Abstract
Disease-causing microbes utilize various strategies to modify their environment in order to create a favorable location for growth and survival. Gram-negative bacterial pathogens often use specialized secretion systems to translocate effector proteins directly into the cytosol of the eukaryotic cells they infect. These bacterial proteins are responsible for modulating eukaryotic cell functions. Identification of the bacterial effectors has been a critical step toward understanding the molecular basis for the pathogenesis of the bacteria that use them. Chlamydiae are obligate intracellular bacterial pathogens that have a type III secretion system believed to translocate virulence effector proteins into the cytosol of their host cells. Selective permeabilization of the eukaryotic cell membrane was used in conjunction with metabolic labeling of bacterial proteins to identify chlamydial proteins that localize within the cytosol of infected cells. More than 20 Chlamydia trachomatis and C. pneumoniae proteins were detected within the cytoplasmic compartment of infected cells. While a number of cytosolic proteins were shared, others were unique to each species, suggesting that variation among cytosolic chlamydial proteins contributes to the differences in the pathogenesis of the chlamydial species. The spectrum of chlamydial proteins exported differed concomitant with the progress of the developmental cycle. These data confirm that a dynamic relationship exists between Chlamydia and its host and that translocation of bacterial proteins into the cytosol is developmentally dependent.
Pathogenic bacteria frequently modify the tissues they colonize or the cells they infect. Creating an environment conducive to microbial survival and replication often involves the directed secretion of specific bacterial products via specialized secretion systems. A hallmark of many secretion systems is that bacterial proteins are translocated into the cells the bacteria parasitize. Although of bacterial origin, these effector proteins are responsible for modulating signal transduction systems and cellular functions within the eukaryotic host (1, 27). Crucial to understanding the pathogenesis of the microbes that have these secretion systems has been identification of the bacterial effector proteins being secreted into the cell cytosol and elucidation of their function (27).
Chlamydia spp. are medically important pathogens responsible for causing a number of diseases in humans ranging from blinding trachoma, pelvic inflammatory disease, and infertility to community-acquired pneumonia and heart disease (41). Sequencing chlamydial genomes revealed the presence of a type III secretion system by identifying the complement of genes that encode homologues to the structural proteins that comprise type III secretion systems in other pathogens (29, 45). Transcriptome analysis demonstrates that these genes are expressed throughout the chlamydial developmental cycle, suggesting that the chlamydial type III secretion system is functional (4, 16, 35). Unlike genes encoding the structural components of type III secretion systems, the genes encoding type III secretion effector proteins are generally not conserved between pathogens, making identification of these virulence factors problematic (27). In Chlamydia, this issue is exacerbated by the inability to genetically modify the bacteria or grow these pathogens in cell-free culture. Nonetheless, several putative effector proteins have been identified by expressing chlamydial genes in other bacteria possessing type III secretion systems and probing with antigen-specific antisera. Examples include a family of chlamydial proteins that localize to the inclusion membrane called Incs (3, 38), TARP (9), a protein that localizes at the plasma membrane just beneath attaching Chlamydia, and CT847 (8), a chlamydial protein that interacts with human GCIP. Expression of chlamydial proteins in a heterologous type III secretion system, however, has yielded little insight into the spectrum of chlamydial proteins that may reside in the cell cytosol. In contrast, immune detection approaches have led to the identification of five chlamydial proteins that localize within the cell cytosol (10, 32, 47, 48, 55). Functional characterization has been assessed for one of these proteins. Chlamydial proteasome-like activity factor (CPAF) is a protease that has been shown to degrade a number of cellular proteins (12, 37, 56, 57). CPAF demonstrates that chlamydiae have evolved to engage and modify their hosts using at least one protein that is exported into the cytoplasmic compartment of the eukaryotic cell. Although immunolocalization approaches have proven successful, this method requires antigen-specific antisera and likely lacks the sensitivity needed to detect effector proteins secreted at low abundance.
Given its obligate intracellular niche and the coevolution of chlamydiae within the host cell for nearly one billion years (21, 44), we hypothesize that chlamydiae secrete a select number of proteins into the host cell cytosol that are necessary for the modulation of cellular functions at different developmental stages. Our objective was to characterize the constellation of chlamydial proteins that localize within the host cell cytosolic compartment and to determine whether this population of proteins changes during the developmental cycle of the bacteria. The cholesterol-dependent cytolysin perfringolysin O (PFO) was used to selectively permeabilize the plasma membrane of Chlamydia-infected cells in order to obtain a soluble cytosolic protein fraction. Chlamydial proteins were distinguished from those of the host by metabolic labeling under conditions that prevented eukaryotic cell protein synthesis. Resolution of the soluble fraction by two-dimensional gel electrophoresis revealed more than 20 chlamydial proteins residing in the cytosol of infected cells. Moreover, the population of cytosolic proteins changed over the course of an infection, suggesting that they play developmental stage-specific roles for the growth and development of chlamydiae.
MATERIALS AND METHODS
Tissue culture and bacterial culture conditions.
Chlamydia trachomatis serovar L2 (L2/434/Bu) was cultured in HeLa or L929 cells as previously described (31). C. pneumoniae (CWL029) was grown on Hep2 cells by centrifuging inoculum onto monolayers at 900 × g for 1 h at 24°C. After a 1-h incubation at 37°C, RPMI (Invitrogen, Carlsbad, CA) containing 5% fetal bovine serum (FBS; HyClone, Logan, UT) was added to each flask. Infections were allowed to progress to the designated time at 37° with 5% CO2.
Wild-type Listeria monocytogenes 10403S (5) and Hly− mutant L. monocytogenes strain DP-L2161 (28) were grown in 2 ml of Luria-Bertani broth (Fisher Scientific, Fair Lawn, NJ) overnight at 30°C without agitation. J774 mouse macrophage-like cells were grown in DME (Invitrogen) containing 7.5% FBS and 2 mM l-glutamine (Invitrogen). Tissue cultures were grown at 37°C with 5% CO2.
Mycoplasma testing of bacterial strains and tissue culture.
PCR was used to test all cell culture and bacterial stocks for the presence of mycoplasma. Sample DNA was eluted from miniprep spin columns (Qiagen, Valencia, CA) using 50 μl of distilled H2O. A 2-μl sample DNA was added to the following buffer mixture to yield a 30-μl reaction: 3 mM MgCl2, 0.2 mM deoxynucleoside triphosphate, 1 μM concentrations of both the forward and reverse primers, and 1.5 U of TacI polymerase (Fermentas, Inc., Hanover, MD). Reactions were carried out under the following conditions: 2 min at 90°C, followed by 30 cycles of denaturation at 94°C, annealing at 55°C, and extension at 72°C. The primer pairs used for Mycoplasma spp. were 5′-TGCACCATCTGTCACTCTGTTAACCTC-3′ and 5′-GGGAGCAAACAGGATTAGATACCCT-3′ (49, 50). Analysis of amplified products was determined by electrophoresis on 1% agarose gels.
