Abstract
Aromatic amines constitute one of the most extensively studied classes of chemical carcinogens. Although monocyclic aromatic amines are generally regarded as weak carcinogens, a recent epidemiologic study of bladder cancer found that the arylamine 3,5-dimethylaniline (3,5-DMA) may play a significant role in the etiology of this disease in man. Investigations using experimental animals also strongly suggested that DNA adducts—of indeterminate structure—formed by 3,5-DMA might account for its presumptive activity. The present study was undertaken to determine the structures of the major DNA adducts formed in vitro by the known, and possibly carcinogenic, N-hydroxylated metabolite. Calf thymus DNA (ct-DNA) was modified by reaction with N-acetoxy-3,5-dimethylaniline (N-AcO-3,5-DMA). After enzymatic hydrolysis of DNA to individual 2'-deoxyribonucleosides, adduct profiles were determined using HPLC/MS. 3,5-DMA formed four major DNA adducts, one to 2’-deoxyguanosine (dG), two to 2’-deoxyadenosine (dA), and one to 2’-deoxycytidine (dC). Reactions of N-AcO-3,5-DMA with dG, dA, and dC produced the same adducts as reaction with ct-DNA with very similar profiles. Adducts were isolated chromatographically and unambiguously characterized as N-(deoxyguanosin-8-yl)-3,5-dimethylaniline (dG-C8−3,5-DMA), 4-(deoxyadenosin-N6-yl)-3,5-dimethylaniline (dA-N6-3,5-DMA), N-(deoxyadenosin-8-yl)-3,5-dimethylaniline (dA-C8−3,5-DMA), and N-(deoxycytidin-5-yl)-3,5-dimethylaniline (dC-C5−3,5-DMA) by high-resolution mass spectra (HR-MS) and NMR spectroscopy including 1H-NMR, 13C-NMR, and two-dimensional NMR. This report includes the first detailed description of a dC adduct of an aromatic amine. The present results provide chemical support for a carcinogenic mechanism of action by 3,5-DMA based on N-hydroxylation and the intermediacy of a nitrenium ion in the formation of DNA adducts.
Introduction
Aromatic amines constitute one of the most extensively studied classes of chemical carcinogens (1-3). Some of these compounds, specifically 4-aminobiphenyl, 2-naphthylamine, and benzidine, are established human bladder carcinogens. A common feature of the strongly carcinogenic arylamines is that they are composed of more than one aromatic ring system. Monocyclic aromatic amines are generally considered to be less biologically active, since experimental animal models usually demonstrate only weak carcinogenic activity for most of them. However, occupational studies have implicated o-toluidine as a human bladder carcinogen in a case of reasonably well-defined exposure (4). Further indication that monocyclic aromatic amine exposure might constitute a human health risk comes from a recent non-occupational epidemiologic study (5) of ethyl- and dimethylaniline exposure and bladder cancer risk in Los Angeles County that found that three of the nine isomeric amines, 3,5-DMA1, 2,6-dimethylaniline (2,6-DMA), and 3-ethylaniline (3-EA), were significantly and independently associated with bladder cancer incidence. When compared to the other two, 3,5-DMA was found to have the highest DNA binding index in experimental animals (6). At least one study has found that 3,5-DMA exhibits mutagenic activity in the presence of metabolic activation (7), but others have failed to confirm this finding (8, 9).
Like other aromatic amines (10,11), 3,5-DMA has the potential to undergo metabolic transformation through cytochrome P450-mediated N-hydroxylation. The observation that synthetic N-hydroxy-3,5-DMA is a direct-acting mutagen (12) supports the hypothesis that N-hydroxylation is an activating step. O-acetylation of the hydroxylamine produces a reactive derivative of 3,5-DMA that yields the DNA adduct dG-C8−3,5-DMA in vitro (12). Our previous study with experimental animals demonstrates that 3,5-DMA has the ability to bind to DNA in vivo (6) but did not provide information regarding the structure of the adducts formed. The present study was undertaken to determine the structures of all major adducts formed in DNA through a mechanism involving the intermediacy of a nitrenium ion, using N-AcO-3,5-DMA as the source of electrophilic reactant, with the ultimate goal of comparing those adducts to the ones formed in vivo.
Materials and methods
Caution
The following chemicals are hazardous and should be handled carefully: 3,5-DMA, pyruvonitrile, triethylamine.
Chemicals and Enzymes
Alkaline phosphatase (AP) from calf intestine was obtained from Roche (Indianapolis, IN). Nuclease P1 (NP1) was obtained from Calbiochem (San Diego, CA). Deoxyribonucleosides (dG, dA, and dC), ct-DNA, and other chemicals and enzymes were purchased from Sigma-Aldrich (St. Louis, MO).
Instrumentation
The MS/MS analyses were performed on an Agilent 1200 series XCT Ultra ion-trap mass spectrometer with a Chip-Cube interface. The pumping system included a G1376A binary nano pump and G2226 binary capillary pump, a G1379B vacuum degasser, a G I 327A auto-sampler, and the Agilent ChemStation data system (Agilent Technologies, San Francisco, USA). A G4240−62001 HPLC-Chip (ZORBAX 80SB-C18, Agilent Technologies) was used with a loading flow rate of 4 ML/minute from the capillary pump and an elution flow rate of 0.4 ML/minute from the nano pump. The HPLC mobile phases consisted of water (A) and acetonitrile (B) with a gradient starting at 95% A:5% B for 5 min followed by a linear increase of solvent B to 60% from 5 to 30 minutes. Loading was with an isocratic mobile phase of 97% A:3% B.
