Abstract
Some clinical isolates of Staphylococcus aureus produce the superantigenic toxic shock syndrome toxin 1 (TSST-1), encoded by tst, located on pathogenicity islands. The expression of tst is complex and is influenced by environmental conditions such as pH, CO2, and glucose. We identified a putative catabolite-responsive element (cre) in the promoter regions of all known tst genes, indicating that tst transcription may be regulated by the catabolite control protein CcpA. By introducing tst genes under the control of their native promoters or tst promoter-reporter gene fusions in wild-type strain Newman, we showed that glucose was able to repress tst transcription and TSST-1 production, whereas glucose repression was abolished in the corresponding ΔccpA mutant. Stabilizing the pH ruled out a pH effect due to acid production during glucose catabolism. CcpA thus directly regulates tst transcription, linking carbohydrate utilization to virulence gene expression in S. aureus.
Toxic shock syndrome (TSS) is an acute and potentially fatal illness caused by a group of bacterial superantigens, such as toxic shock syndrome toxin 1 (TSST-1), staphylococcal enterotoxins, and streptococcal pyrogenic toxins. Superantigens, unlike conventional peptide antigens, bind to invariant regions of the major histocompatibility complex class II molecules at the surfaces of antigen-presenting cells, outside the classical antigen-binding groove, and also to invariant regions of the T-cell receptor. This leads to activation of T cells at orders of magnitude above that for antigen-specific activation, resulting in massive cytokine release, which in turn leads to capillary leakage and is believed to be responsible for hypotension, shock, and finally, death (19). Especially in the early 1980s, there was a major interest in TSS because there were several staphylococcal TSS cases reported for otherwise healthy young women (7). This rise in TSS cases was associated with high-absorbency tampons, and the disease was named menstrual TSS. In most cases, menstrual TSS is associated with TSST-1, but cases of nonmenstrual TSS have also been associated with this toxin (8, 9), which is encoded by tst, a gene found on several staphylococcal pathogenicity islands (SaPIs) (reviewed in reference 25).
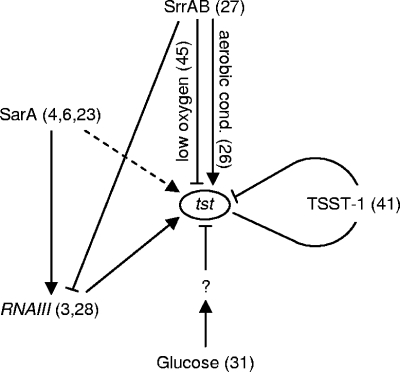
The regulation of TSST-1 is known to be complex, and several environmental conditions, including salt, oxygen, carbon dioxide, growth rate, temperature, glucose, and pH, were reported to modulate its production (3, 29, 31, 46). Global regulators shown to be involved in the regulation of TSST-1 are SarA (4, 6, 23), RNAIII of the agr system (3, 28), and SrrAB (45) (Fig. 1). However, there is some contradiction about the role of SarA in tst expression, since Chan and Foster (4) described that repression of tst by SarA was agr independent, while Novick (23) found that tst repression was dependent on agr. SrrAB, which is also a repressor of RNAIII (27), was previously shown to repress tst transcription under limited oxygen pressure (45). More recently, Pragman et al. (26) proposed that SrrAB, in addition to its downregulatory function under low-oxygen conditions, enhances the level of tst under aerobic conditions. More complexity is added by the fact that tst has autoregulatory functions, with TSST-1 acting as a repressor of tst transcription (41) (Fig. 1).
FIG. 1.
Regulation of tst transcription. Arrows represent upregulation, bars represent downregulation, dashed arrows indicate controversial findings, and numbers in parentheses indicate references. The question mark corresponds to CcpA.
We recently analyzed the impact of the carbon catabolite protein A (CcpA) of Staphylococcus aureus on carbon metabolism, virulence determinant expression, and biofilm formation (34, 35). CcpA induces or represses the transcription of genes by binding to so-called catabolite-responsive elements (cre) (33). These cis-acting DNA sequences, which consist of 14 to 18 bp, have been studied extensively in Bacillus subtilis (17, 20, 21, 38, 44, 47). Binding of CcpA to the cre sites is induced by complex formation with HPr, a component of the phosphoenolpyruvate-dependent phosphotransferase transport system (32, 37, 43). In the presence of glucose or other rapidly metabolized carbon sources, HPr is phosphorylated on serine-46 and binds to CcpA. But CcpA can apparently also regulate gene expression without binding to cre sites (12-14, 22). It was proposed that CcpA can directly interact with and inhibit RNA polymerase, in a process that is stimulated by NADP or NADPH, independent of the redox state of the cell (14).
