Abstract
The protein trafficking machinery of eukaryotic cells is employed for protein secretion and for the localization of resident proteins of the exocytic and endocytic pathways. Protein transit between organelles is mediated by transport vesicles that bear integral membrane proteins (v-SNAREs) which selectively interact with similar proteins on the target membrane (t-SNAREs), resulting in a docked vesicle. A novel Saccharomyces cerevisiae SNARE protein, which has been termed Vti1p, was identified by its sequence similarity to known SNAREs. Vti1p is a predominantly Golgi-localized 25-kDa type II integral membrane protein that is essential for yeast viability. Vti1p can bind Sec17p (yeast SNAP) and enter into a Sec18p (NSF)-sensitive complex with the cis-Golgi t-SNARE Sed5p. This Sed5p/Vti1p complex is distinct from the previously described Sed5p/Sec22p anterograde vesicle docking complex. Depletion of Vti1p in vivo causes a defect in the transport of the vacuolar protein carboxypeptidase Y through the Golgi. Temperature-sensitive mutants of Vti1p show a similar carboxypeptidase Y trafficking defect, but the secretion of invertase and gp400/hsp150 is not significantly affected. The temperature-sensitive vti1 growth defect can be rescued by the overexpression of the v-SNARE, Ykt6p, which physically interacts with Vti1p. We propose that Vti1p, along with Ykt6p and perhaps Sft1p, acts as a retrograde v-SNARE capable of interacting with the cis-Golgi t-SNARE Sed5p.
INTRODUCTION
Protein transport and membrane flow in eukaryotic cells is mediated by vesicles that bud from one membrane-bounded compartment and then target to and fuse with the appropriate acceptor compartment (Palade, 1975; Rothman, 1994). Biochemical and genetic analyses in organisms as diverse as yeast, flies, worms, and mammals have produced a partial inventory of the components involved in membrane traffic and an outline of the molecular mechanisms involved.
Vesicle budding is mediated by coat proteins that assemble on nascent buds under the direction of small GTPases (Schekman and Orci, 1996). Cargo proteins are incorporated into budding vesicles either passively or by binding to “cargo receptors” that concentrate the itinerant proteins (Rothman and Wieland, 1996). After vesicle budding, uncoating ensues, and the vesicle docks with the accepter compartment in a process that involves members of three distinct protein families (Pfeffer, 1996). A family of integral membrane proteins, termed soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) (Söllner et al., 1993a), lies at the heart of the docking apparatus: vesicles carry v-SNAREs that specifically interact with cognate t-SNAREs displayed on the target compartment. The t-SNAREs are maintained in an inactive state (Pevsner et al., 1994; Lupashin and Waters, 1997) by association with members of the Sec1/Sly1 protein family (Halachmi and Lev, 1996) until they undergo activation through the displacement of the Sec1/Sly1 protein by a member of the Rab-GTPase family (Lupashin and Waters, 1997). The characteristic localization of the SNARE, Rab, and Sec1/Sly1 families to distinct intracellular compartments is thought to delineate the road map for vesicular trafficking and mediate its extraordinary specificity.
The final phase of vesicular transport is initiated after assembly of a v/t-SNARE docking complex when two other components, termed SNAP (Clary et al., 1990) and N-ethylmalemide-sensitive factor (NSF) (Block et al., 1988), which are employed at multiple transport steps (Graham and Emr, 1991; Rothman, 1994), bind to the v/t-SNARE complex (Söllner et al., 1993a,b). NSF is an ATPase (Tagaya et al., 1993; Whiteheart et al., 1994) that harvests the energy of ATP hydrolysis to disassemble the v/t-SNARE complex (Söllner et al., 1993b), thereby initiating the process of membrane fusion. Recently, an additional role for NSF has been uncovered in yeast vacuole-vacuole fusion. In that system, NSF acts to “prime” the vacuoles prior to docking, perhaps by dissociating SNARE complexes and thereby allowing them to engage in subsequent interactions (Mayer et al., 1996; Mayer and Wickner, 1997). After vesicle docking is complete, membrane bilayer fusion ensues, and describing this process represents one of the major challenges ahead.
It is clear that vesicular transport is not a unidirectional process. It has long been appreciated that delivery of newly made secretory proteins or intracellular proteins that reside in the secretory pathway requires vesicular transport from their site of synthesis on the endoplasmic reticulum (ER) to their final destinations (Palade, 1975), a process termed anterograde transport. Transport in the opposite direction, retrograde transport, is also clearly required if cells are to maintain the requisite surface area and volume of their intracellular organelles. Retrograde transport is also necessary for the recovery of components of the transport apparatus, such as SNAREs (Rothman and Warren, 1994) or cargo receptors (Marcusson et al., 1994; Schimmöller et al., 1995; Fiedler et al., 1996), if they are to be utilized more than once. Finally, retrograde transport provides the cell with a mechanism to salvage proteins that have escaped from their resident compartment (Dean and Pelham, 1990; Jackson et al., 1993; Nothwehr et al., 1993; Schutze et al., 1994; Harris and Waters, 1996). To fully appreciate the intracellular vesicular traffic pattern then, both anterograde and retrograde traffic must be understood.
Recently, the realization that SNAREs are central to vesicle docking and that they are arrayed on specific compartments (Bennett, 1995) has prompted efforts to identify and localize the full complement of SNAREs. The recent elucidation of the complete yeast genome sequence should allow the full complement of yeast SNAREs to be identified based on protein sequence homology. Identification of all of the yeast SNAREs, coupled with localization and functional analyses, may provide an ideal model system to develop a high resolution map of the secretory pathway encompassing both anterograde and retrograde traffic.
In this report, we describe a novel yeast SNARE protein, Vti1p, likely to function in retrograde transport to the cis-Golgi compartment. This protein has also been examined recently by Fischer von Mollard et al. (1997).
MATERIALS AND METHODS
Media, Plasmids, and Strains
YPD, YNB, and sporulation media were prepared as described (Rose et al., 1990). YPGal and YPGlu/Gal media contain 1% yeast extract, 2% Bacto-peptone, 2% galactose, and 0.1% or 1% glucose, respectively. Transformation of yeast was performed as described (Elble, 1992); all other standard genetic techniques were done as indicated (Rose et al., 1990). The Escherichia coli strain XL1-Blue (Stratagene, La Jolla, CA), which was used for all molecular genetic manipulations, was grown on standard media (Miller, 1972) and transformed according to Hanahan (1983).
The plasmids used in this study are listed in Table 1 and were constructed as follows. VTI1 [open reading frame (ORF) YMR197c] was amplified from genomic DNA by pfu of DNA polymerase- (Stratagene) based polymerase chain reaction (PCR) placing a SacII site 535 bp upstream of the initiator methionine codon of VTI1 and a XhoI site 299 bp downstream of the stop codon (5′ primer, 5′-CGCCCGCGGCTTATGTCAACGAGTTTCAAATGCCAAAAC-3′; 3′ primer, 5′-CGCCTCGAGCTAAAAGATAAGGTGAAAAATATGAAAG-3′). The PCR product was digested with SacII and XhoI and ligated either into Bluescript II KS− (Stratagene) to generate pPI1 or into pRS426 (Sikorski and Hieter, 1989) to generate pPI2. Plasmid pPI3 was generated by removal of the 750-bp internal HindIII-XbaI fragment of VTI1 from plasmid pPI1 and replacement with the 2.1-kb HindIII-XbaI fragment from pMR2253 (from M. Rose, Princeton University), which contains a functional LEU2 gene. To construct pPI6, the plasmid encoding the glutathione S-transferase (GST)-Vti1 fusion protein, the VTI1 ORF was amplified by PCR from genomic DNA, placing BamHI and SacI sites upstream to the sequence encoding the second codon of VTI1 and a XhoI site 299 bp downstream of the stop codon (5′ primer, 5′-GCGAGCTCGGATCCAGTTCCCTATTAATATCATACG-3′; 3′ primer, same as above for pPI1). The BamHI- and XhoI-digested PCR product was ligated into pEMBLyex4 (Baldari et al., 1987) digested with the same enzymes. pPI7 and pPI8 were generated by amplification of the VTI1 ORF by PCR, placing a SacI site upstream of the second codon of VTI1 and a XhoI site 300 bp downstream of the stop codon with the same sets of primers as for pPI6. The PCR products were digested with SacI and XhoI and ligated into similarly digested pYATAG200 (from M. Latterich, Salk Institute, La Jolla, CA), which encodes the hemagglutinin (HA)-epitope tag behind the CUP1 promoter (PCUP), to generate pPI7, or into pYATAG100 (from M. Latterich), to generate pPI8. To construct pPI9, HA-VTI1 was excised from pPI7 as a 1.4-kb PvuII-EcoRI fragment and ligated into EcoRI and SmaI cleaved pSM578 (from S. Michaelis, Johns Hopkins University School of Medicine, Baltimore, MD). Plasmids pVLhPI1 and pVLmPI2, bearing the cDNA encoding a human and mouse sequences related to Vti1p (hVti1-rp1 and mVti1-rp2) under GAL1 promoter (PGAL) control, was constructed by ligation of the 600-bp EcoRI-NotI fragment of expressed sequence tag (EST) AA056932 (for pVLhPI1) or the 800-bp EcoRI-NotI fragment of EST AA016379 (for pVLmPI2) into a pYES2 (Invitrogen, Carlsbad, CA) vector that had been digested with the same enzymes. To construct the plasmid pGST-Sec17, encoding the GST-Sec17 fusion protein, the SEC17 ORF was amplified by PCR, placing a BamHI site adjacent to its 12th codon and an EcoRI site 3′ to the stop codon (5′ primer, 5′-CGGGATCCGCTGAGAAGAAGGGTGTTCC-3′; 3′ primer, 5′-CGGAATTCTCTAAGTCATCGCTATATG-3′). The PCR product was digested with BamHI and EcoRI and ligated into similarly digested pGEX-2T (Pharmacia Biotech, Piscataway, NJ). The first 17 amino acid residues of Sec17p are not essential for function (Griff et al., 1992).
Table 1.