Expression and purification of His-tagged PFO from Escherichia coli.
Full-length, recombinant PFO was overexpressed and purified from strain DP-4167 as previously described (20).
PFO-mediated permeabilization of plasma membrane.
Selective permeabilization of the plasma membrane of Chlamydia-infected or Listeria-infected cells was achieved by using PFO. Monolayers were chilled on ice for 10 min, followed by a single wash with cold Hanks balanced salt solution (HBSS; Invitrogen). Purified recombinant PFO was diluted in cold HBSS and added to the appropriate flask. After a 10-min incubation on ice, monolayers were washed three times with cold HBSS. Fresh HBSS (room temperature) was added, and monolayers were incubated at 37°C for 30 min. Supernatants were transferred to conical tubes and centrifuged at 900 × g for 10 min at 4°C. Supernatants were collected and subsequently microfuged at maximum speed for 3 min. Cleared supernatants were stored at −80°C prior to subsequent analysis.
PFO-mediated delivery of fluorophores to infected cells.
Cell monolayers grown on coverslips in the bottom of 24-well plates were infected with C. trachomatis serovar L2 as previously described (30). At the appropriate time after infection was initiated, monolayers were permeabilized with PFO as described above. Lysine-fixable 10-kDa fluorescein-conjugated dextran (Invitrogen) was diluted in HBSS to 50 μM and added to the monolayers, followed by a 1-h incubation at room temperature. Monolayers were washed three times with HBSS and fixed for 20 min with 4% formaldehyde. Cells were permeabilized by a brief incubation with methanol. Where indicated, cellular nuclei were visualized by staining with DAPI (Invitrogen). The presence of Chlamydia was determined by probing with a monoclonal antibody to major outer membrane protein (MOMP) (46) and detected with an anti-mouse secondary antibody conjugated to fluorescein (Invitrogen) or Alexa Fluor 594 (Invitrogen).
Determination of CFU in L. monocytogenes-infected cells.
J774 macrophage-like cells were grown on coverslips in the bottom of 24 wells plates. Wild-type or Hly− L. monocytogenes grown overnight at 30°C (see above) were washed with HBSS, suspended in DME to yield a final multiplicity of infection of 4, and inoculated onto cells. Bacteria were allowed to infect cells for 30 min at 37°C, after which monolayers were washed three times in HBSS. DME containing 50 μg of gentamicin (Invitrogen)/ml was added to the cultures to kill any extracellular bacteria. Plates were incubated 30 min at 37°C and then washed three times with HBSS. Plates were chilled on ice in preparation for PFO treatment as described above. After permeabilization, DME containing 50 μg of gentamicin/ml was added to the cultures, and the plates were incubated for 30 min at 37°C. After the final incubation, monolayers were washed three times with HBSS. The number of viable bacteria remaining after treatment was quantified by plating the bacteria harvested from each coverslip. Coverslips were transferred to 15-ml conical tubes containing 1 ml of sterile water and vortex mixed to lyse the cells. Dilutions of each culture were plated on Luria-Bertani plates and incubated overnight at 37°C. The next day the colonies were counted. This number was multiplied by the appropriate dilution factor to yield the CFU. All treatment groups were performed in duplicate for each experiment, and all experiments were performed a minimum of three times.
Metabolic labeling of chlamydial proteins.
Infected monolayers grown in flasks were cultivated as described above until the designated time postinfection. Monolayers were then pulse-labeled with 35S-Translabel (MP Biomedicals, Solon, OH) under conditions adapted from Brundage et al. (6). The growth medium was removed and replaced with labeling medium: methionine- and cysteine-deficient RPMI (MP Biomedicals) containing 2 mM l-glutamine (Invitrogen), 5% FBS (HyClone), 225 μg of cycloheximide (Sigma)/ml, 30 μg of anisomycin (Sigma)/ml, 50 μM lactacystin (A. G. Scientific, Inc., San Diego, CA), and 5 μM LLnL (Calbiochem, La Jolla, CA). Flasks were incubated for 30 min at 37°C. The medium was removed from each flask, and new labeling medium was added that contained 100 μCi of 35S-Translabel/ml. Radioactive amino acids were allowed to incorporate into bacterial proteins for 2 h at 37°C. After labeling, monolayers were prepared for PFO-mediated permeabilization as described above.
Immunodetection.
Equal volumes of each sample were resolved on sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to nitrocellulose. Nitrocellulose sheets were incubated for 1 h at room temperature in a phosphate-buffered saline-Tween (PBS-T) solution containing 5% nonfat dried milk. Samples were probed with mouse anti-GAPDH diluted 1:1,000 (Ambion, Austin, TX), mouse anti-MOMP diluted 1:6,000 (R. S. Stephens), or mouse anti-CPAF undiluted (a gift from G. Zhong) in PBS-T containing 5% nonfat dried milk for 1 h and then washed three times in PBS-T. Immune reactions were detected with goat anti-mouse antibody conjugated to horseradish peroxidase (Sigma) diluted in PBS-T, incubated for 1 h at room temperature, and washed three times in PBS-T before detection of immune reactions by chemiluminescence (ECL kit; GE Healthcare).
Two-dimensional gel analysis and autoradiography.
Proteins were precipitated from each sample by using the methanol-chloroform protocol described by Wessel and Flugge (53). Samples were initially vortex mixed with methanol in microcentrifuge tubes. Chloroform was then added to each sample and vortex mixed, followed by the addition of water and a final 10-s vortexing step. Samples were microfuged for 10 min at maximum speed. After the aqueous phase was removed, the remaining sample was combined with methanol, vortex mixed, and microfuged at maximum speed for 10 min. Supernatants were discarded, and the resulting pellet was air dried before resuspension in sample rehydration buffer (8 M urea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 0.5% IPG buffer pH 4-7, 0.002% bromophenol blue, and 40 mM dithiothreitol [all from GE Healthcare]). Immobiline drystrips pH 4-7 (133 cm; GE Healthcare) were rehydrated with sample in rehydration buffer for 12 h at room temperature. Isoelectric focusing was performed using an Ettan IGPhor isoelectric focusing system (GE Healthcare) under the following conditions: 1 h at 500 V, 1 h at 1,000 V, and 2 h at 8,000 V. After focusing, strips were incubated for 15 min in sodium dodecyl sulfate (SDS) equilibration buffer solution (6 M urea, 75 mM Tris-HCl [pH 8.8; Sigma], 29.3% glycerol Σ, 2% SDS Σ, 0.002% bromophenol blue) containing 10 mg of dithiothreitol (Sigma)/ml and then 25 mg of iodoacetamide (GE Healthcare)/ml, respectively. Equilibrated strips were placed on 10% SDS-polyacrylamide gels for resolution of the second dimension under the following conditions: 30 min at 20 μA (constant) and 6 h at 40 μA (constant). Proteins were fixed into place by incubation in 40% methanol (Fisher Scientific, Fairlawn, NJ) and 10% acetic acid (Fisher Scientific). After equilibration in 20% methanol, 4% glycerol and a subsequent 30 min incubation in Enlightning Autoradiography Enhancer (Perkin-Elmer, Wellesley, MA) gels were dried in a gel drier (Bio-Rad, Hercules, CA) for 9 h with a maximum temperature of 60°C. Kodak BioMax MR film (Fisher Scientific) was exposed to dried gels at −80°C.