Since TSST-1 production was shown to be repressed by glucose (31) and since our in silico analysis of the sequenced S. aureus genomes identified putative cre sites in the tst promoter regions, we investigated whether CcpA was the mediator of this regulatory phenomenon. We therefore analyzed the effects of glucose on tst transcription and TSST-1 production and compared them to the effects in a ccpA null mutant. Our results indicate that repression of tst by glucose is mediated directly by the carbon catabolite protein CcpA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains, relevant phenotypes, and plasmids used in this study are listed in Table 1. All strains generated for this study were confirmed by pulsed-field gel electrophoresis of total genomic SmaI digests (42) and by Southern blot analysis according to standard protocols, using CcpA- and RNAIII-specific digoxigenin (DIG)-labeled probes, which were generated using previously published primers (35). S. aureus was routinely grown in Luria-Bertani (LB) medium buffered with 50 mM HEPES (pH 7.5), with a flask volume/culture volume ratio of 5:1, at 37°C and 200 rpm. The medium was supplemented with 50 μg ml−1 kanamycin and 10 μg ml−1 tetracycline, if appropriate.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype and/or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus strains | ||
| RN4220 | NCTC8325-4 r− m+ (restriction negative, modification positive) | 15 |
| RN6911 | NCTC8325-4 agr::tet(M) Tcr | 40 |
| Newman | ATCC 25904; clinical isolate, CP5 producer | 10 |
| MST14 | Newman ccpA::tet(L) Tcr | 35 |
| N315 | Multiresistant clinical methicillin-resistant S. aureus isolate, TSST-1 producer | 16 |
| RF122 | Wild-type strain from bovine mastitis, TSST-1 producer | 11 |
| RN450 | NCTC8325-4 | 24 |
| KS87 | Newman/pSKA12 (tstpN315::luc+) Kanr | This study |
| KS64 | Newman ccpA::tet(L)/pSKA12 (tstpN315::luc+) Tcr Kanr | This study |
| KS179 | Newman/pSKA20 (tstN315+) Kanr | This study |
| KS180 | MST14/pSKA20 (tstN315+) Tcr Kanr | This study |
| KS181 | Newman/pSKA21 (tstRF122+) Kanr | This study |
| KS182 | MST14/pSKA21 (tstRF122+) Tcr Kanr | This study |
| KS186 | Newman agr::tet(M) Tcr | This study |
| KS187 | Newman agr::tet(M)/pSKA12 (tstpN315::luc+) Tcr Kanr | This study |
| KS188 | Newman agr::tet(M)/pSKA20 (tstN315+) Tcr Kanr | This study |
| KS189 | Newman agr::tet(M)/pSKA21 (tstRF122+) Tcr Kanr | This study |
| KS197 | RN450/pSKA20 (tstN315+) Kanr | This study |
| KS198 | RN450/pSKA21 (tstRF122+) Kanr | This study |
| E. coli strain | ||
| DH5α | Restriction-negative strain for cloning | Invitrogen |
| Plasmids | ||
| pSPluc+ | Luciferase fusion plasmid, ColE1 replication origin; Apr | Promega |
| pCN34 | E. coli-S. aureus shuttle plasmid, ColE1 replication origin; pT181 cop-wt repC; Ampr Kanr | 5 |
| pAW17 | E. coli-S. aureus shuttle plasmid, ColE1 and pAMα1 replication origins; Kanr | 30 |
| pSKA12 | pCN34 containing a 2.5-kb tstpN315::luc+ fusion; Apr Kanr | This study |
| pSKA20 | pAW17 with a 1-kb PCR fragment covering tst of strain N315 and its proposed promoter; Kanr | This study |
| pSKA21 | pAW17 with a 1-kb PCR fragment covering tst of strain RF122 and its proposed promoter; Kanr | This study |
Tcr, tetracycline resistant; Kanr, kanamycin resistant; Apr, ampicillin resistant.
Construction of plasmid tstpN315::luc+-pCN34 (pSKA12).
A 0.8-kb fragment containing the tst promoter region was amplified by PCR from chromosomal DNA of S. aureus N315 by the use of primers TstPAsp718+ (GCGCCATGGTTAATTCTCCTTCATTCAAA), including an Asp718 linker (underlined), and TstPNcoI− (GCGGGTACCTTCGAGAGGCAGATTACTCC), including an NcoI linker (underlined). The PCR product was cloned in front of the luciferase gene of plasmid pSPluc+. From this plasmid, a 2.4-kb Asp718-EcoRI fragment, including the tst promoter region fused to the luciferase coding region, was cloned into plasmid pCN34, generating plasmid pSKA12. The identity of the construct was confirmed by sequence analysis and comparison to the respective N315 sequence in the NCBI database (accession no. NC_002745). The plasmid was electroporated into strain RN4220 and subsequently transduced into strain Newman and its isogenic ΔccpA mutant (MST14). The plasmid was also introduced into strain Newman Δagr (KS186), which was obtained by transduction of agr::tet(M) from strain RN6911.
Construction of plasmids tstN315-pAW17 (pSKA20) and tstRF122-pAW17 (pSKA21).