The mass spectrometer was operated in the positive ion mode with an ionization voltage of −1700 V and drying gas temperature of 320°C. Nitrogen was used as drying gas at a flow rate of 6 L/min. Helium was used as the collision gas for the tandem mass spectrometric experiments. Fragmentation was induced with resonant excitation amplitude of 1.0−1.2 V, following isolation of the ion of interest over a mass window of 4 Da. The MSn experiments were performed using an ion charge control (ICC) facility to automatically adjust the accumulation time as the ion abundance changed.
High-resolution mass spectrometry experiments were performed on an Agilent 1200 series LC-TOF mass spectrometer with a standard ESI interface with dual nebulizers for simultaneous introduction of analyte stream and reference masses. The mass spectrometer was operated in positive ion mode at an ionization voltage of −4000 V and source temperature of 325 °C. Nitrogen was used as nebulizer gas at 36 psi and drying gas at a flow rate of 12 L/min.
The NMR spectra were measured on a Varian 500 or Bruker 400 MHz spectrometer. The samples were dissolved in Me2SO-d6, and the solvent was used as an internal reference standard. All chemical shifts are given in ppm.
Synthesis of Nucleoside Adducts
The nucleoside adducts from 3,5-DMA were prepared by reacting dG, dA, or dC with N-AcO-3,5-DMA as described for other alkylaniline adducts (12). Briefly, equivalent molar ratios of N-OH-3,5-DMA (823 mg, 6 mmol) and triethylamine (830 μL, 6 mmol) were dissolved in THF (25 mL) at −45 °C. An equivalent of pyruvonitrile (420 μL, 6 mmol) was added to yield N-AcO-3,5-DMA, which was not isolated. After 90 min, dG (668 mg, 2.5 mmol), dA (628 mg, 2.5 mmol), or dC (568 mg, 2,5 mmol) in water (25 mL) and triethylamine (345 μL, 2.5 mmol) was added and incubated at 37°C for overnight. The reactions were repeated several times as needed to acquire enough product for NMR analysis. Based on the amount of product finally purified the yields are estimated at 2%, 0.3%, 0.5%, and 0.3% for dG-C8−3,5-DMA, dA-N6-3,5-DMA, dA-C8−3,5-DMA, and dC-C5−3,5-DMA, respectively.
Purification was performed with combined reaction mixtures that were evaporated and reconstituted in water. After extraction with methylene chloride, the aqueous phase was fractionated by column chromatography on reversed phase C18 modified silica, using a 10 to 100% step gradient (in 10% steps) of aqueous methanol. The 30−50% methanol fractions were combined and subjected to Sephadex LH-20 (GE Healthcare, Piscataway, USA) column chromatography using a step gradient of methanol in water (10−100%) as before. The adducts derived from dA and dC were further purified by preparative reversed phase HPLC, which was performed on a 5 μm, 2.1 × 250 mm reversed phase C18 column (LEJNA 5 C18, Phenomenex). The column was coupled to the Agilent 1100 HPLC system.
DNA Modification
The modification of DNA with 3,5-DMA was conducted by reacting ct-DNA with N-AcO-3,5-DMA as described previously (13). DNA concentration was calculated from absorbance at 260 nm (1 OD = 50 μg/mL), and purity by UV ratios of 260/280 nm (1.7−2.0 OD indicating low contamination).
Enzymatic digestion of DNA was carried out as described (14). The final volume of the reaction was ∼200 μL. After incubation, the sample was centrifuged at 12000 × g for 5 min and the supernatant was applied to a preconditioned Waters Sep-Pak C18 cartridge. The column was washed with a mixture of 5% methanol in water and eluted with methanol. The methanolic eluent was collected and evaporated under a stream of nitrogen and the residue was reconstituted with methanol for HPLC/MS.
Results
N-AcO-3,5-DMA was used as an electrophilic synthon to generate 3,5-DMA adducts from dG, dA, dC, and DNA. The adducts formed upon reaction with deoxyribonucleosides were isolated by chromatography in sufficient amounts and in sufficient purity to produce high quality spectral data. High resolution ESI-MS, 1H-NMR (collected in Table 1), 13C-NMR (Table 2), DEPT, COSY, HSQC and HMBC NMR analyses served as the basis for structure elucidation. The adducts formed upon reaction of N-AcO-3,5-DMA with DNA were determined using HPLC with selected ion monitoring MS after enzymatic hydrolysis of the modified DNA to deoxyribonucleosides as shown in Figure 1. Adducts formed by individual deoxyribonucleosides were also monitored by HPLC/MS during the purification process at which times MS2 experiments were conducted. These experiments confirmed that the ions observed in all the MS spectra at [M+H-116]+ derived from fragmentation of the quasi-molecular ion.
Table 1.