Plasmids used in this work
| Plasmid | Description | Source |
|---|---|---|
| pPI1 | VTI1 in Bluescript II KS− | This study |
| pPI2 | VTI1 in pRS426 (2μ, URA3) | This study |
| pPI3 | VTI1Δ::LEU2 in pRS304 (YIp, TRP1) | This study |
| pPI6 | PGAL-GST-VTI1 in pEMBLyex4 (2μ, URA3) | This study |
| pPI7 | PCUP-HA-VTI1 in pYATAG200 (CEN, TRP1) | This study |
| pPI8 | PCUP-HA-VTI1 in pYATAG100 (YIp, TRP1) | This study |
| pPI9 | PGAL-HA-VTI1 in pSM578 (CEN, TRP1) | This study |
| pVLhPI1 | hVti1-rp1 (EcoRI-NotI fragment of EST AA056932) in pYES2 (2μ, URA3) | This study |
| pVLmPI2 | mVti1-rp2 (EcoRI-NotI fragment of EST AA016379) in pYES2 (2μ, URA3) | This study |
| pGST-Sec17 | GST-SEC17 (aa 11–291) in pGEX2T | This study |
| pSK54 | SLY1-20 in pRS426 (2μ, URA3) | This laboratory |
| pSK60 | YKT6 in pRS426 (2μ, URA3) | This laboratory |
| pSED5 | SED5 in YEP352 (2μ, URA3) | H. Pelham |
| pSFT1 | 2μ, SFT1 URA3 | H. Pelham |
| pJG103 | 2μ, SEC22 URA3 | S. Ferro-Novick |
| pSFN2d | 2μ, BET1 URA3 | S. Ferro-Novick |
The S. cerevisiae strains used in this study are listed in Table 2. To construct the GWY150 strain, pPI3 was linearized by PvuII and used to transform the diploid strain GWY30. Leu+ transformants were purified; the deletion was confirmed by PCR utilizing primers in LEU2 and flanking the deletion. GWY151 and GWY152 strains were obtained by transformation of GWY150 with pPI2 and pPI6, respectively, followed by sporulation, tetrad dissection, and selection of Leu+ Ura+ segregants. GWY153 was obtained by the plasmid shuffle method from GWY151. GWY154 was obtained by transformation of GWY151 with linearized pPI8, followed by a plasmid shuffle. GWY155 was obtained by transformation of GWY150 with pPI9, followed by sporulation, tetrad dissection and selection for Leu+ Trp+ segregants. GWY156 was isolated from a cross between RSY271 and GWY152 and GWY157 from a cross between RSY271 and GWY154.
Table 2.
S. cerevisiae strains used in this work
| Strain | Genotype | Source |
|---|---|---|
| RSY255 | MATα ura3-52 leu2-3,-112 | R. Schekman |
| RSY271 | MATα sec18-1 ura3-52 his4-612 | R. Schekman |
| RSY272 | MATa sec18-1 ura3-52 his4-612 | R. Schekman |
| GWY30 | MATa/α ura3-52/ura3-52 leu2Δ1/leu2Δ1 trp1Δ63/trp1Δ63 | This laboratory |
| GWY147 | MATα ura3-52 his3Δ200 leu2-3,-112 trp1Δ901 sft1Δ::LEU2 psft1-1 (CEN, HIS3) | H. Pelham |
| GWY150 | MATa/α ura3-52/ura3-52 leu2Δ1/leu2Δ1 trp1Δ63/trp1Δ63 vti1Δ::LEU2/VTI1 | This study |
| GWY151 | MATa ura3-52 leu2Δ1 trp1Δ63 vti1Δ::LEU2 pPI2 (2μ, URA3, VTI1) | This study |
| GWY152 | MATa ura3-52 leu2Δ1 trp1Δ63 vti1Δ::LEU2 pPI6 (2μ, URA3, PGAL-GST-VTI1 | This study |
| GWY153 | MATa ura3-52 leu2Δ1 trp1Δ63 vti1Δ::LEU2 pPI7 (CEN, TRP1, PCUP-HA-VTI1) | This study |
| GWY154 | MATa ura3-52 leu2Δ1 vti1Δ::LEU2 PCUP-HA-VTI1 | This study |
| GWY155 | MATα ura3-52 leu2Δ1 trp1Δ63 vti1Δ::LEU2 pPI9(CEN, TRP, PGAL-HA-VTI1) | This study |
| GWY156 | MATα ura3-52 trp1Δ63 vti1Δ::LEU2 sec18-1 pPI6 (2μ, URA3, PGAL-GST-VTI1) | This study |
| GWY157 | MATα ura3-52 vti1Δ::LEU2 sec18-1 PCUP-HA-VTI1 | This study |
| GWY158 | MATa ura3-52 leu2Δ1 trp1Δ63 vti1Δ::LEU2 pPIts3 (CEN, TRP1, PCUP-HA-vti1-21) | This study |
| GWY159 | MATa ura3-52 leu2Δ1 trp1Δ63 vti1Δ::LEU2 pPIts4 (CEN, TRP1, PCUP-HA-vti1-22) | This study |
| GWY160 | MATa ura3-52 leu2Δ1 trp1Δ63 vti1Δ::LEU2 pPIts6 (CEN, TRP1, PCUP-HA-vti1-23) | This study |
| GWY161 | MATa ura3-52 leu2Δ1 trp1Δ63 vti1Δ::LEU2 pPIts7 (CEN, TRP1, PCUP-HA-vti1-24) | This study |
| GWY162 | MATa ura3-52 leu2Δ1 trp1Δ63 vti1Δ::LEU2 pPIts16 (CEN, TRP1, PCUP-HA-vti1-25) | This study |
Generation of the vti1 Temperature-conditional Strains
Mutations in VTI1 were generated by mutagenic PCR (Fromant et al., 1995). The same set of primers used to generate pPI6 were used to PCR amplify a 1.8-kb fragment containing VTI1 from pPI7 using Taq polymerase (Perkin Elmer-Cetus Corp., Norwalk, CT) under standard conditions except for the presence of 0.5 mM MnCl2. The PCR product was digested with SacI and XhoI and ligated into similarly digested pYATAG200. GWY151 [vti1Δ::LEU2, pPI2] cells were transformed with the resulting plasmid library, and transformants were selected on SC-Trp plates at 25°C. Trp+ colonies were replica plated onto plates containing 5-fluoroorotic acid, which selects against URA3-containing plasmids (Boeke et al., 1984), to remove the plasmid bearing unmutagenized VTI1. 5-Fluoroorotic acid-resistant colonies were replica plated to YPD and incubated at either 25°C or 37.5°C. Temperature-conditional mutants (GWY158 through GWY162) that grew at 25°C but not 37.5°C were isolated.
Subcellular Fractionation
A 100-ml culture of GWY154 expressing HA-Vti1p was grown in YPD to an OD600 of 0.6. The cells were collected by centrifugation, washed in distilled water, and resuspended at 30 OD/ml in ice-cold lysis buffer [20 mM HEPES/KOH, pH 7.0, 100 mM KOAc, 2 mM Mg(OAc)2, 1 mM dithiothreitol (DTT), 2 mM EGTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), 5 mM 1,10-phenanthroline, 2 μM pepstatin A, 2 μg/ml aprotinin, 0.5 μg/ml leupeptin]. One-half sample volume of acid-washed glass beads (0.45 mm diameter, Thomas Scientific, Swedesboro, NJ) was added to the cell suspension in a glass tube, which was then vortexed five times for 30 s with 1-min incubations on ice between each burst. The crude lysate was centrifuged at 500 × g for 5 min at 4°C in a Sorvall SA600 rotor to remove unlysed cells. The supernatant (Lysate) was diluted to 5 mg protein/ml in lysis buffer and centrifuged at 10,000 × g for 15 min at 4°C in the same rotor to generate supernatant (S10) and pellet (P10) fractions. The S10 was then centrifuged at 150,000 × g for 60 min at 4°C in a Beckman TLA100.2 rotor to generate high-speed supernatant (S150) and pellet (P150) fractions.
For the membrane extraction experiments (Figure 3B), the lysate was treated on ice for 15 min with lysis buffer or lysis buffer containing either 1% Triton X-100, 0.1 M Na2CO3, final pH 11.5, or 1 M NaCl and then separated into S150 and P150 fractions by centrifugation as above.
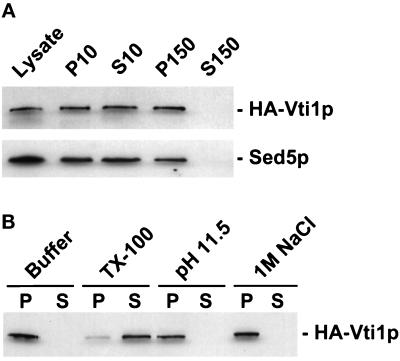
Figure 3.
Vti1p is membrane associated. (A) The GWY154 strain (HA-VTI1) was grown to logarithmic phase, lysed with glass beads, and centrifuged at 500 × g for 3 min. The supernatant (Lysate) was centrifuged at 10,000 × g for 15 min, yielding the P10 pellet and S10 supernatant fractions, and the S10 was centrifuged again at 150,000 × g for 60 min to generate the P150 pellet and S150 supernatant fractions. Aliquots (derived from 0.2 OD600 units of cells) of each fraction were separated by SDS-PAGE and immunoblotted with the anti-HA epitope monoclonal antibody or anti-Sed5p affinity-purified antisera. (B) Aliquots of lysate were extracted with buffer, 1% Triton X-100, 0.1 M Na2CO3, or 1 M NaCl. After centrifugation at 150,000 × g, HA-Vti1p was detected in pellet (P) and supernatant (S) fractions via immunoblotting.
The sucrose flotation gradient was constructed as follows. Clarified glass bead cell lysate (0.5 ml) from strain GWY154 made in D2O buffer (20 mM HEPES/KOH, pH 7.0, 100 mM KOAc, 1 mM DTT, 1 mM PMSF, 5 mM 1, 10-phenanthroline, 2 μM pepstatin A, 2 μg/ml aprotinin, 0.5 μg/ml leupeptin in D2O) was mixed with two volumes of 60% sucrose in D2O buffer, placed at the bottom of a 12-ml SW41 (Beckman Instruments, Fullerton, CA) tube, and overlaid with 1.5 ml each of 45%, 37%, 33%, 29%, 23%, 17%, and 10% sucrose, respectively, in D2O buffer. The gradient was centrifuged at 150,000 × g for 24 h in a SW41 rotor. Twenty-four 0.45-ml fractions were collected from the top of the tube and analyzed using immunoblotting. Western blots were quantified by analyzing films with a scanning densitometer (Microtek International, Taiwan) and National Institutes of Health Image software.
Separations of the S10 fraction by sucrose density centrifugation was performed as described (Becherer et al., 1996).
Immunofluorescence and Immunoblotting
Indirect immunofluorescence was performed as described by Hardwick and Pelham (1992) with some modifications. Fifty milliliters of log phase GWY154 cells grown in YPD were fixed for 10 min by addition of 1/10th final volume of 37% formaldehyde to the culture. Cells were collected by centrifugation, resuspended in 40 mM KPi (pH 6.5), 0.5 mM Mg(OAc)2, and 3.7% formaldehyde and incubated for 60 min. Cells were washed three times with 40 mM KPi (pH 6.5), once with 40 mM KPi (pH 6.5), and 1.2 M sorbitol and then spheroplasted in the same buffer containing 0.05 mg/ml Zymolyase 100T (Seikagako Kogyo Co., Tokyo, Japan) for 30 min at 30°C. The spheroplasts were washed with 40 mM KPi (pH 6.5) and 1.2 M sorbitol, pipetted onto polylysine-coated microscope slides, and allowed to adhere for 10 min. Slides were plunged into −20°C methanol for 6 min, transferred to −20°C acetone for 1 min, air dried, and incubated in blocking solution [phosphate-buffered saline (PBS), 0.5% bovine serum albumin, 0.5% ovalbumin, 0.2% gelatin, 0.1% Tween 20) for 60 min. Samples were incubated for 2 h at room temperature in a 1:1000 dilution of the anti-Kar2p polyclonal antibodies (from R. Schekman), a 1:200 dilution of the affinity-purified antiSed5p antibodies (Lupashin and Waters, 1997), or a 1:200 dilution of the 12CA5 anti-HA epitope monoclonal antibody (Wilson et al., 1984) in blocking solution. The samples were washed four times with blocking solution, incubated for 1 h at room temperature in a 1:200 dilution of fluorescein isothiocyanate-conjugated goat anti-rabbit or goat anti-mouse immunogloblin (ICN/Cappel, Costa Mesa, CA) in blocking solution, washed again with PBS, and mounted in 90% glycerol and 0.1% p-phenylenediamine. Images were obtained using a Bio-Rad MRC600 laser scanning microscope.
