Abstract
A subset of invasive nontypeable Haemophilus influenzae (NTHI) strains has evidence of IS1016, an insertion element associated with division I H. influenzae capsule serotypes. We examined IS1016-positive invasive NTHI isolates collected as part of Active Bacterial Core Surveillance within the Georgia Emerging Infections Program for the presence or absence of hmw1 and hmw2 (two related adhesin genes that are common in NTHI but absent in encapsulated H. influenzae) and hia (homologue of hsf, an encapsulated H. influenzae adhesin gene). Isolates were serotyped using slide agglutination, confirmed as NTHI strains using PCR capsule typing, and biotyped. Two hundred twenty-nine invasive NTHI isolates collected between August 1998 and December 2006 were screened for IS1016; 22/229 (9.6%) were positive. Nineteen of 201 previously identified IS1016-positive invasive NTHI isolates collected between January 1989 and July 1998 were also examined. Forty-one IS1016-positive and 56 randomly selected IS1016-negative invasive NTHI strains were examined. The hia adhesin was present in 39 of 41 (95%) IS1016-positive NTHI strains and 1 of 56 (1.8%) IS1016-negative NTHI strains tested; hmw (hmw1, hmw2, or both) was present in 50 of 56 (89%) IS1016-negative NTHI isolates but in only 5 of 41 (12%; all hmw2) IS1016-positive NTHI isolates. IS1016-positive NTHI strains were more often biotype V (P < 0.001) or biotype I (P = 0.04) than IS1016-negative NTHI strains, which were most often biotype II. Pulsed-field gel electrophoresis revealed the expected genetic diversity of NTHI with some clustering based on IS1016, hmw or hia, and biotypes. A significant association of IS1016 with biotypes V and I and the presence of hia adhesins was found among invasive NTHI. IS1016-positive NTHI strains may represent a unique subset of NTHI strains, with characteristics more closely resembling those of encapsulated H. influenzae.
Haemophilus influenzae is a gram-negative exclusive human pathogen whose pathogenic potential may vary, in part, depending on the presence or absence of a polysaccharide capsule. Encapsulated (typeable) H. influenzae bacteria are often associated with systemic invasive diseases, such as meningitis, whereas nonencapsulated H. influenzae (nontypeable H. influenzae [NTHI]) bacteria are usually associated with disease at respiratory mucosal sites, such as the sinuses, lung, or middle ear. Although invasive pediatric H. influenzae disease has declined with routine infant use of H. influenzae serotype b (Hib) conjugate vaccines, serious disease caused by non-type b encapsulated and NTHI strains remains important in children and adults (1, 25, 46). In the United States in 2006, 63% of invasive H. influenzae disease in children 0 to 17 years old and 65% of invasive H. influenzae disease in adults of ≥18 years old was due to NTHI (Active Bacterial Core Surveillance Report, Emerging Infections Network, Haemophilus influenzae, http://www.cdc.gov/ncidod/dbmd/abcs/survreports.htm) (18). Among children in the developed world, NTHI is a leading cause of bacterial ear infections (16, 51), with significant socioeconomic consequences (3, 4), and NTHI strains are responsible for a substantial number of pediatric deaths caused by pneumonia in the developing world (27).
Encapsulated H. influenzae strains possess cap genes necessary for production and transport of their respective polysaccharide capsules (39). In division I encapsulated H. influenzae strains, the cap locus is flanked by direct repeats of the IS1016 insertion element; division II encapsulated H. influenzae strains contain one or more IS1016 elements that are unassociated with cap genes. A number of investigators have shown that while IS1016 elements are generally absent in NTHI, they may be present in a small (but varying) proportion of NTHI isolates from the respiratory tract and invasive disease (53, 59, 60). It has been suggested that the presence of IS1016 in NTHI may be a marker for a more recent evolution from encapsulated H. influenzae (59, 60).
NTHI may differ from encapsulated H. influenzae in ways other than the lack of capsule, including a variety of adhesive structures. In studies designed to understand the mechanisms of respiratory tract colonization by NTHI, two high-molecular-weight adhesins, HMW1 and HMW2, expressed by the majority of NTHI strains and found to be immunogenic in natural infection, were identified (6, 10, 58). HMW1 adhesins mediate high levels of attachment to most human epithelial cell lines via a sialylated glycoprotein receptor (10, 58, 61); HMW2 adhesins mediate attachment to only a subset of human epithelial cell lines via an unknown receptor (29). Both are generally absent in encapsulated strains. Hia, a unique autotransporter adhesin protein that mediates attachment to cultured human epithelial cells (6, 57), is found in many non-Hib encapsulated H. influenzae (serotypes a, e, and f) and NTHI strains that lack HMW adhesins (52). hia is a homologue of the hsf gene that is ubiquitous among Hib strains, again suggesting a closer relationship between hia-containing NTHI and encapsulated H. influenzae (60).
Efforts to develop vaccines targeting NTHI disease have focused on a variety of surface-exposed antigens known to be targets of the immune system, including bacterial adhesion proteins (4); such efforts have been hampered by the heterogeneity of NTHI strains. Despite the heterogeneity, in a recent study using multilocus sequence typing to characterize 322 NTHI isolates, Erwin et al. demonstrated well-defined phylogenetic groups of NTHI that differ in genetic content (21). Better characterization of clinically relevant NTHI strains with shared features will improve understanding of NTHI and may inform vaccine development. In this study, using a large, well-characterized isolate collection, we sought to identify additional IS1016-positive, invasive NTHI isolates and to compare the adhesin gene profiles, biotypes, and restriction patterns in IS1016-positive and IS1016-negative NTHI isolates to assess relatedness to each other and to patterns previously recognized among encapsulated strains.
MATERIALS AND METHODS
Surveillance and case characteristics.
