Abstract
Staphylococcus epidermidis is one of the most common causes of infections of prosthetic heart valves (prosthetic valve endocarditis [PVE]) and an increasingly common cause of infections of native heart valves (native valve endocarditis [NVE]). While S. epidermidis typically causes indolent infections of prosthetic devices, including prosthetic valves and intravascular catheters, S. epidermidis NVE is a virulent infection associated with valve destruction and high mortality. In order to see if the differences in the course of infection were due to characteristics of the infecting organisms, we examined 31 S. epidermidis NVE and 65 PVE isolates, as well as 21 isolates from blood cultures (representing bloodstream infections [BSI]) and 28 isolates from nasal specimens or cultures considered to indicate skin carriage. Multilocus sequence typing showed both NVE and PVE isolates to have more unique sequence types (types not shared by the other groups; 74 and 71%, respectively) than either BSI isolates (10%) or skin isolates (42%). Thirty NVE, 16 PVE, and a total of 9 of the nasal, skin, and BSI isolates were tested for virulence in Caenorhabditis elegans. Twenty-one (70%) of the 30 NVE isolates killed at least 50% of the worms by day 5, compared to 1 (6%) of 16 PVE isolates and 1 (11%) of 9 nasal, skin, or BSI isolates. In addition, the C. elegans survival rate as assessed by log rank analyses of Kaplan-Meier survival curves was significantly lower for NVE isolates than for each other group of isolates (P < 0.0001). There was no correlation between the production of poly-β(1-6)-N-acetylglucosamine exopolysaccharide and virulence in worms. This study is the first analysis suggesting that S. epidermidis isolates from patients with NVE constitute a more virulent subset within this species.
Coagulase-negative staphylococci (CoNS) are a leading cause of infections of cardiac valve prostheses (prosthetic valve endocarditis [PVE]) and are increasingly recognized as a cause of infections of native cardiac valves (native valve endocarditis [NVE]). Recent reports from the International Collaboration on Endocarditis identified CoNS as a cause of 31% of the cases of PVE occurring in five countries between 1984 and 1999 (18) and 7.8% of the cases of NVE occurring in 28 countries between 2000 and 2006 (5). Most of the CoNS causing both PVE and NVE have been identified as Staphylococcus epidermidis (15, 26). Past data have characterized S. epidermidis PVE as a relatively indolent disease that involves the valve sewing ring, presumably caused by bacteria inoculated into the ring area from the skin or surgical field at the time of surgery (15). In contrast, data from both older (3) and recent (5) studies have demonstrated the aggressive nature of NVE caused by CoNS, leading to valve destruction, heart failure, and death at a rate comparable to that seen with NVE due to S. aureus. In addition, while PVE isolates are mostly nosocomial and methicillin resistant (15), 51% of NVE isolates were reported previously to be community acquired and 59% were found to be methicillin susceptible (5). These clinical, epidemiological, and susceptibility data would suggest that there is something different about the two S. epidermidis populations that cause these diseases. The recent development of sensitive typing systems, international typing databases, and high-throughput screens for relative bacterial virulence has made it possible to test the hypothesis that there are different S. epidermidis populations causing NVE and PVE.
In this study, we examined the virulence of S. epidermidis isolates from patients with NVE and PVE, as well as both blood culture isolates and commensals, using Caenorhabditis elegans as a model (29). We also determined the genotypes of these isolates by multilocus sequence typing (MLST) (22, 31) in order to identify any unique types that were associated with endocarditis.
MATERIALS AND METHODS
Bacterial strains.