1H-NMR Chemical Shifts of Adducts dG-C8−3,5-DMA, dA-C8−3,5-DMA, dA-N6-3,5-DMA, and dC-C5−3,5-DMA in DMSO-d6a,b
| dG-C8−3,5-DMA | dA-N6-3,5-DMA | dA-C8−3,5-DMA | dC-C5−3,5-DMA | |
|---|---|---|---|---|
| H-1′ | 6.30 (1H, m) | 6.37 (1H, m) | 6.52 (1H, m) | 6.30 (1H, m) |
| H-2′ | 2.50 (1H, m) | 2.79 (1H, m) | 2.71 (1H, m) | 2.13 (1H, o) |
| H-2″ | 1.98 (1H, m) | 2.27 (1H, m) | 2.16 (1H, m) | 1.95 (1H, m) |
| H-3′ | 4.40 (1H, bs) | 4.42 (1H, bs) | 4.45 (1H, bs) | 4.19 (1H, bs) |
| H-4′ | 3.90 (1H, m) | 3.89 (1H, m) | 3.95 (1H, m) | 3.77 (1H, m) |
| H-5′ | 3.74 (1H, o) | 3.63 (1H, m) | 3.72 (1H, o) | 3.50 (1H, o) |
| H-5″ | 3.74 (1H, o) | 3.52 (1H, m) | 3.72 (1H, o) | 3.50 (1H, o) |
| OH-3′ | 5.33 (1H, m) | 5.34 (1H, bs) | 5.39 (1H, bs) | 5.23 (1H, m) |
| OH-5′ | 5.90 (1H, m) | 5.29 (1H, bs) | 6.03 (1H, bs) | 4.95 (1H, m) |
| N1H | 10.58 (1H, s) | - | - | - |
| N2-H2 | 6.35 (2H, s) | - | - | - |
| H-2 | - | 8.06 (1H, s) | 8.02 (1H, s) | - |
| H-8 | - | 8.39 (1H, s) | - | - |
| N6-H/N6-H2 | - | 8.94 (1H, s) | 6.91 (2H, s) | - |
| H-6 | - | - | - | 7.74 (1H, s) |
| N4-H′ | - | - | - | 6.49 (1H, bs) |
| N4-H″ | - | - | - | 7.36 (1H, bs) |
| ArH-2,6 | 7.32 (2H, s) | 6.30 (2H, s) | 7.52 (2H, s) | 6.17 (2H, s) |
| ArH-4 | 6.53 (1H, s) | - | 6.59 (1H, s) | 6.29 (1H, s) |
| ArCH3 | 2.22 (6H, s) | 1.95 (6H, s) | 2.25 (6H, s) | 2.13 (6H, o) |
| ArNH/ArNH2 | 8.45 (1H, s) | 4.88 (2H, s) | 8.87 (1H, s) | 6.81 (1H, s) |
m, multiplet; s, singlet; bs, broad singlet; o, overlapped peaks.
Spectra recorded on a Bruker 400 NMR spectrometer.
Table 2.
13C-NMR Chemical Shifts of Adducts dG-C8−3,5-DMA, dA-C8−3,5-DMA, dA-N6-3,5-DMA, and dC-C5−3,5-DMA in DMSO-d6a
| dG-C8−3,5-DMA | dA-N6-3,5-DMA | dA-C8−3,5-DMA | dC-C5−3,5-DMA | |
|---|---|---|---|---|
| C-1′ | 82.6 | 84.2 | 83.2 | 85.1 |
| C-2′ | 38.2 | 39.8 | 38.3 | 40.6 |
| C-3′ | 71.1 | 71.1 | 71.4 | 70.1 |
| C-4′ | 87.0 | 88.1 | 87.5 | 87.3 |
| C-5′ | 61.2 | 62.0 | 61.5 | 61.5 |
| C-2 | 152.7 | 152.4 | 149.9 | 154.4 |
| C-4 | 149.4 | 148.4 | 148.5 | 164.4 |
| C-5 | 112.2 | 119.6 | 116.4 | 109.3 |
| C-6 | 155.7 | 154.4 | 153.3 | 138.1 |
| C-8 | 140.7 | 139.6 | 146.5 | - |
| ArC-1 | 143.3 | 147.1 | 140.2 | 147.2 |
| ArC-2,6 | 115.0 | 113.2 | 116.1 | 111.1 |
| ArC-3,5 | 137.3 | 136.3 | 137.6 | 137.9 |
| ArC-4 | 122.2 | 124.5 | 123.1 | 119.5 |
| ArCH3 | 21.2 | 18.4 | 21.2 | 21.3 |
Spectra recorded on a Bruker 400 NMR spectrometer. Resonances were assigned on the basis of HSQC spectra and by comparison with known chemical shifts for normal deoxyribonucleotides.
Figure 1.