A total of 734 invasive H. influenzae isolates were collected as part of the Active Bacterial Core Surveillance (54) of the Georgia Emerging Infections Program from January 1989 through December 2006; 437 were PCR-confirmed NTHI isolates, 430 of which were available for further testing. All isolates were obtained from normally sterile body sites, such as blood and cerebrospinal fluid. Basic clinical and demographic information, including patient age, race, gender, clinical syndrome, and disease outcome, was collected for each case.
Bacterial strains and growth.
Characteristics (biotype, IS1016 screening, and selected pulsed-field gel electrophoresis [PFGE] typing) of 201 NTHI strains collected between January 1989 through July 1998 have been previously reported (53). The remaining 229 NTHI isolates collected from August 1998 through December 2006 were PCR capsule typed as previously described (53) with the following exception: new bexA primers were designed based on additional sequence information (bexA101-121, 5′-CAATTTTGAGYTAMAAAAAGG-3′; bexA646-627, 5′-TCCATRTCTTCAAAATGRTG-3′, where Y is C or T, M is A or C, and R is A or G). All bacteria were grown on chocolate II solid medium or in brain heart infusion broth supplemented with 10 μg/ml hemin and 2 μg/ml NAD (Becton Dickinson Microbiology Systems, Cockeysville, MD).
Southern blot hybridization.
Chromosomal DNA was extracted from 229 PCR-confirmed NTHI strains collected between August 1998 and December 2006 (9) and hybridized with digoxigenin-labeled pUO38 (39), pBR322 (8), or IS1016 as previously described (53).
Screen for hmw and hia adhesin genes.
All IS1016-postive NTHI strains from 1989 to 2006 and randomly selected IS1016-negative NTHI strains were screened for the presence of hmw1, hmw2, and hia using PCR on genomic DNA or single colony lysates as described by Erwin et al. (21) with updated primer sequences (A. L. Erwin et al., personal communication). Primers hmw1-F (5′CAAAGCCATCAGGTTGTTGTGC-3′) and hmw1-R (5′-CCTATTTGGTCTTGCTACGAGTGG-3′) correspond to Rd genome coordinates 1746545 to 1751277, and when hmw1 is present, these primers amplify a 13,928-bp product. Primers hmw2-F (5′-CCGCACTTTCTTCTCGTTCTTCT-3′) and hmw2-R (5′-GCTATTCGGTTAGGTAATGCAGATCC-3′) correspond to Rd genome coordinates 1665719 to 1667371, and when hmw2 is present, these primers amplify a 12,717-bp product (26). hia flanking primers Hia F (5′-CCGAAAGCACAAGGATATGGACG-3′) and Hia R (5′-CAGATAAATCCTGACCTCGCTCTC-3′) are located between Rd genome coordinates 1804356 to 1810590 and amplify a product of 6,235 bp if hia is present (26). PCR results, both negative and positive, were confirmed with Southern blot hybridization using hmw1A, hmw2A, and hia probes as previously described (19).
PFGE and composite cluster analysis.
PFGE was performed as previously described (53). Cluster analysis was applied to a composite data set combining the PFGE fingerprint data, biotypes, and the categorical data (with and without IS1016, hmw, and hia). A similarity matrix of the composite data was created and displayed on a dendrogram by averaging similarity matrices from the individual experiments, keeping internal weights equal and using the unweighted pair group method of arithmetic averages (UPGMA) (BioNumeric method manual 5.0, Applied Maths, Kortrijk, Belgium).
Biotyping.
Biochemical assays to determine biotype were performed at either the CDC or the Georgia Public Health Laboratory (Atlanta, GA) using the method of Kilian (37) and API 20E biochemical test strips purchased from bioMerieux (St. Louis, MO) (35).
Statistics.
Frequency data were evaluated by the chi-square test or, where appropriate, Fisher's exact probability test, and means were evaluated by Student's t test using SAS 9.1 (The SAS Institute, Inc., Cary, NC). A P value of <0.05 was considered statistically significant.
RESULTS
Four hundred thirty PCR-confirmed, invasive NTHI isolates collected between 1989 and 2006 in metropolitan Atlanta, GA, were available for testing. Results of biotyping, IS1016 screening, and selected PFGE typing have been previously reported for 201 isolates collected between 1989 and 1998 (53). In this study, 229 isolates were biotyped, screened for IS1016, and PFGE typed, and all 430 isolates were screened for hia and hmw. A combined dendrogram was generated.
IS1016-positive NTHI.
Southern blot hybridization with pUO38, pBR322, and IS1016 was performed on the 229 PCR-confirmed NTHI isolates (collected August 1998 to December 2006) to look for residual capsule-specific DNA as previously described (53). Plasmid pUO38 contains a complete cap locus from a division I Hib strain, including a copy of the insertion sequence IS1016 and the beta-lactamase (bla) gene from the plasmid backbone, pBR322 (39). No capsule-specific DNA was found among the 229 NTHI isolates probed with pUO38, 69% were bla positive, and 22/229 (9.6%) of the strains hybridized to IS1016 with a single band ranging between ∼5 and 10 kb (data not shown). The proportion of invasive NTHI isolates containing IS1016 was nearly identical to the 9.5% (19/201) rate noted previously among isolates collected between January 1989 and July 1998 (53); the combined IS1016-positive total was 41/430 (9.5%).
Biotypes.
The biotypes of all NTHI isolates collected between 1989 and 2006 have been combined and are shown in Table 1. Among the invasive NTHI isolates with known biotypes, 51/421 (12.1%) were biotype I, 216/421 (51.3%) were biotype II, 100/421 (23.7%) were biotype III, 22/421 (5.2%) were biotype IV, 20/421 (4.8%) were biotype V, 8/421 (1.9%) were biotype VI, and 4/421 (l%) were biotype VII. More than half of the IS1016-positive NTHI isolates were biotypes I and V, and compared with IS1016-negative NTHI, a significant association was noted between the presence of IS1016 and biotype V (P < 0.001) and biotype I (P = 0.04). Eighty percent (16/20) of isolates with the uncommon biotype V were IS1016 positive.
