Abstract
In addition to two known staphylococcal enterotoxin-like genes (selj and selr), two novel genes coding for two superantigens, staphylococcal enterotoxins S and T (SES and SET), were identified in plasmid pF5, which is harbored by food poisoning-related Staphylococcus aureus strain Fukuoka 5. This strain was implicated in a food poisoning incident in Fukuoka City, Japan, in 1997. Recombinant SES (rSES) specifically stimulated human T cells in a T-cell receptor Vβ9- and Vβ16-specific manner in the presence of major histocompatibility complex (MHC) class II+ antigen-presenting cells (APC). rSET also stimulated T cells in the presence of MHC class II+ APC, although its Vβ skewing was not found in reactive T cells. Subsequently, we examined the emetic activity of SES and SET. We also studied SElR to determine emetic activity in primates. This toxin was identified in previous studies but was not examined in terms of possession of emetic activity for primates. rSES induced emetic reactions in two of four monkeys at a dose of 100 μg/kg within 5 h of intragastric administration. In one monkey, rSET induced a delayed reaction (24 h postadministration) at a dose of 100 μg/kg, and in the other one, the reaction occurred 5 days postadministration. rSElR induced a reaction in two of six animals within 5 h at 100 μg/kg. On this basis, we speculate that the causative toxins of vomiting in the Fukuoka case are SES and SER. Additionally, SES, SER, and SET also induced emesis in house musk shrews as in the monkeys.
Staphylococcus aureus produces a variety of superantigenic toxins (SAGTs), which selectively activate a vast number of T cells, depending on Vβ elements in the β chain of a T-cell receptor (TCR), in direct association with major histocompatibility complex (MHC) class II molecules on antigen-presenting cells (APC) (14, 31). Staphylococcal SAGTs can be divided into three large groups and one minor group on the basis of similarity of amino acid sequences (31). Most toxins of the three groups, including staphylococcal enterotoxins A and B (SEA and SEB), exhibit strong emetic activity in primates (4, 16, 25); toxic shock syndrome toxin-1, grouped as the minor group, does not possess emetic activity in primates (14, 31). It is noteworthy that toxins designated SE-like toxins, such as SElP and SElR, which either have not been examined for emetic activity or have been reported not to have emetic activity, have been discovered in S. aureus strains (12, 13, 20, 27). S. aureus strain Fukuoka 5 was isolated from food as the causative microbe in a food poisoning outbreak in Fukuoka City, Japan, in 1997, although this strain did not carry any well-recognized SAGT genes with emetic activity (19). Subsequently, Omoe et al. (19) discovered, using a plaque hybridization with a seg probe, that 2.8 kbp of the EcoRI fragment of plasmid pF5, carried by Fukuoka 5, carries two genes, a novel SAGT gene designated selr and a previously reported gene, selj (33).
We undertook research to explore, using a PCR walking technique, whether there were additional SAGT genes on plasmid pF5, because many mobile genetic elements carry various SAGT genes (2, 10, 11, 18, 33). As predicted, pF5 carries two novel toxin genes, designated ses and set, in addition to selj and selr. These two new toxins act as superantigens and exhibit emetic activity in primates. In parallel, we found that SElR also exhibits emetic activity in primates. As a result, we propose changing the name of SElR to SER. We also discuss the causative toxin of the Fukuoka case and the emetic activity of staphylococcal toxins designated SEl-type toxins.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are shown in Table 1. Fukuoka 5, Fukuoka 6, and Fukuoka 7, which harbored the plasmid pF5, were isolated from foods sampled during the outbreak of food poisoning that occurred in Fukuoka City, Japan. S. aureus strains were grown in two media: Trypticase soy broth (Nissui, Tokyo, Japan), kept at 37°C and maintained with aeration for total DNA isolation, and brain heart infusion broth (Difco Laboratories, Detroit, MI) supplemented with 1% yeast extract (Difco) at 37°C with aeration for RNA isolation. SE proteins were produced by S. aureus strains grown in brain heart infusion broth supplemented with 1% yeast extract at 37°C for 48 h with aeration (4). Escherichia coli strains were grown in LB broth (Sigma, St. Louis, MO) containing 100 μg/ml of ampicillin (Wako Chemicals, Osaka, Japan) for plasmid isolation.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | SE genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| Fukuoka 5 | selj selr ses set | 19 |
| Fukuoka 6 | selj selr ses set | 19 |
| Fukuoka 7 | selj selr ses set | 19 |
| E. coli | ||
| DH5α | SE negative | Toyobo |
| BL21 | SE negative | Stratagene |
| Plasmids | ||
| pGEM-Easy | Apr; cloning vector | Promega |
| pGEX 6P-1 | Apr; GST fusion expression vector | Pharmacia |
| pKSX1 | Apr; pGEX 6P-1 with ses | This study |
| pKTX1 | Apr; pGEX 6P-1 with set | This study |
DNA isolation.
Total DNA of S. aureus was purified using a QIAamp DNA minikit (Qiagen, Germantown, MD). E. coli plasmids were purified with a QIAprep spin miniprep kit (Qiagen) following the manufacturer's instructions.
PCR walking of pF5.