One-kilobase fragments containing tst and its promoter region were amplified by PCR from chromosomal DNAs of S. aureus N315 and RF122, using primers TstXbaI+ (GTTGTCTAGAACTCACACTTTGTTTTTTGC), including an XbaI linker (underlined), and TstPstI− (GTTGCTGCAGGTTTACTAATTCACCCTAGC), including a PstI linker (underlined). The PCR products were cloned into pAW17, generating pSKA20 and pSKA21, respectively. The identities of the constructs were confirmed by sequence analysis and comparison to the respective sequences in the NCBI database (for N315, accession no. NC_002745; and for RF122, accession no. NC_007622). The plasmids were electroporated into RN4220 and subsequently transduced into strain Newman, its respective ΔccpA mutant (MST14), strain Newman Δagr (KS186), and strain RN450.
Luciferase assays.
For glucose impulse experiments, the cultures were inoculated at an optical density at 600 nm (OD600) of 0.05 and were grown for 5 h without glucose. At this time point, corresponding to the early stationary growth phase, the cultures were split and different amounts of glucose were added. Samples were taken 0, 30, and 60 min after the glucose impulse. To follow the tst promoter-luciferase reporter activity over time, the medium was inoculated with an overnight culture to an OD600 of 0.05 and the bacteria were grown for 8 h. The medium contained either 10 mM glucose or no glucose. Sampling was performed on an hourly basis, starting 2 h after inoculation. Luciferase activity was measured as described earlier, using a luciferase assay substrate and a Turner Designs TD-20/20 luminometer (Promega) (1). All luciferase assays were performed at least three times in independent experiments.
Northern blots.
Sampling for Northern blot analysis was performed basically as described for the glucose impulse experiments in the luciferase assays. Ten millimolar glucose was added to one-half of the culture, while the other half remained without glucose. Samples were centrifuged for 2 min at 12,000 × g, and cell sediments were snap-frozen in liquid nitrogen. RNA isolation and Northern blotting were performed as described earlier (18). Primers DIGtst+ (TAAGCCTTTGTTGCTTGCG) and DIGtst− (CACTTTGATATGTGGATCCG) were used to generate DIG-labeled tst probes. All Northern blot analyses were performed at least twice with independently isolated RNA samples.
Exoprotein analysis and Western blots.
For the determination of TSST-1 levels in the supernatants, cells were grown for 3 or 5 h either with or without 10 mM glucose. Supernatants were adjusted to the same volume for all samples, using sterile medium. Identical amounts of bovine serum albumin were added to samples as an internal control for semiquantitative detection of TSST-1. Proteins were precipitated with trichloroacetic acid at a final concentration of 10%, samples were placed on ice for 30 min and centrifuged for 15 min at 16,000 × g, and sediments were washed twice with 5 ml acetone. After being air dried, sediments were resuspended in loading buffer and samples corresponding to a final OD600 of 0.25 (1.25 for strain N315) were analyzed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Gels were either stained with Coomassie brilliant blue or blotted on nitrocellulose membranes. TSST-1 detection was performed using rabbit polyclonal anti-TSST-1 antibodies (LucernaChem, Switzerland) after blocking with human immunoglobulin G and visualized using SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
For the determination of exoprotein patterns, cells were sampled at an OD600 of 3.5 (∼5 h). Proteins were precipitated as described above, and samples corresponding to a final OD600 of 1.0 were analyzed by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue or blotted for TSST-1 detection as described above.
Determination of glucose levels.
Two-milliliter aliquots of bacterial cultures were harvested at the indicated time points and centrifuged for 2 min at 16,000 × g. The supernatants were incubated at 80°C for 15 min and stored at −20°C until use. Glucose levels were determined with kits from R-Biopharm (Darmstadt, Germany) according to the manufacturer's directions.
RESULTS AND DISCUSSION
The tst promoter region contains a cre site.
By screening strain N315 with the consensus sequence proposed by Miwa et al. (21), we identified a putative cre site 22 bp upstream of the translational start of tst on SaPIn1 (Fig. 2). Because tst is located on different pathogenicity islands (SaPIs) in different strains (25), we compared the promoter regions of the tst genes of the published SaPIs (SaPIn1 [strain N315], GenBank accession no. NC_002745; SaPm1 [strain Mu50], NP_372535; SaPI1 [strain RN4282], U93688; SaPI2 [strain RN3984], EF010993; and SaPbov1 [strain RF122], NC_007622). We found identical cre sites on SaPI1, SaPIm1, SaPIn1, and SaPI2 (data not shown). With the exception of 1 bp, the cre sites were palindromic and were located downstream of the transcriptional start site suggested by Vojtov et al. (41). The pathogenicity island SaPIbov1 of the bovine mastitis isolate RF122 had a cre site which differed at 2 bp from the cre sites of the other tst genes. However, it was still palindromic with the exception of 1 bp (Fig. 2).
FIG. 2.
Alignment of the cloned tst promoter regions of strains N315 (GenBank accession no. NC_002745; nucleotides 2,060,871 to 2,061,078) and RF122 (GenBank accession no. NC_007622; nucleotides 400,656 to 400,452) used for the construction of pSKA20 and pSKA21. The ribosome binding site (rbs) suggested by Blomster-Hautamaa et al. (2) is shown. Nucleotides fitting with the cre consensus of B. subtilis suggested by Miwa et al. (21) are highlighted in bold. Inverted repeats are indicated by arrows. Differences in nucleotides are highlighted by gray boxes.
tst expression is strongly repressed in the presence of glucose.