Samples for immunoblotting were separated by SDS-PAGE (12% acrylamide, except where noted), electrotransferred to nitrocellulose, and probed with the appropriate primary antibodies according to standard protocols (Harlow and Lane, 1988). Monoclonal antibodies against the HA epitope (12CA5) and Dpm1p (Molecular Probes, Eugene, OR) were used at a dilution of 1:2000. Affinity-purified antibodies against Sed5p, Sec22p, Sly1p, and GST (Lupashin and Waters, 1997) and sera against gp400/hsp150 (Lupashin et al., 1992), Kar2p (from R. Schekman), Pep12p (from E. Jones), Sso1p (from P. Brennwald), and Ykt6p were all used at a dilution of 1:2000. Horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibody (Bio-Rad, Hercules, CA) were used at a dilution of 1:3000. Immunoblots were developed with the Renaissance chemiluminescent detection kit (DuPont New England Nuclear, Boston, MA).
Metabolic Labeling and Immunoprecipitation of CPY and Invertase
Strains were grown at room temperature in SC-MET media to an OD600 of 0.5. For each strain, 6 OD units of cells were collected by centrifugation, resuspended in 1.0 ml of SC-MET media, and preshifted to 38°C for 30 min. Cells were radiolabeled for 5 min with 100 μCi of Tran35S-label (ICN Radiochemicals, Irvine, CA), followed by a 30-min chase initiated by the addition of 1/25th volume of chase mix (250 mM cysteine, 250 mM methionine). The reactions were stopped by addition of an equal volume of 20 mM sodium azide and placed on ice for 10 min. Cell lysis and immunoprecipitation of CPY were performed as described previously (Sapperstein et al., 1996).
For the experiment described in Figure 7B, cells were grown at room temperature in SC-MET media to an OD600 of 0.5, collected by centrifugation, resuspended in 1.0 ml of SC-MET media with 0.1% glucose, incubated for 30 min at room temperature, and then for 30 min at 25°C or 38°C as indicated. The cells were labeled as above, chased for 20 min, and spheroplasted for 20 min by addition of 1/3 final volume of 0.3 mg/ml Zymolyase 100T in 2.8 M sorbitol. The spheroplasts were isolated by centrifugation at 3000 × g for 5 min and lysed on ice in 1 ml of 0.2 M NaOH and 0.5% β-mercaptoethanol. Proteins from the lysed spheroplasts and the spheroplasting media were precipitated with 10% trichloroacetic acid, resuspended in 2% SDS and 50 mM Tris-Cl (pH 8.0) and diluted in 60 mM Tris-HCl (pH 7.4), 190 mM NaCl, 6 mM EDTA, and 1.25% Triton X-100. Invertase was immunoprecipitated from the lysed spheroplasts (I, for intracellular) and from the spheroplasting media (E, for extracellular). To detect outer chain mannosyl residues, the radiolabeled invertase and CPY were immunoprecipitated, eluted, and reimmunoprecipitated with anti α-1,6-mannose linkage-specific antisera as described (Franzusoff and Schekman, 1989).
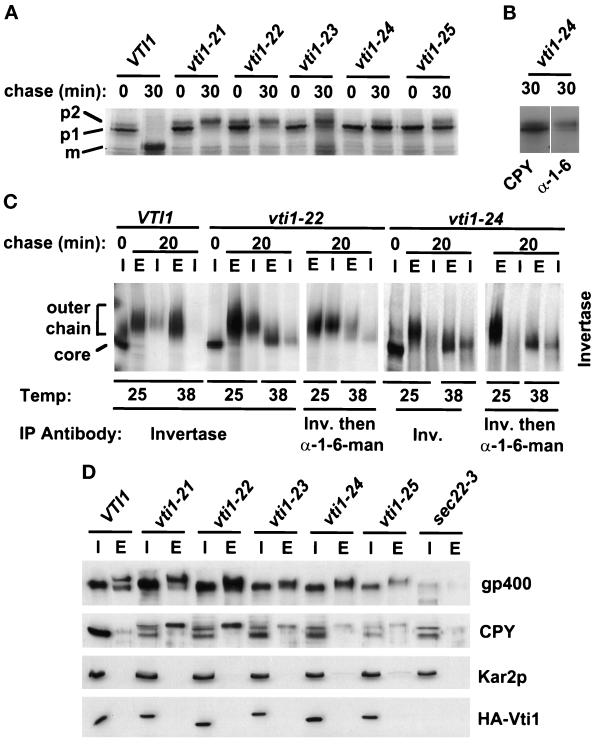
Figure 7.
vti1 ts mutants are defective in the Golgi sorting and glycosylation, but secretion is not severely compromised. (A) vti1 ts mutants block CPY transport at different stages. Autoradiograph of CPY immunoprecipitations from an igsΔ strain bearing HA-VTI1 (GWY153) or one of five HA-Vti1 ts mutant alleles (GWY158-GWY162). Cells were shifted to the restrictive temperature of 38°C for 30 min, pulse labeled for 5 min, and chased for 30 min at the same temperature. (B) The immunoprecipitated CPY was eluted and reprecipitated with anti-CPY and anti-α-1,6-mannose-specific antiserum and analyzed by SDS-PAGE and autoradiography. (C) Invertase is secreted in the underglycosylated form. Autoradiograph of invertase immunoprecipitations from an igsΔ strain bearing the HA-VTI1 (GWY153), HA-vti1–22 ts (GWY159), or HA-vti1–24 ts mutant alleles (GWY161). Cells were preincubated in low-glucose media for 30 min, shifted to the restrictive temperature of 38°C for 30 min, pulse labeled for 5 min, and chased for 20 min at the same temperature. Cells were converted to spheroplasts, and invertase was immunoprecipitated from lysed spheroplasts (I) and spheroplasting media (E). The immunoprecipitated invertase was eluted and reprecipitated with anti-invertase and anti-α-1,6-mannose-specific antiserum and analyzed by SDS-PAGE and autoradiography (D) vti1-ts cells secrete gp400/hsp150 and CPY but not ER-resident protein Kar2p at a semipermissive temperature. Mutant cells were grown at 35°C for 5 h in fresh YPD medium. The extracellular medium (E) and total cell lysates (I) were subjected to SDS-PAGE (10% acrylamide) followed by immunoblotting with anti-gp400/hsp150, anti-CPY, anti-Kar2p, and anti-HA epitope antibodies. We determined that the small amount of Kar2p detectable in the culture media of vti1–24 and vti1–25 cell was due to the cell lysis (our unpublished results).
For the experiment described in Figure 6, cells were grown at 30°C in 1% glucose/1% galactose containing SC-MET media to an OD600 of 0.2, after which they were transferred to SC-MET media containing 5% glucose or 0.1% glucose/2% galactose and allowed to grow for 6 h. Cells were pulse labeled for 5 min and chased in the media containing 5% glucose for the indicated time periods as described above. Antibodies against invertase, CPY, and α-1,6-mannose-specific linkage were kindly provided by M. Rose (Princeton University) and R. Schekman (University of California, Berkeley, CA).
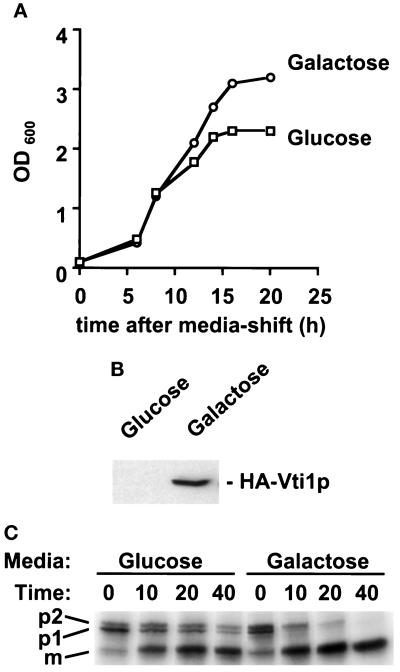
Figure 6.
Depletion of HA-Vti1p results in the accumulation of the Golgi forms of CPY. (A) Growth curve of the GWY155 (vti1Δ strain carrying PGAL-HA-VTI1) grown in galactose-containing media (Galactose) and glucose-containing media (Glucose). (B) Immunoblot detection of HA-Vti1p in whole-cell extracts after 6 h of growth on each medium shown in A. Note that upon long exposure of the immunoblot, HA-Vti1p is detectable in glucose-grown cells. (C) Autoradiograph of pulse-chase analysis of CPY processing after 6 h of growth on each media shown in A. Cells were pulse labeled for 5 min and chased in the glucose-containing media for times indicated. CPY was isolated from cell extracts by immunoprecipitation and resolved by SDS-PAGE (8% acrylamide). The different forms of CPY are indicated on the left. p1 indicates the ER and cis-Golgi form, p2 denotes the outer-chain glycosylated medial/trans-Golgi form, and m marks the mature, vacuolar form.
Suppression Analysis
Strains GWY158 to GWY162 were transformed with the following plasmids: pPI2, pRS426, pJG103, pSFN2d, pSK60, pSK54, pSED5, pSFT1, pVLhPI1, and pVLmPI2. Purified transformants were grown to stationary phase at 23°C in synthetic media. Suppression was assessed by inoculation of cells from these saturated cultures to YPD or YPGal plates and examination of growth after incubation for 3 d at 37.5°C.
Purification of Recombinant Proteins from E. coli
GST and GST-Sec17p were expressed in XL1-Blue from pGEX-2T and pGEX-Sec17, respectively. Protein expression was induced in 500-ml cultures grown at 37°C to an OD600 of 0.5 by the addition of isopropylthiogalactoside to 1.0 mM. After 4 h the cells were harvested (7000 × g for 10 min at 4°C in a Sorvall SA600 rotor), resuspended in 25 ml PBS with 0.1 mg/ml lysozyme, incubated on ice for 15 min, and lysed by sonication with three 30-s bursts at half-maximum intensity with a Branson Sonifer 450 (VWR Scientific Products Co., Bridgeport, NJ) separated by 1-min incubations on ice. Triton X-100 was added to 1%, the lysates were rocked at 4°C for 30 min, clarified by centrifugation (12,000 × g for 10 min at 4°C in an SA600 rotor), and incubated with 2 ml of glutathione-Sepharose 4B beads (Pharmacia Biotech) with gentle end-over-end mixing for 2 h at 4°C. The beads were washed twice with 25 ml PBS/1% Triton X-100 and loaded into a 2-ml chromatography column. The fusion proteins were eluted with 5 ml of 20 mM reduced glutathione in 50 mM Tris-Cl (pH 8.0) and dialyzed against PBS.