TABLE 1.
Distribution of biotypes of PCR-confirmed NTHI isolates from 1989 to 2006 and association with IS1016a
| Isolate | No. of NTHI isolates with the following biotype:
|
||||||
|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | |
| NTHI with IS1016 | 9 | 2 | 7 | 4 | 16 | 2 | 1 |
| NTHI without IS1016 | 40 | 213 | 92 | 15 | 4 | 6 | 3 |
| NTHI IS1016 N/Ab | 2 | 1 | 1 | 3 | 0 | 0 | 0 |
| Total | 51 | 216 | 100 | 22 | 20 | 8 | 4 |
Correlation of HMW and Hia adhesins with IS1016 in NTHI.
All 41 IS1016-positive NTHI isolates collected from 1989 to 2006 and 56 randomly selected IS1016-negative invasive NTHI isolates (also collected between 1989 and 2006) were examined for the presence or absence of the adhesin genes hmw1, hmw2, and hia by PCR and Southern blot hybridization. None of the 41 IS1016-positive NTHI strains had evidence of hmw1, and 5 of 41 (12%) were hmw2 positive (Table 2). The majority (39/41 [95%]) of IS1016-positive NTHI strains contained hia (Table 2). In contrast, most (50/56 [89%]) IS1016-negative NTHI isolates tested contained hmw, 10/56 (18%) isolates tested contained hmw1, 11/56 (19%) isolates tested contained hmw2, and 29/56 (52%) isolates tested contained both hmw1 and hmw2 (Table 2). The presence of hia in IS1016-negative NTHI was rare (1/56 [1.8%]) (Table 2). The majority (36/41 [87.8%]) of IS1016-positive NTHI isolates were hmw negative and hia positive, and the majority (49/56 [87.5%]) of IS1016-negative NTHI were hmw positive and hia negative; 6 strains were negative for both hmw and hia, and 4 were positive for both by PCR (Table 3). The association of hia with IS1016-positive NTHI was significant (P < 0.001).
TABLE 2.
Prevalence of hmw and hia adhesin genes in PCR-confirmed NTHI isolates and association with IS1016
| Isolate | No. of strains possessing the following gene(s)/total no. of strains (%)
|
||||
|---|---|---|---|---|---|
| hmw1 only | hmw2 only | Both hmw1 and hmw2 | hmw1 or hmw2 or both | hia | |
| NTHI with IS1016 | 0/41 | 5/41 (12) | 0/41 | 5/41 (12) | 39/41 (95.1) |
| NTHI without IS1016 | 10/56 (18) | 11/56 (19) | 29/56 (52) | 50/56 (89) | 1/56 (1.8) |
TABLE 3.
hmw and hia adhesin gene profiles in PCR-confirmed NTHI isolates and association with IS1016
| Isolate | No. of NTHI strains with the adhesin gene(s)/total no. of strains (%)
|
|||
|---|---|---|---|---|
| hmw negative and hia negative | hmw negative and hia positive | hmw positive and hia negative | hmw positive and hia positive | |
| NTHI with IS1016 | 0/41 | 36/41 (87.8) | 2/41 (4.9) | 3/41 (7.3) |
| NTHI without IS1016 | 6/56 (10.7) | 0/56 | 49/56 (87.5) | 1/56 (1.3) |
Epidemiological characteristics of IS1016-positive NTHI.
For the combined group of 430 cases of invasive NTHI disease, patients ranged in age between 0 and 100 years (mean age of 46.5 years; 75.8% adults were ≥18 years old). Cases of invasive NTHI disease due to IS1016-positive (n = 41) and IS1016-negative isolates (n = 389) were compared for significant differences in patient age, race, gender, clinical syndrome, and disease outcome (Table 4). A statistically significant association was observed between younger age and the presence of IS1016 in invasive NTHI disease; the association between older age (≥18 years) and the absence of IS1016 in invasive NTHI disease was nearly significant (P = 0.05). Among the 41 NTHI isolates positive for IS1016, one case occurred in a neonate (<1 month of age), 10 cases in children aged 1 month to 4 years old, 4 cases in children 5 to 17 years old, and 26 cases in adults 18 years old or older. Ten of 41 (24.4%) of the IS1016-positive NTHI strains were from children 1 month to 4 years of age compared to 10.6% of IS1016-negative NTHI (P = 0.01).
TABLE 4.
Epidemiological characteristics of PCR-confirmed NTHI and association with IS1016
| Variable | % NTHI isolates
|
P value | |
|---|---|---|---|
| With IS1016 (n = 41) | Without IS1016 (n = 389) | ||
| Mean age (yr) | 38 | 47 | 0.06 |
| Age groups | |||
| <1 mo | 2.4 | 6.2 | 0.21 |
| 1 mo-4 yrs | 24.4 | 10.6 | 0.01a |
| 5-17 yrs | 9.8 | 5.7 | 0.14 |
| ≥18 yrs | 63.4 | 77.5 | 0.05 |
| Race | |||
| White | 63.4 | 56.3 | 0.38 |
| Black | 31.7 | 40.4 | 0.28 |
| Other | 4.9 | 3.3 | 0.26 |
| Gender | |||
| Male | 49 | 45 | 0.67 |
| Female | 51 | 55 | 0.67 |
| Clinical syndrome | |||
| Meningitis | 5 | 9 | 0.50 |
| Bacteremia | 68 | 66 | 0.82 |
| Pneumonia | 27 | 25 | 0.86 |
| Disease outcome (survived) | 93 | 82 | 0.09 |
The percentage of NTHI isolates with IS1016 compared to the percentage of NTHI isolates without IS1016 was significantly different for 1-month- to 4-year-old children.