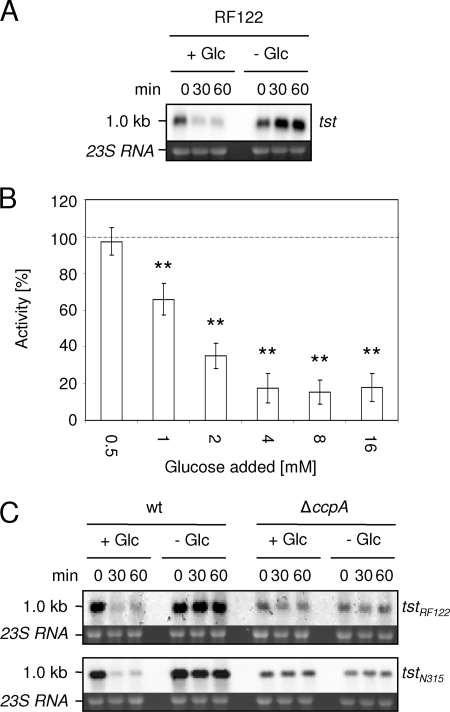
The presence of cre sites in the different tst genes together with previous findings showing that glucose decreased TSST-1 production (31) indicated that CcpA might mediate glucose-induced tst repression. To confirm that glucose also had an effect on tst transcription, we performed a Northern blot analysis in a glucose impulse experiment. We chose strains N315 and RF122 to compare the two different cre variants. Because tst is known to be expressed in early stationary growth phase (39), we cultured the cells in buffered medium for 5 h, split the cultures, and added glucose to one-half of each culture. In line with the literature, we found strong tst transcription in RF122 during this growth stage, while the addition of glucose led to a clear repression of tst (Fig. 3A). Surprisingly, in strain N315, tst expression was much weaker and smears appeared instead of a distinct band, impeding an interpretation of the glucose effect (data not shown). To see if the glucose-dependent activation of CcpA was functional in this particular strain, we tested the glucose-mediated repression of pckA, a gene previously reported to be under the control of CcpA (35). We observed a strong glucose-dependent repression of pckA, indicating that N315 possessed a functional CcpA protein (data not shown).
FIG. 3.
Influence of glucose on tst expression. (A) Northern blot analysis of tst expression in response to glucose in strain RF122 after the addition of 10 mM glucose to a culture in the early stationary growth phase. Ethidium bromide-stained 23S rRNA indicates RNA loading. (B) Luciferase activity of strain KS87 in response to different glucose concentrations. The luciferase activity at time zero was set to 100% (dashed line), and relative activity after 60 min was assessed. The averages of three independent measurements are shown. Error bars indicate the standard deviations of the mean activities. **, P < 0.01 for supplemented versus unsupplemented cultures. (C) tst transcription in response to glucose in strains Newman (wt) and MST14 (ΔccpA) carrying pSKA20 (tstN315) or pSKA21 (tstRF122) after the addition of 10 mM glucose to a culture in the early stationary growth phase. Ethidium bromide-stained 23S rRNA indicates RNA loading. The exposure time of tstN315 had to be adjusted in comparison to that of tstRF122.
To determine the influence of the genetic background on tst expression in N315, we fused the promoter region of the tst gene of this strain to the firefly luciferase gene, yielding plasmid tstpN315::luc+-pCN34 (pSKA12), and transduced the construct into strain Newman. The luciferase activity in the transformed Newman derivative reached high values of between 30,000 and 50,000 relative light units. By testing the impact of the glucose concentration on tst repression, we observed that the addition of 1 mM (0.018%) glucose reduced the luciferase activity to 66% (±8.5%), 2 mM (0.036%) glucose decreased the activity to 35% (±7.0%), and concentrations of >4 mM decreased the activity to <18% (±8.0%) (Fig. 3B). This indicated that transcription of the tst promoter of N315 was highly sensitive to low concentrations of glucose and suggested that the low tst expression in N315 was likely due to the genetic background.
Glucose-dependent repression of tst transcription requires CcpA.