In Vitro GST-Sec17p Binding Reaction
GWY157 spheroplasts were shifted to 37°C for 1 h and lysed in the presence of 1% Triton X-100, as described above for subcellular fractionation. Solubilized protein (0.5 mg) was incubated with purified GST or GST-Sec17p (2 μg each) in buffer B (20 mM HEPES-KOH, 150 mM KOAc, 0.5 mM DTT, 0.05% Tween 20) for 1 h at 4°C and centrifuged at 15,000 × g for 15 min to remove aggregated material. The supernatant was incubated for 1 h at 4°C with 40 μl of glutathione-Sepharose 4B beads equilibrated with buffer B. The beads were washed three times with buffer B and bound proteins were eluted with 50 μl of 10 mM reduced glutathione in 50 mM Tris-Cl (pH 7.5), separated by SDS-PAGE (10% acrylamide), and immunoblotted with anti-Kar2p, anti-HA epitope, or anti-Sed5p antibodies.
Analysis of GST-Vti1p Interactions in Whole-Yeast Detergent Extracts
GWY156 cells were grown at room temperature in YPGlu/Gal media to an OD600 of 0.5–1.0, collected by centrifugation, resuspended in fresh YPGlu/Gal media, incubated at the permissive (25°C) or restrictive (37°C) temperature for 1 h, and then transferred to ice. Cells were lysed with glass beads in the presence of 1% Triton X-100 as described above. Clarified detergent lysates (∼2 mg of protein) were incubated for 1 h at 4°C with 300 μl of glutathione-Sepharose 4B equilibrated with buffer B. The beads were transferred into 1-ml chromatography columns, washed with buffer B, and bound proteins were eluted with 10 mM reduced glutathione in 50 mM Tris-Cl (pH 7.5). Fractions (300 μl each) were collected, proteins were separated by SDS-PAGE followed by immunoblotting with anti-GST, anti-Sed5p, anti-Sso1p, anti-Ykt6p, or anti-Sec22p antibodies.
Immunoprecipitation from Detergent Extracts of Yeast Spheroplasts
GWY157 cells were grown to OD600 of 0.5 in YPD media. Cultures (50 ml) were centrifuged for 5 min at 3500 rpm in a Beckman TJ-6 centrifuge, resuspended in 100 mM Tris-SO4 (pH 9.4)/10 mM DTT at 5 OD/ml, and placed on ice for 10 min. The cells were collected by centrifugation as above, resuspended in the same volume of YPD, 0.8 M sorbitol, and 25 mM HEPES/KOH (pH 7.0). Zymolyase 20T (Seikagak Kogyo Co.) was added to 100 μg/ml, and the cells were incubated at 25°C for 30 min. Spheroplasts were washed in YPD, 0.8 M sorbitol, and 25 mM HEPES/KOH (pH 7.0), resuspended in the same buffer, and incubated for 20 min at 25°C with gentle rocking. The regenerated spheroplasts were collected by centrifugation, resuspended in 20 mM HEPES/KOH (pH 7.0), 200 mM KCl, 1 mM DTT, 1 mM MgCl2, 1 mM PMSF, 0.5 mM 1:10 phenanthroline, 2 μM pepstatin A, 0.5 μg/ml leupeptin, 2 μg/ml aprotinin, and 2% Triton X-100 (Søgaard et al., 1994) at 10 OD/ml, and lysed by repeated pipetting. Debris was removed by centrifugation at 10,000 × g for 10 min at 4°C in an SA600 rotor and the cleared extracts were transferred to fresh tubes. Immunoprecipitations were done essentially as described (Sapperstein et al., 1996; Lupashin and Waters, 1997) except that each reaction contained 500 μg of extract protein and 40 μl of a 50% slurry of protein A-Sepharose (Pharmacia Biotech) bearing affinity-purified antibodies to Sed5p, Sec22p, or Sly1p that had been crosslinked to the beads with dimethylpimelimidate as described (Harlow and Lane, 1988).
RESULTS
Identification of Novel Yeast SNARE Proteins
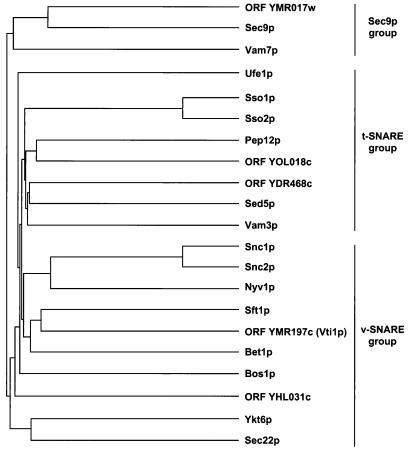
In an effort to identify the full complement of yeast SNARE proteins, we used known SNAREs as query sequences for BLAST (Altschul et al., 1990) and FASTA (Pearson and Lipman, 1988) searches of the complete S. cerevisiae genome database (Cherry, Adler, Ball, Dwight, Chervitz, Jia, Juvik, Weng, and Botstein, Saccharomyces Genome Database, http://genome-www.stanford.edu/Saccharomyces). Predicted proteins were considered SNAREs if 1) they yielded significant BLAST or FASTA scores (p value < 10−2); 2) the predicted topology corresponded to a type II integral membrane protein with a very short lumenal domain; and 3) they bore heptad repeats capable of forming a coiled coil in the membrane-proximal region as predicted by the COILS algorithm [with a window size of 21 and the MTDIK matrix (Lupas et al., 1991)]. The Sec9p (SNAP-25 related) group (Weimbs et al., 1997) and Ykt6p (Søgaard et al., 1994; McNew et al., 1997) are exceptions to these criteria because they do not possess transmembrane domains. The results of the searches are presented in Figure 1A as a dendrogram. We found one new member of the Sec9p group of sequences (ORF YMR017w), two new potential members of the Sso1p-Sed5p (t-SNARE) family (ORFs YOL018c and YDR468c), and two new members of the Snc1p/Bet1p (v-SNARE) protein family (ORFs YMR197c and YHL031c).
Figure 1.
Sequence analysis of yeast SNARE proteins. (A) Nearest-neighbor dendrogram of known and putative yeast SNARE molecules. Common protein names or ORF numbers are indicated. The dendrogram was generated with the PILEUP program of the GCG sequence analysis package. (B) Multiple sequence alignment of S. cerevisiae Vti1p (denoted S.c.Vti1p), and Vti1p-related proteins from S. pombe (S.p.Vti1), human (hVti1-rp1 and hVti1-rp2), and mouse (mVti1-rp1 and mVti1-rp2). Blackened boxes denote identical residues, and gray shading denotes conserved residues. Optimal alignment was produced with the PILEUP program. S.p.Vti1 is encoded by cDNA clone D89116 and mVti1-rp1, mVti1-rp2, hVti1-rp1, and hVti1-rp2 are encoded by EST AA056932, EST AA016379, EST AA105524, and EST T70362, respectively. In some cases, primary sequence data were obtained from the Washington University Genome Sequencing Center and used to extend the EST sequences to encompass the complete ORFs.
We have analyzed ORF YMR197c, a new member of the putative v-SNARE family, in detail. Comparison of the YMR197c predicted protein sequence using the GAP and BESTFIT programs from the GCG sequence analysis package revealed similarity (32% identity) with the previously described yeast intra-Golgi retrograde v-SNARE Sft1p (Banfield et al., 1995). The next nearest relative of YMR197c, at 23% identity, is the ER-to-Golgi v-SNARE, Bet1p/Sly12p (Newman et al., 1990; Dascher et al., 1991). When we extended our analysis beyond S. cerevisiae, we found that the mammalian v-SNARE GOS-28/GS28, which is involved in ER-to-Golgi and/or intra-Golgi traffic (Nagahama et al., 1996; Subramaniam et al., 1996), is also related (21% identity) to YMR197c. We originally termed this new SNARE Igs1p (intra-Golgi SNARE 1 protein), based on it role in intracellular protein traffic. After submission of this work, however, Fischer von Mollard et al. (1977) published a report on the same protein. Thus, we have adopted the name they used, Vti1p.
The predicted protein sequence of Vti1p comprises 217-amino acid residues with a predicted molecular mass of 24.7 kDa (Figure 1B). GenBank and dbEST database searches with Vti1p indicated that several proteins are related to Vti1p: a putative protein encoded by Schizosaccharomyces pombe cDNA clone D89116, two hypothetical mouse proteins, mVti1-rp1 (mouse Vti1p-related protein 1) and mVti1-rp2, and two hypothetical human proteins, hVti1-rp1 and hVti1-rp2. The aligned amino acid sequences of the putative Vti1p-related proteins are shown in Figure 1B, and their pairwise sequence identities are shown in Table 3. The human and mouse orthologous pairs of proteins (i.e., the hVti1-rp1/mVti1-rp1 pair and the hVti1-rp2/mVti1-rp2 pair) are very similar (87 and 90% identity, respectively). The human paralogues are more distantly related to each other and to their yeast orthologues. The existence of two distinct mammalian homologues is interesting since a similar situation has been demonstrated for the yeast v-SNARE Sec22p (Hay et al., 1996, 1997; Paek et al., 1997). These observations may reflect the difference between a more streamlined yeast secretory pathway and the more highly elaborated mammalian protein transport machinery. It is also possible that the yeast protein is involved in more than one transport step, as recently proposed (Fischer von Mollard et al., 1997), whereas in mammalian eukaryotic cells these distinct functions could be performed by related, yet distinct proteins.
Table 3.
Relative sequence identity of Vti1p-related proteinsa
| S.p.Vti1p | hVti1-rp1 | hVti1-rp2 | mVti1-rp1 | mVti1-rp2 | |
|---|---|---|---|---|---|
| S.c.Vti1p | 38 | 29 | 35 | 27 | 32 |
| S.p.Vti1p | 33 | 36 | 30 | 34 | |
| hVti1-rp1 | 21 | 87 | 27 | ||
| hVti1-rp2 | 27 | 90 | |||
| mVti1-rp1 | 30 |
The percentage of amino acid identity between yeast, human, and mouse Vti1p isoforms are displayed. Optimal alignments were produced by the BESTFIT program from the GCG package.
VTI1 Encodes an Essential Yeast Golgi Membrane Protein
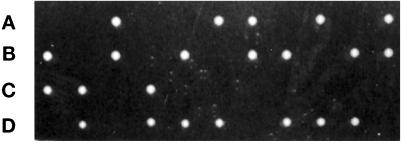
To determine whether VTI1 is an essential gene, the viability of haploid igsΔ disruptants was examined. The VTI1 ORF was replaced by the sequence of LEU2 in a diploid strain, the presence of both the wild-type and disrupted VTI1 alleles in the heterozygous diploid was confirmed by PCR, the strain was sporulated, and tetrads were dissected. All 24 tetrads examined displayed 2+:2− segregation for viability at 25°C (Figure 2). The same 2+:2− segregation pattern was evident when the spores were incubated at 30°C (our unpublished results). Microscopic examination of the inviable segregants revealed microcolonies of ∼20 cells, indicating that the vti1Δ spores can germinate. All viable spores were Leu−, and therefore contained the wild-type VTI1 allele. These data indicate that VTI1 is essential for vegetative growth. Transformation of the vti1Δ/VTI1 heterozygous diploid strain with a low-copy number centromere- (CEN) based plasmid-bearing VTI1 prior to sporulation and dissection complemented the lethality of the vti1Δ haploid segregants (our unpublished results). In addition, the vti1Δ strain could be rescued by expression of HA epitope-tagged Vti1p (HA-Vti1p) or a GST-Vti1p fusion protein (GST-Vti1p) expressed from CEN-based plasmids (our unpublished results), indicating that these fusion proteins, which were used in experiments described below, are functional.