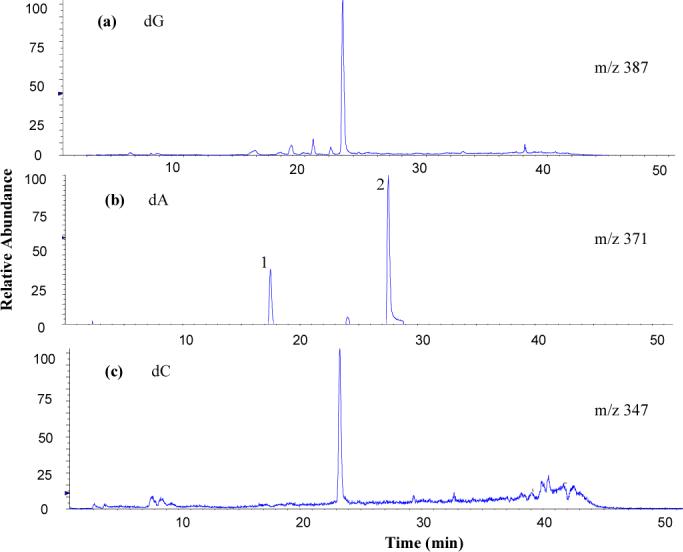
Extracted ion chromatogram of the [M+H]+ adduct ions of (a) dG-3,5-DMA, (b) dA-3,5-DMA, and (c) dC-3,5-DMA derived from digested ct-DNA modified in vitro with N-AcO-3,5-DMA at m/z 387, m/z 371, and m/z 347, respectively. Major peak area ratio is 67:9:21:3 (dG:dA1:dA2:dC).
Adducts from reaction with dG (dG-C8−3,5-DMA)
The HPLC/MS profile from N-AcO-3,5-DMA dG indicated the presence of one major adduct. The HR-ESI-MS showed the quasi-molecular ion [M+H]+ at m/z 387.1784 (calcd. 387.1781, mass difference of 0.8 ppm), corresponding to the molecular formula C18H23N6O4, as expected from an adduct of dG with 3,5-DMA. In addition, the detection of an intense ion at m/z 271.1309 (calcd. 271.1307) is consistent with loss of the deoxyribosyl fragment from the parent ion. The adduct was identified as dG-C8−3,5-DMA, the product of C8 substitution through the arylamine nitrogen, by comparing 1H-NMR and 13C-NMR data with those reported in the literature (12).
Formation of other, relatively minor, adducts was apparent when the reaction products were analyzed by HPLC/MS. Their low abundance precluded isolating sufficient amounts for NMR spectroscopic analysis and no further effort was made to identify them.
Adducts from reaction with dA
HPLC/MS of the dA reaction mixture indicated the presence of two major adducts. They were characterized as N-(deoxyadenosin-8-yl)-3,5-dimethylaniline (dA-C8−3,5-DMA, peak 1 in Figure 1) and 4-(deoxyadenosin-N6-yl)-3,5-dimethylaniline (dA-N6-3,5-DMA, peak 2) based on the following analysis:
(1) dA-N6-3,5-DMA (dA adduct 1)
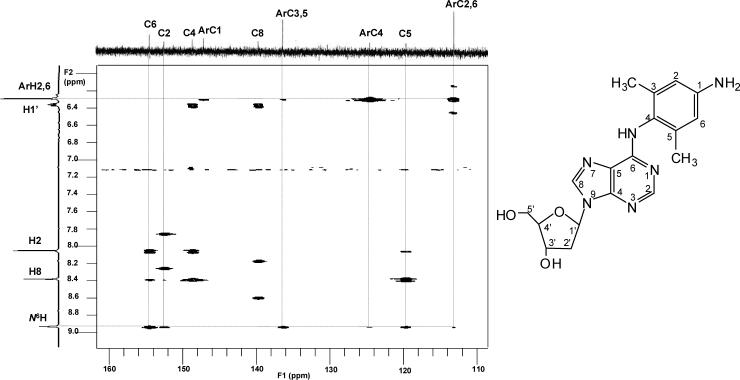
The HR-ESI-MS showed the quasi-molecular ion [M+H]+ at m/z 371.1828 (calcd. 371.1832, mass difference of −1.1 ppm), corresponding to the molecular formula C18H23N6O3, as expected from an adduct of dA with 3,5-DMA. In addition, the detection of an intense ion [M-116+H]+ at m/z 255.1351 (calcd. 255.1358) was consistent with fragment loss of the deoxyribose moiety. The assignment of this formula was further supported by the 1H-NMR and 13C-NMR data. In the 1H–NMR spectrum, both purine C-H resonances were present, as well as a 1-proton resonance at δ 8.94 assigned to the exocyclic N6-H. The arylamine ring proton resonances ArH-2 and ArH-6 were observed but the resonance attributable to ArH-4 was not. Meanwhile, the presence of a two-proton singlet at δ 4.88, assigned to the arylamine NH2, indicated that the arylamine nitrogen remained unsubstituted. The only connectivity consistent with these observations is between the exocyclic nitrogen of the adenosine ring and the arylamine C-4. This conclusion was further confirmed from the 1H-1H COSY and HMBC results. In the 1H-1H COSY spectrum, the signal of N6H (δ 8.94, 1H, s) showed correlations with the signals of H-2 (δ 8.06, 1H, s), ArH-2,6 (δ 6.30, 2H, s), and Ar-CH3 (δ 1.95, 6H, s). In the HMBC spectrum (Figure 2), the signal of N6H had peaks correlated with C-6 (δ 154.4), C-2 (δ 152.4), ArC-4 (δ 136.3), ArC-3,5 (δ 124.5), and C-5 (δ 119.6). From the above evidence, dA adduct 1 was determined to be 4-(deoxyadenosin-N6-yl)-3,5-dimethylaniline. The full assignments of carbon and proton signals are summarized in Tables 1 and 2.
Figure 2.
Partial HMBC spectrum of dA adduct 1, dA-N6-3,5-DMA.