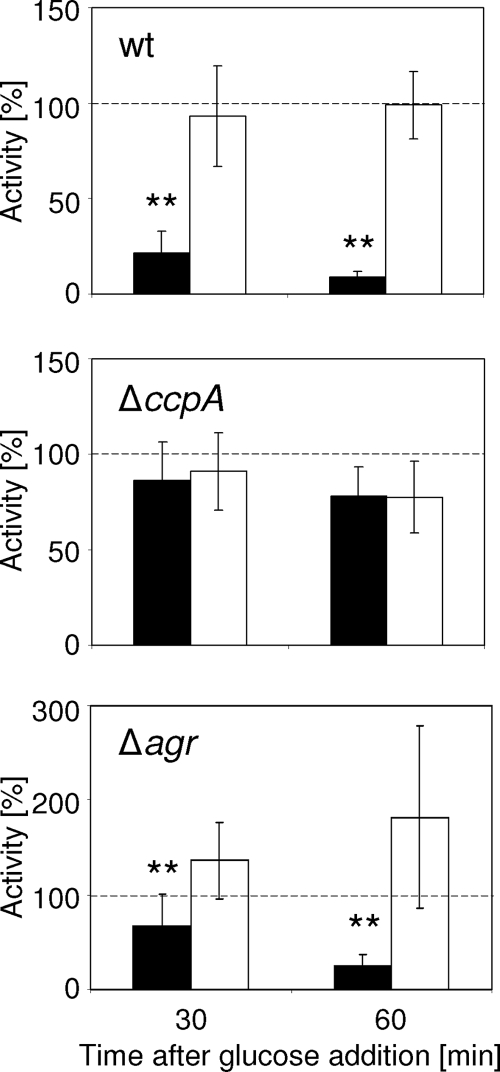
To assess the impact of CcpA on the glucose-dependent repression of tst transcription, we transduced the tstpN315::luc+-pCN34 construct into the ΔccpA mutant of strain Newman and followed the glucose effect over time in the wild-type and mutant strains. In the wild type, repression of luciferase activity was already clearly detectable 30 min after glucose addition, and activity decreased further until 1 h after glucose addition (Fig. 4). The pH remained stable throughout the time course followed, ruling out a possible pH effect on tst expression (31). In the mutant, no changes in activity were observed upon glucose addition, strongly suggesting that CcpA was involved in glucose-mediated tst repression (Fig. 4).
FIG. 4.
Relative luciferase activities of strains KS87 (wt), KS64 (ΔccpA), and KS187 (Δagr). Glucose (10 mM) was added to one-half of a culture in the early stationary growth phase (black bars), while the other half remained without glucose (white bars). Activity at time zero was set to 100% (dashed line), and relative activities after 30 and 60 min were assessed. The averages of three independent measurements are shown. Error bars indicate the standard deviations of the mean activities. **, P < 0.01 for supplemented versus unsupplemented cultures.
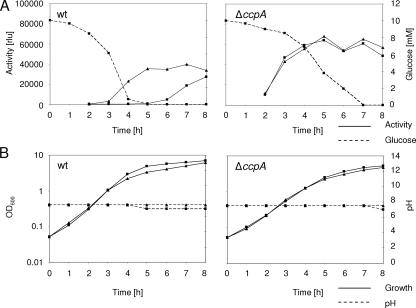
To observe the long-term effects of glucose on tst promoter activity during growth, the wild-type and ΔccpA mutant strains were grown for 8 h in either the presence or absence of glucose, and luciferase activity was followed at 1-h intervals. Without glucose, activity in the wild type started to rise after 3 h, reached its maximum after 5 h, and remained stable from then on (Fig. 5A). In the presence of glucose, activity remained very low (about 1% of the activity without glucose) and started to rise only slowly after 5 h, when glucose was depleted from the medium, reaching a similar value to that for cells grown without glucose after 8 h (Fig. 5A and B). Despite HEPES buffering, in the presence of glucose, the pH dropped slightly after 5 h (Fig. 5B). For the mutant, luciferase activities were similar in the presence and absence of glucose, and the pH remained stable under both conditions for up to 7 h (Fig. 5A and B). Compared with that of the wild type, luciferase activity in the mutant rose faster and tended to be higher.
FIG. 5.
tst promoter activity and growth characteristics of KS87 (wt) and KS64 (ΔccpA) in the presence (squares) and absence (triangles) of glucose. (A) Luciferase activity and glucose consumption. rlu, relative light units. (B) Growth and pH. Data are representative of three independent measurements.
The luciferase assays left the question open of whether and how CcpA would repress tst transcription in the presence of an intact tst gene. Unfortunately, we were not successful in transducing the ΔccpA mutation into strain N315, strain RF122, or other clinical TSST-1-expressing isolates from our strain collection. We therefore introduced the tst genes of strains N315 and RF122 (tstN315 and tstRF122, respectively) in trans into strain Newman and its ΔccpA mutant. The plasmid-borne tst transcripts were the same size (1 kb) as the chromosomal tst transcripts of strains N315 and RF122 (data not shown). Even though the higher copy number of the plasmid-carried tstN315 and tstRF122 genes may blur a CcpA effect in strains KS181 and KS179, tst transcription in the respective Newman ΔccpA mutants KS182 and KS180 was lower than that in the wild type, indicating that CcpA might exert some regulatory function on tst expression even in the absence of glucose (Fig. 3C). The overall apparently stronger tstN315 transcription, although it was cloned into the same plasmid backbone as tstRF122 and expressed in the same Newman background, may be due to small sequence differences in the operator region (Fig. 2) or possibly also within the coding or terminator region (not shown).