PCR walking of the pF5 selj and selr flanking region was performed using a Genome Walker Universal kit (Clontech Laboratories, Mountain View, CA) according to the manufacturer's instructions. This method is used to amplify regions of unknown DNA sequences flanking a region of known DNA sequence. The PCR fragments obtained were subcloned to pGEM-Easy (Promega Corporation, Madison, WI) and sequenced with an ABI3100 Avant DNA sequencer (Applied Biosystems, Foster City, CA). The DNA sequences obtained were assembled by Genetyx-Mac software, version 13 (Genetyx, Tokyo, Japan). Identification of open reading frames (ORFs) was performed with ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), and the identified ORFs were annotated by a Basic Local Alignment Search Tool (BLAST) search of the DNA Data Bank of Japan (DDBJ) (http://blast.ddbj.nig.ac.jp/top-j.html). The N-terminal signal peptide sequences of SES and SET were predicted using the online signal peptide prediction software SignalP (http://www.cbs.dtu.dk/services/SignalP) (17). Multiple alignments and the construction of the phylogenetic tree for SEs and SEls were performed using ClustalW software (28).
Expression of rSES and rSET in a GST fusion system.
To construct the recombinant SES (rSES) and rSET expression plasmids, PCR primers including the BamHI and SalI sites were designed to amplify the ses and set gene fragments corresponding to their mature toxin sequences (Table 2). The gene fragments of ses and set were amplified by PCR using Pyrobest DNA polymerase (Takara, Shiga, Japan). After digestion with BamHI and SalI, the PCR products were subcloned into pGEX6P-1 glutathione S-transferase (GST) fusion expression vector and designated pKSX (which included ses) and pKTX (which included set), respectively. Nucleotide sequences were verified using an ABI3100 Avant DNA sequencer (Applied Biosystems). Expression, purification of the GST-fused recombinant proteins, and cleavage and removal of the GST tag from rSES and rSET were performed by the methods previously described (19, 22). The resulting rSES and rSET had five additional amino acid residues, GPLGS, at the N termini of the mature forms of SES and SET. Preparation of rSEA and rSElR was as previously described (6, 19).
TABLE 2.
Primer sequences and predicted sizes of PCR products
| Purpose | Gene | Primer | Oligonucleotide sequence (5′→3′) | PCR product (bp) |
|---|---|---|---|---|
| Cloning | ses | ORF6GSTF | CCCCGGATCCGATGAATCTAGACCTAAAATAG | 794 |
| ORF6GSTR | CCCCGTCGACTTATTGGGAATAAAC | |||
| set | ORF5GSTF | CCCCGGATCCGATTCTCGTGAAGGTTTAAAAG | 671 | |
| ORF5GSTR | CCCCGTCGACCTATTTTTCCATATATATATC | |||
| RT-PCR | ses | ORF6F | TTCAGAAATAGCCAATCATTTCAA | 195 |
| ORF6R | CCTTTTTGTTGAGAGCCGTC | |||
| set | ORF5F | GGTGATTATGTAGATGCTTGGG | 170 | |
| ORF5R | TCGGGTGTTACTTCTGTTTGC | |||
| femA | femA1 | AAAAAAGCACATAACAAGCG | 134 | |
| femA2 | GATAAAGAAGAAACCAGCAG |
Mitogenic activity of toxins.
Human peripheral blood mononuclear cells (PBMCs) were obtained from three healthy donors and processed by Ficoll-Conray density gradient centrifugation. The PBMCs were incubated for 72 h in 96-well flat-bottomed tissue culture plates with different concentrations of rSES or rSET and then assayed to test for uptake of [3H]thymidine. Data (in counts per minute) are presented as means ± standard errors of triplicate determinations, as previously described (30).
Analysis of requirement for MHC class II molecules in activation of T cells by toxins.
T cells were obtained by the S-2-aminoethylisothiouronium-treated sheep red blood cell rosette method. They were further enriched by removal of CD16−, CD14−, CD19−, and HLA-DR+ cells, using monoclonal antibodies (MAbs) to those antigens and anti-mouse immunoglobulin-coated magnetic beads (Dynabeads; Dynal, Oslo, Norway) (8, 9). L cells transfected with the DR4 gene (8124 L cells) and control L cells (8400 cells) were prepared as previously described (30). L cells were then treated with mitomycin C, irradiated with an MBR-1404R X-ray generator (Hitachi, Tokyo, Japan) to block proliferation, and used as accessory cells for T-cell activation by rSES, rSET, and SEA. We then measured interleukin-2 (IL-2) production from stimulated T cells to measure T-cell activation. IL-2 activity in culture supernatants was determined with IL-2-dependent CTLL-2 (9, 30). The assays were performed in triplicate for samples and standards, and the data obtained are presented as units of IL-2 per milliliter. In parallel, we examined the effects of the antibody to HLA-DR on T-cell response (30).
Analysis of TCR Vβ repertoires of SES- or SET-reactive human T cells.
rSES-, rSET-, or anti-CD3-induced T-cell blasts were obtained by stimulating PBMCs with 20 ng of rSES or rSET/ml, or with 5 μg of MAbs to anti-CD3/ml, for 3 days and expanding harvested blasts for 4 days in the presence of 100 U of recombinant human IL-2/ml (Shionogi, Osaka, Japan). The T-cell blasts obtained were stained with MAbs to TCR Vβ elements (IOTest Beta Mark kit; Beckman Coulter, Miami, FL). Samples were analyzed on an EPICS XL cytometer (Beckman Coulter) with FlowJo software, as previously described (22, 26). The Vβ frequencies of the T-cell preparations were expressed as percentages on CD3+ T cells. We determined SE-specific reactive T cells when an increased percentage of a certain Vβ element-positive T cells was observed in all donors. TCR Vα expression in rSET- or anti-CD3-induced T-cell blasts was analyzed by reverse transcription-PCR (RT-PCR) (22, 23).
Emesis assay.