Of the 145 isolates investigated, 31 were from patients with NVE, 65 were from patients with PVE, 4 were from specimens indicating nasal carriage (hereinafter referred to as nasal isolates), 24 were from blood cultures but were ascribed to contamination from skin (hereinafter referred to as skin isolates), and 21 were from blood cultures and were felt to represent true bloodstream infections (BSI). Fifty PVE isolates were from patients diagnosed with infections in the 1970s and have been described previously (33). The remaining 15 PVE isolates, as well as 22 NVE isolates, were from patients with cases of infective endocarditis as defined by the Duke criteria (9) and belong to the International Collaboration on Endocarditis collection of isolates recovered from patients between 1999 and 2006. These isolates were obtained in previous multicenter cohort studies and have been extensively clinically characterized elsewhere (5, 18). The remaining nine NVE isolates were from patients hospitalized at the Medical College of Virginia between 1975 and 1979. Nasal isolates were obtained from individuals whose nares were swabbed in an attempt to document S. aureus nasal carriage. Skin isolates were from the Virginia Commonwealth University health system clinical microbiology laboratory. Each isolate was grown from the contents of only one of four or more blood culture bottles containing blood samples obtained at the same time from a single patient. BSI isolates were obtained from patients hospitalized at the Duke University Medical Center. These isolates were categorized as nonendocarditis bacteremia isolates based on the presence of at least two sets of S. epidermidis-positive blood cultures (four total blood cultures) from samples drawn on separate days in the presence of an intravascular catheter, along with either an echocardiogram that was negative for valvular vegetations or the resolution of bacteremia upon the removal of the intravascular catheter. All isolates were confirmed to be S. epidermidis by the use of 16S rRNA sequencing (11).
Drug susceptibility testing.
All isolates were tested for drug susceptibility by spreading 0.1 ml of an overnight brain heart infusion (BHI) broth culture onto a Mueller-Hinton agar plate containing 6 μg of oxacillin/ml. Any growth after 72 h of incubation at 35°C indicated resistance to oxacillin.
DNA extraction, PCR amplification, and sequencing.
Genomic DNA for MLST was extracted using the QIAamp DNA mini kit (Qiagen, Hilden, Germany). Lysis buffer contained lysostaphin (200 μg/ml; Sigma) in a mixture of 20 mM Tris-HCl, pH 8.0, 2 mM EDTA, and 1.2% Triton. PCR was performed with 50-μl reaction mixtures by using 100 pmol of a primer with an annealing temperature of 55°C before the visualization of the products upon a 1.5% acrylamide gel stained with ethidium bromide. All reaction mixtures were cleaned up using the Qiagen PCR cleanup kit, and sequences were obtained using the previously reported primers at 10 pmol and an annealing temperature of 55°C with BigDye fluorescent terminators on an ABI Prism 3700 instrument (Applied Biosystems, Foster City, CA).
Genotyping analysis.
MLST was performed as described previously (31) using primers for internal portions of the following genes: arcC, aroE, gtr, mutS, pyr, tpi, and yqiL. MLST alleles were classified using the S. epidermidis MLST website at http://sepidermidis.mlst.net. MLST eBURST was performed as described previously (10, 22) using the S. epidermidis MLST website.
Virulence assays.
Virulence assays were performed as described previously (29). Thirty microliters of 1:10 dilutions of overnight cultures of S. epidermidis grown in BHI (Becton Dickinson, Sparks, MD) broth were spread onto the surface of BHI agar supplemented with 5 μg of nalidixic acid/ml and were grown for 4 h at 35°C. Twenty-five to 30 C. elegans worms in a larval stage (larval stage 4) were added to each plate, and worms were counted at 24-h intervals for the first 2 days and at 12-h intervals for the remaining 3 days. Worms were assessed for movement by the agitation of the plate and physical stimulation with a platinum wire. Lack of movement indicated death. Virulence was measured in two ways: by the total number of C. elegans worms surviving at each time point and by the time taken to kill ≥50% of the worms.
Biofilm and exopolysaccharide assays.
The production of biofilms was measured as described previously (4). Briefly, 5 μl of an overnight BHI broth culture of each isolate to be analyzed was inoculated into 200 μl of fresh BHI broth in one well of a 96-well tissue culture plate, and the plate was incubated at 35°C for 18 h. The broth was then removed, the wells were washed with phosphate-buffered saline, and the well contents were stained with crystal violet. Two independent observers then assessed the stained wells, rating each biofilm as weak, medium, or strong compared to those of test controls: S. aureus MN8M (strong, 3+), S. epidermidis RP62A (medium, 2+), S. aureus MN8MΔica (weak, 1+), and BHI (negative).