Figure 2.
VTI1 is an essential gene. The VTI1/vti1Δ::LEU2 heterozygous deletion strain was sporulated, tetrads were dissected, and the segregants incubated at 25°C for 4 d on YPD plates. All viable segregants were Leu−. A–D indicate the segregants within each tetrad.
We used the vti1Δ strain carrying genomic HA-Vti1p to initiate a biochemical analysis of Vti1p. Immunoblot analysis with an anti-HA epitope monoclonal antibody (Wilson et al., 1984) specifically recognized one protein with an apparent molecular mass of ∼28 kDa, which was absent in the wild-type parent strain (our unpublished results). The intracellular localization of HA-Vti1p was examined by subcellular fractionation via differential centrifugation (Figure 3A). HA-Vti1p-expressing cells were lysed, unbroken cells were removed, and the lysate was centrifuged at 10,000 × g, yielding pellet (P10) and supernatant (S10) fractions. The S10 was further centrifuged at 150,000 × g to generate high-speed pellet (P150) and supernatant (S150) fractions. The proteins in each fraction were resolved by SDS-PAGE and immunoblotted to localize HA-Vti1p and, as a control, the cis-Golgi t-SNARE Sed5p. HA-Vti1p behaves as an integral or peripheral membrane protein, partially sedimenting at 10,000 × g and completely sedimenting at 150,000 × g (Figure 3A).
To test the prediction that Vti1p is an integral membrane protein, we extracted cell lysates under several conditions that perturb membrane proteins and examined the sedimentation behavior of Vti1p (Figure 3B). Neither 1 M NaCl nor 0.1 M Na2CO3 (pH 11.5) treatments, which are expected to release peripheral membrane proteins (Fujiki et al., 1982), extracted Vti1p from membranes, whereas 1% Triton X-100, which solubilizes membranes, did solubilize Vti1p, suggesting that Vti1p is an integral membrane protein. Furthermore, fusion of myc epitope-tagged yeast vacuolar alkaline phosphatase to the carboxyl-terminus of Vti1p produced a protein of ∼90 kDa that was glycosylated, as shown by interaction with concanavalin A-Sepharose beads (our unpublished results), suggesting that the carboxyl-terminus of Vti1p is lumenally oriented.
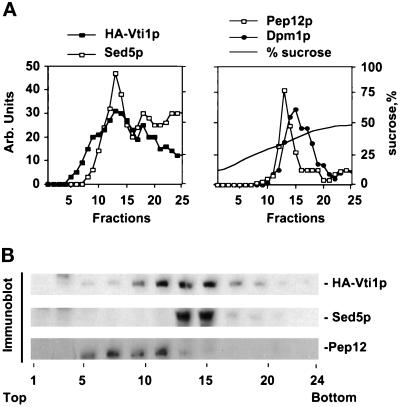
To further examine the intracellular location of HA-Vti1p, a cell lysate was subjected to isopycnic flotation through a sucrose/D20 density gradient, the gradient was fractionated, and aliquots were subjected to immunoblotting (Figure 4A). Markers of the endoplasmic reticulum (Dpm1p), cis-Golgi (Sed5p), and prevacuolar (Pep12p) compartments were used as internal controls. HA-Vti1p was present in two peaks whose distribution was most like that of the cis-Golgi marker Sed5p. The more buoyant HA-Vti1p peak (fraction 13) also coincided with the prevacuolar marker Pep12p (Becherer et al., 1996).
Figure 4.
Vti1p partially colocalizes with the Golgi t-SNARE Sed5p and prevacuolar t-SNARE Pep12p on sucrose gradients. (A) A clarified glass bead cell lysate (500 × g supernatant) from GWY154 was mixed with two volumes of 60% sucrose/D2O, placed at the bottom of a 10–45% sucrose step gradient, and subjected to centrifugation at 150,000 × g for 24 h. Fractions were collected and subjected to SDS-PAGE, and the locations of HA-Vti1p, the Golgi marker Sed5p, prevacuolar marker Pep12p, and ER marker Dpm1p were determined by immunoblotting. Sucrose density was measured with a refractometer. (B) Immunoblots of fractions from sucrose density centrifugation of the S10 fraction on a shallow sucrose gradient.
To better resolve the cis-Golgi from the prevacuolar compartment, we separated a 10,000 × g supernatant of lysed spheroplasts by sedimentation through a shallow sucrose gradient (Figure 4B). Under these conditions the Sed5p and Pep12p peaks were separated. HA-Vti1p could be found in both the Sed5p and the Pep12p regions of the gradient. Recently, Fischer von Mollard et al. (1997) have shown colocalization of Vti1p with both prevacuolar and TGN markers.
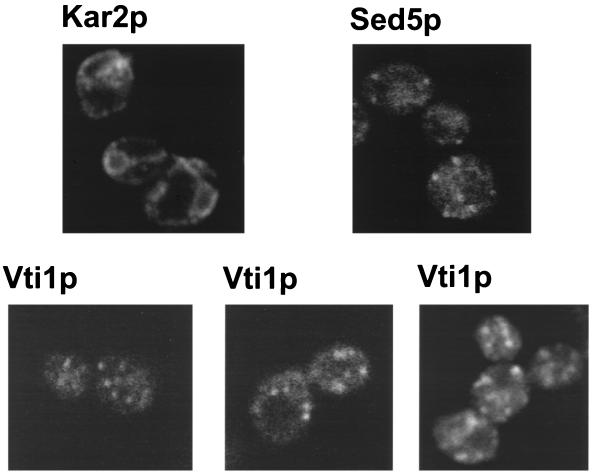
We also carried out an immunofluorescence study in S. cerevisiae cells expressing HA-Vti1p as the sole source of Vti1 protein. We observed a small number of punctate structures (Figure 5, Vti1p) that did not coincide with the plasma membrane or vacoule nor with nuclear envelope or ER, which were revealed by anti-Kar2p staining (Figure 5, Kar2p). The staining observed for Vti1p resembled that of Sed5p (Figure 5, Sed5p): numerous punctate structures were found throughout the cytoplasm.
Figure 5.
Vti1p is localized to punctate structures throughout the cytoplasm. The indicated proteins were detected in strain GWY154 by indirect immunofluorescence.
Our observations, along with those of Fischer von Mollard et al. (1997), suggest that Vti1p is a type II integral membrane protein of the Golgi apparatus and the prevacuolar compartment.
Vti1p Is Required for Intra-Golgi Protein Transport
To investigate the function of Vti1p, we tested the effect of its depletion on cell growth and protein transport. HA-VTI1 was placed behind the galactose-inducible/glucose-repressible GAL1 promoter (PGAL) on a low-copy number CEN plasmid. vti1Δ cells bearing the PGAL-HA-VTI1 construct were grown in medium containing galactose, where they grew as rapidly as wild-type cells in the same media (our unpublished results), indicating that the HA-Vti1p was functional. However, when HA-Vti1p expression was repressed by transferring the cells to glucose-containing medium, the strain gradually ceased to grow (Figure 6A). To examine the role of Vti1p in protein transit through the secretory pathway, we analyzed the kinetics of maturation of vacuolar CPY 6 h after shift to high-glucose media, a time point when the level of HA-Vti1p was significantly reduced (Figure 6B), but growth was not inhibited (Figure 6A). CPY is a useful probe of secretory pathway function, because it is sequentially modified as it transits the secretory pathway: upon translocation into the ER, it receives N-linked oligosaccharides resulting in the p1 form, subsequent transport to the medial Golgi, and finally, to the vacuole, generates the p2 and mature (m) forms, respectively. Cells grown in the galactose-containing medium efficiently transported CPY to the vacuole; most of the CPY was matured within 10 min of synthesis, and no p1 or p2 forms remained after 40 min (Figure 6C, Galactose). In contrast, cells grown in glucose-containing medium, where the amount of HA-Vti1p was greatly reduced (Figure 6B), matured only about half of the CPY within 10 min of synthesis, and some p1 and p2 forms were still found after 40 min (Figure 6C, Glucose). These data suggest that movement of the ER/cis-Golgi p1 form of CPY to the compartments that generate the p2 (medial/trans-Golgi) and m forms (vacoule) is compromised by depletion of Vti1p activity. The persistence of both the p1 and p2 forms of CPY when the level of Vti1p is low suggests that transport is defective at multiple steps within the Golgi. This result, however, does not by itself allow us to determine whether Vti1p acts in anterograde or retrograde protein transport, since intracellular transport of the CPY is very sensitive to defects in either of these processes (Stevens et al., 1982; Banfield et al., 1995; Gaynor and Emr, 1997).
Golgi Function Is Disrupted in vti1 Temperature-Sensitive Mutants
To further analyze the role of Vti1p in intracellular protein transport, we prepared temperature-sensitive (ts) alleles of HA-VTI1 by PCR-mediated mutagenesis (Fromant et al., 1995) and studied their secretion phenotypes at the restrictive temperature. We obtained five ts mutants of VTI1, all of which ceased growth at 37.5°C but differed from each other by their growth rates at lower temperatures (e.g., vti1–23 grows as well as wild type at 30°C, but very slowly at 20°C; vti1–25 grows poorly at 30°C). All five ts mutants harbor approximately equivalent amount of HA-Vti1p after prolonged incubation at semipermissive conditions, suggesting that stability of the mutant proteins was not compromised (Figure 7D, lowest panel). We did note, however, that the electrophoretic mobility of some of the mutant forms of HA-Vti1p was slightly aberrant .
To test the effect of the ts mutations on the intracellular transport of CPY, we used vti1Δ cells bearing either VTI1 or one of the vti1 ts alleles on CEN plasmids. The cells were shifted to the restrictive temperature for 30 min, pulse labeled for 5 min, chased for 30 min, and CPY was immunoprecipitated and analyzed by SDS-PAGE and autoradiography (Figure 7A). After 5 min of pulse label, most of the CPY in the wild-type cells and the vti1–21, vti1–22, and vti1–23 mutants was present as the ER/cis-Golgi p1 form with a small amount of the medial/trans-p2 form detectable; in vti1–24 and vti1–25, the p1 form was found exclusively. After 30 min of chase, neither p1 nor p2 forms of CPY were detectable in the wild-type cells, since all of the CPY had been processed to the mature (m) form. In the vti1–21, vti1–22, and vti1–23 strains, CPY was detected after the chase predominantly in the medial/trans-Golgi p2 form with a small amount of p1 form present. In contrast, in the vti1–24 and vti1–25 strains the ER/cis-Golgi p1 form of CPY was predominant, with a small amount of p2 form present (Figure 7A). Some of the p1 and most of the p2 form of CPY that accumulated in vti1–24 mutants was modified with Golgi-specific α-1,6-mannose glycosylation (Figure 7B), suggesting that delivery of CPY from the ER to the Golgi was not significantly compromised. All vti1 mutants were able to deliver CPY to the vacuole at the permissive temperature (our unpublished results).