In this study we conducted two types of emesis assays. First, experiments using cynomolgus monkeys (Macaca fascicularis) were conducted at the Tsukuba Primate Research Center of the National Institute of Biomedical Innovation of Japan (Tsukuba City, Ibaragi) under the approval of the Animal Ethics Committee of Iwate University and Tsukuba Primate Research. Monkeys used in the experiments were individually housed in stainless steel cages in rooms kept at 23 to 27°C and 50 to 70% humidity, using a 12-h/12-h light/dark cycle. An emesis assay using a primate model was performed according to Bergdoll's monkey feeding test (3) with a slight modification. rSEA, rSElR, rSES, and rSET were dissolved in 10 ml of sterile distilled water and fed by nasogastric intubation without anesthesia to young (4-year-old) female cynomolgus monkeys at a dose of 100 μg/kg. The monkeys were kept under continuous observation for 5 h after intragastric administration of the toxin. If monkeys did not appear to exhibit emetic reactions during the first 5 h, the monkey cages were checked for the presence of vomited material every morning for 2 weeks. In addition, all monkeys were subjected to routine observation every morning, and their appetites and stools were observed for abnormalities throughout the experimental period.
The emesis assay with house musk shrews was performed by a method described elsewhere (6, 7), under the approval of the Animal Ethics Committee at Iwate University. Healthy adult (1.5- to 14-month-old) house musk shrews (Suncus murinus; Nihon Clea, Tokyo, Japan) were kept at 22 to 25°C in a room lit for 12 h from 7:00 a.m. to 7:00 p.m. Purified rSElR, rSES, or rSET was diluted in 0.01 M phosphate-buffered saline (pH 7.2). Two hundred microliters of rSElR, rSES, or rSET appropriately diluted was injected intraperitoneally into each house musk shrew. The animals were observed for emesis for 3 h after intraperitoneal administration. The number of vomiting episodes, the time of each vomiting episode, the length of time before the first vomiting episode, and behavioral changes were recorded.
RT-PCR.
Total RNA was extracted from S. aureus cultures using an RNeasy spin column (Qiagen) according to the manufacturer's instructions. Purified total RNA was treated with DNase I (Roche Diagnostics K.K., Basel, Switzerland) to degrade contaminating genomic DNA. cDNA was synthesized with SuperScript II reverse transcriptase (Gibco BRL, Grand Island, NY) and random primer (Gibco BRL). As a control for genomic DNA contamination, total RNA was also subjected to PCR but without the RT step. The ses and set cDNAs were detected with the primer sets shown in Table 2. femA, a cytoplasmic protein gene involved in the biosynthesis of staphylococcal cell walls, was used as the control for RNA isolation and RT-PCR (15).
Preparation of specific antibodies and detection of SES and SET in S. aureus cultures.
Anti-rSES and anti-rSET rabbit sera were prepared by immunizing rabbits with purified rSES or rSET, as previously reported (24). Titers of antiserum were monitored by enzyme-linked immunosorbent assay. Specific antibodies were purified from hyperimmune sera using a MAbTrap kit (GE Healthcare UK Ltd., Buckinghamshire, England). Using these specific antibodies, production of SES and SET from S. aureus isolates harboring pF5 was determined. Culture supernatants of these isolates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (Bio-Rad, Richmond, CA) by the method described by Towbin et al. (29). Reactive signals were detected using a horseradish peroxidase-labeled anti-rabbit immunoglobulin G goat antibody (Bio-Rad) and an ECL Plus system (GE Healthcare UK Ltd.) in accordance with the manufacturers’ instructions.
Nucleotide sequence accession number.
The nucleotide sequence of flanking SEl genes in plasmid pF5 was submitted to the GenBank, EMBL, and DDBJ databases and assigned accession number AB330135.
RESULTS
Identification of two staphylococcal superantigen-like sequences in Fukuoka 5.
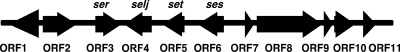
The nucleotide sequence flanking selr and selj on pF5 was determined by PCR walking. We obtained an 11,335-bp nucleotide sequence containing novel two genes, both having significant sequence similarity to staphylococcal SAGT genes in ORF5 and ORF6, in addition to selr in ORF3 and selj in ORF4 (Fig. 1). The remaining genes in seven ORFs did not show sequence similarity with staphylococcal SAGT genes.
FIG. 1.
Location of the two new superantigen-like genes in the selj and selr flanking regions. An 11,335-bp nucleotide sequence of the selr (ORF3) and selj (ORF4) flanking region was determined, and two new superantigen-like genes, ses (ORF6) and set (ORF5), were identified.
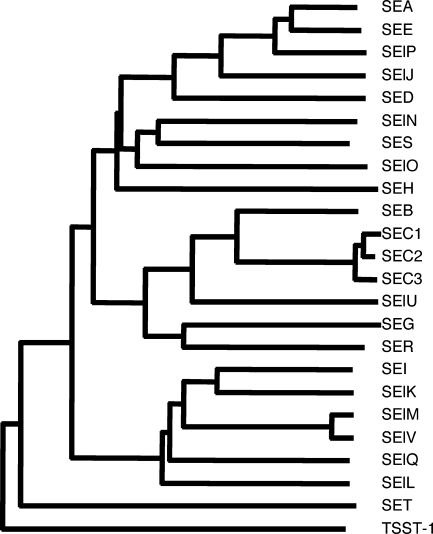
ORF5, tentatively designated set, encoded a polypeptide of 216 amino acids in length. We predicted that the mature form of SET would have a molecular weight of 22,614. Deducing the amino acid sequence of SET, we determined that it is most closely related (38% sequence similarity) to that of putative exotoxin SAB2421c, identified in bovine mastitis isolate RF122 (GenBank/EMBL/DDBJ accession number AJ938182). Also, SET showed sequence similarity to streptococcal pyrogenic toxin type K (SpeK) (27.0% sequence similarity). ORF6, tentatively designated ses, encoded a polypeptide of 257 amino acids in length. We predicted that the mature form of SES would have a molecular weight of 26,217. Its deduced amino acid sequence is most closely related to that of SElN (48.0% sequence similarity). Phylogenetic analysis showed that SES belongs to the same group as SEA, while SET is distinct from other SAGTs (Fig. 2).