The presence of the major component of biofilms, poly-β(1-6)-N-acetylglucosamine (PNAG) exopolysaccharide, was also evaluated using the following assay, similar to the method described by Cramton et al. (6). A single bacterial colony was picked from agar, inoculated into 5 ml of BHI broth, and grown at 37°C overnight. The bacteria were centrifuged at 3,000 × g for 10 min, the pellet was resuspended in 250 μl of 0.5 M EDTA, pH 8.0, and the resuspended sample was boiled for 5 min. This sample was centrifuged again at 16,000 × g for 10 min, the supernatant was treated with proteinase K at 1.6 μg/ml, and this sample was incubated at 56°C for 30 min. After each sample was boiled for an additional 5 min, 200 ml was loaded into a slot blot apparatus containing a nitrocellulose membrane wetted with 10% methanol. Samples of the strong, medium, and weak biofilm producer control strains described above were also prepared and loaded in a similar manner. All samples were diluted 1:5 except for that of the strong biofilm producer control (MN8M), which was serially diluted from 1:1 to 1:1,000.
Following slot blotting, the nitrocellulose membrane was blocked overnight in 1× phosphate-buffered saline containing 5% bovine serum albumin and then washed in 1× Tris-buffered saline (TBS; 2.9 g of NaCl, 0.24 g of Tris base to reach pH 7.5 in 1 liter of distilled H2O) containing 0.05% Tween 20 three times for 20 min each with gentle shaking at room temperature. The primary antibody (anti-PNAG produced in rabbits as reported previously [21] and supplied by Tomás Maira-Litrán, Harvard Medical School, Boston, MA), diluted 1:1,000, and secondary antibody (goat anti-rabbit immunoglobulin G [whole molecule]-peroxidase [A9169; Sigma, St Louis, MO]), diluted 1:10,000, both in 1× TBS containing 0.05% Tween 20 and 5% bovine serum albumin, were added to the membrane, and the membrane was washed three times in the same solution used for previous washes and shaken gently at room temperature. The membrane was visualized with a 3,3′-diaminobenzidine peroxidase substrate kit (SK-4100) per the instructions of the manufacturer (Vector Labs, Burlingame, CA). The S. epidermidis biofilm samples were visually scored as strong (3+), medium (2+), or weak (1+) compared to the strong, medium, and weak controls listed above.
Statistical analyses.
Virulence data were analyzed as follows. Within-group Kaplan-Meier (KM) survival estimates (the total number of worms surviving out of the total number inoculated for each group) were computed for each time interval in order to build the KM survival curves for each group. The log rank test was used to perform between-group comparisons, testing the equivalence of the KM survival curves for a pair of groups. Several additional analyses were carried out to check for the effects of censoring and sample size and the stability of the distribution functions, including robust estimates and alternative test statistics analyses (8, 34). However, the standard log rank test was considered to provide an adequate representation of the differences between the groups.
The comparison of the numbers of isolates from the different groups with unique sequence types (STs) was performed by chi-square analysis.
Origin of isolates.
Isolates were classified as being nosocomial or health care associated if the patients had been hospitalized within the previous 3 months or had contact with a health care facility within the previous year. The latter criterion was required because of the long latency period that may occur between the implantation of organisms into the valve ring at surgery and the clinical presentation of PVE (15). Those isolates that were neither nosocomial nor health care associated in origin were classified as community acquired.
RESULTS
MLST.
A summary of the MLST data is found in Table 1. Typing data on each isolate can be found in Table S2 in the supplemental material. The most common ST was ST2, the group in which 22 (15%) of the 145 isolates were found. This number included 13% of the NVE isolates, 12% of the PVE isolates, 38% of the BSI isolates, 13% of the skin isolates, and none of the nasal isolates. Interestingly, 23 (74%) of the 31 NVE isolates and 46 (71%) of the 65 PVE isolates were unique. That is, none of the skin, BSI, or nasal isolates (a total of 49 isolates) shared the STs of these isolates. In addition, only 19% of NVE isolates shared STs with PVE isolates, and only 21% of PVE isolates shared STs with NVE isolates. This result is in contrast to the finding that only 10 skin isolates (42%) and 2 BSI isolates (10%) had unique STs. All four nasal isolates were unique. The difference between the numbers of unique NVE and PVE isolates and unique BSI isolates was significant (P < 0.005), and that between the numbers of unique NVE and PVE isolates and unique skin isolates was nonsignificant (P > 0.05). eBURST analysis found all isolates to be highly clonal, with 116 (80%) of the isolates falling into a single clonal complex, as has been reported previously (22) (data not shown). eBURST data are found in Fig. S3 in the supplemental material.
