1999. IFN-gamma knockout mice show Th2-associated delayed-type hypersensitivity and the inflammatory cells fail to localize and control chlamydial infection. Eur. J. Immunol. 293782-3792. [DOI] [PubMed] [Google Scholar]
- 75.Watkins, N. G., W. J. Hadlow, A. B. Moos, and H. D. Caldwell. 1986. Ocular delayed hypersensitivity: a pathogenetic mechanism of chlamydial-conjunctivitis in guinea pigs. Proc. Natl. Acad. Sci. USA 837480-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watson, R. R., A. B. MacDonald, E. S. Murray, and F. Z. Modabber. 1973. Immunity to chlamydial infections of the eye. 3. Presence and duration of delayed hypersensitivity to guinea pig inclusion conjunctivitis. J. Immunol. 111618-623. [PubMed] [Google Scholar]
- 77.Wintergerst, E. S., S. Maggini, and D. H. Hornig. 2007. Contribution of selected vitamins and trace elements to immune function. Ann. Nutr. Metab. 51301-323. [DOI] [PubMed] [Google Scholar]
- 78.Yamamoto, K., K. Takeshita, T. Shimokawa, H. Yi, K. Isobe, D. J. Loskutoff, and H. Saito. 2002. Plasminogen activator inhibitor-1 is a major stress-regulated gene: implications for stress-induced thrombosis in aged individuals. Proc. Natl. Acad. Sci. USA 99890-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang, X., J. Gartner, L. Zhu, S. Wang, and R. C. Brunham. 1999. IL-10 gene knockout mice show enhanced Th1-like protective immunity and absent granuloma formation following Chlamydia trachomatis lung infection. J. Immunol. 1621010-1017. [PubMed] [Google Scholar]
- 80.Yang, Z. P., P. K. Cummings, D. L. Patton, and C. C. Kuo. 1994. Ultrastructural lung pathology of experimental Chlamydia pneumoniae pneumonitis in mice. J. Infect. Dis. 170464-467. [DOI] [PubMed] [Google Scholar]
- 81.Zheng, W., and R. A. Flavell. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89587-596. [DOI] [PubMed] [Google Scholar]
- 82.Zhou, L.-J., D. C. Ord, S. A. Omori, and T. F. Tedder. 1992. Structure of the genes encoding the CD19 antigen of human and mouse B lymphocytes. Immunogenetics 35102-111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.