In performing glucose impulse experiments with the strains carrying the tst-expressing plasmids, we found strong repression of tst transcription by glucose in strain Newman, independent of whether it carried the N315 or RF122 tst variant. However, no such effect was observed in the ΔccpA mutant (Fig. 3C). The finding that both tst genes were repressed by glucose in a CcpA-dependent manner suggests that the minor difference in their cre sites did not affect CcpA activity.
tst does not affect exoprotein patterns in LB medium.
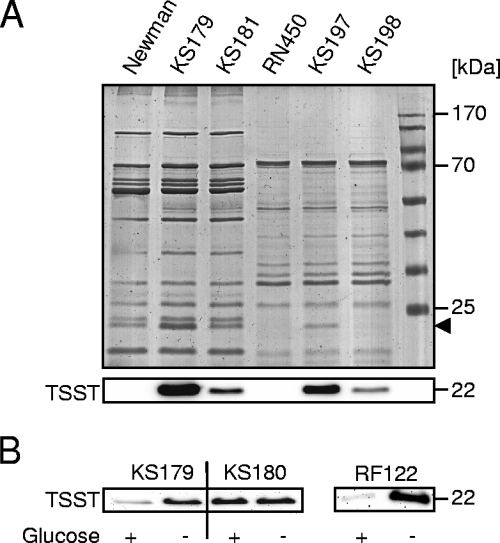
To assess TSST-1 production and the impact of TSST-1 on the exoprotein patterns of the different strains, we analyzed supernatants of post-exponential-growth-phase cultures. Consistent with its low tst transcription, strain N315 produced significantly smaller amounts of TSST-1 than those in RF122 or Newman carrying the tst plasmids after 5 h of growth (data not shown). The production of larger TSST-1 amounts in KS179 (Newman tstN315) than in KS181 (Newman tstRF122) (Fig. 6A) was in line with the stronger tstN315 transcription in this strain.
FIG. 6.
Influence of TSST-1 production on exoprotein patterns and influence of ccpA deletion on TSST-1 production in response to glucose. (A) Exoprotein patterns and TSST-1 production of strains Newman, KS179 (Newman tstN315), KS181 (Newman tstRF122), RN450, KS197 (RN450/pSKA20 tstN315), and KS198 (RN450/pSKA21tstRF122) at an OD600 of 3.5. The arrow indicates the position of TSST-1. (B) Comparison of TSST-1 production in response to glucose in strains KS179 (Newman tstN315), KS180 (Newman ΔccpA tstN315), and RF122 after 3 h of growth.
Unlike Vojtov et al. (41), who found a strong difference in exoprotein production between TSST-1-producing isolates and their isogenic non-TSST-1-producing strains in CYGP medium (Casamino Acids [10 g/liter]; yeast extract [10 g/liter], NaCl [5 g/liter], 20% glucose, 1.5 M phosphoglycerate), we did not observe such differences between the TSST-1-producing derivatives of strain Newman and the parental strain in LB medium (Fig. 6A). Transducing the tst-expressing plasmids into strain RN450 (NCTC8325-4 agr+), a background similar to the one used by Vojtov et al. (41), did not change the exoprotein pattern in comparison to that of the wild type (Fig. 6A), suggesting that this effect may be medium dependent. As already found in the Newman background, tst transcription and TSST-1 production (Fig. 6A) were stronger when RN450 carried the tstN315 gene than when it carried tstRF122.
Glucose affects TSST-1 production in a CcpA-dependent manner.
To confirm that the observed glucose-mediated repression of tst transcription also affected TSST-1 production, we determined the TSST-1 contents in the supernatants of KS179 (Newman tstN315) and KS180 (Newman ΔccpA tstN315) cell cultures after 3 h of growth in the presence and absence of 10 mM glucose. In the presence of glucose, KS179 reached a higher OD600 than that in the absence of glucose, while no such difference in OD600 was found for strain KS180. In line with the transcriptional data, we found a clear decrease in TSST-1 in the supernatant of strain KS179 grown in the presence of glucose, while the TSST-1 levels in the supernatants of the ΔccpA mutant KS180 were almost identical in the presence and absence of glucose (Fig. 6B). Glucose also led to a higher OD600 in strain RF122 and repressed TSST-1 production (Fig. 6B).
Repression of tst expression by CcpA in an agr-null mutant.
Strain N315 was previously reported to have defective RNAIII production (36). Since RNAIII is a known inducer of tst transcription (3, 28), this may explain the low tst transcription levels in N315. We confirmed that agr inactivation in strain Newman strongly reduced tstN315 and tstRF122 transcription (data not shown). The low level of tst transcription observed in N315, which harbors a functional CcpA system, might therefore be due, at least in part, to the lack of RNAIII. Nevertheless, by luciferase assays, we observed glucose-mediated repression of tst promoter-reporter gene activity in the Δagr mutant of strain Newman (Fig. 4), indicating that the glucose-mediated repression by CcpA remained functional in the Δagr mutant. This suggests that expression of tst depends on dual, direct RNAIII stimulation and glucose-mediated CcpA repression and on indirect CcpA-mediated control of RNAIII levels. However, we cannot rule out that additional factors may influence tst transcription, as N315 differs in terms of metabolism from strain Newman by being unable to catabolize acetate in the post-exponential growth phase (36). This loss of secondary metabolite catabolism might have consequences on tst expression. Also, regulatory circuits which may be present in strain N315 but not in the other strains could be the reason for the low tst expression in N315.