TABLE 1.
Distribution of MLST sequence types among groups of S. epidermidis isolates
| Sequence type(s) | Isolate group(s) (no. of isolates) |
|---|---|
| 1 | PVE (5) |
| 2 | PVE (8), NVE (4), BSI (7), skin (3) |
| 3 | PVE (7) |
| 4 | PVE (2) |
| 5 | PVE (4), skin (3), BSI (2), NVE (1) |
| 6 | PVE (1), NVE (1), skin (1) |
| 7 | PVE (3), skin (2), BSI (1) |
| 8 to 20 | PVE (15) |
| 22 | PVE (1), BSI (3) |
| 23 to 27 | PVE (6) |
| 29 and 30 | PVE (2) |
| 30 | PVE (1) |
| 35 | Skin (2) |
| 43 | NVE (3) |
| 44 | PVE (1) |
| 54 | NVE (1), BSI (1) |
| 57 | Skin (3), BSI (2) |
| 59 | PVE (1), skin (1) |
| 83 | Nasal (1) |
| 85 | BSI (1), NVE (1) |
| 88 | Nasal (1) |
| 89 | NVE (1) |
| 115 to 122 | NVE (8) |
| 123 and 124 | Skin (2) |
| 125 | NVE (1), skin (1) |
| 126 to 129 | PVE (4) |
| 130 to 138 | NVE (9) |
| 139 to 143 | Skin (5) |
| 144 | PVE (1), BSI (2) |
| 145 | Nasal (1) |
| 146 | PVE (1) |
| 147 | BSI (1) |
| 160 | Skin (1) |
| 161 and 162 | PVE (2) |
| 163 | NVE (1) |
| 164 | BSI (1) |
| 165 | Nasal (1) |
Oxacillin susceptibility.
As reported in previous studies (15), most of the PVE isolates (58 of 65; 89%) were oxacillin resistant. In contrast, only 16 (51%) of the 31 NVE isolates were oxacillin resistant. Among the commensals (nasal and skin isolates), 15 of 27 (55.5%) were oxacillin resistant while 14 (67%) of 21 BSI isolates grew on oxacillin plates.
Virulence.
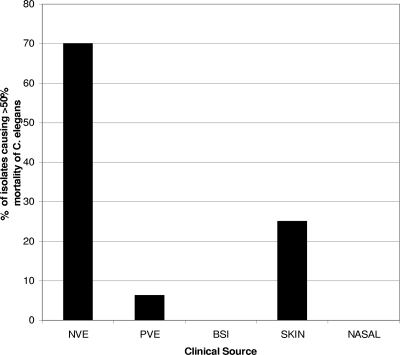
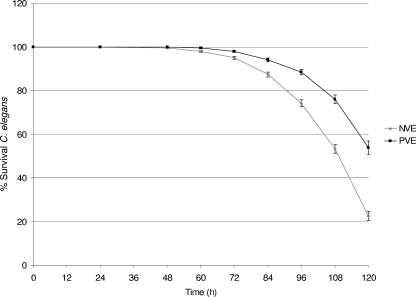
Fifty-five isolates were assessed in the C. elegans model of virulence: 30 NVE, 16 PVE, 4 skin, 3 BSI, and 2 nasal isolates. Only 23 isolates (42%), 21 NVE isolates, 1 PVE isolate, and 1 skin isolate, were able to kill ≥50% of the C. elegans worms by day 5. The lethal isolates represented 70% (21 of 30) of the NVE, 6% (1 of 16) of the PVE, and 25% (1 of 4) of the skin isolates tested for virulence. These data are depicted in Fig. 1. The isolates killed C. elegans between 60 and 108 h, with a mean time of 96 h. When the numbers of worms surviving at each time point (expressed as the percent survival) were compared for NVE and PVE isolates by the KM function, as shown in Fig. 2, the results of the log rank test for differences in survival estimates indicated that NVE isolates were significantly more lethal than PVE isolates (P < 0.0001). The differences in survival rates for NVE isolates and each of the other groups (BSI, skin, and nasal isolates) were also significant (P < 0.0001). Finally, on day 5 of the experiment, the NVE isolates corresponded to an average rate of survival of C. elegans worms of only 22.3% (95% confidence interval [CI], 20.3 to 24.7%), while the PVE isolates corresponded to a survival rate of 53.8% (95% CI, 50.9 to 56.9%). Data on the virulence of each isolate toward worms are found in Table S2 in the supplemental material.