FIG. 2.
Phylogenetic tree of SEs and SEls, including SES and SET. The phylogenetic tree was constructed by the neighbor-joining method based on amino acid sequences. Five distinct groups can be observed. SES is most closely related to SElN and belongs to the SEA group, while SET is distinct from the SEA, SEB, and SEI groups.
Superantigenic activities of rSES and rSET.
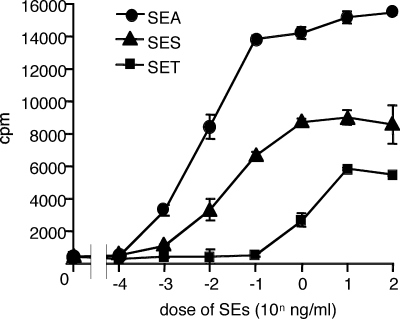
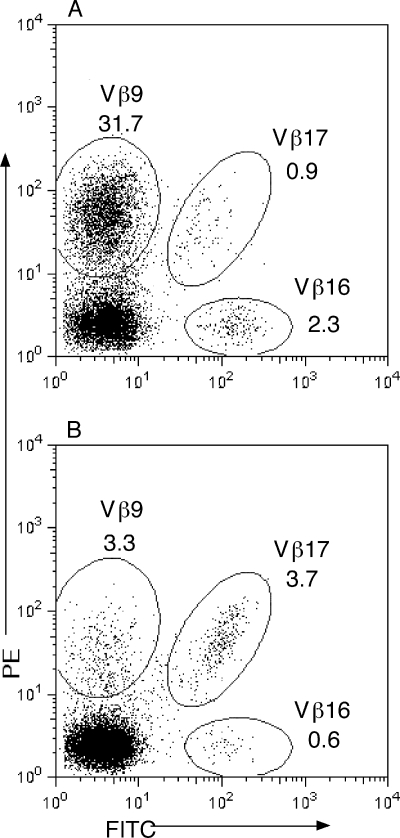
Recombinant proteins of SES and SET were examined for SAGT activity. First, rSES and rSET were tested for mitogenic activity to human PBMCs, and that activity was compared with the mitogenic activity of SEA. Representative results for three experiments are shown in Fig. 3. Lymphocyte proliferation was observed at an SES concentration of 1 pg/ml or more, and the strength of mitogenic activity was slightly lower in SES than in SEA. The mitogenic activity of SET was very weak in comparison with that of SES. SET required a concentration 2 orders of magnitude higher than that of SES to achieve substantial lymphocyte proliferation. Second, we examined the requirements for MHC class II molecules on APC for activation of human T cells by SES and SET. Table 3 shows representative results for three different donors. Both rSES and rSET induced production of substantial levels of IL-2 from T cells in the presence of DR-transfected L cells (8124 cells), but not in the presence of control L cells (8400 cells). Anti-DR MAb markedly inhibited rSES- and rSET-induced IL-2 production from T cells in the presence of 8124 L cells. Anti-mouse MHC class I MAb (anti-H-2Kk) showed no effect on the T-cell response induced by rSES or rSET. Third, we analyzed the TCR Vβ specificity of SES- and SET-induced activation of human T cells by flow cytometry. Table 4 and Fig. 4 show results for one of three different donors with similar results. T cells bearing TCR Vβ9 responded with the highest level of expansion (from 3.3% [in the normal range] to 31.7% in activated T cells) by rSES stimulation. T cells bearing Vβ16 seem to have responded to rSES, although the level of response was very low. The percentage of Vβ16-positive cells increased from 0.6%, in the normal range, to 2.3% in the activated T cells. No obvious TCR Vβ skewing was observed in T-cell activation using SET (Table 4). SEH is unique in that it stimulates T cells bearing Vα10 but not those bearing Vβ (23). We analyzed the TCR Vα specificity of SET-induced activation of human T cells by RT-PCR. Frequencies of T cells bearing all Vα elements tested were similar in SET-induced T-cell blasts and anti-CD3-induced T-cell blasts (data not shown).
FIG. 3.
Mitogenic activity of SES and SET. PBMCs were isolated from human blood samples and incubated with a number of concentrations of rSEA, rSES, and rSET. Means and standard errors for triplicate wells from a single experiment are shown.
TABLE 3.
MHC class II molecule requirement for T-cell activation by SES and SET
| APCa | Antibody | IL-2 production (U/ml)b with:
|
|||
|---|---|---|---|---|---|
| SES (20 ng/ml) | SET (20 ng/ml) | SEA
|
|||
| 0 ng/ml | 10 ng/ml | ||||
| 8400 | None | <0.1 | <0.1 | <0.1 | <0.1 |
| 8124 | None | 13.7 | 11.2 | <0.1 | 84.3 |
| 8124 | Anti-DR | 1.6 | 0.9 | <0.1 | 23.5 |
| 8124 | anti-H-2Kk | 18.0 | 8.5 | <0.1 | 89.1 |
8400, control L cells; 8124, HLA-DR4+ L cells.
