Abstract
We have created a panel of recombinant HIV-1 infectious clones containing common patterns of reverse transcriptase (RT) mutations responsible for resistance to each of the currently available nucleoside reverse transcriptase inhibitors (NRTI), and we have submitted the panel to the National Institutes of Health AIDS Research and Reference Reagent Programme. Testing the activity of new antiretroviral compounds against this panel of drug-resistant clones will determine their relative activity against many clinically relevant NRTI-resistant viruses.
Although the large number of drug-resistant HIV-1 mutations makes the development of new non-cross-resistant antiretroviral inhibitors challenging, HIV-1 strains from heavily treated individuals often develop common, co-linear combinations of these mutations [1]. We have created a panel of recombinant infectious molecular clones containing combinations of mutations that confer resistance to multiple nucleoside reverse transcriptase inhibitors (NRTI). We hypothesize that NRTI that maintain full activity against the clones in this panel will also be active against the much larger number of NRTI-resistant variants currently found in individuals failing therapy.
In previous studies, we identified common combinations of NRTI-resistance mutations by determining the extent of co-variation between all pairs of NRTI-resistance mutations [2], by determining the frequency of specific NRTI-resistance mutations in a clinical database [3], and by using statistical clustering approaches [4]. We used the data from these studies and from recent clinical trials [5–7] to select cryopreserved plasma samples containing previously sequenced HIV-1 isolates with specific patterns of NRTI-resistance mutations.
HIV-1 RNA was extracted using the QIAamp Viral RNA Kit (QIAGEN Inc., Valencia, CA, USA) from 500 μl of cryopreserved plasma and amplified by reverse transcriptase—polymerase chain reaction to create an amplicon encompassing RT codons 23–312 (871 bp). Amplified RT fragments were ligated into an RT-deleted pNL4-3 vector (pNLPFB digested with Msc1 and PflM1) [8] and transformed with competent Escherichia coli Top10 cells (Invitrogen, Carlsbad, CA, USA). These recombinant clones were transfected into C8166 cells using Lipofectin (Invitrogen). When syncytia were observed, the C8166 cells were co-cultured with SupT1 cells to create high-titer virus stocks as measured by p24 enzyme-linked immunosorbent assay (Perkin Elmer, Boston, MA, USA) and endpoint dilution. The mutations present in each of the clones were then confirmed by sequencing. Virus stocks were then submitted for HIV-1 drug susceptibility testing using the PhenoSense assay (ViroLogic, South San Francisco, CA, USA). In 2002 and 2004, the clones and their associated virus stocks were submitted to the National Institutes of Health AIDS Research and Reference Reagent Programme (Rockville, MD, USA; www.aidsreagent.org, catalog #7384-7395) for use without restriction.
The mutations in the 12 clones and their associated drug susceptibilities are shown in Table 1. Eight clones have reduced susceptibility to each of the six NRTI tested; four have reduced susceptibility to three to five inhibitors. Susceptibilities were not available for emtricitabine, which has a drug-resistance profile identical to lamivudine. Median reductions in susceptibility were greater than 300-fold to lamivudine, 161-fold to zidovudine, 7.6-fold to abacavir, 4.4-fold to stavudine, 3.1-fold to tenofovir, and 2.7-fold to didanosine.
Table 1.
Nucleoside reverse transcriptase inhibitor resistance mutations and drug susceptibility results of the 12 infectious molecular reverse transcriptase clones.a
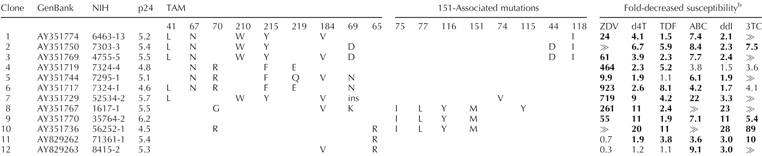 |
ABC, Abacavir; ddI, didanosine; d4T, stavudine; NIH, National Institutes of Health; p24, logarithm of the p24 antigen concentration (pg/ml) of the initial virus stock; TAM, thymidine analog mutation; TDF, tenofovir; 3TC, lamivudine; ZDV, zidovudine.