RESULTS
Selective permeabilization of the eukaryotic cell plasma membrane.
Chlamydiae spend their entire growth cycle within the mammalian cell. Therefore, it is reasonable to propose that these bacteria modify cytosolic components of their eukaryotic host in order to create an environment conducive to survival and replication. The goal of this research was to describe the spectrum of chlamydial proteins that reside within the host cell cytosol. Identification of the range of possible chlamydial proteins that are secreted or localized to the cell cytosol has not been previously reported because it has not been possible to separate the cytosolic compartment of Chlamydia-infected cells from the bulk of chlamydial proteins localized within the inclusion and embedded within the inclusion membrane. An important experimental design issue is the apparently high sensitivity of the chlamydial inclusion to lysis after the manipulation of infected cells (25).
In order to gain access to the host cell cytosol of Chlamydia-infected cells without disrupting the integrity of the inclusion membrane, PFO was utilized to create transient pores in the cell plasma membrane. PFO is an exotoxin produced by Clostridium perfringens with functional characteristics that make it compatible for the task of selective permeabilization. As a member of the cholesterol-dependent cytolysins, 40 to 50 PFO monomers bind to cholesterol-containing membranes and oligomerize to form pores ∼300 Å in diameter (36). The large pores enable transfer of molecules up to 100 kDa across the permeabilized membranes (51). Permeabilization can be controlled by temperature such that monomers bind to cell surfaces at low temperature, but insertion into the membrane and subsequent pore formation do not occur until temperatures reach 15°C or higher (43). Moreover, the permeabilization process is reversible in that pores heal, allowing cells to recuperate and to continue growing after treatment (51, 52).
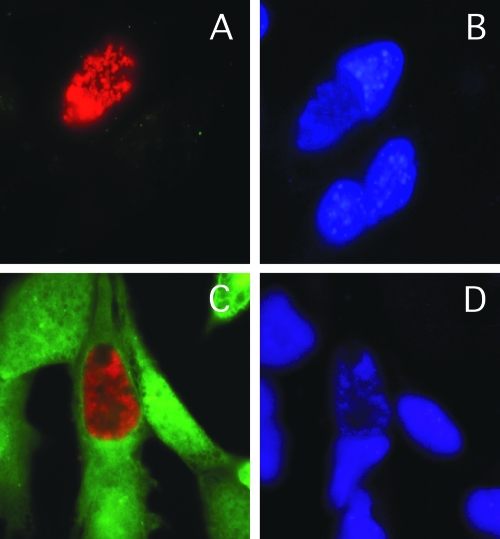
Heinzen and Hackstadt (25) previously reported that the chlamydial inclusion is not passively permeable to molecules greater than 520 Da, the smallest molecule tested. We reasoned that if PFO were able to enter the cell and permeabilize the inclusion, then the large fluorophores normally excluded would gain access and be visible in this compartment. In addition, if PFO treatment resulted in disruption of the integrity of the vacuole, then the inclusion should collapse and acquire fluorophores, as has been reported with the use of microinjection (25). Fibroblasts were infected with C. trachomatis, and the infection was allowed to progress to the mid-developmental cycle. By this stage the inclusions are large enough to be clearly visible within the cell. Monolayers were then permeabilized with PFO and incubated in the presence of a 10-kDa fluorescein-conjugated dextran to ascertain whether the chlamydial inclusion had also been permeabilized during toxin treatment. The 10-kDa dextran was observed to localize within the cell cytosol but not within the lumen of the chlamydial inclusion despite quantitative delivery to the cytoplasm of the eukaryotic cell (Fig. 1). In addition, inclusions maintained their typical round and turgid appearance similarly in the presence or absence of toxin treatment (Fig. 1). This demonstrates that inclusion membrane integrity was not compromised in the presence of toxin.
FIG. 1.
Selective permeabilization of the eukaryotic cell membrane by PFO treatment. C. trachomatis-infected L929 cells were treated with buffer alone (A and B) or with buffer containing PFO (C and D). After treatment, monolayers were incubated in the presence of 10-kDa fluorescein-conjugated dextran (green). Chlamydiae were detected using mouse anti-MOMP antisera and a secondary anti-mouse antibody conjugated to Texas Red (A and C). Monolayers were stained with DAPI (4′,6′-diamidino-2-phenylindole) (blue) to locate cellular nuclei (B and D).
An alternative test of the specificity of PFO for the plasma membrane and for probing membrane permeability is to use a biologically functional probe. The aminoglycoside gentamicin is often used in microbiological experimental designs to specifically kill extracellular bacteria while sparing intracellular bacteria because it is not permeable to eukaryotic cell membranes. Gentamicin is bactericidal; thus, a biologic readout can be obtained, facilitating quantitation of bacterial survival after treatment of infected cells. The use of selective permeabilization by PFO to deliver membrane-impermeable gentamicin to the cell cytosol should provide a sensitive measure for determining whether internal membranes have become permeabilized subsequent to PFO treatment and therefore whether bacteria sequestered within vacuoles have access to the cytosolic compartment.
Listeria monocytogenes was used as a model system to test for bacterial sensitivity to gentamicin delivered to the cell cytosol by PFO treatment. L. monocytogenes is a facultative intracellular bacterium that is taken into phagocytic cells where it resides for a short period of time within a phagosome. Wild-type bacteria express a number of factors, including a hemolysin called listeriolysin O (LLO) that facilitate their escape from the phagosome and into the cytosol of the cell. Mutants of L. monocytogenes that do not express LLO (Hly−) are unable to escape the phagosome and remain trapped within the intracellular vacuole (19).