Data are representative of the results for three different donors.
TABLE 4.
TCR Vβ specificities of SES and SET
| Vβ | % in T cells stimulated witha:
|
||
|---|---|---|---|
| Anti-CD3 | SES | SET | |
| 1 | 4.2 | 1.9 | 5.1 |
| 2 | 6.6 | 0.2 | 4.9 |
| 3 | 14.1 | 2.6 | 7.0 |
| 4 | 0.6 | 0.0 | 0.3 |
| 5.1 | 6.2 | 0.9 | 7.0 |
| 5.2 | 0.5 | 1.5 | 0.6 |
| 5.3 | 0.5 | 0.8 | 0.6 |
| 7.1 | 2.8 | 3.4 | 1.8 |
| 7.2 | 0.7 | 0.0 | 0.3 |
| 8 | 3.6 | 0.7 | 6.2 |
| 9 | 3.3 | 31.7* | 2.4 |
| 11 | 1.5 | 0.1 | 0.8 |
| 12 | 1.2 | 0.2 | 2.0 |
| 13.1 | 3.3 | 0.3 | 3.1 |
| 13.2 | 1.3 | 0.1 | 1.2 |
| 13.6 | 1.1 | 0.3 | 2.1 |
| 14 | 7.1 | 2.0 | 4.5 |
| 16 | 0.6 | 2.3* | 1.2 |
| 17 | 3.7 | 0.9 | 4.7 |
| 18 | 0.3 | 0.1 | 0.8 |
| 20 | 3.9 | 1.2 | 3.3 |
| 21.3 | 3.2 | 0.8 | 2.9 |
| 22 | 4.5 | 3.3 | 2.8 |
| 23 | 1.1 | 0.2 | 0.5 |
| Otherb | 24.1 | 44.5 | 33.9 |
Data are representative of the results for three different donors. Increases in comparison with anti-CD3 stimulation observed in three donors are indicated with an asterisk.
Vβ elements that were not covered by the anti-Vβ MAbs available in this study.
FIG. 4.
Representative dot plots of Vβs for CD3-gated events from PBMCs stimulated with SES (A) and anti-CD3 (B). MAbs against Vβ9, Vβ16, and Vβ17 were labeled with phycoerythrin (PE), fluorescein isothiocyanate (FITC), or PE and FITC, respectively. Numbers indicate the percentage of T cells expressing a particular Vβ element.
These results show that SES activates human T cells in association with MHC class II molecules expressed on APC in a manner specific to Vβ9+ and Vβ16+ T cells, indicating that SES acts on T cells as an SAGT. Although Vβ specificity was not found in the activation of SET on T cells, we think that SET also acts on T cells as a superantigen because it stimulates T cells in the presence of MHC class II molecules on APC and has amino acid sequences similar to those of other SAGTs.
Emetic activities of rSElR, rSES, and rSET.
rSES, rSET, and rSElR were examined for emetic activity using the monkey feeding test, which has been recognized as the gold standard to confirm “enterotoxin” activity of staphylococcal SAGTs. It is possible to think that all SEs, including SEl types, have emetic activity in primates, although their emetic activities are not equal. An observation period of 5 h may be insufficient to detect the weak emetic activity of some of the enterotoxins. Thus, we checked the emetic activity of the three SAGTs and rSEA as a positive control in cynomolgus monkeys for 24 h or more.
The results are summarized in Table 5. rSEA induced emetic reactions in six of seven monkeys at a dose of 100 μg/kg within 1 to 4 h after intragastric administration. Each monkey had from 2 to more than 10 emetic episodes. rSES induced emetic reactions in two of four animals at a dose of 100 μg/kg within 1 to 3 h postadministration, and five to seven emetic episodes per monkey were observed. rSElR induced emetic reactions in two of six monkeys at a dose of 100 μg/kg within 2 to 3 h postadministration, and six to eight emetic episodes per monkey were observed. Due to its clear emetic activity, we propose that SElR be designated SER. rSET did not induce an emetic reaction in four monkeys within 5 h at a dose of 100 μg/kg, but in one of the four monkeys, we found vomitus several times on monkey cage floor either within 24 h or during subsequent days. In another monkey, vomitus was observed on the 5th and 12th days postadministration. The delayed emetic reactions seen in SET administration were not observed for administrations of SEA, SER and SES.
TABLE 5.
Emetic response of Macaca fascicularis to SER, SES, and SET
| Toxin (100 μg/kg) | No. of monkeys tested | No. of monkeys with emetic reaction:
|
||
|---|---|---|---|---|
| Within 5 h | Within 24 h | After 24 h | ||
| SEA | 7 | 6 | 0 | 0 |
| SER | 6 | 2 | 0 | 0 |
| SES | 4 | 2 | 0 | 0 |
| SET | 4 | 0 | 1 | 2a |
These monkeys, including one monkey that vomited within 24 h, intermittently vomited for 2 weeks after administration of SET.
Additionally, the emetic activity of SER, SES, and SET was examined by a recently established system using house musk shrews (6, 7). rSES induced emetic reactions in two of three house musk shrews at a dose of 100 μg/animal and in one of three animals at a dose of 20 μg/animal within 80 to 100 min postadministration (Table 6). rSER induced emetic reactions in two of five animals at a dose of 1,000 μg/animal within 100 to 120 min after administration but not at a dose of 200 μg/animal. rSET induced an emetic reaction in one of five animals at a dose of 1,000 μg/animal 130 min postadministration but not in three animals at a dose of 500 μg/animal.
TABLE 6.