The ability of the vti1 ts strains preincubated at the restrictive temperature to accumulate the p1 and p2 Golgi forms of CPY, in conjunction with their inability to generate the vacuolar mature (m) form suggested that transport of CPY through the Golgi toward the vacuole is compromised. In addition, the reduced intensity of the p2 form after the chase relative to the intensity of the p1 form before the chase suggested that some CPY was either degraded intracellularly or secreted from the cells. We found that all tested vti1 ts mutants secreted some p2 CPY at the semirestrictive temperature (Figure 7D). We have observed similar missorting of CPY in mutants of previously described intra-Golgi v-SNAREs Sft1p (Banfield et al., 1995) and Ykt6p (Søgaard et al., 1994; McNew et al., 1997) when they were incubated at semirestrictive conditions (our unpublished results).
To further test the effect of the ts mutations on protein secretion, we analyzed the transport of the yeast periplasmic enzyme invertase. Invertase synthesis was derepressed for 30 min on low-glucose media prior to the 30-min shift to the vti1 ts restrictive temperature, the cells were pulse labeled for 5 min, chased for 20 min, spheroplasted, and then separated into intracellular (I) and extracellular (E) fractions from which invertase was immunoprecipitated. We found that invertase was efficiently secreted from the vti1–22 and vti1–24 mutants at both the permissive and restrictive temperatures (Figure 7C) and only a small fraction of protein was retained intracellularly (about 10% or 35% of the total invertase at the restrictive temperature, respectively). Invertase secreted by both mutants at the restrictive temperature migrated faster in SDS-PAGE than the hyperglycosylated forms secreted from wild-type cells or from mutant cells at 25°C (Figure 7C), suggesting that it was underglycosylated. To observe post-ER glycosyl modifications in the vti1–22 and vti1–24 strains, we reimmunoprecipitated the invertase with an antisera against the α-1,6-mannose linkages, which are added in the cis- and medial-Golgi (Franzusoff and Schekman, 1989). Both the hyperglycosylated forms secreted at 25°C and, to a lesser degree, the intermediate form produced at 38°C were modified with α-1,6–mannose (Figure 7C). These results suggest that invertase export, unlike CPY traffic, was not seriously affected in the vti1 ts mutants.
To examine secretion in the vti1 ts mutants further, we harvested the cells, transferred them to fresh media, shifted the mutant strains to a semipermissive temperature (35°C) for 5 h, and analyzed the cells (I) and the extracellular medium (E) for several protein markers (Figure 7D). Similar to invertase, we found that secretion of gp400/hsp150, an O-glycosylated protein that is primarily localized extracellularly (Lupashin et al., 1992; Russo et al., 1992), occurred efficiently in all of the vti1 ts strains. In wild-type cells, gp400/hsp150 is secreted as a heteroligomer (7C and Lupashin et al., 1992) as a result of partial proteolytic cleavage by the Kex2p protease (Russo et al., 1992). We noted that gp400/hsp150 secreted by the vti1 ts strains migrated on SDS-PAGE as a single band, which may indicate defects in the Kex2p processing, glycosylation, or both. The lack of Kar2p in the media (Figure 7D) also suggests that HDEL-dependent retrieval of the ER luminal proteins (Dean and Pelham, 1990) from the cis-Golgi to the ER was not seriously affected by the vti1 mutations. These results indicate that a partial loss of Vti1p function compromises traffic of CPY without seriously affecting the transit of invertase or gp400/hsp150 to the cell surface.
Vti1p Is a Putative Intra-Golgi Retrograde v-SNARE
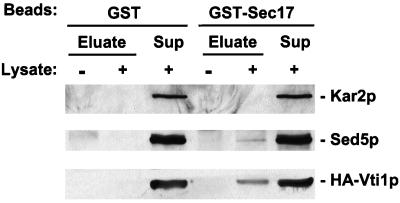
SNAREs, SNAP receptors by definition, are known to physically interact with SNAP molecules, which facilitate subsequent binding of NSF (Clary et al., 1990; Whiteheart et al., 1992; Söllner et al., 1993a,b). Yeast SNAP is encoded by the SEC17 gene (Griff et al., 1992) and NSF is encoded by SEC18 (Eakle et al., 1988; Wilson et al., 1989). To examine the possibility that Vti1p interacts with Sec17p, we incubated purified bacterially expressed GST or GST-Sec17p fusion protein with a cell lysate from a yeast strain expressing HA-Vti1p. GST and GST-Sec17p, as well as any associated proteins, were harvested by glutathione-Sepharose affinity. Bound proteins were eluted, separated by SDS-PAGE, transferred to nitrocellulose, and stained with Ponceau S to ensure that equivalent amounts of GST and GST-Sec17p were recovered from the binding reaction (our unpublished results). The immunoblots (Figure 8) were developed with anti-HA epitope antibodies to detect HA-Vti1p, as well as with anti-Sed5p (as a positive control) and anti-Kar2p (as a negative control) antibodies. Neither HA-Vti1p, Sed5p, nor Kar2p bound detectably to GST. However, both HA-Vti1p and Sed5p were found to be associated with GST-Sec17, whereas Kar2p was not. These data strongly indicate that Vti1p is a bona fide SNARE.
Figure 8.
Vti1p physically interacts with Sec17p. Spheroplasts of GWY154 cells (HA-VTI1) were lysed in the presence of Triton X-100. Solubilized proteins (0.5 mg) were incubated with purified bacterially expressed GST or GST-Sec17p (2 μg each) in buffer B (20 mM HEPES-KOH, 150 mM KOAc, 0.5 mM DTT, 0.05% Tween 20) for 1 h at 4°C and centrifuged at 15,000 × g for 15 min. The supernatant was incubated for 1 h at 4°C with 40 μl of glutathione-Sepharose 4B equilibrated with buffer B. Beads were washed three times with buffer B, and bound proteins were eluted with 50 μl of 10 mM reduced glutathione in 50 mM Tris-Cl (pH 7.5), separated by SDS-PAGE (10% acrylamide) followed by immunoblotting with anti-HA, anti-Sed5p, or anti-Kar2p antibodies. Beads that had not been incubated with yeast lysate were included as a primary antibody specificity control.
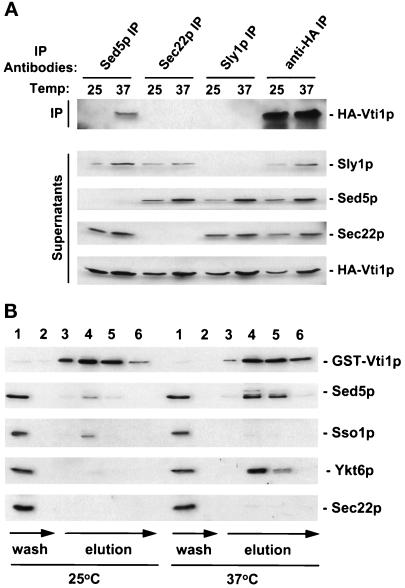
Since Sed5p and Vti1p are both localized to the Golgi (at least partially), we tested whether they can interact with each other in a SEC18-sensitive manner. Lysates were made from a sec18 strain expressing HA-Vti1p after incubation at the permissive and restrictive temperatures, Sed5p was immunoprecipitated, and precipitate was probed for the presence of HA-Vti1p (Figure 9A, Sed5 IP). We found that HA-Vti1p associated with the cis-Golgi t-SNARE Sed5p only at the sec18 restrictive temperature, suggesting that Vti1p is a v-SNARE that participates in vesicular transport to the cis-Golgi. Since Sed5p can interact with the ER-to-Golgi v-SNARE Sec22p and form a separate complex with its negative regulator Sly1p (Søgaard et al., 1994; Lupashin and Waters, 1997), we tested the interaction of Vti1p with each of these proteins. Although the immunoprecipitations of Sec22p and Sly1p were complete (Figure 9A, Supernatants), no interaction of Vti1p with either of these proteins was detected (Figure 9A, Sec22 and Sly1 IP). These data suggest that the t-SNARE Sed5p can enter into at least two distinct v/t-SNARE complexes: one containing Sec22p and another containing Vti1p.
Figure 9.
Vti1p interacts with Sed5p and Ykt6p in a SEC18-sensitive manner. (A) Interaction of Vti1p and Sed5p is specific. Spheroplasts of GWY157 cells (sec18–1 with chromosomal HA-VTI1) were incubated at the permissive (25°C) or restrictive (37°C) temperature for 1 h, lysed in the presence of Triton X-100, and proteins immunoprecipitated (IP) with affinity-purified antibody to Sed5p, Sec22p, Sly1p, or the anti-HA monoclonal antibody (12CA5). The IPs (upper panel) and one-fifth of the corresponding supernatants (lower panel) were immunoblotted to detect HA-Vti1p, Sed5p, Sec22p, and Sly1p. (B) GST-Vti1p interactions. GWY156 cells (sec18–1 with GST-VTI1) were incubated at the permissive (25°C) or restrictive (37°C) temperature for 1 h, chilled on ice, and lysed with glass beads in the presence of 1% Triton X-100. Clarified detergent lysates (∼2 mg of protein) were incubated for 1 h at 4°C with 300 μl of glutathione-Sepharose 4B equilibrated with buffer B (see MATERIALS AND METHODS). Beads were transferred into 1-ml chromatography columns, washed with buffer B (wash), and bound proteins were eluted (elution) with 10 mM reduced glutathione in 50 mM Tris-Cl (pH 7.5). Fractions (300 μl each) were collected and proteins were separated by SDS-PAGE followed by immunoblotting with anti-GST, anti-Sed5p, anti-Sso1, anti-Ykt6p, or anti-Sec22p antibodies.
In an effort to confirm this data, we attempted to immunoprecipitate HA-Vti1p with the aim of identifying interacting factors. Unfortunately, the recovery of HA-Vti1p was incomplete (Figure 9A, anti-HA IP and Supernatant). We therefore examined copurification of several factors with GST-Vti1p (Figure 9B) in a sec18 strain. The GST-Vti1p-expressing sec18 cells were grown to midlogarithmic phase, spheroplasted, and incubated for 1 h at the sec18 permissive or restrictive temperature. After incubation, lysates were prepared and GST-Vti1p was recovered on glutathione-Sepharose beads. Eluates from the beads were analyzed for their Sed5p, Sso1p, Ykt6p, or Sec22p content (Figure 9B). Ykt6p was discovered as a component of the cis-Golgi v/t-SNARE complex (Søgaard et al., 1994), and is thought to be a v-SNARE acting in ER-to-Golgi transport (McNew et al., 1997). Sso1p is a plasma membrane t-SNARE (Aalto et al., 1993). Interestingly, we found that the inactivation of Sec18p resulted in accumulation of Vti1p/Sed5p and Vti1p/Ykt6p complexes. No Vti1p/Sec22p or Vti1p/Sso1p complexes accumulated at the sec18 restrictive temperature.