FIG. 1.
Percentage of isolates examined for each clinical category in this investigation that were able to cause the death of ≥50% of C. elegans worms in the experimental model of virulence.
FIG. 2.
KM plot illustrating the relative virulence of NVE and PVE isolates examined in this investigation. The percent survival of C. elegans at each time point for the PVE and NVE isolates is shown. The 95% CIs are indicated by bars above and below each datum point.
Biofilms.
Exopolysaccharide production has been identified as a staphylococcal virulence factor in the C. elegans model (2). Therefore, we assessed 51 of the 55 isolates examined in the virulence assay for both biofilm formation and specific PNAG exopolysaccharide production. In general, more isolates (35 of 51 [69%]) were graded as strong (3+) biofilm producers than strong PNAG producers (8 of 51 [16%]). Most isolates (39 of 51 [76%]) produced medium (2+) levels of PNAG. There was no evidence that PNAG or biofilm production was correlated with virulence: 18 (62%) of 29 avirulent and 17 (77%) of 22 virulent isolates produced 3+ biofilms, while 20 (68%) of 29 avirulent and 19 (86%) of 22 virulent isolates produced 2+ PNAG levels. Four isolates (two PVE, one NVE, and one skin isolate; 8%) produced weak PNAG levels, and all were avirulent. Data on each isolate are found in Table S2 in the supplemental material.
Origin of isolates.
Nineteen (61%) of the 31 NVE isolates were community acquired, while only 12 (39%) were health care associated. In contrast, 57 (88%) of the 65 PVE isolates were health care associated and only 8 (12%) were community acquired.
DISCUSSION
CoNS in general and S. epidermidis in particular are generally considered to be microorganisms with low levels of virulence for humans, usually requiring the presence of a foreign body, such as an intravenous catheter or prosthetic device, to establish an infection. While genes for toxins such as enterotoxins and toxic shock syndrome toxin 1 can be found in some CoNS species (7), they are uncommon in S. epidermidis and rarely associated with disease syndromes (14, 16, 24, 28, 32). However, numerous investigators have attempted to identify CoNS that are more virulent in animal models. Claims for the increased virulence of S. epidermidis versus that of other CoNS and for the increased virulence of specific S. epidermidis isolates have been made using the rat endocarditis (1), mouse intraperitoneal inoculation (23), mouse weight retardation (12), mouse catheter (25), and mouse intravenous 50% lethal dose (13) models. The clinical sources of isolates evaluated in these studies were not always clear, and in most cases, there was no difference in the clinical significance of isolates with high and low levels of virulence. No specific factors or genes were proposed as virulence candidates in any of these studies, and a recent study found no difference in the presence or absence of postulated S. epidermidis virulence factors between isolates causing infections and commensals (27). Therefore, our study differs from these earlier reports in that we identified S. epidermidis isolates obtained under clearly defined clinical conditions and found a marked increase in virulence among those isolates causing an infection, NVE, that is differentiated from other S. epidermidis infections by its more aggressive and virulent clinical course.