Emetic response of Suncus murinus to SER, SES, and SET
| Toxin | Dose (μg/animal) | No. of animals tested | No. of animals that vomited |
|---|---|---|---|
| SER | 1,000 | 5 | 2 |
| 200 | 5 | 0 | |
| SES | 100 | 3 | 2 |
| 20 | 3 | 1 | |
| 4 | 3 | 0 | |
| SET | 1,000 | 5 | 1 |
| 500 | 3 | 0 |
Although we lacked sufficient numbers of animals to conduct a quantitative analysis of emetic activity, our results suggest that SER and SES have similar levels of emetic activity in monkeys, although the levels of activity are slightly lower than that for SEA. In the analysis using house musk shrews, emetic \activity was highest for SES, medium for SER, and lowest for SET.
Production of SES and SET in S. aureus strains isolated in the Fukuoka outbreak.
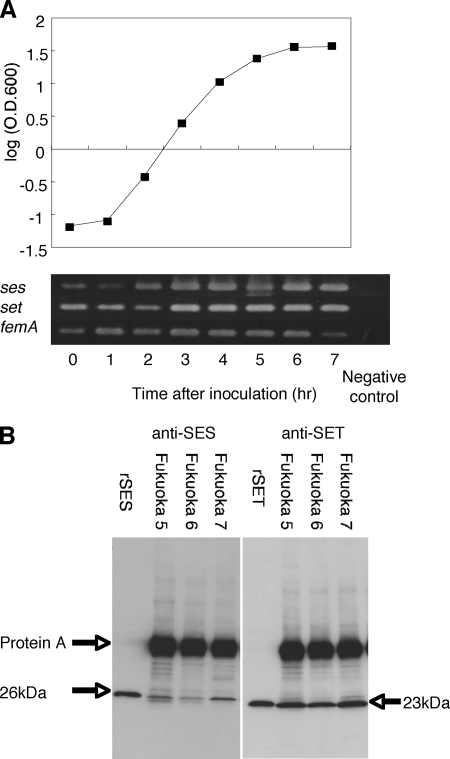
In order to check whether SES and SET are produced by the S. aureus strains isolated from the Fukuoka outbreak, we conducted two experiments. First, total RNA isolated from Fukuoka 5 was subjected to RT-PCR analysis for ses and set mRNA transcription for a number of time points during bacterial growth. Figure 5A shows the Fukuoka 5 strain growth curve and the results of RT-PCR at each point. ses and set mRNAs were transcribed at all phases of growth of S. aureus Fukuoka 5. Second, Western blot analysis was employed to investigate whether SES and SET proteins were present in the culture supernatants of S. aureus isolates (Fukuoka 5, Fukuoka 6, and Fukuoka 7). The results showed that significant amounts of SES and SET were detected in all culture supernatants from these bacteria (Fig. 5B).
FIG. 5.
Expression of ses and set. (A) Detection of ses and set mRNAs by RT-PCR. S. aureus Fukuoka 5 was cultured at 37°C and collected for RNA isolation at 0 to 7 h after inoculation. The Fukuoka 5 growth curve was graphed by determining the optical density at 600 nm (O.D.600). (B) Detection of SES and SET in culture supernatants of S. aureus strains harboring plasmid pF5 (Fukuoka 5, Fukuoka 6, and Fukuoka 7) by Western blotting.
DISCUSSION
In this study, we examined both emetic and superantigenic activities of two novel SAGTs, SES and SET, produced by S. aureus strain Fukuoka 5. Emetic activity of SER, previously designated SElR and also produced by Fukuoka 5, was examined in parallel. We discuss several aspects of the emetic and superantigenic activities of these toxins. In the toxins of the SE/SEl groups, the naming of SES and SET used between SER and SElU has not yet been. In recent years, genome analyses of several S. aureus strains revealed clusters of multiple genes encoding superantigen-like proteins designated staphylococcal exotoxin-like toxins (SETs) (2, 11, 32). Studies have shown, however, that SETs have no capacity to stimulate T cells (1). Later, researchers recommended renaming this protein family the “staphylococcal superantigen-like” (SSL) proteins (13). Therefore, in the present study we designated the two novel SAGTs SES and SET.
Our study revealed that SER and SES induced emetic reactions in cynomolgus monkeys at a dose of 100 μg/kg in two to six animals and in two to four animals, respectively, within the first 5 h after SES administration (under the standard examination). SEA induced emetic reactions in six or seven animals. Other reports indicate that the 50% effective dose of SEA in rhesus monkeys (Macaca mulatta) is 5 μg/animal (3, 4); therefore, it seems likely that SER and SES are weaker in emetic activity than SEA. It is noteworthy that SET induced an atypical emetic reaction in cynomolgus monkeys. Two of four animals examined exhibited emetic reactions only after significant time lapses: at 24 h postadministration in one animal and at 5 days postadministration in another animal. We consider that the mechanisms of the SE-induced emetic responses are fundamentally the same and that the prolonged incubation time for the SET-induced emetic response reflected its weak emetic activity compared with SEA, SER, and SES. However, it seems that further analysis is necessary to obtain a conclusion. As no well-recognized SEs with strong emetic activity, such as SEA to SEE and SEG to SEI, were detected in the causative bacteria from the Fukuoka outbreak, it seems likely that SER, SES, and/or SElJ was the causative toxin of this outbreak. One of these toxins, or any of these three toxins acting additively with one another, would have produced vomiting.