Genetic Interactions of VTI1
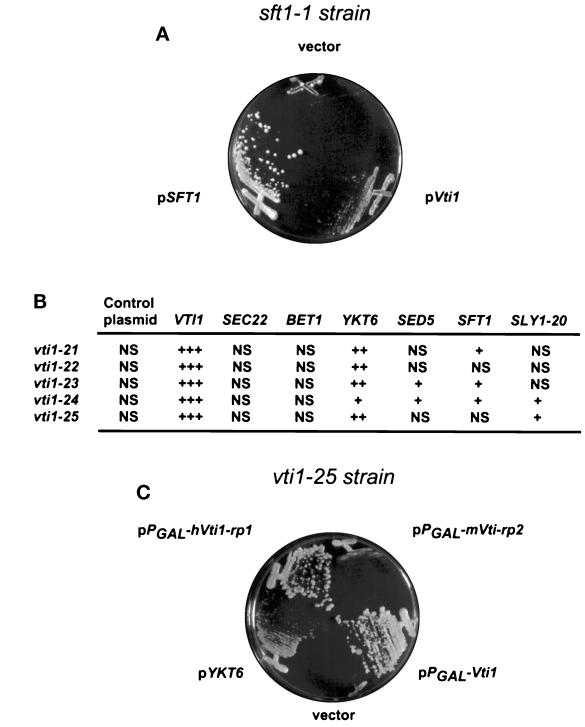
The protein–protein interaction studies and the analyses of the effects of Vti1p depletion or mutation on protein trafficking suggested that Vti1p is required for intra-Golgi protein traffic. To gain more insight into the role of Vti1p, we undertook an analysis of the genetic interactions of VTI1 with other genes involved in ER-to-Golgi and intra-Golgi traffic. High levels of Vti1p were expressed from a 2-μm plasmid in each of the following ts strains, but suppression of the growth defect at the restrictive temperatures was not evident: sec22–3, bos1–1, bet1–1 (ER-to-Golgi v-SNAREs), uso1–1 (ER-to-Golgi transport factor), sly1ts (t-SNARE-associated regulatory protein), sec19–1 (GDP dissociation inhibitor for Ypt1p and Sec4p), sec7–1 (ER-to-Golgi and intra-Golgi docking factor), ypt1–3 (ER-to-Golgi Rab-like GTPase), ypt6ts (intra-Golgi Rab-like GTPase), sec4–8 (Golgi-to-plasma membrane Rab-like GTPase), sec21–1 and sec21–2 (γ-COP), sec27–1 (β′-COP), and ret1–1 (α-COP), sec35–1, and sec34–2 (novel ER-to-Golgi transport factors). In contrast, high levels of Vti1p expression could partially suppress the sft1–1 temperature-sensitive growth defect (Figure 10A).
Figure 10.
Multicopy suppression of sft1–1 and vti1 ts growth defect. (A) Vti1p overexpression suppresses the growth defect of the sft1–1. Growth at 35°C on YPD plates of the sft1–1 strain carrying multicopy plasmids bearing VTI1, SFT1, or no insert as indicated. (B) Growth at (37.5°C) on YPD plates of the indicated mutants carrying 2μ multicopy plasmids bearing the indicated genes. +++, wild-type growth; ++, good growth; +, poor growth; NS, no suppression. (C) Human Vti1p homologue (hVti1-rp1) expression suppresses the growth defect of the vti1–25 mutant allele. Growth at 37°C on YPGal plates of the vti1–25 strain carrying multicopy PGAL plasmids bearing VTI1, hVti1-rp1, mVti1-rp2, or no insert as indicated. YKT6 was not under PGAL control.
We also tested whether any of several known genes could act as multicopy suppressors of the growth defects of the vti1 ts alleles. As shown in Figure 10B, the temperature sensitivity of all tested vti1 ts strains could be completely or partially suppressed by overexpression of the SNARE YKT6 from a 2-μm plasmid. In addition, the vti1–23 and vti1–24 temperature-sensitive growth defect was partially suppressed by a multicopy plasmid carrying genes encoding the putative intra-Golgi retrograde t-SNARE SFT1 (Banfield et al., 1995), the ER-to-Golgi t-SNARE SED5 (Hardwick and Pelham, 1992), or a dominant mutation of SLY1, termed SLY1–20 (Ossig et al., 1991). SEC22 and BET1 (Newman et al., 1990; Ossig et al., 1991), v-SNAREs involved in ER-to-Golgi transport, were also tested for the ability to suppress vti1 ts mutants, but these genes did not improve the growth of vti1 ts strains at the restrictive temperature.
Finally, to test whether any of the mammalian sequences that encode Vti1p homologues (Figure 1B) were capable of complementing a Vti1p defect, we tested the ability of the human homologue, hVti1-rp1, and the mouse homologue mVti1-rp2 to suppress the growth defects of the vti1 ts alleles. Expression of hVti1-rp1 under control of PGAL completely suppressed the growth defect of the vti1–25 (Figure 10C) and partially suppressed the growth defect of other vti1 ts alleles (our unpublished results). In contrast, suppression by the mouse homologue, mVti1-rp2, was very weak.
DISCUSSION
In this study, we used protein homology searches of the complete yeast genome database to identify five ORFs that could encode novel SNARE-like proteins. We have performed a functional analysis of one of these, named Vti1p. This 24.7-kDa type II transmembrane protein is most closely related to the putative intra-Golgi retrograde v-SNARE Sft1p (Banfield et al., 1995) and is essential for vegetative growth of yeast cells. Subcellular fractionation and immunofluorescence suggested that Vti1p is localized to the Golgi apparatus and perhaps to the prevacuolar compartment as well (see also Fischer Von Mollard et al., 1997). We have confirmed that Vti1p is a SNARE by showing that it physically interacts with yeast SNAP (Sec17p).
Reduction of the cellular level of Vti1p, expressed under the control of the glucose-repressible GAL1 promoter, resulted in a defect in the intra-Golgi transport of CPY prior to cessation of cell growth. Both the p1 (ER/cis-Golgi) and p2 (medial/trans-Golgi) forms of CPY accumulated intracellularly upon loss of Vti1p, suggesting that this SNARE is required, either directly or indirectly, for CPY transport through multiple compartments of the Golgi.
To further define the role of this Golgi SNARE, we generated a set of temperature-conditional vti1 mutants and studied their secretion phenotypes. Pulse-chase analysis of CPY transport in these strains indicated that the mutants accumulate the p1 and/or p2 forms of CPY at the restrictive temperature. In addition, we found that some p2 CPY was secreted in the vti1 ts mutants. In a similar pulse-chase experiment, we found that the secretion of invertase was not significantly compromised, but that the secreted invertase lacked the extensive outer-chain glycosylation which is normally added in the Golgi. Another extracellular protein, termed gp400/hsp150, was also secreted by the vti1 ts strains.
This combination of secretion phenotypes is unusual. For example, the accumulation of p1 and p2 suggests an intra-Golgi block, whereas secretion of p2 CPY indicates that anterograde transport to the cell surface can still occur and that vacuolar protein sorting may be defective. The continued secretion of invertase and gp400/hsp150 in the vti1 ts mutants also indicates that anterograde transport is operational, but the finding that the secreted invertase is underglycosylated suggests a defect in Golgi glycosylation.
In an effort to understand how a loss of Vti1p function could lead to this spectrum of secretion phenotypes, we identified the SNAREs with which Vti1p interacts. Since Vti1p is localized, at least partially, to the Golgi apparatus and it is most homologous to Sft1p (Banfield et al., 1995) and Bet1p (Newman et al., 1990), two v-SNAREs known to interact with the cis-Golgi t-SNARE Sed5p (Søgaard et al., 1994; McNew et al., 1997), we tested whether Vti1p could interact with Sed5p. Indeed, Vti1p physically interacts with Sed5p in a complex that accumulates in the absence of NSF (Sec18p) activity. This suggests that Vti1p is a v-SNARE that can target vesicles to the cis-Golgi. Interestingly, the Vti1p/Sed5p complex is distinct from the well-characterized Sec22p/Sed5p complex (Søgaard et al., 1994) that targets ER-derived vesicles to the cis-Golgi. It has previously been proposed, on the basis of genetic evidence, that Sed5p can act as the cis-Golgi docking site for both anterograde ER-to-Golgi vesicles as well as retrograde intra-Golgi vesicles (Banfield et al., 1995). This concept has recently been supported by the observation that the mammalian homologue of Sed5p, syntaxin 5, can enter into at least two unique v/t-SNARE complexes (Hay et al., 1997). Our protein–protein interaction data demonstrate that a similar situation exists in yeast for Sed5p and also suggests that Vti1p is a retrograde v-SNARE.
Support for the idea that Vti1p is a retrograde cis-Golgi-directed was obtained by analysis of the multicopy suppression profile of the vti1 ts mutants. First, we found that overexpression of Sed5p, or Sly1–20p, which physically interacts with Sed5p and facilitates its function (Dascher et al., 1991, Søgaard et al., 1994, Grabowski and Gallwitz, 1997), could suppress vti1 mutants in an allele specific manner. This finding is consistent with the physical interaction of Vti1p with Sed5p. In contrast, overexpression of the ER-to-Golgi v-SNARE Sec22 did not suppress any of the vti1 ts mutants, reminiscent of its lack of physical interaction with Vti1p. Interestingly, overexpression of Sft1p, which is a putative retrograde intra-Golgi v-SNARE (Banfield et al., 1995), could weakly suppress several vti1 ts alleles; likewise overexpression of Vti1p suppressed sft1–1. Finally, high levels of Ykt6p, an isoprenylated v-SNARE involved in early Golgi traffic (McNew et al., 1997), efficiently rescued all of the vti1 ts mutants.
In agreement with this genetic suppression data, we found that Vti1p physically interacts with Ykt6p. To summarize then, Vti1p can physically interact with the cis-Golgi t-SNARE Sed5p, and the v-SNARE Ykt6p, but not with the anterograde ER-to-Golgi v-SNARE Sec22p. VTI1 also genetically interacts with the putative intra-Golgi v-SNARE SFT1.
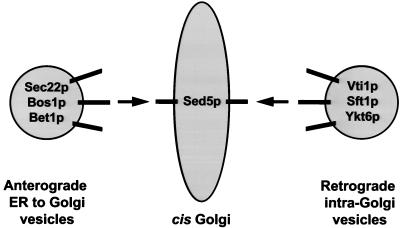
These protein–protein interaction and genetic analyses suggest a model for the role of Vti1p in vesicular traffic (Figure 11). We propose that Sed5p marks the cis-Golgi docking site for ER-to-Golgi vesicles bearing the anterograde v-SNAREs Sec22p, Bet1p, and Bos1p as well for retrograde intra-Golgi vesicles that display the v-SNAREs Vti1p, Sft1p, and Ykt6p.
Figure 11.
Model for Vti1p function.