(2) dA-C8−3,5-DMA (dA adduct 2)
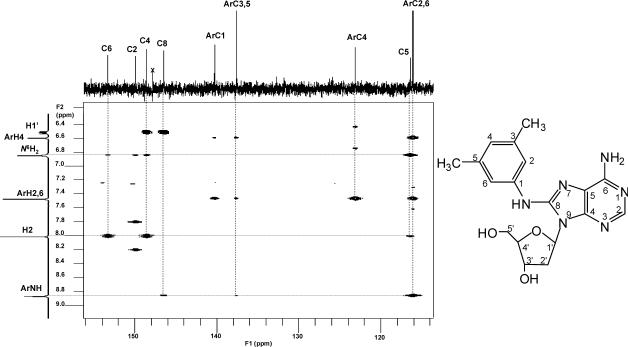
The HR-ESI-MS showed the quasi-molecular ion [M+H]+ at m/z 371.1829 (calcd. 371.1832, mass difference of −0.8 ppm), corresponding to the molecular formula C18H23N6O3, as expected from an adduct of dA with 3,5-DMA. In addition, the detection of an intense ion [M-116+H]+ at m/z 255.1351 (calcd. 255.1358) was consistent with fragment loss of the deoxyribose moiety. The assignment of this formula was further supported by the 1H-NMR and 13C-NMR data. In the 1H-NMR spectrum, the purine H-2 resonance was present but the resonance expected for H-8 was not. Meanwhile, all the arylamine ring proton resonances were observed, as well as a 1-proton resonance at δ 8.87 assigned to the arylamine NH. 13C-NMR spectra showed the presence of all the expected carbon resonances. From the DEPT spectrum it was clear that the C-8 nucleus, which was shifted downfield from δ 139.6 in dA to δ 146.5, was no longer bound to a proton. These results indicate the formation of a covalent bond between the arylamine nitrogen and the C-8 atom of the adenosine ring. This conclusion was further confirmed from the 1H-1H COSY and HMBC results. In the 1H-1H COSY spectrum, the proton signal of ArH-2,6 (δ 7.52, 2H, s) showed correlations with the proton signals of ArH-4 (δ 6.59, 1H, s) and Ar-CH3 (δ 2.25, 6H, s), while the signal of ArH-4 also correlated with the signal of Ar-CH3. In the HMBC spectrum (Figure 3), the signal of Ar-NH (δ 8.87, 1H, s) had correlations with C-8 (δ 146.5) and ArC-2,6 (δ 116.1). Based on the above data, dA adduct 2 was determined to be N-(deoxyadenosin-8-yl)-3,5-dimethylaniline. The full assignments of carbon and proton signals are summarized in Table 1 and 2.
Figure 3.
Partial HMBC spectrum of dA adduct 2, dA-C8−3,5-DMA.
Adducts from the reaction with dC
The HPLC/MS profile of products from reaction of N-AcO-3,5-DMA with dC indicated the presence of one adduct. It was characterized as N-(deoxycytidin-5-yl)-3,5-dimethylaniline (dC-C5−3,5-DMA) based on the following spectroscopic evidence.
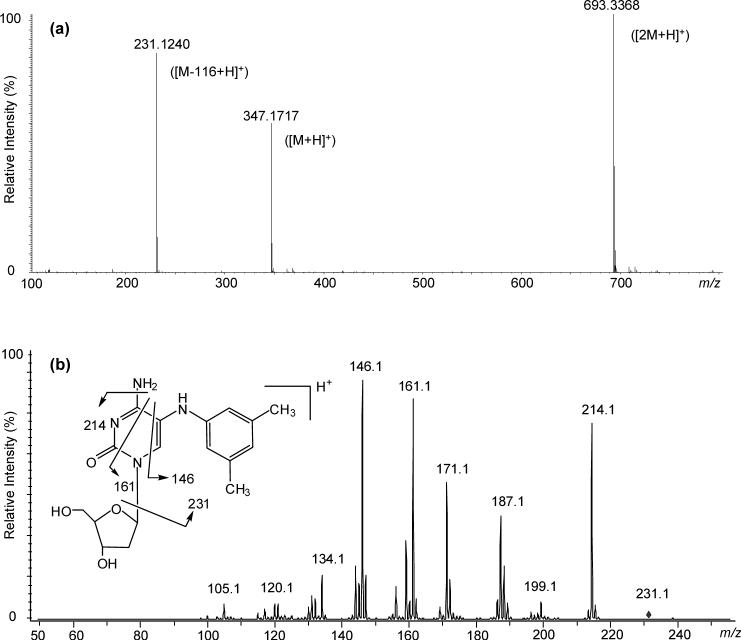
The HR-ESI-MS showed the quasi-molecular ion [M+H]+ at m/z 347.1717 (calcd. 347.1719, mass difference of −0.6 ppm) corresponding to the molecular formula C17H23N4O4, as expected from an adduct of dC with 3,5-DMA. In addition, the detection of an intense ion [M-116+H]+ at m/z 231.1240 (calcd. 231.1246) is consistent with fragment loss of the deoxyribose moiety (Figure 4a). The assignment of this formula was further supported by the 1H-NMR and 13C-NMR data. In the 1H-NMR spectrum, all the arylamine ring proton resonances were observed, as well as a 1-proton exchangeable resonance at δ 6.81 assigned to the arylamine NH. Meanwhile, the pyrimidine H-6 was present but the resonance at δ 6.05 (in deoxycytidine) corresponding to the nonexchangeable H-5 was not. D2O exchange showed that, in addition to the protons at δ 6.81 (Ar-NH), δ 5.23 (OH-3’), and δ 4.95 (OH-5’), there are two more exchangeable protons at δ 7.36 and δ 6.49. Their assignment and the implications thereof for determination of the tautomeric structure of the pyrimidine ring are described below.