SE/SEl-type toxins exhibit similarities in their amino acid sequences. In most cases, emetic reactions in primates induced by staphylococcal SAGTs with strong emetic activity have been observed within 5 h of toxin ingestion (4). This length of observation time seems insufficient for observing emetic activities caused by staphylococcal SAGTs with low emetic activity. We assume that a longer observation period will reveal emetic activity caused by newly identified SAGTs. As expected, emetic reactions induced by SET were observed after the routine observation period was over. Because many S. aureus strains isolated in food poisoning cases carry multiple genes for the SEl-type toxins, in addition to genes for the classical SEs (5, 21), it seems important to examine the possible emetic activity of SEl-type toxins. We have started to investigate this subject, to illuminate the complete picture of the emetic activity of staphylococcal SAGTs in association with staphylococcal food poisoning.
Our study showed that a major portion of human T cells reactive to SES are TCR Vβ9+ and that TCR Vβ16+ T cells are a minor reactive fraction of these T cells. This TCR Vβ skewing resembles that of SElN, which is phylogenetically related to SES and has been shown to selectively stimulate human T cells harboring TCR Vβ9 (10). Vβ skewing similarity between phylogenetically related SAGTs has been well established (31). The assay system used in the present study to analyze the Vβ repertoire in SAGT-reactive human T cells, however, was not complete. The panel of MAbs used in the present study did not cover all TCR Vβ elements in human T cells. As shown in Table 4, the percentage of Vβ elements in T cells stimulated by SES shown as “other,” which were not covered by the anti-Vβ MAbs available, was 44.5%, much higher than the control value (24.1%), suggesting that the assay system failed to detect another SES-reactive fraction. The percentage of “other” Vβ elements reactive to SET was 33.9%, much higher than the control value (Table 4), suggesting the presence of a SET-reactive fraction which was missed the present assay system. Alternatively, SET may have activated T cells in TCR Vα-specific manner. Another possibility is that SET may have activated T cells in a polyclonal manner, which is not restricted by particular sets of TCR Vβ and TCR Vα specificities. Further analysis is needed to elucidate the nature of the superantigenic activity of SET in detail.
We employed house musk shrews as an experimental animal model to test emetic reactions caused by SAGTs, in addition to the primate model. Although the monkey feeding assay is a valid method to examine human food poisoning by SEs, it seems important to make an attempt to develop a suitable assay system using small animals to investigate emetic reactions induced by staphylococcal SAGTs. In the emesis assay using house musk shrews, intraperitoneal injection of SER or SES induced various levels of emetic reactions. It seems that SEs can induce emetic reactions through two potentially different mechanisms, their enterotoxic activity and their superantigenic activity. At present, we cannot say that this system can replace the system using monkeys to examine SE-induced food poisonings. Recently, Hu et al. showed that intraperitoneal injection of SEA increased 5-hydroxytryptamine (5-HT) release in the small intestine and induced emetic reaction through stimulation of the 5-HT type 3 receptor on vagal afferent neurons in house musk shrews (7). Currently, we cannot define which mechanism, superantigenic activity of SEA or enterotoxigenic activity of SEA per se, triggered the 5-HT release in the small intestine in SEA-injected house musk shrews. Further study is needed to clarify which mechanism is responsible for the emetic reactions induced by injection of staphylococcal SAGTs in house musk shrews.
Acknowledgments
We thank Keiichi Hiramatsu at Juntendo University for advice on SE nomenclature.
This work was supported in part by grants-in-aid for scientific research from the Japan Society for the Promotion of Science (grants 16380205, 18580304, and 18590439).
Editor: V. J. DiRita
Footnotes
Published ahead of print on 18 August 2008.
REFERENCES
- 1.Arcus, V. L., R. Langley, T. Proft, J. D. Fraser, and E. N. Baker. 2002. The three-dimensional structure of a superantigen-like protein, SET3, from a pathogenicity island of the Staphylococcus aureus genome. J. Biol. Chem. 27732274-32281. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 3591819-1827. [DOI] [PubMed] [Google Scholar]
- 3.Bergdoll, M. S. 1988. Monkey feeding test for staphylococcal enterotoxin. Methods Enzymol. 165324-333. [DOI] [PubMed] [Google Scholar]
- 4.Bergdoll, M. S. 1989. Staphylococcus aureus, p. 463-523. In M. P. Doyle (ed.), Foodborne bacterial pathogens. Marcel Dekker, Inc., New York, NY.