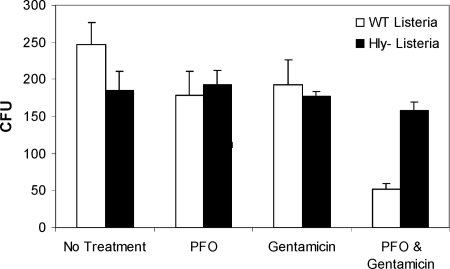
To demonstrate the specificity of PFO-dependent permeabilization for the eukaryotic cell plasma membrane, and not internal membranes, J774 macrophage-like cells were infected with wild-type and Hly− L. monocytogenes. After bacteria were taken into cells, monolayers were incubated in the presence of gentamicin to kill extracellular bacteria. Antibiotic was removed, and the cells were washed before proceeding with the PFO permeabilization protocol utilized for Chlamydia-infected cells. When monolayers were treated with HBSS only, PFO only, or gentamicin only, the number of CFU obtained from both wild-type and Hly− bacteria was equivalent between each treatment group (Fig. 2). In contrast, when gentamicin was delivered to the cytosol of cells by treatment with PFO, the number of wild-type bacteria remaining viable after treatment dropped significantly compared to vacuole-bound Hly− bacteria (Fig. 2). Because wild-type bacteria escape the phagosome within 30 min after entering a cell, these data suggest that cytosolic L. monocytogenes is sensitive to cytosolically delivered gentamicin. Moreover, the fact that Hly− L. monocytogenes, which remains trapped within the phagosome, was not killed by the presence of gentamicin in the cytoplasm demonstrates that these bacteria remain effectively compartmentalized within an intact phagosome throughout the PFO treatment protocol.
FIG. 2.
Listeriae trapped within the phagosome were not sensitive to cytosolic gentamicin. J774 macrophage-like cells were infected with wild-type (□) or hemolysin-deficient (Hly−) (▪) L. monocytogenes. Monolayers were subjected to one of four treatments: no treatment, PFO permeabilization only, antibiotic treatment only, or PFO permeabilization and antibiotic treatment. CFU were quantified after treatment. Each treatment was performed in duplicate, and the entire experiment was repeated three times. Error bars represent the standard error of the mean.
Altogether, these data establish that PFO can be used to selectively permeabilize the eukaryotic cell plasma membrane without disrupting internal membranes, including the chlamydial inclusion.
PFO permeabilization and selective metabolic labeling enable identification of chlamydial proteins within the host cell cytosol.
Because the pores formed by PFO facilitated the delivery of large molecules to the cell (51), we reasoned that a similar procedure would reciprocally contribute to the release of soluble molecules from the cell cytosol. Therefore, PFO-mediated permeabilization was used in conjunction with a selective metabolic radiolabeling protocol to distinguish bacterial proteins from host cell proteins that reside within the eukaryotic cell cytosolic compartment.
Infected and uninfected cells were labeled with [35S]methionine-cysteine for 2 h in the presence of cycloheximide and anisomycin to inhibit eukaryotic protein synthesis. After the short pulse-labeling period, monolayers were permeabilized with PFO. Numerous radiolabeled proteins were detected in lysates of Chlamydia-infected monolayers, whereas mock-infected monolayers contained no detectable isotope (Fig. 3A). This analysis showed the specific labeling of only chlamydial proteins. In addition, cells not treated with PFO produced supernatants devoid of detectable radioactivity (Fig. 3A). Thus, infected cells remained intact during the treatment procedure, and bacterial proteins were not released. Selective permeabilization of infected host cell membranes with PFO, however, resulted in supernatants containing a spectrum of bacterial proteins ranging in size from ∼20 to ∼100 kDa (Fig. 3A). The predominant bands in the SDS-treated whole-cell lysates were different from the predominant bands visualized within the PFO-treated sample supernatants (Fig. 3A). Analogous to the discovery of inclusion membrane proteins (39), these results demonstrated that PFO permeabilization resulted in a soluble fraction enriched for a subset of chlamydial, eukaryotic cell-associated proteins that were distinct from the population of total chlamydial proteins.
FIG. 3.
PFO permeabilization facilitates the release of chlamydial proteins from the cytosol of infected cells. Supernatants were collected from mock-infected HeLa cells (Not Inf) treated with SDS (SDS) or C. trachomatis-infected HeLa cells (Infected) treated with either HBSS alone (HBSS), PFO-diluted in HBSS (PFO), or SDS (SDS). (A) Autoradiogram of samples harvested after infected monolayers were incubated in the presence of [35S]methionine-cysteine and eukaryotic protein synthesis inhibitors. Samples were loaded into wells as follows: HBSS treated (100 μl/well), PFO treated (100 μl/well), or SDS treated (10 μl/well) and resolved by SDS-polyacrylamide gel electrophoresis. The gel was dried and exposed to film. (B) Western blot of samples after being probed with mouse anti-GAPDH (GAPDH), mouse anti-MOMP (MOMP), or mouse anti-CPAF (CPAF). Equal volumes of supernatants from all treatment groups were loaded in each well.
The soluble fraction obtained from PFO permeabilization of Chlamydia-infected cells was evaluated for host cell cytosolic proteins to test the origin of this fraction. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is localized exclusively within the cell cytoplasm and serves as a specific marker for this cellular compartment. GAPDH was detected in abundance in samples obtained from cells treated with PFO or solubilized by SDS, while very little was found in the buffer-treated control cell supernatants (Fig. 3B). In contrast, the quantitatively predominant bacterial protein of Chlamydia (MOMP) was found in the infected monolayers solubilized with SDS, but it was not detected in the samples treated with buffer alone or in monolayers permeabilized with PFO (Fig. 3B). This suggests that inclusions remained intact during toxin treatment, preventing bacteria and other contents of the lumen of the inclusion from contaminating the cytosolic fraction. CPAF, a Chlamydia protein shown to localize in the cell cytosol (14), was released upon PFO permeabilization, confirming the ability of the assay to enrich for cytosolic proteins of low abundance (Fig. 3B). Consistent with the known function of PFO to permeabilize cell plasma membranes, this analysis showed that PFO treatment resulted in the quantitative and selective release of the soluble contents of the cell cytosol.
Chlamydial proteins released from cell cytosol upon PFO permeabilization.
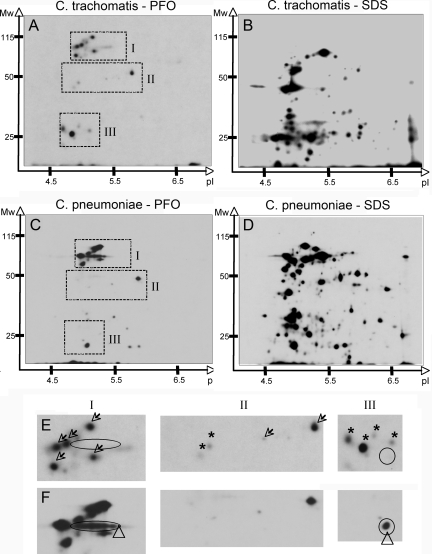
The population of chlamydial proteins localized within the host cell cytosolic fraction was characterized by proteomic analyses. More than 20 protein spots were reproducibly detected in the fraction obtained from PFO permeabilization of C. trachomatis-infected cells (Fig. 4A). There was an acidic bias in the isoelectric point of this population of proteins since all spots focused between a pH of 4 and a pH of 7. The estimated molecular mass of these proteins ranged in size from ∼25 to ∼115 kDa corresponding to the migration of bands visualized by one-dimensional gel electrophoresis (Fig. 3A and 4A). Comparison of the constellation of proteins obtained from the cytosol of infected cells (Fig. 4A) to that of whole-cell lysates (Fig. 4B) suggested that there was a profound enrichment for cytosolic proteins of low abundance upon permeabilization of infected cells with PFO since many of the most predominant spots from the toxin-treated samples were not seen in the samples obtained by solubilization with SDS despite loading twice as much PFO-treated sample on the gel (Fig. 4A and B).