Figure 4.
(a) HR-ESI-MS spectrum of dC-C5−3,5-DMA, (b) MS3 (347→231) spectrum of dC-C5−3,5-DMA.
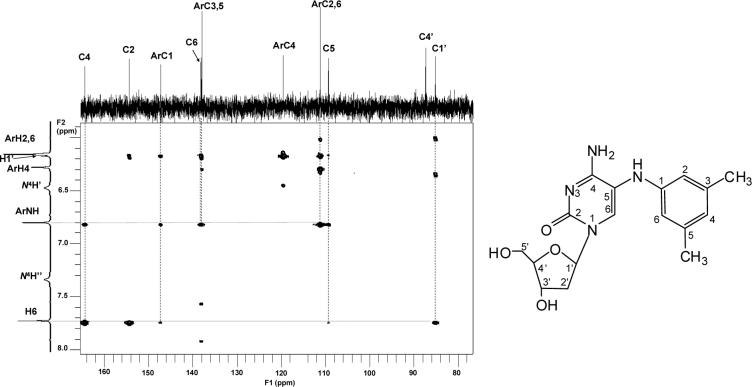
The results given above indicated the formation of a covalent bond between the arylamine nitrogen and the C-5 atom of the cytidine ring. The MS3 spectrum (Figure 4b), is fully consistent with this conclusion, particularly with respect to the amine-nucleobase linkage. The structure was further confirmed by 1H-1H COSY and HMBC results. In the 1H-1H COSY spectrum, the signal of ArH-4 (δ 6.29, 1H, s) showed correlations with the signals of ArH-2,6 (δ 6.17, 2H, s) and Ar-CH3 (δ 2.13, 6H, overlapped), while Ar-NH (δ 6.81, 1H, s) showed correlations with H-6 (δ 7.74, 1H, s). The signals at δ 6.49 and δ 7.36 were correlated. In the HMBC spectrum (Figure 5), the signal of Ar-NH had peaks correlated with C-4 (δ 164.4), ArC-1 (δ 147.2), C-6 (δ 138.1), ArC-3,5 (δ 137.9), ArC-2,6 (δ 111.1), and C-5 (δ 109.3), while H-6 correlated with C-4 (δ 164.4), C-2(154.4), ArC-1 (δ 147.2), C-5 (δ 109.3) and C-1’ (δ 85.1). Based on the above analyses, the dC adduct was determined to be N-(deoxycytidin-5-yl)-3,5-dimethylaniline. The full assignments of carbon and proton signals are summarized in Table 1 and 2.
Figure 5.
Partial HMBC spectrum of dC adduct, dC-C5−3,5-DMA.
The structure shown in Figures 4 and 5 represents one of two possible proton tautomers, the other having an imine group at position 4 of the cytosine ring and a proton on N3. The alternate imine tautomer offers a ready explanation for the presence of two exchangeable proton resonances at 6.49 and 7.36 but the amine tautomer could also give rise to two distinct resonances if the two protons are non-equivalent. To distinguish between the two tautomers, a 1H-15N HSQC spectrum was recorded. In that spectrum, both proton resonances were correlated with a single nitrogen resonance, a result that essentially rules out the imine tautomer. Non-equivalence of the two protons of the amino group presumably arises through hydrogen bonding of one to the arylamine nitrogen.
Adducts from the reaction with DNA
N-AcO-3,5-DMA was used to react with ct-DNA in vitro. Following solvent extractions and enzymatic hydrolysis of the DNA to deoxyribonucleosides, the HPLC/MS profiles of the reaction products showed the same pattern as those obtained from reaction of NAcO-3,5-DMA with dG, dA, and dC.
Discussion
Metabolic activation of aromatic amines is often presumed to occur by the well understood pathway of P450-mediated N-hydroxylation followed by phase II conjugation of the hydroxyl group. In this context, we undertook to determine the adducts formed when N-AcO-3,5-DMA reacted with DNA, choosing the acyloxyamine as a plausible and likely product of the foregoing metabolic pathway. In common with previous studies of reactive alkylaniline derivatives, the adduct formed by linkage of the aniline amino group with C-8 of dG was a major product (12, 15). This adduct structure can be understood as the result of formation, by heterolysis of the acyloxy-amine bond, of an electrophilic nitrenium ion with subsequent attack on a nucleophilic center of the nucleic acid. In reaction with dA, we found that N-AcO-3,5-DMA also formed a C-8 adduct through the amino group. Formation of the other dA adduct, in which a bond is formed between the exocyclic amino group of adenine and the para-carbon atom of 3,5-DMA, demonstrates that electrophilicity of the intermediate nitrenium ion is not centered exclusively on the nitrogen atom but is actually delocalized to a considerable extent. Formation of the analogous adduct by 2,6-DMA has also been reported (15).