The complete sequences, list of mutations, and list of drug susceptibility results (including for the non-nucleoside reverse transcriptase inhibitors) for each clone can be found on the NIH AIDS Research and Reference Reagent Programme website (http://www.aidsreagent.org) or on the Stanford Drug Resistance Database (http://hivdb.stanford.edu).
Zalcitibine results are not shown because this drug is rarely used and its resistance profile is similar to that of ddI. Although susceptibilities to emtricitabine, the most recently approved nucleoside reverse transcriptase inhibitor, were not determined, its resistance profile is considered identical to that of 3TC. Results in bold are above the PhenoSense assay clinical cut-off. ⪢indicates that the fold resistance (reduction in drug susceptibility) was greater than the upper limit of assay detection, which is 300-fold for 3TC and approximately 1000-fold for ZDV. Non-nucleoside reverse transcriptase inhibitor resistance mutations at position 103 (K103N) were present in clones 2, and 10, and at position 190 (G190C and G190A) in clones 7 and 11. The ‘ins’ at position 69 for clone 7 indicates the presence of a double amino acid (SS) insertion following a T69S substitution at position 69.
The dynamic range in susceptibility to stavudine, didanosine, and tenofovir is much lower than that to zidovudine, lamivudine, and even abacavir. With the PhenoSense assay, reductions in susceptibility of greater than 1.5-fold to stavudine, didanosine, and tenofovir occur only in isolates with drug-resistance mutations [9,10], and reductions of three to fourfold are associated with markedly decreased antiretroviral activity in vivo [11–13].
The panel contains HIV-1 isolates with four previously described mechanisms of multiple NRTI resistance [14,15]: (i) multiple thymidine analog mutations (TAM) + M184V ± T69D/N (clones 1–6); (ii) multiple TAM + T69 insertion (clone 7); (iii) Q151M-mediated multi-NRTI resistance ± K65R ± M184V (clones 8–10); and (iv) K65R ± M184V ± (clones 11–12). Because the first pattern ±is the most common, it is represented by the greatest number of clones. Three are characterized by the common mutation triad M41L, L210W, and T215Y, and four by D67N, K70R, T215F, and K219Q/E. These four mechanisms of multi-NRTI resistance are partly overlapping — T69 insertions nearly always occur with multiple TAM, and K65R often occurs with Q151M. However, Q151M and particularly K65R rarely occur in combination with TAM [2,16].
New compounds that demonstrate in-vitro antiretroviral activity are usually tested on a range of drug-resistant clinical isolates. However, because no standard sets of clinical drug-resistant isolates have been identified, it is usually not possible to determine the activity of a new compound relative to other experimental compounds and approved drugs. Testing the activity of new compounds against this panel will allow researchers from different laboratories to standardize their results against a set of reference viruses. Infectious molecular clones provide an advantage over primary HIV-1 isolates because primary isolates often contain mixtures of wild-type and mutant viruses that may evolve independently during in-vitro passage and because molecular clones are amenable to biochemical and biophysical studies. The panel we have described will be continuously updated as new NRTI are developed and new patterns of RT mutations associated with multi-NRTI resistance are observed.
Acknowledgments
Sponsorship: E.J., M.J.G., K.M.D, M.A.W., and R.W.S. were partly supported by a grant from the NIAID (National Institutes of Health) AI-46148-03.