PFGE and cluster analysis of composite data set.
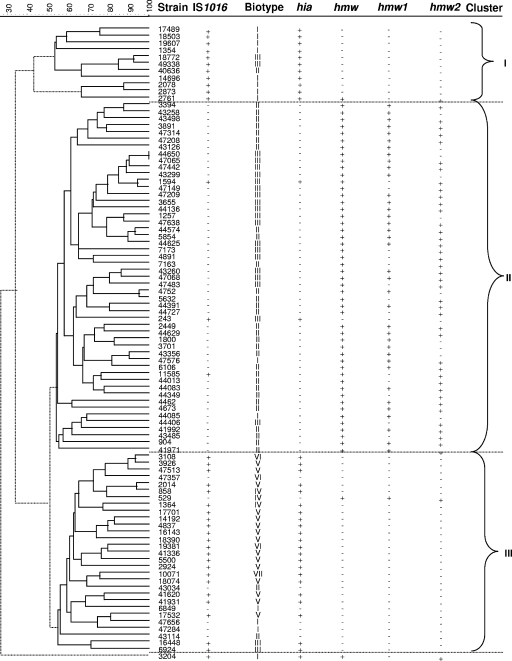
We compared the PFGE patterns of genomic DNA from 37 IS1016-positive and 55 IS1016-negative NTHI isolates. As expected, genetic diversity among the NTHI strains was relatively high, and 90 distinct restriction patterns were identified among the 92 strains available for PFGE (data not shown). However, when a composite data set was examined, clustering with respect to PFGE patterns, biotypes, and the presence or absence of IS1016, hmw, and hia was noted (Fig. 1). Using a dendrogram generated by BioNumerics' Cluster Analysis program, combined with the Cluster Cutoff method using the composite data set (PFGE, biotype, and presence/absence of IS1016, hmw, and hia), strains were grouped into three clusters. Clusters I and III contained the majority (92%) of IS1016-positive strains; 100% of cluster I strains and 75% (22/29) of cluster III strains were IS1016 and hia positive (Fig. 1). In addition, 11/13 biotype I strains fell within clusters I and III, and all biotype V strains were in cluster III.
FIG. 1.
Evolutionary analysis of invasive NTHI strains. The dendrogram was generated by BioNumerics software showing the genetic relationships between 17 IS1016-positive and 19 IS1016-negative NTHI strains. The dendrogram was constructed by cluster analysis of PFGE patterns (data not shown) obtained after digestion of chromosomal DNA with SmaI enzyme, using the unweighted pair group method of arithmetic averages. The strain designation, capsule type by PCR, biotype, and presence (+) or absence (−) of IS1016, hmw, and hia are shown. Three different clusters of branches of PFGE patterns are identified.
DISCUSSION
Since the introduction of the H. influenzae type b conjugate vaccine, the epidemiological characteristics of invasive Haemophilus influenzae disease have changed. Serious pediatric disease due to serotype b has been largely eliminated, while adult disease remains and is dominated by nontypeable strains. In addition to the significant burden of adult invasive NTHI disease, NTHI can occasionally cause bacteremia and meningitis in otherwise healthy newborns and older children (11, 15, 46, 48). NTHI bacteria remain important causes of localized respiratory tract infections (44).
Efforts to better understand the virulence factors responsible for NTHI disease pathogenesis have been increasing (23, 33, 62), and current evidence suggests that NTHI bacteria rely on multiple factors to colonize the human mucosal surfaces and ultimately spread to other sites (56). Genomic DNA-based microarray and bioinformatics approaches were used to compare the genomes of 1885MEE and 86-02NP, middle ear and nasopharyngeal NTHI isolates from children with otitis media, with the genome of Rd. These analyses suggested that the genomes of the two NTHI strains are more similar to each other than to Rd, a formerly encapsulated strain. A number of genes were identified in the NTHI strains that were absent in Rd, including proteins with possible roles in adhesion, biofilm formation, pH regulation, protection against reactive oxygen species, and a member of a family of virulence-associated autotransporters (43).
Although extensive heterogeneity among NTHI isolates compared with Hib and non-Hib encapsulated strains has been well described (41, 45), nonrandom associations of a number of genetic loci have been noted in a subset of NTHI strains (21, 22). H. influenzae strains differ in protein adhesins (reviewed in reference 57). Most NTHI strains express one or both of two high-molecular-weight adhesins, HMW1 and HMW2. St. Geme et al. reported that IS1016 was often present in hmw-negative NTHI strains and uniformly absent in hmw-positive NTHI strains and speculated that hmw-negative NTHI strains evolved more recently from an encapsulated ancestor (57). Furthermore, NTHI strains that are hmw negative often express the adhesin Hia, a homologue of the Hib adhesin, Hsf. Although the prevalence and distribution of hmw and hia genes do not define distinct divisions among NTHI, the hmw-hia dichotomy suggests that hmw-deficient strains may form subgroups within the larger NTHI population (21, 52, 60). Because hia is present in a number of non-Hib encapsulated H. influenzae strains and is a homologue of the Hib gene hsf, it has also been speculated that hia-containing NTHI strains may have evolved more recently from an encapsulated ancestor.