An unexpected feature of the present study is the observation of a dC adduct. To our knowledge, no adduct of an aromatic amine with dC has previously been described although there is a report of closely related adducts formed by acetamidostilbene (16). Of further interest is the adduct structure, which results from electrophilic attack of the alkylaniline amino group at C-5 of cytosine. Addition reactions at the C5−C6 bond of cytosine are well known but are almost unprecedented with respect to electrophilic drug or other xenobiotic metabolites. The only example found in the literature is that of chlorambucil, which was reported to form a C5 adduct in very low yield compared to the N3 adduct (17). In contrast, we found that the C5 adduct was predominant, which may be an indication that the two reactants have very different electrophilic properties.
The relative yields of each adduct from reactions of N-AcO-3,5-DMA with DNA were not determined quantitatively, but it was apparent from the synthesis reactions that this dC adduct, as well as the dA adducts, were formed in considerably lower yields than the dG adduct when the nucleophilic partner was a free deoxyribonucleoside. The same conclusion applies to relative adduct yields from DNA if the mass spectrometer response to the different adducts is assumed to be essentially constant. This adduct spectrum is significantly different from that observed for reaction of N-AcO-2,6-DMA with DNA, in which case the dG-C8 adduct is the least abundant (15). Whether the difference reflects differences in reactivity or results from steric differences is unclear at present.
The observation here that N-AcO-3,5-DMA produces multiple adducts in DNA supports the previous conclusion based on experiments with N-AcO-2,6-DMA that nitrenium ions derived from single-ring amines may react rather indiscriminately with DNA as compared to nitrenium ions from multi-ring amines (15). The significance of these findings for the biological plausibility of nitrenium ion intermediates from alkylanilines being the critical mediators of genotoxicity in vivo remains unclear but, generally, more selective electrophilic intermediates tend to be more biologically active. Results unequivocally demonstrate that alkylaniline nitrenium ions are capable of forming DNA adducts entirely analogous to those produced by carcinogenic aromatic amines. Nevertheless, there is as yet no evidence that these same adducts are formed in vivo and whether they are remains a critical issue regarding biological activity of alkylanilines.
Resolving this issue was a principal factor in undertaking the present study. Previously, using 14C-labeled alkylanilines we found that 3,5-DMA, as well as 2,6-DMA and 3-ethylaniline, became bound to DNA of several tissues when the amines were administered to mice at relatively low doses (ca. 100 Mg/kg). A higher dose study of 2,6-DMA similarly found isotope bound to liver DNA and DNA in certain epithelial tissues of the nasal cavity (18). Neither study provides evidence regarding the structure of the adducts formed because quantitation of isotope bound to purified DNA was the analytical endpoint of each. With adducts of known structure as standards, it will now be possible to chromatographically analyze digests of DNA from animals given [14C]3,5-DMA and confirm that adducts different from the standards are present therein. Chromatographic identity with the standards would suggest that the same adducts are present, but such findings would require further proof of identity.
Supplementary Material
Acknowledgments
This work was supported by grants PO1-ES006052 and P30-ES002109 from NIEHS. We thank Agilent Technologies for access to the XCT Ultra ion trap mass spectrometer, and Dr. Julie Marr (Agilent) for helpful discussions.
Footnotes
The abbreviations used are: 3,5-DMA, 3,5-dimethylaniline; 2,6-DMA, 2,6-dimethylaniline; dG, 2’-deoxyguanosine; dA, 2’-deoxyadenosine; dC, 2’-deoxycytidine; N-AcO-3,5-DMA, N-acetoxy-3,5-dimethylaniline; ct-DNA, calf thymus DNA; dG-C8−3,5-DMA, N-(deoxyguanosin-8-yl)-3,5-dimethylaniline; dA-C8−3,5-DMA, N-(deoxyadenosin-8-yl)-3,5-dimethylaniline; dA-N6-3,5-DMA, 4-(deoxyadenosin-N6-yl)-3,5-dimethylaniline; dC-C5−3,5-DMA, N-(deoxycytidin-5-yl)-3,5-dimethylaniline; HR-MS, high resolution mass spectrometry; ESI-MS, electrospray ionization mass spectrometry; DEPT, distortionless enhancement by polarization transfer; HSQC, two-dimensional heteronuclear single-quantum correlation; COSY, 1H-1H two-dimensional correlation spectroscopy; HMBC, heteronuclear multiple-bond correlation.