FIG. 4.
Numerous Chlamydia proteins localize within the eukaryotic cell cytosol. Autoradiogram of the soluble fraction obtained from C. trachomatis-infected HeLa cells (A, B, and E) or C. pneumoniae-infected Hep2 cells (C, D, and F) after treatment with PFO (A, C, E, and F) or SDS (B and D). Monolayers were metabolically labeled with [35S]methionine-cysteine in the presence of eukaryotic protein synthesis inhibitors. After appropriate treatment, supernatants were precipitated with methanol and chloroform. Protein was precipitated from two times the volume of the PFO-treated supernatant compared to the SDS-treated supernatant. Three sections (I to III) from each of the two-dimensional autoradiograms were enlarged to compare the constellation of spots obtained from C. trachomatis-infected (E, top) and C. pneumoniae-infected (F, bottom) samples. Open arrows indicate proteins shared by both chlamydial species, ovals indicate Pmp-like protein found in C. pneumoniae, triangles indicate C. pneumoniae-specific proteins, and asterisks indicate C. trachomatis-specific proteins.
C. pneumoniae proteins within the host cell cytosol.
Comparative analysis of the C. pneumoniae and C. trachomatis genomes revealed that 80% of the annotated genes within C. pneumoniae have orthologous genes encoded in the C. trachomatis genome, demonstrating functional conservation of a large number of proteins. Nevertheless, the genome from each species also encodes numerous proteins not found in the other (29). Given the essential and specific roles cytosolic microbial proteins play in the virulence of intracellular pathogenic bacteria (1, 27), the spectrum of C. pneumoniae proteins that localize within the cellular cytosol was compared to those for C. trachomatis (Fig. 4A). Proteomic analysis of the soluble fraction obtained after PFO-mediated cell permeabilization revealed more than 20 C. pneumoniae proteins within the cell cytosol (Fig. 4C), many of which shared gel coordinates with the proteins from C. trachomatis. Similar to C. trachomatis proteins in the host cell cytosol, the cytosolic C. pneumoniae proteins have acidic isoelectric points and have estimated molecular masses ranging from <25 kDa to nearly 115 kDa (Fig. 4A and C).
Cytosolic proteins from both species tend to cluster within 3 groups. Group I and group III proteins represent the high-molecular-weight and low-molecular-weight protein clusters, respectively, while group II proteins are of intermediate mass (Fig. 4). Group I contains a cluster of at least five proteins that have identical migration patterns, suggesting that these proteins are likely encoded by genes shared by the two bacterial species (Fig. 4E and F, arrows). Others display unique gel coordinates (Fig. 4E and F, arrowhead). One group of proteins (Fig. 4E and F, oval) is characteristic of the gel profile of a member of the C. pneumoniae polymorphic membrane proteins (Pmp) (22). The group II cluster contains two proteins that were shared (Fig. 4E and F, arrows) and two that were only detected in the sample from C. trachomatis (Fig. 4E and F, asterisks). Group III is characterized by species-specific proteins since those found in the C. trachomatis sample were not seen in the C. pneumoniae sample (Fig. 4E and F, asterisks), and an additional protein found in the C. pneumoniae sample was not seen in the C. trachomatis sample (Fig. 4E and F, triangle).
Developmental stage-specific changes in chlamydial proteins localized to the cell cytosolic compartment.
Chlamydiae are obligate intracellular bacterial pathogens characterized by a unique developmental cycle. The developmental cycle includes entry of the infectious elementary body (EB) into the cell, reorganization of the EB to the metabolically active reticulate body form (RB), and RB differentiation into EB with subsequent cell lysis. These processes take ca. 48 to 72 h to complete, and all take place within the chlamydial inclusion. It is during this time that Chlamydia requires and obtains many nutrients from the cell (33, 34) and avoids immune surveillance long enough to survive and replicate (7). This biological framework suggests that chlamydiae exert some level of control over cellular functions despite physical sequestration within the inclusion. Given the length of time the bacteria reside within a single cell, it is possible that manipulating cellular functions occurs in a stepwise fashion corresponding to the specific needs of the bacterial population at progressive times in the developmental cycle. This hypothesis is supported by characterization of CPAF that is secreted to the cell cytoplasm only late during infection (26).
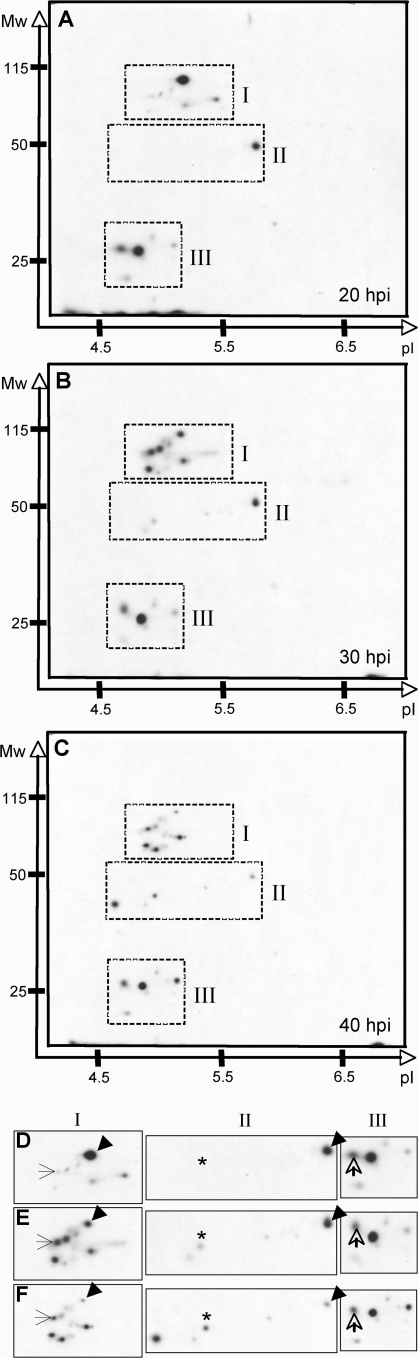
Given the relatively large number of chlamydial proteins discovered in the host cell cytosol, it was tested whether there are changes in the temporal distribution of the spectrum of chlamydial proteins localizing within this compartment. The cytosolic fraction was isolated from Chlamydia-infected cells at 20, 30, and 40 h postinfection (hpi). Fractions obtained at 10 hpi did not yield labeled proteins (data not shown). Although it is possible that Chlamydia do not release proteins into the cytosol or that secreted proteins were not synthesized at this time, it is more likely that there was not sufficient metabolic activity to produce detectable protein labeling within 2 h because there are fewer metabolically active organisms present at this early time point. Because of the selective and differential metabolic labeling requirement of the experimental system, 20 hpi was the earliest time point tested for which labeled proteins were obtained. At this time chlamydial RBs have initiated replication and are ramping up their metabolic demands on the host cell. The 30 hpi represents a midpoint in which there remains great demand for metabolism but many RBs have begun their reorganization to the EB. By 40 hpi much of the developmental program is focused on RB transformation back to EB; however, this is early enough to avoid cellular lysis, a result that could complicate the detection of cytosolically localized proteins.