Conclusion.
The expression of S. aureus virulence genes is regulated by complex networks in which global regulators such as the agr system and the sarA family play a central role. We recently showed that CcpA is also involved in the regulation of major virulence determinants by influencing the expression of hla, spa, and RNAIII (35). By demonstrating that the expression of tst is affected by CcpA as well, we add another important virulence factor to the group of CcpA-regulated genes/operons. The significance of glucose-mediated TSST-1 expression in vivo is unknown and remains to be evaluated using appropriate S. aureus constructs in wild-type and diabetic animal pathogenesis models, with the latter elaborating elevated blood glucose levels.
Acknowledgments
This study was supported by Swiss National Science Foundation grant 31-117707.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 1835171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blomster-Hautamaa, D., B. Kreiswirth, J. Kornblum, R. Novick, and P. Schlievert. 1986. The nucleotide and partial amino acid sequence of toxic shock syndrome toxin-1. J. Biol. Chem. 26115783-15786. [PubMed] [Google Scholar]
- 3.Chan, P. F., and S. J. Foster. 1998. The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiology 1442469-2479. [DOI] [PubMed] [Google Scholar]
- 4.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 1806232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charpentier, E., A. I. Anton, P. Barry, B. Alfonso, Y. Fang, and R. P. Novick. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 706076-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. L., A. S. Bayer, G. Zhang, H. Gresham, and Y.-Q. Xiong. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 401-9. [DOI] [PubMed] [Google Scholar]
- 7.Davis, J. P., P. J. Chesney, P. J. Wand, and M. LaVenture. 1980. Toxic-shock syndrome: epidemiologic features, recurrence, risk factors, and prevention. N. Engl. J. Med. 3031429-1435. [DOI] [PubMed] [Google Scholar]
- 8.Descloux, E., T. Perpoint, T. Ferry, G. Lina, M. Bes, F. Vandenesch, I. Mohammedi, and J. Etienne. 2008. One in five mortality in non-menstrual toxic shock syndrome versus no mortality in menstrual cases in a balanced French series of 55 cases. Eur. J. Clin. Microbiol. Infect. Dis. 2737-43. [DOI] [PubMed] [Google Scholar]
- 9.Dinges, M. M., P. M. Orwin, and P. M. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 1316-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duthie, E., and L. L. Lorenz. 1952. Staphylococcal coagulase; mode of action and antigenicity. J. Gen. Microbiol. 695-107. [DOI] [PubMed] [Google Scholar]
- 11.Fitzgerald, J. R., P. J. Hartigan, W. J. Meaney, and C. J. Smyth. 2000. Molecular population and virulence factor analysis of Staphylococcus aureus from bovine intramammary infection. J. Appl. Microbiol. 881028-1037. [DOI] [PubMed] [Google Scholar]
- 12.Gösseringer, R., E. Küster, A. Galinier, J. Deutscher, and W. Hillen. 1997. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J. Mol. Biol. 266665-676. [DOI] [PubMed] [Google Scholar]
- 13.Kim, J.-H., M. I. Voskuil, and G. H. Chambliss. 1998. NADP, corepressor for the Bacillus catabolite control protein CcpA. Proc. Natl. Acad. Sci. USA 959590-9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, J.-H., Y.-K. Yang, and G. H. Chambliss. 2005. Evidence that Bacillus catabolite control protein CcpA interacts with RNA polymerase to inhibit transcription. Mol. Microbiol. 56155-162. [DOI] [PubMed] [Google Scholar]
- 15.Kreiswirth, B. N., S. Löfdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305709-712. [DOI] [PubMed] [Google Scholar]
- 16.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K.-I. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R.-I. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 3571225-1240. [DOI] [PubMed] [Google Scholar]
- 17.Martin-Verstraete, I., J. Stülke, A. Klier, and G. Rapoport. 1995. Two different mechanisms mediate catabolite repression of the Bacillus subtilis levanase operon. J. Bacteriol. 1776919-6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCallum, N., H. Karauzum, R. Getzmann, M. Bischoff, P. Majcherczyk, B. Berger-Bächi, and R. Landmann. 2006. In vivo survival of teicoplanin-resistant Staphylococcus aureus and fitness cost of teicoplanin resistance. Antimicrob. Agents Chemother. 502352-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCormick, J. K., J. M. Yarwood, and P. M. Schlievert. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 5577-104. [DOI] [PubMed] [Google Scholar]
- 20.Miwa, Y., and Y. Fujita. 2001. Involvement of two distinct catabolite-responsive elements in catabolite repression of the Bacillus subtilis myo-inositol (iol) operon. J. Bacteriol. 1835877-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miwa, Y., A. Nakata, A. Ogiwara, M. Yamamoto, and Y. Fujita. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 281206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 391366-1381. [DOI] [PubMed] [Google Scholar]