Supporting Information Available. 1H-1H COSY spectra of the dA and dC adducts and partial 1H-15N HSQC of the dC adduct. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Woo YT, Lai DY. Aromatic amino and nitro-amino compounds and their halogenated derivatives. In: Bingham E, Cohrssen B, Powell CH, editors. Patty's Toxicology. Wiley; New York: 2001. pp. 969–1105. [Google Scholar]
- 2.Talaska G. Aromatic amines and human urinary bladder cancer: exposure sources and epidemiology. J. Environ. Sci. Health C Environ. Carcinog. Ecotoxicol. Rev. 2003;21:29–43. doi: 10.1081/GNC-120021372. [DOI] [PubMed] [Google Scholar]
- 3.Vineis P, Pirastu R. Aromatic amines and cancer. Cancer Causes Control. 1997;8:346–355. doi: 10.1023/a:1018453104303. [DOI] [PubMed] [Google Scholar]
- 4.Markowitz SB, Levin K. Continued epidemic of bladder cancer in workers exposed to ortho-toluidine in a chemical factory. J. Occup. Environ. Med. 2004;46:154–160. doi: 10.1097/01.jom.0000111602.76443.15. [DOI] [PubMed] [Google Scholar]
- 5.Gan J, Skipper PL, Gago-Domingues M, Arakawa K, Ross RK, Yu MC, Tannenbaum SR. Alkylaniline-hemoglobin adducts and risk of non-smoking-related bladder cancer. J. Natl. Cancer Inst. 2004;96:1425–1431. doi: 10.1093/jnci/djh274. [DOI] [PubMed] [Google Scholar]
- 6.Skipper PL, Trudel LJ, Kensler TW, Groopman JD, Egner PA, Liberman RG, Wogan GN, Tannenbaum SR. DNA adduct formation by 2,6-dimethyl-, 3,5-dimethyl, and 3-ethylaniline in vivo in mice. Chem. Res. Toxicol. 2004;19:1086–1090. doi: 10.1021/tx060082q. [DOI] [PubMed] [Google Scholar]
- 7.Zeiger E, Anderson B, Haworth S, Lawlor T, Mortelmans K. Salmonella mutagenicity test: IV. Results from the testing of 300 chemicals. Environ. Mol. Mutagen. 1988;11(suppl 12):1–157. [PubMed] [Google Scholar]
- 8.Zimmer D, Mazurek J, Petzold G, Bhuyan BK. Bacterial mutagenicity and mammalian cell DNA damage by several substituted anilines. Mutation Res. 1980;77:317–326. doi: 10.1016/0165-1218(80)90003-8. [DOI] [PubMed] [Google Scholar]
- 9.Nohmi T, Yoshikawa K, Nakadate M, Miyata R, Ishidate M., Jr. Mutations in Salmonella typhimurium and inactivation of Bacillus subtilis transformating DNA induced by phenylhydroxylamine derivatives. Mutation Res. 1984;136:159–168. doi: 10.1016/0165-1218(84)90049-1. [DOI] [PubMed] [Google Scholar]
- 10.Kadlubar FF. DNA adducts of carcinogenic aromatic amino. In: Hemminki K, Dipple A, Shuker DEG, Kadlubar FF, Segerback D, Bartsch H, editors. DNA Adducts Identification and Biological Significance. IARC Scientific Publication; Lyon, France: 1994. pp. 199–216. [PubMed] [Google Scholar]
- 11.Gan J, Skipper PL, Tannenbaum SR. Oxidation of 2,6-Dimethylaniline by recombinant human cytochrome P450 and human liver microsomes. Chem. Res. Toxicol. 2001;14:672–677. doi: 10.1021/tx000181i. [DOI] [PubMed] [Google Scholar]
- 12.Marques MM, Mourato LLG, Amorim MT, Santos MA, Melchior WB, Jr., Beland FA. Effect of substitution site upon the oxidation potentials of alkylanilines, the mutagenicities of N-hydroxyalkylanilines, and the conformations of alkylaniline-DNA adducts. Chem. Res. Toxicol. 1997;10:1266–1274. doi: 10.1021/tx970104w. [DOI] [PubMed] [Google Scholar]
- 13.Jones CR, Sabbioni G. Identification of DNA adducts using HPLC/MS/MS following in vitro and in vivo experiments with arylamines and nitroarenes. Chem. Res. Toxicol. 2003;16:1251–1263. doi: 10.1021/tx020064i. [DOI] [PubMed] [Google Scholar]
- 14.Zayas B, Stillwell SW, Wishnok JS, Trudel LJ, Skipper PL, Yu MC, Tannenbaum SR, Wogan GN. Detection and quantification of 4-ABP adducts in DNA from bladder cancer patients. Carcinogenesis. 2007;28:342–349. doi: 10.1093/carcin/bgl142. [DOI] [PubMed] [Google Scholar]
- 15.Gonçalves LL, Beland FA, Marques MM. Synthesis, characterization, and comparative 32P-postlabeling efficiencies of 2,6-dimethylaniline-DNA adducts. Chem. Res. Toxicol. 2001;14:165–174. doi: 10.1021/tx0002031. [DOI] [PubMed] [Google Scholar]
- 16.Scribner NK, Scribner JD, Smith DL, Schram KH, McCloskey JA. Reactions of the carcinogen N-acetoxy-4-acetamidostilbene with nucleosides. Chem.-Biol. Interact. 1979;26:27–46. doi: 10.1016/0009-2797(79)90091-7. [DOI] [PubMed] [Google Scholar]
- 17.Florea-Wang D, Haapala E, Mattinen J, Hakala K, Vilpo J, Hovinen J. Reactions of N,N-bis(2-chloroethyl)-p-aminophenylbutyric acid (Chloroambucil) with 2’deoxycytidine, 2’-deoxy-5-methylcytidine, and thymidine. Chem. Res. Toxicol. 2004;17:383–391. doi: 10.1021/tx034233q. [DOI] [PubMed] [Google Scholar]
- 18.Short CR, Joseph M, Hardy ML. Covalent binding of [14C]-2,6-dimethylaniline to DNA of rat liver and ethmoid turbinate. J. Toxicol. Environ. Health. 1989;27:85–94. doi: 10.1080/15287398909531280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.