- 5.Fueyo, J. M., M. C. Mendoza, and M. C. Martin. 2005. Enterotoxins and toxic shock syndrome toxin in Staphylococcus aureus recovered from human nasal carriers and manually handled foods: epidemiological and genetic findings. Microbes Infect. 7187-194. [DOI] [PubMed] [Google Scholar]
- 6.Hu, D. L., K. Omoe, Y. Shimoda, A. Nakane, and K. Shinagawa. 2003. Induction of emetic response to staphylococcal enterotoxins in the house musk shrew (Suncus murinus). Infect. Immun. 71567-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu, D. L., G. Zhu, F. Mori, K. Omoe, M. Okada, K. Wakabayashi, S. Kaneko, K. Shinagawa, and A. Nakane. 2007. Staphylococcal enterotoxin induces emesis through increasing serotonin release in intestine and it is downregulated by cannabinoid receptor-1. Cell. Microbiol. 92267-2277. [DOI] [PubMed] [Google Scholar]
- 8.Imanishi, K., K. Seo, H. Kato, T. Miyoshi-Akiyama, R. H. Zhang, Y. Takahashi, Y. Imai, and T. Uchiyama. 1998. Post-thymic maturation of migrating human thymic single-positive T cells: thymic CD1a-CD4+ T cells are more susceptible to anergy induction by toxic shock syndrome toxin-1 than cord blood CD4+ T cells. J. Immunol. 160112-119. [PubMed] [Google Scholar]
- 9.Imanishi, K., H. Kato, H. Fujii, and T. Uchiyama. 2003. Maturation of adult peripheral blood CD38+CD4+ T cells demonstrated by cytokine production in response to a superantigen, TSST-1. Cell. Immunol. 22289-96. [DOI] [PubMed] [Google Scholar]
- 10.Jarraud, S., M. A. Peyrat, A. Lim, A. Tristan, M. Bes, C. Mougel, J. Etienne, F. Vandenesch, M. Bonneville, and G. Lina. 2001. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J. Immunol. 166669-677. [DOI] [PubMed] [Google Scholar]
- 11.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 3571225-1240. [DOI] [PubMed] [Google Scholar]
- 12.Letertre, C., S. Perelle, F. Dilasser, and P. Fach. 2003. Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J. Appl. Microbiol. 9538-43. [DOI] [PubMed] [Google Scholar]
- 13.Lina, G., G. A. Bohach, S. P. Nair, K. Hiramatsu, E. Jouvin-Marche, and R. Mariuzza. 2004. Standard nomenclature for the superantigens expressed by Staphylococcus. J. Infect. Dis. 1892334-2336. [DOI] [PubMed] [Google Scholar]
- 14.McCormick, J. K., J. M. Yarwood, and P. M. Schlievert. 2001. Toxic shock syndrome and bacterial superantigens: an update. Annu. Rev. Microbiol. 5577-104. [DOI] [PubMed] [Google Scholar]
- 15.Mehrotra, M., G. Wang, and W. M. Johnson. 2000. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 381032-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Munson, S. H., M. T. Tremaine, M. J. Betley, and R. A. Welch. 1998. Identification and characterization of staphylococcal enterotoxin type G and I from Staphylococcus aureus. Infect. Immun. 663337-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 101-6. [DOI] [PubMed] [Google Scholar]
- 18.Novick, R. P. 2003. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 4993-105. [DOI] [PubMed] [Google Scholar]
- 19.Omoe, K., D. L. Hu, H. Takahashi-Omoe, A. Nakane, and K. Shinagawa. 2003. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect. Immun. 716088-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Omoe, K., K. Imanishi, D. L. Hu, H. Kato, H. Takahashi-Omoe, A. Nakane, T. Uchiyama, and K. Shinagawa. 2004. Biological properties of staphylococcal enterotoxin-like toxin type R. Infect. Immun. 723664-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Omoe, K., D. L. Hu, H. Takahashi-Omoe, A. Nakane, and K. Shinagawa. 2005. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiol. Lett. 246191-198. [DOI] [PubMed] [Google Scholar]
- 22.Omoe, K., K. Imanishi, D. L. Hu, H. Kato, Y. Fugane, Y. Abe, S. Hamaoka, Y. Watanabe, A. Nakane, T. Uchiyama, and K. Shinagawa. 2005. Characterization of novel staphylococcal enterotoxin-like toxin type P. Infect. Immun. 735540-5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersson, K., H. Pettersson, N. J. Skartved, B. Walse, and G. Forsberg. 2003. Staphylococcal enterotoxin H induces Vα-specific expansion of T cells. J. Immunol. 1704148-4154. [DOI] [PubMed] [Google Scholar]
- 24.Shinagawa, K., M. Ishibashi, H. Yamamoto, N. Kunita, and K. Hisa. 1974. A consideration to immune doses of staphylococcal enterotoxin B to rabbits. Jpn. J. Med. Sci. Biol. 27309-314. [DOI] [PubMed] [Google Scholar]
- 25.Su, Y. C., and A. C. L. Wong. 1995. Identification and purification of a new staphylococcal enterotoxin, H. Appl. Environ. Microbiol. 611438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi, N., H. Kato, K. Imanishi, K. Miwa, S. Yamanami, H. Nishida, and T. Uchiyama. 2000. Immunopathophysiological aspects of an emerging neonatal infectious disease induced by a bacterial superantigen. J. Clin. Investig. 1061409-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas, D. Y., S. Jarraud, B. Lemercier, G. Cozon, K. Echasserieau, J. Etienne, M. Gougeon, G. Lina, and F. Vandenesch. 2006. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect. Immun. 744724-4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 764350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uchiyama, T., T. Miyoshi-Akiyama, H. Kato, W. Fujimaki, K. Imanishi, and X. J. Yan. 1993. Superantigenic properties of a novel mitogenic substance produced by Yersinia pseudotuberculosis isolated from patients manifesting acute and systemic symptoms. J. Immunol. 1514407-4413. [PubMed] [Google Scholar]
- 31.Uchiyama, T., K. Imanishi, T. Miyoshi-Akiyama, and H. Kato. 2006. Staphylococcal superantigens and the diseases they cause, p. 830-843. In J. E. Alouf and M. R. Popoof (ed.), Comprehensive sourcebook of bacterial protein toxins, 3rd ed. Academic Press, Burlington, MA.
- 32.Williams, R. J., J. M. Ward, B. Henderson, S. Poole, B. P. O'Hara, M. Wilson, and S. P. Nair. 2000. Identification of a novel gene cluster encoding staphylococcal exotoxin-like proteins: characterization of the prototypic gene and its protein product, SET1. Infect. Immun. 684407-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, S., J. J. Iandolo, and G. C. Stewart. 1998. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej). FEMS Microbiol. Lett. 168227-233. [DOI] [PubMed] [Google Scholar]