The findings that the majority of our IS1016-positive NTHI strains were hmw negative and hia positive and the majority of those without IS1016 were hmw positive and hia negative suggest that the presence or absence of IS1016 in NTHI may be a linked marker for two distinct lineages. It is important to note that since hmw and hia were detected by PCR amplification and Southern blot hybridization, we cannot be certain that these strains actually express HMW or Hia adhesin. Although it is very uncommon, we found four strains positive for both hmw and hia. Erwin et al. and O'Neill et al. each identified a single NTHI isolate positive for both hmw and hia (21, 48). We also observed six NTHI strains that lacked hmw1 and hmw2 as well as hia by PCR and Southern blotting. Although it is possible that strains found to be negative by PCR may be due to variation within the sequence at the primer binding sites, the results of Southern blot hybridization were in agreement with the PCR results in all cases. Rao et al. reported that 25% of NTHI strains lack proteins belonging to HMW1/HMW2 family of adhesins, yet nearly all such strains remain capable of efficient in vitro adherence, independent of piliation, suggesting the existence of other nonpilus adhesins (50).
An appreciable number of NTHI isolates show partial hybridization with cap locus sequences, most notably cap-associated IS1016, suggesting a relationship to encapsulated ancestors (36, 53, 60). Residual IS1016 sequence was found in a highly virulent NTHI strain, R2866 (INT1) (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi) isolated from a child with meningitis (20, 21, 46). This invasive, IS1016-positive, NTHI demonstrated unusual resistance to killing by normal human serum, and DNA isolated from strain INT1 was able to transform a nonvirulent NTHI strain and cause bacteremia and meningitis in infant rats (46, 64). Also, strain R2866 has several genetic loci not found in strain Rd KW20 (21). These include the gene for an autotransporter, lav (17), the lysogenic bacteriophage HP2 (63), the tna cluster (40), and a 53-kb plasmid similar to other large integrative plasmids of Haemophilus spp. (42).
The complete genome sequences of several NTHI strains isolated from various sites have demonstrated that each of these strains contains 30 to 40 genetic loci not present in the previously sequenced Rd KW20 strain (34). We chose to focus our characterization of IS1016-positive NTHI on the association with hmw and hia adhesin genes. Future studies to evaluate the association of other NTHI genetic features with the presence of IS1016 will be of interest. However, alternatively, the presence of IS1016 in NTHI may instead be a marker for genetic characteristics that more closely resemble encapsulated H. influenzae, such as lipopolysaccharide biosynthesis, iron uptake, and other genes identified using a position-based scanning (7) on the Hib genome and lacking in the laboratory-passaged, unencapsulated, and avirulent Rd KW20.
Seven of the eight currently recognized H. influenzae biotypes were present in both IS1016-positive and -negative strains. Since the development of Kilian's assay for biotyping (37), several laboratories have examined the relationship between biotype and other characteristics, such as anatomic site of isolation, serotype, or pattern of antimicrobial susceptibility (2, 30, 38, 47). In past surveys, most encapsulated H. influenzae (serotypes a, b, and f) strains were biotypes I and II, serotype e strains were biotype IV, and NTHI strains were biotypes II and III (37). Biotype distribution among invasive disease and noninvasive disease isolates has been well documented. However, the relationship between biotype and pathogenicity has produced some conflicting conclusions (32). Although previous studies showed that the majority of invasive disease isolates from cases of meningitis and septicemia were biotype I and noninvasive isolates were biotype II, the data were derived from the pre-Hib vaccine era and likely reflect the predominance of biotype I Hib clonal groups in systemic disease.
We previously noted the association of IS1016-positive NTHI with biotype V, an otherwise very uncommon H. influenzae biotype (53). The more recent invasive H. influenzae isolates (1998 to 2006) also show the same significant association of biotype V with the presence of IS1016. The highly virulent, IS1016-positive NTHI strain INT1/R2866 belongs to biotype V and is also hmw negative (20, 46). Biotype V strains have been most frequently reported as occurring in association with acute otitis media or as respiratory tract commensals (37, 45). Although uncommon, a small proportion of Hib strains belong to biotype V, and at least one clinical study found that an unusually large percentage (14/28 [50%]) of NTHI strains were biotype V and nearly 50% of these caused invasive disease (31).
An obvious question is whether IS1016-positive NTHI strains differ in pathogenic potential or in the characteristics of clinical disease. Despite the association of IS1016-containing NTHI with certain biochemical and adhesin characteristics of encapsulated H. influenzae, no association between the presence of IS1016 and clinical syndrome, race, or disease outcome were noted in our comparison. However, a significant association was observed between the presence of IS1016 and younger patient age. Although of unclear significance, it is of interest that the capsule-associated IS1016 element was more often present in NTHI disease affecting children in the age group previously at highest risk for Hib disease. Host factors may play a significant role in the pathogenesis of invasive NTHI disease in adults, and microbiologic characteristics important to invasive disease may differ from those important for respiratory tract disease. A particular strength of this study is the analysis of a large series of invasive NTHI cases and isolates collected by active, population-based surveillance over a 19-year period. However, the findings may reflect regional characteristics that may not be generalizable to all invasive NTHI disease.
Since the introduction of the 7-valent pneumococcal vaccine, NTHI has emerged as a leading cause of acute otitis media in children (12). The NTHI antibody response that develops during the course of infection has been associated with a decrease in bacteria in middle ear fluid (24) and more efficient resolution of infection (55), suggesting a vaccine with NTHI bacterial components may be feasible (13). Outer membrane proteins are the major focus of NTHI vaccine development efforts (14, 49). However, despite excellent preclinical studies, the optimal antigen(s) for vaccine development remains unclear (28). HMW proteins are important targets of serum antibody in children who have had otitis media (5), and antibodies produced following immunization of chinchillas with HMW proteins are opsonophagocytic (65). A report from the Eighth International Otitis Media Research Conference recommended continued preclinical evaluation of this family of proteins (28). It has been estimated that HMW proteins are present in approximately 75% of unrelated NTHI strains (59). Our data would suggest that this number might be somewhat higher among invasive NTHI.