The model that we used to compare the degrees of virulence of S. epidermidis isolates assessed the abilities of individual isolates to kill the worm C. elegans. The C. elegans virulence model was developed by Mahajan-Miklos et al. (20) to allow them to screen large numbers of isogenic mutants of Pseudomonas aeruginosa that differed by the presence or absence of specific putative virulence genes. The model has been used successfully by Sifri et al. to assess virulence determinants in S. aureus (29). A recent review (30) has detailed the use of C. elegans in the study of the virulence of human microbial pathogens. More than 20 bacterial and fungal pathogens have been fed to C. elegans worms and found to result in the premature death of the worms. Killing has been associated with the accumulation of bacteria in the worms' intestines and by the production of toxins; bacteria do not appear to invade the worm body from the intestine, nor do they invade gut-lining cells. Thus, the mechanisms underlying worm killing are not well-understood. One study associated worm death after the ingestion of S. aureus or S. epidermidis with the production of exopolysaccharide (2) and hypothesized that the exopolysaccharide protected bacteria from C. elegans innate immune factors and allowed the bacteria to accumulate. However, in the present study, 69% of S. epidermidis isolates produced abundant (3+) biofilms, 76% made moderate (2+) amounts of exopolysaccharide (PNAG [21]), and there was no correlation between either biofilm or exopolysaccharide production and worm virulence. Thus, the differences in worm death caused by NVE isolates and other isolates were not likely to be related to the production of PNAG. A major defense mechanism of C. elegans, which has no professional phagocytic cells, is the secretion of antimicrobial peptides into the intestines (30). Thus, the number and type of antimicrobial peptide defense systems within the worm intestine may be associated with different rates of survival. S. epidermidis has been shown to regulate its response to human antimicrobial peptides and to produce a number of factors that help protect the pathogen from the lethal effect of the peptides (17, 19). In addition, the ability of NVE isolates to cause valve destruction may be related to the production of exoenzymes not previously identified as virulence factors. In fact, one of the factors induced in S. epidermidis upon exposure to an antimicrobial peptide is a potent protease (17). These same factors may also cause intestinal toxicity in C. elegans.
Our study also used MLST to find out if S. epidermidis isolates of documented clinical significance could be grouped on the basis of their STs. MLST is the typing technique most commonly used for large-scale studies of the evolutionary relationships of S. aureus genomes (10). This technique assigns STs on the basis of single nucleotide polymorphisms among a group of stable housekeeping genes located throughout the genome. An international MLST database has been established for S. epidermidis by using a set of genes, some of which are different from those used for S. aureus, chosen for their random genomic distribution (31). MLST has been used successfully to infer a population structure for S. epidermidis (22), validating the usefulness of this method in establishing clonal relationships. However, the isolates that we describe in this report differ from the international isolates included in the typing study reported by Miragaia et al. (22). Even though ST2 was the most common type in both studies, it comprised 31% of the isolates in the study by Miragaia et al. but only 15% of our isolates. Furthermore, 50% of the STs assigned to the isolates in our study were not found among the isolates in the collection of Miragaia et al. This distinction may reflect the relatively small number of isolates that have been typed by the new MLST scheme, or it may be an indication that the two sets of isolates came from different clinical sources. The international set was composed of patient isolates, but their sources were unclear. They were largely nosocomial isolates, and 82% were methicillin resistant. In contrast, our isolates were from defined sources, and 96 were specifically from patients with infective endocarditis, either PVE or NVE. The sources from which these endocarditis isolates originated reflected characteristics reported in the literature: 88% of the PVE isolates were nosocomial and 89% were oxacillin resistant, while only 52% of the NVE isolates were oxacillin resistant and 61% were community acquired. Thus, the different sources of these isolates may explain the unique STs found in our isolates compared to those of Miragaia et al. They may also explain the lack of overlap in STs between our PVE and NVE isolates. Of the 96 total NVE and PVE isolates, only 18 (19%) had STs in common, and 74% of NVE and 71% of PVE isolates had unique STs. In contrast, even though skin and BSI isolates were also, like PVE isolates, largely health care associated, only 12 (27%) of 45 had unique STs. Thus, while MLST analysis did not identify a group of STs that would define the clinical source of an S. epidermidis isolate, it suggested that S. epidermidis isolates causing NVE and PVE may be different from each other and from nosocomial isolates routinely recovered from blood culture bottles.
In summary, this study has provided evidence, using both molecular typing and high-throughput virulence testing with C. elegans, that there may be strains of S. epidermidis that are uniquely able to cause a specific human infection. Analyses of these isolates may reveal novel virulence genes for this generally avirulent species.
Supplementary Material
Acknowledgments
This work was supported by NIH grant 5R01AI35705 from the NIAID (G.L.A.) and AHA grant 0675027N from the American Heart Association (V.H.C.).