The C. trachomatis proteins found within the cell cytosol changed both quantitatively and qualitatively over the course of the developmental cycle (Fig. 5). A number of chlamydial proteins were found in the cytosol at equivalent intensities at all three time points, suggesting that these bacterial proteins are responsible for maintaining interaction with cellular components that must be modulated for the growth phase of the developmental cycle (Fig. 5D, E, and F, open arrows). In contrast, many cytosolic chlamydial proteins appear to be temporally regulated as the concentration of protein present changed over the course of the infection. Several proteins increased or appeared de novo during the time course (Fig. 5D E, and F, arrows and asterisks), whereas others decreased (Fig. 5D E, and F, arrowheads). Together, these data demonstrate that the Chlamydia-cell interaction is dynamic and that chlamydial modulation of the cytosolic space is regulated in a temporal fashion.
FIG. 5.
Developmental stage-specific localization of chlamydial proteins in the host cell cytosol. Autoradiogram of the soluble fraction obtained from C. trachomatis-infected HeLa cells after treatment with PFO at 20 hpi (A), 30 hpi (B), or 40 hpi (C). Monolayers were metabolically labeled with [35S]methionine-cysteine in the presence of eukaryotic protein synthesis inhibitors. After PFO permeabilization, supernatants were precipitated with methanol and chloroform. Three sections (I to III) from each of the two-dimensional autoradiograms were enlarged to compare constellations of spots obtained at each time point: panel D, 20 hpi; panel E, 30 hpi; and panel F, 40 hpi. Open arrows indicate proteins whose abundance remains unchanged throughout the developmental cycle, and arrowheads indicate proteins that decrease throughout the course of the developmental cycle. Proteins that appear de novo (thin arrows) or increase over time (asterisks) are also indicated.
DISCUSSION
Obligate intracellular bacteria have evolved numerous strategies to exploit the cells they inhabit. One approach is to translocate bacterial proteins into the host cell to modulate signal transduction cascades involved in various cellular activities. This is commonly exemplified by the type III and type IV secretion systems of gram-negative pathogens, wherein bacterial effector proteins interact with and modify eukaryotic proteins (1, 27). In most cases, disruption of the secretion system or single secreted effector protein results in an avirulent phenotype (27). This phenomenon underscores the important role that the secretion systems and their effector proteins play in microbial virulence.
There are many aspects of Chlamydia biology that indicate that these intracellular bacteria are usurping cellular processes. In addition to exploiting host cell functions for chlamydial attachment and uptake, the inclusion is nonfusogenic with endosomal and lysosomal compartments (42, 54) and yet intercepts cholesterol- and sphingomyelin-containing vesicles originating from the Golgi body (23, 24). Chlamydia-infected cells are resistant to apoptotic stimuli (11, 15, 37), demonstrating bacterial control of cell death induction pathways. The cellular immune response is also modulated as major histocompatibility complex expression is markedly downregulated in Chlamydia-infected cells (14, 55, 57). Altogether, this indicates that the Chlamydia occupy a unique niche within the cell. Although many of the molecules responsible for mediating the host-microbe interaction have yet to be identified, six chlamydial proteins have been shown to localize to the cytosol of infected cells: CPAF (14, 55), Cpn0797 (10), CCA00037 (47), Cpn0796 (50), Cpn0809, and Cpn1020 (32). A definitive role for these proteins in the biology of a Chlamydia infection has yet to be established.
Considering the intimate relationship with its host, we hypothesized that Chlamydia translocate many bacterial proteins beyond the inclusion and into the cytosol of the cell. A comprehensive examination of the chlamydial proteins that reside in the cell cytosol has not previously been documented, most likely due to the technical difficulties associated with working with the chlamydial system. Therefore, an important contribution of the present study is the development of a protocol that allows for the recovery of a soluble cytosolic fraction from Chlamydia-infected cells in conjunction with the ability to distinguish the presence of bacterial proteins in a complex mixture, including proteins from the eukaryotic host.
Although we were able to demonstrate that more than 20 chlamydial proteins localize within the cytosolic compartment of infected cells, the experimental design has a few limitations that must be taken into consideration. The proteins elucidated in the present study are limited to soluble cytosolic bacterial proteins and thus do not represent the total population of bacterial proteins that may be translocated beyond the inclusion membrane. It is possible that chlamydial proteins may remain trapped within the cell after PFO treatment because they interact with cytoskeletal or nuclear components or other membrane-bound or integral membrane proteins. In addition, the proteins detected here are only the bacterial proteins that contain the amino acids methionine and/or cysteine since metabolic labeling was performed with [35S]methionine-cysteine. Consequently, it is possible that other soluble chlamydial proteins localize within the cytoplasmic fraction of infected cells but were not detected because they did not incorporate the labeled amino acids. Finally, the fact that the bacterial proteins were present in small quantities does not render them directly amenable to identification by mass spectrometry. This, however, underscores the exquisite sensitivity of this assay and demonstrates its utility as a strong analytical tool allowing the detection of cytosolic chlamydial proteins, including those of relatively low abundance.
Independent of the overall similarity in the constellation of proteins identified from both chlamydial species, a number of differences were detected. Because we were unable to determine the specific proteins that comprise each spot we cannot rule out the possibility that differences in protein migration are due merely to differences in protein processing. Although it is possible that proteins with similar function are encoded differently in the two genomes, the dissimilarity in migration patterns of these proteins between species suggests that each Chlamydia species interacts with its host in a species-specific manner. As a result, chlamydial proteins translocated to the host cytosolic compartment may define a molecular basis for the differences in pathogenesis between C. trachomatis and C. pneumoniae. In fact, two of the chlamydial proteins previously shown to localize within the host cell cytosol are specific to C. pneumoniae (10, 48).