In summary, we found that a subset of invasive NTHI isolates possessed DNA homologous to the insertion sequence IS1016, an element often associated with the H. influenzae capsule gene cluster. The significant association of IS1016 with biotypes V and I, the presence of hia adhesin and the absence of hmw adhesins in NTHI may represent a unique subgroup of NTHI with characteristics more closely resembling those of encapsulated H. influenzae. Future characterization and comparison of systematically collected isolates from invasive disease, respiratory disease, and nasopharyngeal carriage may provide a more complete understanding of the prevalence and clinical relevance of IS1016-positive NTHI.
Acknowledgments
This work was supported in part by a VA Merit Grant to M.M.F. and the CDC-sponsored Emerging Infections Program.
We thank Bill Cheek (Georgia Public Health Laboratory), John Elliott and Richard Facklam (CDC) for biotyping and serotyping, and Wendy Baughman and Paul Malpiedi for statistical support.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 15 September 2008.
REFERENCES
- 1.Adderson, E. E., C. L. Byington, L. Spencer, A. Kimball, M. Hindiyeh, K. Carroll, S. Mottice, E. K. Korgenski, J. C. Christenson, and A. T. Pavia. 2001. Invasive serotype a Haemophilus influenzae infections with a virulence genotype resembling Haemophilus influenzae type b: emerging pathogen in the vaccine era? Pediatrics 108E18. [DOI] [PubMed] [Google Scholar]
- 2.Albritton, W. L., S. Penner, L. Slaney, and J. Brunton. 1978. Biochemical characteristics of Haemophilus influenzae in relationship to source of isolation and antibiotic resistance. J. Clin. Microbiol. 7519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alsarraf, R., C. J. Jung, J. Perkins, C. Crowley, N. W. Alsarraf, and G. A. Gates. 1999. Measuring the indirect and direct costs of acute otitis media. Arch. Otolaryngol. Head Neck Surg. 12512-18. [DOI] [PubMed] [Google Scholar]
- 4.Barenkamp, S. J. 2004. Rationale and prospects for a nontypable Haemophilus influenzae vaccine. Pediatr. Infect. Dis. J. 23461-462. [DOI] [PubMed] [Google Scholar]
- 5.Barenkamp, S. J., and F. F. Bodor. 1990. Development of serum bactericidal activity following nontypable Haemophilus influenzae acute otitis media. Pediatr. Infect. Dis. J. 9333-339. [DOI] [PubMed] [Google Scholar]
- 6.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 191215-1223. [DOI] [PubMed] [Google Scholar]
- 7.Bergman, N. H., and B. J. Akerley. 2003. Position-based scanning for comparative genomics and identification of genetic islands in Haemophilus influenzae type b. Infect. Immun. 711098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 295-113. [PubMed] [Google Scholar]
- 9.Bruant, G., S. Watt, R. Quentin, and A. Rosenau. 2003. Typing of nonencapsulated Haemophilus strains by repetitive-element sequence-based PCR using intergenic dyad sequences. J. Clin. Microbiol. 413473-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buscher, A. Z., K. Burmeister, S. J. Barenkamp, and J. W. St. Geme III. 2004. Evolutionary and functional relationships among the nontypeable Haemophilus influenzae HMW family of adhesins. J. Bacteriol. 1864209-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campos, J., M. Hernando, F. Roman, M. Perez-Vazquez, B. Aracil, J. Oteo, E. Lazaro, and F. de Abajo. 2004. Analysis of invasive Haemophilus influenzae infections after extensive vaccination against H. influenzae type b. J. Clin. Microbiol. 42524-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casey, J. R., and M. E. Pichichero. 2004. Changes in frequency and pathogens causing acute otitis media in 1995-2003. Pediatr. Infect. Dis. J. 23824-828. [DOI] [PubMed] [Google Scholar]
- 13.Cripps, A. W., and J. Kyd. 2003. Bacterial otitis media: current vaccine development strategies. Immunol. Cell Biol. 8146-51. [DOI] [PubMed] [Google Scholar]
- 14.Cripps, A. W., and D. C. Otczyk. 2006. Prospects for a vaccine against otitis media. Expert Rev. Vaccines 5517-534. [DOI] [PubMed] [Google Scholar]
- 15.Cuthill, S. L., M. M. Farley, and L. G. Donowitz. 1999. Nontypable Haemophilus influenzae meningitis. Pediatr. Infect. Dis. J. 18660-662. [DOI] [PubMed] [Google Scholar]
- 16.Dagan, R., A. Hoberman, C. Johnson, E. L. Leibovitz, A. Arguedas, F. V. Rose, B. R. Wynne, and M. R. Jacobs. 2001. Bacteriologic and clinical efficacy of high dose amoxicillin/clavulanate in children with acute otitis media. Pediatr. Infect. Dis. J. 20829-837. [DOI] [PubMed] [Google Scholar]
- 17.Davis, J., A. L. Smith, W. R. Hughes, and M. Golomb. 2001. Evolution of an autotransporter: domain shuffling and lateral transfer from pathogenic Haemophilus to Neisseria. J. Bacteriol. 1834626-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dworkin, M. S., L. Park, and S. M. Borchardt. 2007. The changing epidemiology of invasive Haemophilus influenzae disease, especially in persons ≥65 years old. Clin. Infect. Dis. 44810-816. [DOI] [PubMed] [Google Scholar]