We are grateful to Tomás Maira-Litrán (Harvard Medical School, Boston, MA) for providing the antibody used in the PNAG assays and to Kim Jefferson (Department of Microbiology and Immunology, Virginia Commonwealth University, Richmond, VA) for helpful advice and supervision in the performance of both the biofilm and PNAG assays.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 15 September 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Baddour, L. M., G. D. Christensen, M. G. Hester, and A. L. Bisno. 1984. Production of experimental endocarditis by coagulase-negative staphylococci: variability in species virulence. J. Infect. Dis. 150721-727. [DOI] [PubMed] [Google Scholar]
- 2.Begun, J., J. M. Gaiani, H. Rohde, D. Mack, S. B. Calderwood, F. M. Ausubel, and C. D. Sifri. 2007. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 3e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caputo, G. M., G. L. Archer, S. B. Calderwood, M. J. DiNubile, and A. W. Karchmer. 1987. Native valve endocarditis due to coagulase-negative staphylococci. Clinical and microbiologic features. Am. J. Med. 83619-625. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, G. D., A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu, V. H., C. W. Woods, J. M. Miro, B. Hoen, C. H. Cabell, P. A. Pappas, J. Federspiel, E. Athan, M. E. Stryjewski, F. Nacinovich, F. Marco, D. P. Levine, T. S. Elliott, C. Q. Fortes, P. Tornos, D. L. Gordon, R. Utili, F. Delahaye, G. R. Corey, V. G. Fowler, Jr., and the International Collaboration on Endocarditis-Prospective Cohort Study Group. 2008. Emergence of coagulase-negative staphylococci as a cause of native valve endocarditis. Clin. Infect. Dis. 15232-242. [DOI] [PubMed] [Google Scholar]
- 6.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Götz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 675427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.da Cunha, M. D. L. R. S., R. A. O. Calsolari, and J. P. Araújo, Jr. 2007. Detection of enterotoxin and toxic shock syndrome toxin 1 genes in Staphylococcus, with emphasis on coagulase-negative staphylococci. Microbiol. Immunol. 51381-390. [DOI] [PubMed] [Google Scholar]
- 8.Dorey, F. J., and E. L. Korn. 1987. Effective sample size for confidence intervals for survival probabilities. Stat. Med. 6679-687. [DOI] [PubMed] [Google Scholar]
- 9.Durack, D. T., A. S. Lukes, D. K. Bright, et al. 1994. New criteria for diagnosis of infective endocarditis: utilization of specific echocardiographic findings. Am. J. Med. 96200-209. [DOI] [PubMed] [Google Scholar]
- 10.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 997687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forsman, P., A. Tilsala-Timisjarvi, and T. Alatossava. 1997. Identification of staphylococcal and streptococcal causes of bovine mastitis using 16S-23S rRNA spacer regions. Microbiology 1433491-3500. [DOI] [PubMed] [Google Scholar]
- 12.Gunn, B. A. 1989. Comparative virulence of human isolates of coagulase-negative staphylococci tested in an infant mouse weight retardation model. J. Clin. Microbiol. 27507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herndona, B. L., H. Bialkowska-Hobrzanska, and L. Dall. 2000. Molecular characterization and LD50 identify virulent isolates of Staphylococcus epidermidis from adult sepsis. Diagn. Microbiol. Infect. Dis. 3675-80. [DOI] [PubMed] [Google Scholar]
- 14.Jaulhac, B., M. L. De Buyser, F. Dilasser, G. Prevost, and Y. Piedmont. 1991. Screening of staphylococci for the toxic shock syndrome toxin-1 (TSST-1) gene. Lett. Appl. Microbiol. 1390-92. [DOI] [PubMed] [Google Scholar]
- 15.Karchmer, A. W., G. L. Archer, and W. E. Dismukes. 1983. Staphylococcus epidermidis causing prosthetic valve endocarditis: microbiologic and clinical observations as guides to therapy. Ann. Intern. Med. 98447-455. [DOI] [PubMed] [Google Scholar]
- 16.Kreiswirth, B. N., P. M. Schlievert, and R. P. Novick. 1987. Evaluation of coagulase-negative staphylococci for ability to produce toxic shock syndrome toxin 1. J. Clin. Microbiol. 252028-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai, Y., A. E. Villaruz, M. Li, D. J. Cha, D. E. Sturdevant, and M. Otto. 2007. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol. Microbiol. 63497-506. [DOI] [PubMed] [Google Scholar]