Many of the cytosolic chlamydial proteins identified in the mid-developmental cycle stage were found in samples obtained from both C. trachomatis- and C. pneumoniae-infected cells. This suggests that these proteins are encoded by genes found within both genomes. Because all members of the genus Chlamydia share a eukaryotic host and thus the same fundamental host biology, it seems reasonable that many of the effector proteins would be highly conserved between chlamydial species. Therefore, there are components of the host-microbe interaction that are shared across the chlamydiae on a molecular level and likely affect basic needs for chlamydial growth.
A unique characteristic of the chlamydiae is their biphasic developmental cycle. Given the amount of time Chlamydia require to complete their developmental cycle and the changes in metabolic demands of the bacteria, it seems reasonable to hypothesize that the molecular mediators of the host-Chlamydia interaction change over time as well. Analysis of the cytosolic fraction obtained from Chlamydia-infected cells bears this out since there were both qualitative and quantitative differences in the proteins present at each stage. The proteins detected at each time point represent proteins synthesized during the short pulse-labeling period rather than an accumulation of the protein product during that stage of the developmental cycle. The asynchronous nature of the developmental cycle, however, precludes normalization of the cytosolic fraction obtained from Chlamydia-infected cells between time points, making it difficult to speculate how much protein is being produced by each individual bacterium at any given time.
Both C. trachomatis- and C. pneumoniae-infected cells secreted numerous bacterial proteins into the cytosol of their host cells. Considering the small size of the chlamydial genomes, and adding the large number of Inc family proteins (2), this suggests that a significant fraction of the genome is devoted to genes encoding proteins which function to modulate the eukaryotic cell. Perhaps it is not surprising that the biology of Chlamydia is inextricably linked to that of the cell given the long evolutionary relationship that exists between Chlamydia and its host. The number of cytosolic chlamydial proteins detected in the present study is comparable to the number of type III secretion effector proteins identified from pathogenicity island I in Salmonella enterica serovar Typhimurium (18). Although it is attractive to speculate that the secreted chlamydial proteins are type III secretion effectors, it is important to note that of the six cytosolic chlamydial proteins previously identified, three are apparently not substrates of type III secretion since they contain sec-dependent secretion signals (10, 50, 55). While the sec-dependent secretion pathway is involved in the translocation of proteins out of the bacterial cell and into the surrounding environment (13, 17, 40), in the chlamydial system these bacterial proteins would be expected to be localized within the lumen of the inclusion. Simple diffusion cannot be used to explain the location of these bacterial proteins since the inclusion does not contain large enough pores to allow the passive diffusion of molecules of this size (25). How chlamydial proteins are translocated across the inclusion membrane is unknown at this time.
Acknowledgments
We thank Guangming Zhong for the anti-CPAF antibody and Pam Schnupf and Ian Glomski for numerous conversations about working with toxins and for generously sharing PFO stocks. We especially thank Dan Portnoy for insights that contributed to the direction of this work.
This research was supported by National Institutes of Health grants AI042156, HL071730, and AI32943.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Backert, S., and T. F. Meyer. 2006. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr. Opin. Microbiol. 9207-217. [DOI] [PubMed] [Google Scholar]
- 2.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 235-47. [DOI] [PubMed] [Google Scholar]
- 3.Bannantine, J. P., D. D. Rockey, and T. Hackstadt. 1998. Tandem genes of Chlamydia psittaci that encode proteins localized to the inclusion membrane. Mol. Microbiol. 281017-1026. [DOI] [PubMed] [Google Scholar]
- 4.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 1008478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bishop, D. K., and D. J. Hinrichs. 1987. Adoptive transfer of immunity to Listeria monocytogenes: the influence of in vitro stimulation on lymphocyte subset requirements. J. Immunol. 1392005-2009. [PubMed] [Google Scholar]
- 6.Brundage, R. A., G. A. Smith, A. Camilli, J. A. Theriot, and D. A. Portnoy. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. USA 9011890-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunham, R. C., F. A. Plummer, and R. S. Stephens. 1993. Bacterial antigenic variation, host immune response, and pathogen-host coevolution. Infect. Immun. 612273-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chellas-Gery, B., C. N. Linton, and K. A. Fields. 2007. Human GCIP interacts with CT847, a novel Chlamydia trachomatis type III secretion substrate, and is degraded in a tissue-culture infection model. Cell. Microbiol. 92417-2430. [DOI] [PubMed] [Google Scholar]
- 9.Clifton, D. R., K. A. Fields, S. S. Grieshaber, C. A. Dooley, E. R. Fischer, D. J. Mead, R. A. Carabeo, and T. Hackstadt. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. USA 10110166-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, F., R. Flores, D. Chen, J. Luo, Y. Zhong, Z. Wu, and G. Zhong. 2006. Localization of the hypothetical protein Cpn0797 in the cytoplasm of Chlamydia pneumoniae-infected host cells. Infect. Immun. 746479-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong, F., M. Pirbhai, Y. Xiao, Y. Zhong, Y. Wu, and G. Zhong. 2005. Degradation of the proapoptotic proteins Bik, Puma, and Bim with Bcl-2 domain 3 homology in Chlamydia trachomatis-infected cells. Infect. Immun. 731861-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, F., H. Su, Y. Huang, Y. Zhong, and G. Zhong. 2004. Cleavage of host keratin 8 by a Chlamydia-secreted protease. Infect. Immun. 723863-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driessen, A. J., P. Fekkes, and J. P. van der Wolk. 1998. The Sec system. Curr. Opin. Microbiol. 1216-222. [DOI] [PubMed] [Google Scholar]
- 14.Fan, P., F. Dong, Y. Huang, and G. Zhong. 2002. Chlamydia pneumoniae secretion of a protease-like activity factor for degrading host cell transcription factors required major histocompatibility complex antigen expression. Infect. Immun. 70345-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in Chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields, K. A., D. J. Mead, C. A. Dooley, and T. Hackstadt. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol. Microbiol. 48671-683. [DOI] [PubMed] [Google Scholar]
- 17.Filloux, A., G. Michel, and M. Bally. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22177-198. [DOI] [PubMed] [Google Scholar]
- 18.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 2841322-1328. [DOI] [PubMed] [Google Scholar]