- 19.Ecevit, I. Z., K. W. McCrea, M. M. Pettigrew, A. Sen, C. F. Marrs, and J. R. Gilsdorf. 2004. Prevalence of the hifBC, hmw1A, hmw2A, hmwC, and hia genes in Haemophilus influenzae isolates. J. Clin. Microbiol. 423065-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erwin, A. L., S. Allen, D. K. Ho, P. J. Bonthuis, J. Jarisch, K. L. Nelson, D. L. Tsao, W. C. Unrath, M. E. Watson, Jr., B. W. Gibson, M. A. Apicella, and A. L. Smith. 2006. Role of lgtC in resistance of nontypeable Haemophilus influenzae strain R2866 to human serum. Infect. Immun. 746226-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erwin, A. L., K. L. Nelson, T. Mhlanga-Mutangadura, P. J. Bonthuis, J. L. Geelhood, G. Morlin, W. C. Unrath, J. Campos, D. W. Crook, M. M. Farley, F. W. Henderson, R. F. Jacobs, K. Muhlemann, S. W. Satola, L. van Alphen, M. Golomb, and A. L. Smith. 2005. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect. Immun. 735853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erwin, A. L., S. A. Sandstedt, P. J. Bonthuis, J. L. Geelhood, K. L. Nelson, W. C. Unrath, M. A. Diggle, M. J. Theodore, C. R. Pleatman, E. A. Mothershed, C. T. Sacchi, L. W. Mayer, J. R. Gilsdorf, and A. L. Smith. 2008. Analysis of genetic relatedness of Haemophilus influenzae isolates by multilocus sequence typing. J. Bacteriol. 1901473-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erwin, A. L., and A. L. Smith. 2007. Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol. 15355-362. [DOI] [PubMed] [Google Scholar]
- 24.Faden, H., L. Brodsky, J. Bernstein, J. Stanievich, D. Krystofik, C. Shuff, J. J. Hong, and P. L. Ogra. 1989. Otitis media in children: local immune response to nontypeable Haemophilus influenzae. Infect. Immun. 573555-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farley, M. M., D. S. Stephens, P. S. Brachman, Jr., R. C. Harvey, J. D. Smith, J. D. Wenger, and CDC Meningitis Surveillance Group. 1992. Invasive Haemophilus influenzae disease in adults. A prospective, population-based surveillance. Ann. Intern. Med. 116806-812. [DOI] [PubMed] [Google Scholar]
- 26.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269496-512. [DOI] [PubMed] [Google Scholar]
- 27.Funkhouser, A., M. C. Steinhoff, and J. Ward. 1991. Haemophilus influenzae disease and immunization in developing countries. Rev. Infect. Dis. 13(Suppl. 6)S542-S554. [DOI] [PubMed] [Google Scholar]
- 28.Giebink, G. S., Y. Kurono, L. O. Bakaletz, J. M. Kyd, S. J. Barenkamp, T. F. Murphy, B. Green, P. L. Ogra, X. X. Gu, J. A. Patel, T. Heikkinen, S. I. Pelton, M. Hotomi, and P. Karma. 2005. Recent advances in otitis media. 6. Vaccine Ann. Otol. Rhinol. Laryngol. Suppl. 19486-103. [PubMed] [Google Scholar]
- 29.Giufrè, M., M. Muscillo, P. Spigaglia, R. Cardines, P. Mastrantonio, and M. Cerquetti. 2006. Conservation and diversity of HMW1 and HMW2 adhesin binding domains among invasive nontypeable Haemophilus influenzae isolates. Infect. Immun. 741161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Golberg, R., and J. A. Washington II. 1978. The taxonomy and antimicrobial susceptibility of Haemophilus species in clinical specimens. Am. J. Clin. Pathol. 70899-904. [DOI] [PubMed] [Google Scholar]
- 31.Granato, P. A., E. A. Jurek, and L. B. Weiner. 1983. Biotypes of Haemophilus influenzae: relationship to clinical source of isolation, serotype, and antibiotic susceptibility. Am. J. Clin. Pathol. 7973-77. [DOI] [PubMed] [Google Scholar]
- 32.Harper, J. J., and M. H. Tilse. 1991. Biotypes of Haemophilus influenzae that are associated with noninvasive infections. J. Clin. Microbiol. 292539-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison, A., D. W. Dyer, A. Gillaspy, W. C. Ray, R. Mungur, M. B. Carson, H. Zhong, J. Gipson, M. Gipson, L. S. Johnson, L. Lewis, L. O. Bakaletz, and R. S. Munson, Jr. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J. Bacteriol. 1874627-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hogg, J. S., F. Z. Hu, B. Janto, R. Boissy, J. Hayes, R. Keefe, J. C. Post, and G. D. Ehrlich. 2007. Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains. Genome Biol. 8R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holmes, R. L., L. M. DeFranco, and M. Otto. 1982. Novel method of biotyping Haemophilus influenzae that uses API 20e. J. Clin. Microbiol. 151150-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlsson, E., and A. Melhus. 2006. Nontypeable Haemophilus influenzae strains with the capsule-associated insertion element IS1016 may mimic encapsulated strains. APMIS 114633-640. [DOI] [PubMed] [Google Scholar]
- 37.Kilian, M. 1976. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J. Gen. Microbiol. 939-62. [DOI] [PubMed] [Google Scholar]
- 38.Kilian, M., I. Sorensen, and W. Frederiksen. 1979. Biochemical characteristics of 130 recent isolates from Haemophilus influenzae meningitis. J. Clin. Microbiol. 9409-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kroll, J. S., I. Hopkins, and E. R. Moxon. 1988. Capsule loss in H. influenzae type b occurs by recombination-mediated disruption of a gene essential for polysaccharide export. Cell 53347-356. [DOI] [PubMed] [Google Scholar]
- 40.Martin, K., G. Morlin, A. Smith, A. Nordyke, A. Eisenstark, and M. Golomb. 1998. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J. Bacteriol. 180107-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meats, E., E. J. Feil, S. Stringer, A. J. Cody, R. Goldstein, J. S. Kroll, T. Popovic, and B. G. Spratt. 2003. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J. Clin. Microbiol. 411623-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohd-Zain, Z., S. L. Turner, A. M. Cerdeno-Tarraga, A. K. Lilley, T. J. Inzana, A. J. Duncan, R. M. Harding, D. W. Hood, T. E. Peto, and D. W. Crook. 2004. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 1868114-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munson, R. S., Jr., A. Harrison, A. Gillaspy, W. C. Ray, M. Carson, D. Armbruster, J. Gipson, M. Gipson, L. Johnson, L. Lewis, D. W. Dyer, and L. O. Bakaletz. 2004. Partial analysis of the genomes of two nontypeable Haemophilus influenzae otitis media isolates. Infect. Immun. 723002-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy, T. F. 2003. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis. 16129-134. [DOI] [PubMed] [Google Scholar]