- 23.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 481429-1449. [DOI] [PubMed] [Google Scholar]
- 24.Novick, R. P. 1967. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology 33155-166. [DOI] [PubMed] [Google Scholar]
- 25.Novick, R. P., and A. Subedi. 2007. The SaPIs: mobile pathogenicity islands of Staphylococcus. Chem. Immunol. Allergy 9342-57. [DOI] [PubMed] [Google Scholar]
- 26.Pragman, A. A., Y. Ji, and P. M. Schlievert. 2007. Repression of Staphylococcus aureus SrrAB using inducible antisense srrA alters growth and virulence factor transcript levels. Biochemistry 46314-321. [DOI] [PubMed] [Google Scholar]
- 27.Pragman, A. A., J. M. Yarwood, T. J. Tripp, and P. M. Schlievert. 2004. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J. Bacteriol. 1862430-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Recsei, P., B. Kreiswirth, M. O'Reilly, P. M. Schlievert, A. Gruss, and R. P. Novick. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agr. Mol. Gen. Genet. 20258-61. [DOI] [PubMed] [Google Scholar]
- 29.Ross, R. A., and A. B. Onderdonk. 2000. Production of toxic shock syndrome toxin 1 by Staphylococcus aureus requires both oxygen and carbon dioxide. Infect. Immun. 685205-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi, J., M. Bischoff, A. Wada, and B. Berger-Bächi. 2003. MsrR, a putative cell envelope-associated element involved in Staphylococcus aureus sarA attenuation. Antimicrob. Agents Chemother. 472558-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlievert, P. M., and D. A. Blomster. 1983. Production of staphylococcal pyrogenic exotoxin type C: influence of physical and chemical factors. J. Infect. Dis. 147236-242. [DOI] [PubMed] [Google Scholar]
- 32.Schumacher, M. A., G. S. Allen, M. Diel, G. Seidel, W. Hillen, and R. G. Brennan. 2004. Structural basis for allosteric control of the transcription regulator CcpA by the phosphoprotein HPr-Ser46-P. Cell 118731-741. [DOI] [PubMed] [Google Scholar]
- 33.Seidel, G., M. Diel, N. Fuchsbauer, and W. Hillen. 2005. Quantitative interdependence of coeffectors, CcpA and cre in carbon catabolite regulation of Bacillus subtilis. FEBS J. 2722566-2577. [DOI] [PubMed] [Google Scholar]
- 34.Seidl, K., C. Goerke, C. Wolz, D. Mack, B. Berger-Bächi, and M. Bischoff. 2008. The Staphylococcus aureus CcpA affects biofilm formation. Infect. Immun. 762044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seidl, K., M. Stucki, M. Rüegg, C. Goerke, C. Wolz, L. Harris, B. Berger-Bächi, and M. Bischoff. 2006. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 501183-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Somerville, G. A., B. Said-Salim, J. M. Wickman, S. J. Raffel, B. N. Kreiswirth, and J. M. Musser. 2003. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect. Immun. 714724-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stülke, J., and W. Hillen. 1998. Coupling physiology and gene regulation in bacteria: the phosphotransferase sugar uptake system delivers the signals. Naturwissenschaften 85583-592. [DOI] [PubMed] [Google Scholar]
- 38.Stülke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus subtilis. Annu. Rev. Microbiol. 54849-880. [DOI] [PubMed] [Google Scholar]
- 39.Timmins, B. S., and K. T. Holland. 1999. Shift-down in growth rate rather than high cell density induces toxic shock syndrome toxin-1 gene expression in Staphylococcus aureus. FEMS Microbiol. Lett. 172173-177. [DOI] [PubMed] [Google Scholar]
- 40.Vandenesch, F., J. Kornblum, and R. P. Novick. 1991. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J. Bacteriol. 1736313-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vojtov, N., H. F. Ross, and R. P. Novick. 2002. Global repression of exotoxin synthesis by staphylococcal superantigens. Proc. Natl. Acad. Sci. USA 9910102-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wada, A., Y. Katayama, K. Hiramatsu, and T. Yokota. 1991. Southern hybridization analysis of the mecA deletion from methicillin-resistant Staphylococcus aureus. Biochem. Biophys. Res. Commun. 1761319-1325. [DOI] [PubMed] [Google Scholar]
- 43.Warner, J. B., and J. S. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weickert, M., and G. Chambliss. 1990. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 876238-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 1831113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yarwood, J. M., and P. M. Schlievert. 2000. Oxygen and carbon dioxide regulation of toxic shock syndrome toxin 1 production by Staphylococcus aureus MN8. J. Clin. Microbiol. 381797-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zalieckas, J. M., L. V. Wray, Jr., and S. H. Fisher. 1998. Expression of the Bacillus subtilis acsA gene: position and sequence context affect cre-mediated carbon catabolite repression. J. Bacteriol. 1806649-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]