This model provides the framework for interpretation of the secretion phenotypes exhibited by the vti1 mutants. First, we suggest that anterograde transport of invertase and gp400/hsp150 is largely unaffected by loss of Vti1p activity because the primary function of this SNARE is in retrograde transport. The inefficient anterograde transport of CPY could be a secondary manifestation of the retrograde transport defect if CPY requires recycling of components that facilitate its forward movement, e.g., Vps10p (Marcusson et al., 1994). This model was also proposed recently by Gaynor and Emr (1997) who found similar transport phenotypes when studying the role of Sec21p, the γ-subunit (γ-COP) of yeast coatomer (Hosobuchi et al., 1992), in retrograde transport from the Golgi to the ER. Whereas mutants in γ-COP exhibited a CPY-specific ER-to-Golgi transport defect, mutants in Vti1p allowed CPY transit from the ER-to-Golgi, but compromised further forward transport. Similar effects on CPY and invertase transport were demonstrated previously in the sft1 mutant (Banfield et al., 1995). Finally, we suggest that the inefficient glycosylation of invertase seen in the vti1 mutants is due to mislocalization of the Golgi glycosyltransferases such as the α-1,6-mannosyltransferase, Och1p, which must be recycled to maintain its cis-Golgi localization (Harris and Waters, 1996).
The recent work of Fischer von Mollard et al. (1997) provides another model that is not mutually exclusive with ours. In their work they showed that Vti1p interacts with both Sed5p and with Pep12p, the t-SNAREs that marks the prevacuolar compartment (Becherer et al. 1996). This suggests that Vti1p may be involved in docking of late Golgi-derived vesicles with the prevacuolar compartment (Fischer von Mollard et al., 1997) as well as retrograde transport to the cis-Golgi.
ACKNOWLEDGMENTS
We thank P. Brennwald, E. Jones, S. Ferro-Novick, M. Latterich, S. Michaelis, H. Pelham, M. Rose, J. Rothman, and R. Schekman for materials. We thank J. Goodhouse for technical assistance and S. Harris, K. Paul, J. Rothman, T. Söllner, and S. VanRheenen for discussions and critical reading of the manuscript. The collegiality of G. Fischer von Mollard and T. Stevens was greatly appreciated. This work was supported by the National Institutes of Health (GM-47853), National Science Foundation (MCB 9631129), and Lucille P. Markey Charitable Trust.
Footnotes
Abbreviations used: CEN, centromere; CPY, carboxypeptidase Y; ER, endoplasmic reticulum; EST, expressed sequence tag; GST, glutathione S-transferase; HA, hemagglutinin; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; PGAL, GAL1 promoter; PCUP, CUP1 promoter.
REFERENCES
- Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Baldari C, Murray JAH, Ghiara P, Cesareni G, Galeotii CL. A novel leader peptide which allows efficient secretion of a fragment of human interleukin 1β in Saccharomyces cerevisiae. EMBO J. 1987;6:229–234. doi: 10.1002/j.1460-2075.1987.tb04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Pelham HRB. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–809. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homolog, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK. SNAREs and the specificity of transport vesicle targeting. Curr Opin Cell Biol. 1995;7:581–586. doi: 10.1016/0955-0674(95)80016-6. [DOI] [PubMed] [Google Scholar]
- Block MR, Glick BS, Wilcox CA, Wieland FT, Rothman JE. Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc Natl Acad Sci USA. 1988;85:7852–7856. doi: 10.1073/pnas.85.21.7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Clary DO, Griff IC, Rothman JE. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell. 1990;61:709–721. doi: 10.1016/0092-8674(90)90482-t. [DOI] [PubMed] [Google Scholar]
- Dascher C, Ossig R, Gallwitz D, Schmitt HD. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N, Pelham HR. Recycling of proteins from the Golgi compartment to the ER in yeast. J Cell Biol. 1990;111:369–377. doi: 10.1083/jcb.111.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eakle KA, Bernstein M, Emr SD. Characterization of a component of the yeast secretion machinery: identification of the SEC18 gene product. Mol Cell Biol. 1988;8:4098–4109. doi: 10.1128/mcb.8.10.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- Fiedler K, Veit M, Stamnes MA, Rothman JE. Bimodal interaction of coatomer with the p24 family of putative cargo receptors. Science. 1996;273:1296–1299. doi: 10.1126/science.273.5280.1396. [DOI] [PubMed] [Google Scholar]
- Fischer von Mollard G, Nothwehr SF, Stevens TH. The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J Cell Biol. 1997;137:1511–1524. doi: 10.1083/jcb.137.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A, Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromant M, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Emr SD. COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J Cell Biol. 1997;136:789–802. doi: 10.1083/jcb.136.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski R, Gallwitz D. High-affinity binding of the yeast cis-Golgi t-SNARE, Sed5p, to wild-type and mutant Sly1p, a modulator of transport vesicle docking. FEBS Lett. 1997;411:169–172. doi: 10.1016/s0014-5793(97)00720-5. [DOI] [PubMed] [Google Scholar]
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18 (NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griff IC, Schekman R, Rothman JE, Kaiser CA. The yeast SEC17 gene product is functionally equivalent to mammalian α-SNAP protein. J Biol Chem. 1992;267:12106–12115. [PubMed] [Google Scholar]
- Halachmi N, Lev Z. The Sec1 family: a novel family of proteins involved in synaptic transmission and general secretion. J Neurochem. 1996;66:889–897. doi: 10.1046/j.1471-4159.1996.66030889.x. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HR. SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- Hay JC, Hirling H, Scheller RH. Mammalian vesicle trafficking proteins of the endoplasmic reticulum and Golgi apparatus. J Biol Chem. 1996;271:5671–5679. doi: 10.1074/jbc.271.10.5671. [DOI] [PubMed] [Google Scholar]
- Hosobuchi M, Kreis T, Schekman R. SEC21 is a gene required for ER to Golgi protein transport that encodes a subunit of a yeast coatomer. Nature. 1992;360:603–605. doi: 10.1038/360603a0. [DOI] [PubMed] [Google Scholar]
- Jackson MR, Nilsson T, Peterson PA. Retrieval of transmembrane proteins to the endoplasmic reticulum. J Cell Biol. 1993;121:317–333. doi: 10.1083/jcb.121.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Lupashin VV, Kononova SV, Ratner YN, Tsiomenko AB, Kulaev IS. Identification of a novel secreted glycoprotein of the yeast Saccharomyces cerevisiae stimulated by heat shock. Yeast. 1992;8:157–169. doi: 10.1002/yea.320080302. [DOI] [PubMed] [Google Scholar]
- Lupashin VV, Waters MG. t-SNARE activation through transient interaction with a Rab-like guanosine triphosphatase. Science. 1997;276:1255–1258. doi: 10.1126/science.276.5316.1255. [DOI] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Mayer A, Wickner W. Docking of yeast vacuoles is catalyzed by the Ras-like GTPase Ypt7p after symmetric priming by Sec18p (NSF) J Cell Biol. 1997;136:307–313. doi: 10.1083/jcb.136.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (α-SNAP) can precede docking and fusion of yeast vacuoles. Cell. 1996;85:83–94. doi: 10.1016/s0092-8674(00)81084-3. [DOI] [PubMed] [Google Scholar]
- McNew, J.A., Søgaard, M., Lampen, N.M., Machida, S., Ye, R.R., Lacomis, L., Tempst, P., Rothman, J.E., and Söllner, T.H. (1997). Ykt6p, a prenylated SNARE essential for ER-Golgi transport. J. Biol. Chem. (in press). [DOI] [PubMed]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Nagahama M, Orci L, Ravazzola M, Amherdt M, Lacomis L, Tempst P, Rothman JE, Söllner TH. A v-SNARE implicated in intra-Golgi transport. J Cell Biol. 1996;133:507–516. doi: 10.1083/jcb.133.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossig R, Dascher C, Trepte HH, Schmitt HD, Gallwitz D. The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1 protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol. 1991;11:2980–2993. doi: 10.1128/mcb.11.6.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek I, Orci L, Ravazzola M, Erdjument-Bromage H, Amherdt M, Tempst P, Söllner TH, Rothman JE. ERS-24, a mammalian v-SNARE implicated in vesicle traffic between the ER and the Golgi. J Cell Biol. 1997;137:1017–1028. doi: 10.1083/jcb.137.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G. Intracellular aspects of the process of protein synthesis (secretion) Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Braun JE, Calakos N, Ting AE, Bennett MK, Scheller RH. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. Transport vesicle docking: SNAREs and associates. Annu Rev Cell Dev Biol. 1996;12:441–461. doi: 10.1146/annurev.cellbio.12.1.441. [DOI] [PubMed] [Google Scholar]
- Rose M, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Rothman JE. Mechanisms of intracellular protein transport. Nature. 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Warren G. Implications of the SNARE hypothesis for intracellular membrane topology and dynamics. Curr Biol. 1994;4:220–232. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Russo P, Kalkkinen N, Sareneva H, Paakkola J, Makarow M. A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc Natl Acad Sci USA. 1992;89:3671–3675. doi: 10.1073/pnas.89.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein SK, Lupashin VV, Schmitt HD, Waters MG. Assembly of the ER to Golgi SNARE complex requires Uso1p. J Cell Biol. 1996;132:755–767. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R, Orci L. Coat proteins and vesicle budding. Science. 1996;271:1526–1533. doi: 10.1126/science.271.5255.1526. [DOI] [PubMed] [Google Scholar]
- Schimmöller F, Singer-KrŸger B, Schröder S, KrŸger U, Barlowe C, Riezman H. The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J. 1995;14:1329–1339. doi: 10.1002/j.1460-2075.1995.tb07119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze M-P, Peterson PA, Jackson MR. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 1994;13:1696–1705. doi: 10.1002/j.1460-2075.1994.tb06434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Søgaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Söllner T. A Rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Söllner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Subramaniam VN, Peter F, Philp R, Wong SH, Hong W. GS28, a 28-kilodalton Golgi SNARE that participates in ER-Golgi transport. Science. 1996;272:1161–1163. doi: 10.1126/science.272.5265.1161. [DOI] [PubMed] [Google Scholar]
- Tagaya M, Wilson DW, Brunner M, Arango N, Rothman JE. Domain structure of an N-ethylmaleimide-sensitive fusion protein involved in vesicular transport. J Biol Chem. 1993;268:2662–2666. [PubMed] [Google Scholar]
- Weimbs T, Low SH, Chapin SJ, Mostov KE, Bucher P, Hofmann K. A conserved domain is present in different families of vesicular fusion proteins: A new superfamily. Proc Natl Acad Sci USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteheart SW, Brunner M, Wilson DW, Wiedmann M, Rothman JE. Soluble N-ethylmaleimide-sensitive fusion attachment proteins (SNAPs) bind to a multi-SNAP receptor complex in Golgi membranes. J Biol Chem. 1992;267:12239–12243. [PubMed] [Google Scholar]
- Whiteheart SW, Rossnagel K, Buhrow SA, Brunner M, Jaenicke R, Rothman JE. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J Cell Biol. 1994;126:945–954. doi: 10.1083/jcb.126.4.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DW, Wilcox CA, Flynn GC, Chen E, Kuang WJ, Henzel WJ, Block MR, Ullrich A, Rothman JE. A fusion protein required for vesicle-mediated transport in both mammalian cells and yeast. Nature. 1989;339:355–359. doi: 10.1038/339355a0. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Niman HL, Houghten RA, Cherenson ML, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]