- 19.Gedde, M. M., D. E. Higgins, L. G. Tilney, and D. A. Portnoy. 2000. Role of listeriolysin O in cell-to-cell spread of Listeria monocytogenes. Infect. Immun. 68999-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glomski, I. J., M. M. Gedde, A. W. Tsang, J. A. Swanson, and D. A. Portnoy. 2002. The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J. Cell Biol. 1561029-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greub, G., and D. Raoult. 2003. History of the ADP/ATP-translocase-encoding gene, a parasitism gene transferred from a Chlamydiales ancestor to plants 1 billion years ago. Appl. Environ. Microbiol. 695530-5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimwood, J., L. Olinger, and R. S. Stephens. 2001. Expression of Chlamydia pneumoniae polymorphic membrane protein family genes. Infect. Immun. 692383-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hackstadt, T., D. D. Rockey, R. A. Heinzen, and M. A. Scidmore. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15964-977. [PMC free article] [PubMed] [Google Scholar]
- 24.Hackstadt, T., M. A. Scidmore, and D. D. Rockey. 1995. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. USA 924877-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinzen, R. A., and T. Hackstadt. 1997. The Chlamydia trachomatis parasitophorous vacuolar membrane is not passively permeable to low-molecular-weight compounds. Infect. Immun. 651088-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heuer, D., V. Brinkmann, T. F. Meyer, and A. J. Szczepek. 2003. Expression and translocation of chlamydial protease during acute and persistent infection of the epithelial HEp-2 cells with Chlamydophila (Chlamydia) pneumoniae. Cell. Microbiol. 5315-322. [DOI] [PubMed] [Google Scholar]
- 27.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, S., and D. A. Portnoy. 1994. Characterization of Listeria monocytogenes pathogenesis in a strain expressing perfringolysin O in place of listeriolysin O. Infect. Immun. 625608-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21385-389. [DOI] [PubMed] [Google Scholar]
- 30.Koehler, J. E., S. Birkelund, and R. S. Stephens. 1992. Overexpression and surface localization of the Chlamydia trachomatis major outer membrane protein in Escherichia coli. Mol. Microbiol. 61087-1094. [DOI] [PubMed] [Google Scholar]
- 31.Koehler, J. E., R. R. Burgess, N. E. Thompson, and R. S. Stephens. 1990. Chlamydia trachomatis RNA polymerase major sigma subunit. Sequence and structural comparison of conserved and unique regions with Escherichia coli sigma 70 and Bacillus subtilis sigma 43. J. Biol. Chem. 26513206-13214. [PubMed] [Google Scholar]
- 32.Lugert, R., M. Kuhns, T. Polch, and U. Gross. 2004. Expression and localization of type III secretion-related proteins of Chlamydia pneumoniae. Med. Microbiol. Immunol. 194163-171. [DOI] [PubMed] [Google Scholar]
- 33.McClarty, G. 1994. Chlamydiae and the biochemistry of intracellular parasitism. Trends Microbiol. 2157-164. [DOI] [PubMed] [Google Scholar]
- 34.Moulder, J. W. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol. Rev. 55143-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicholson, T. L., L. Olinger, K. Chong, G. Schoolnik, and R. S. Stephens. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 1853179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olofsson, A., H. Hebert, and M. Thelestam. 1993. The projection structure of perfringolysin O (Clostridium perfringens theta-toxin). FEBS Lett. 319125-127. [DOI] [PubMed] [Google Scholar]
- 37.Pirbhai, M., F. Dong, Y. Zhong, K. Z. Pan, and G. Zhong. 2006. The secreted protease factor CPAF is responsible for degrading proapoptotic BH3-only proteins in C. trachomatis-infected cells. J. Biol. Chem. 4231495-31501. [DOI] [PubMed] [Google Scholar]
- 38.Rockey, D. D., R. A. Heinzen, and T. Hackstadt. 1995. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol. Microbiol. 15617-626. [DOI] [PubMed] [Google Scholar]
- 39.Rockey, D. D., and J. L. Rosquist. 1994. Protein antigens of Chlamydia psittaci present in infected cells but not detected in the infectious elementary body. Infect. Immun. 62106-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279485-499. [DOI] [PubMed] [Google Scholar]
- 41.Schachter, J. 1999. Infection and disease epidemiology, p. 139-170. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, DC.
- 42.Scidmore, M. A., E. R. Fischer, and T. Hackstadt. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect. Immun. 71973-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shepard, L. A., O. Shatursky, A. E. Johnson, and R. K. Tweten. 2000. The mechanism of pore assembly for a cholesterol-dependent cytolysin: formation of a large prepore complex precedes the insertion of the transmembrane beta-hairpins. Biochemistry 3910284-10293. [DOI] [PubMed] [Google Scholar]
- 44.Stephens, R. S. 2002. Chlamydial evolution: a billion years and counting, p. 3-12. In Chlamyd. Infect. Proc. 10th Int. Symp. Hum. Chlamyd. Infect., Antalya, Turkey, 16 to 21 June 2002.
- 45.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282754-759. [DOI] [PubMed] [Google Scholar]
- 46.Stephens, R. S., M. R. Tam, C. C. Kuo, and R. C. Nowinski. 1982. Monoclonal antibodies to Chlamydia trachomatis: antibody specificities and antigen characterization. J. Immunol. 1281083-1089. [PubMed] [Google Scholar]
- 47.Subtil, A., C. Delevoye, M. E. Balana, L. Tastevin, S. Perrinet, and A. Dautry-Varsat. 2005. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol. Microbiol. 561636-1647. [DOI] [PubMed] [Google Scholar]
- 48.Vandahl, B. B., A. Stensballe, P. Roepstorff, G. Christiansen, and S. Birkelund. 2005. Secretion of Cpn0796 from Chlamydia pneumoniae into the host cell cytoplasm by an autotransporter mechanism. Cell. Microbiol. 7825-836. [DOI] [PubMed] [Google Scholar]
- 49.van Kuppeveld, F. J., J. T. van der Logt, A. F. Angulo, M. J. van Zoest, W. G. Quint, H. G. Niesters, J. M. Galama, and W. J. Melchers. 1993. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl. Environ. Microbiol. 59655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Kuppeveld, F. J., J. T. van der Logt, A. F. Angulo, M. J. van Zoest, W. G. Quint, H. G. Niesters, J. M. Galama, and W. J. Melchers. 1992. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl. Environ. Microbiol. 582606-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walev, I., S. C. Bhakdi, F. Hofmann, N. Djonder, A. Valeva, K. Aktories, and S. Bhakdi. 2001. Delivery of proteins into living cells by reversible membrane permeabilization with streptolysin-O. Proc. Natl. Acad. Sci. USA 983185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walev, I., M. Hombach, W. Bobkiewics, D. Fenske, S. Bhakdi, and M. Husmann. 2002. Resealing of large transmembrane pores produced by streptolysin O in nucleated cells is accompanied by NF-κB activation and downstream events. FASEB J. 16237-239. [DOI] [PubMed] [Google Scholar]
- 53.Wessel, D., and U. I. Flugge. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138141-143. [DOI] [PubMed] [Google Scholar]
- 54.Wyrick, P. B., E. A. Brownridge, and B. E. Ivins. 1978. Interaction of Chlamydia psittaci with mouse peritoneal macrophages. Infect. Immun. 191061-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong, G., T. Fan, and L. Liu. 1999. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J. Exp. Med. 1891931-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong, G., L. Liu, T. Fan, P. Fan, and H. Ji. 2000. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in Chlamydia-infected cells. J. Exp. Med. 1911525-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]