References
- 1.Gonzales MJ, Johnson E, Dupnik KM, Imamichi T, Shafer RW. Colinearity of reverse transcriptase inhibitor resistance mutations detected by population-based sequencing. J Acquir Immune Defic Syndr. 2003;34:398–402. doi: 10.1097/00126334-200312010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzales MJ, Wu TD, Taylor J, Belitskaya I, Kantor R, Israelski D, et al. Extended spectrum of HIV-1 reverse transcriptase mutations in patients receiving multiple nucleoside analog inhibitors. AIDS. 2003;17:791–799. doi: 10.1097/01.aids.0000050860.71999.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhee SY, Liu T, Ravela J, Gonzales MJ, Shafer RW. Distribution of human immunodeficiency virus type 1 protease and reverse transcriptase mutation patterns in 4,183 persons undergoing genotypic resistance testing. Antimicrob Agents Chemother. 2004;48:3122–3126. doi: 10.1128/AAC.48.8.3122-3126.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzales MJ, Belitskaya I, Dupnik KM, Rhee SY, Shafer RW. Protease and reverse transcriptase mutation patterns in HIV type 1 isolates from heavily treated persons: comparison of isolates from Northern California with isolates from other regions. AIDS Res Hum Retroviruses. 2003;19:909–915. doi: 10.1089/088922203322493085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallant J, Rodriguez AE, Weinberg W.Early non-response to tenofovir DF (TDF) + abacavir (ABC) and lamivudine (3TC) in a randomized trial compared to efavirenz (EFV) + ABC and 3TC: ESS30009 43rd Interscience Conference on Antimicrobial Agents and ChemotherapyChicago, IL2003[Abstract 1722a]. [Google Scholar]
- 6.Landman R, Peytavin G, Descamps D, Brun Vezinet F, Benech H, Benalisherif A, et al. Low genetic barrier to resistance is a possible cause of early virologic failures in once-daily regimen of abacavir, lamiduvine, and tenofovir: the Tonus Study 11th Conference on Retroviruses and Opportunistic InfectionsSan Francisco, CA2004[Abstract 52]. [Google Scholar]
- 7.Roge BT, Katzenstein TL, Obel N, Nielsen H, Kirk O, Pedersen C, et al. K65R with and without S68: a new resistance profile in vivo detected in most patients failing abacavir, didanosine and stavudine. Antivir Ther. 2003;8:173–182. [PubMed] [Google Scholar]
- 8.Imamichi T, Berg SC, Imamichi H, Lopez JC, Metcalf JA, Falloon J, Lane HC. Relative replication fitness of a high-level 3‘-azido-3’-deoxythymidine-resistant variant of human immunodeficiency virus type 1 possessing an amino acid deletion at codon 67 and a novel substitution (Thr(Gly) at codon 69. J Virol. 2000;74:10958–10964. doi: 10.1128/jvi.74.23.10958-10964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parkin NT, Hellmann NS, Whitcomb JM, Kiss L, Chappey C, Petropoulos CJ. Natural variation of drug susceptibility in wild-type HIV-1. Antimicrob Agents Chemother. 2004;48:437–443. doi: 10.1128/AAC.48.2.437-443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitcomb JM, Parkin NT, Chappey C, Hellmann NS, Petropoulos CJ. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J Infect Dis. 2003;188:992–1000. doi: 10.1086/378281. [DOI] [PubMed] [Google Scholar]
- 11.Miller MD, Margot N, Lu B, Zhong L, Chen SS, Cheng A, Wulfsohn M. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disoproxil fumarate treatment in antiretroviral-experienced patients. J Infect Dis. 2004;189:837–846. doi: 10.1086/381784. [DOI] [PubMed] [Google Scholar]
- 12.Molina JM, Marcelin AG, Pavie J, Merle C, Troccaz M, Leleu G, Calvez V.Didanosine (ddI) in treatment experienced HIV-infected patients: results from a randomized double-blind study (AI454-176 Jaguar) 43rd Interscience Conference on Antimicrobial Agents and ChemotherapyChicago, IL2003[Abstract H-447]. [Google Scholar]
- 13.Shulman NS, Hughes MD, Winters MA, Shafer RW, Zolopa AR, Hellmann NS, et al. Subtle decreases in stavudine phenotypic susceptibility predict poor virologic response to stavudine monotherapy in zidovudine-experienced patients. J Acquir Immune Defic Syndr. 2002;31:121–127. doi: 10.1097/00126334-200210010-00001. [DOI] [PubMed] [Google Scholar]
- 14.Gallant JE, Gerondelis PZ, Wainberg MA, Shulman NS, Haubrich RH, Clair M, et al. Nucleoside and nucleotide analogue reverse transcriptase inhibitors: a clinical review of antiretroviral resistance. Antivir Ther. 2003;8:489–506. [PubMed] [Google Scholar]
- 15.Shafer RW. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin Microbiol Rev. 2002;15:247–277. doi: 10.1128/CMR.15.2.247-277.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faruki H, Sebastian J, Scott J, Stamp J, Lanier E. Changes in prevalence of NRTI resistance associated mutations among clinical isolates from 1999–2003. Antivir Ther. 2004;9:S91. [Google Scholar]