- 18.Lalani, T., Z. A. Kanafani, V. H. Chu, L. Moore, G. R. Corey, P. Pappas, C. W. Woods, C. H. Cabell, B. Hoen, C. Selton-Suty, T. Doco-Lecompte, C. Chirouze, D. Raoult, J. M. Miro, C. A. Mestres, L. Olaison, S. Eykyn, E. Abrutyn, and V. G. Fowler, Jr. 2006. Prosthetic valve endocarditis due to coagulase-negative staphylococci: findings from the International Collaboration on Endocarditis Merged Database. Eur. J. Clin. Microbiol. Infect. Dis. 25365-368. [DOI] [PubMed] [Google Scholar]
- 19.Li, M., Y. Lai, A. E. Villaruz, D. J. Cha, D. E. Sturdevant, and M. Otto. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. USA 1049469-9474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahajan-Miklos, S., M. W. Tan, L. G. Rahme, and F. W. Ausubel. 1999. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 9647-56. [DOI] [PubMed] [Google Scholar]
- 21.Maira-Litrán, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark III, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 704433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miragaia, M., J. C. Thomas, I. Couto, M. C. Enright, and H. de Lencastre. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 1892540-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molnar, C., Z. Hevessy, F. Rozgonyi, and C. G. Gemmell. 1994. Pathogenicity and virulence of coagulase negative staphylococci in relation to adherence, hydrophobicity, and toxin production in vitro. J. Clin. Pathol. 47743-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsonnet, J., A. E. Harrison, S. E. Spencer, A. Reading, K. C. Parsonnet, and E. H. Kass. 1987. Nonproduction of toxic shock syndrome toxin 1 by coagulase-negative staphylococci. J. Clin. Microbiol. 251370-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patrick, C. C., S. V. Hetherington, P. K. Roberson, S. Henwick, and M. M. Sloas. 1995. Comparative virulence of Staphylococcus epidermidis isolates in a murine catheter model. Pediatr. Res. 3770-74. [DOI] [PubMed] [Google Scholar]
- 26.Petti, C. A., K. E. Simmon, J. M. Miro, B. Hoen, F. Marco, V. H. Chu, E. Athan, S. Bukovski, E. Bouza, S. Bradley, V. G. Fowler, E. Giannitsioti, D. Gordon, P. Reinbott, T. Korman, S. Lang, C. Garcia-de-la-Maria, A. Raglio, A. J. Morris, P. Plesiat, S. Ryan, T. Doco-Lecompte, F. Tripodi, R. Utili, D. Wray, J. J. Federspiel, K. Boisson, L. B. Reller, D. R. Murdoch, C. W. Woods, and the International Collaboration on Endocarditis—Microbiology Investigators. 2008. Genotypic diversity of coagulase-negative staphylococci causing endocarditis: a global perspective. J. Clin. Microbiol. 461780-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohde, H., M. Kalitzky, N. Kroger, S. Scherpe, M. A. Horstkotte, J. K. Knobloch, A. R. Zander, and D. Mack. 2004. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J. Clin. Microbiol. 425614-5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosec, J. P., J. P. Guiraud, C. Dalet, and N. Richard. 1997. Enterotoxin production by staphylococci isolated from foods in France. Int. J. Food Microbiol. 35213-221. [DOI] [PubMed] [Google Scholar]
- 29.Sifri, C. D., J. Begun, F. M. Ausubel, and S. B. Calderwood. 2003. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 712208-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sifri, C. D., J. Begun, and F. M. Ausubel. 2005. The worm has turned—microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 13119-127. [DOI] [PubMed] [Google Scholar]
- 31.Thomas, J. C., M. R. Vargas, M. Miragaia, S. J. Peacock, G. L. Archer, and M. C. Enright. 2007. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 45616-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Eiff, C., G. Peters, and C. Heilmann. 2002. Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2677-685. [DOI] [PubMed] [Google Scholar]
- 33.Wisplinghoff, H., A. E. Rosato, M. C. Enright, M. Noto, W. Craig, and G. L. Archer. 2003. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 473574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, B., Y. Li, and R. A. Betensky. 2006. Effects of unmeasured heterogeneity in the linear transformation model for censored data. Lifetime Data Anal. 12191-203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.