- 45.Musser, J. M., S. J. Barenkamp, D. M. Granoff, and R. K. Selander. 1986. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect. Immun. 52183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nizet, V., K. F. Colina, J. R. Almquist, C. E. Rubens, and A. L. Smith. 1996. A virulent nonencapsulated Haemophilus influenzae. J. Infect. Dis. 173180-186. [DOI] [PubMed] [Google Scholar]
- 47.Oberhofer, T. R., and A. E. Back. 1979. Biotypes of Haemophilus encountered in clinical laboratories. J. Clin. Microbiol. 10168-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Neill, J. M., J. W. St. Geme III, D. Cutter, E. E. Adderson, J. Anyanwu, R. F. Jacobs, and G. E. Schutze. 2003. Invasive disease due to nontypeable Haemophilus influenzae among children in Arkansas. J. Clin. Microbiol. 413064-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poolman, J. T., L. Bakaletz, A. Cripps, P. A. Denoel, A. Forsgren, J. Kyd, and Y. Lobet. 2000. Developing a nontypeable Haemophilus influenzae (NTHi) vaccine. Vaccine 19(Suppl. 1)S108-S115. [DOI] [PubMed] [Google Scholar]
- 50.Rao, V. K., G. P. Krasan, D. R. Hendrixson, S. Dawid, and J. W. St. Geme III. 1999. Molecular determinants of the pathogenesis of disease due to non-typable Haemophilus influenzae. FEMS Microbiol. Rev. 2399-129. [DOI] [PubMed] [Google Scholar]
- 51.Rayner, M. G., Y. Zhang, M. C. Gorry, Y. Chen, J. C. Post, and G. D. Ehrlich. 1998. Evidence of bacterial metabolic activity in culture-negative otitis media with effusion. JAMA 279296-299. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez, C. A., V. Avadhanula, A. Buscher, A. L. Smith, J. W. St. Geme III, and E. E. Adderson. 2003. Prevalence and distribution of adhesins in invasive non-type b encapsulated Haemophilus influenzae. Infect. Immun. 711635-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satola, S. W., J. T. Collins, R. Napier, and M. M. Farley. 2007. Capsule gene analysis of invasive Haemophilus influenzae: accuracy of serotyping and prevalence of IS1016 among nontypeable isolates. J. Clin. Microbiol. 453230-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuchat, A., T. Hilger, E. Zell, M. M. Farley, A. Reingold, L. Harrison, L. Lefkowitz, R. Danila, K. Stefonik, N. Barrett, D. Morse, and R. Pinner for the Active Bacterial Core Surveillance Team of the Emerging Infections Program Network. 2001. Active bacterial core surveillance of the Emerging Infections Program network. Emerg. Infect. Dis. 792-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shurin, P. A., S. I. Pelton, I. B. Tager, and D. L. Kasper. 1980. Bactericidal antibody and susceptibility to otitis media caused by nontypable strains of Haemophilus influenzae. J. Pediatr. 97364-369. [DOI] [PubMed] [Google Scholar]
- 56.St. Geme, J. W., III. 1997. Insights into the mechanism of respiratory tract colonization by nontypable Haemophilus influenzae. Pediatr. Infect. Dis. J. 16931-935. [DOI] [PubMed] [Google Scholar]
- 57.St. Geme, J. W., III. 2002. Molecular and cellular determinants of non-typeable Haemophilus influenzae adherence and invasion. Cell Microbiol. 4191-200. [DOI] [PubMed] [Google Scholar]
- 58.St. Geme, J. W., III, S. Falkow, and S. J. Barenkamp. 1993. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 902875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.St. Geme, J. W., III, V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.St. Geme, J. W., III, A. Takala, E. Esko, and S. Falkow. 1994. Evidence for capsule gene sequences among pharyngeal isolates of nontypeable Haemophilus influenzae. J. Infect. Dis. 169337-342. [DOI] [PubMed] [Google Scholar]
- 61.St. Geme, J. W., III. 1994. The HMW1 adhesin of nontypeable Haemophilus influenzae recognizes sialylated glycoprotein receptors on cultured human epithelial cells. Infect. Immun. 623881-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vitovski, S., K. T. Dunkin, A. J. Howard, and J. R. Sayers. 2002. Nontypeable Haemophilus influenzae in carriage and disease: a difference in IgA1 protease activity levels. JAMA 2871699-1705. [DOI] [PubMed] [Google Scholar]
- 63.Williams, B. J., M. Golomb, T. Phillips, J. Brownlee, M. V. Olson, and A. L. Smith. 2002. Bacteriophage HP2 of Haemophilus influenzae. J. Bacteriol. 1846893-6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams, B. J., G. Morlin, N. Valentine, and A. L. Smith. 2001. Serum resistance in an invasive, nontypeable Haemophilus influenzae strain. Infect. Immun. 69695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winter, L. E., and S. J. Barenkamp. 2006. Antibodies specific for the high-molecular-weight adhesion proteins of nontypeable Haemophilus influenzae are opsonophagocytic for both homologous and heterologous strains. Clin. Vaccine Immunol. 131333-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]