Abstract
The multicopper oxidase (MCO) family of enzymes includes laccases, which oxidize a broad range of substrates including diphenols, and several oxidases with specific substrates such as iron, copper or ascorbic acid. We have identified five putative MCO genes in the genome of Anopheles gambiae and have cloned cDNAs encompassing the full coding region for each gene. MCO1 mRNA was detected in all developmental stages and in all of the larval and adult tissues tested. We observed an increase in MCO1 transcript abundance in the midguts and Malphighian tubules of adult females following a blood meal and in adult abdominal carcasses in response to an immune challenge. Two alternatively spliced isoforms of MCO2 mRNA were identified. The A isoform of MCO2 was previously detected in larval and pupal cuticle where it probably catalyzes sclerotization reactions (He et al., 2007). The B isoform was transcriptionally upregulated in ovaries in response to a blood meal. MCO3 mRNA was detected in the adult midgut, Malpighian tubules, and male reproductive tissues; like MCO1, it was upregulated in response to an immune challenge or a blood meal. MCO4 and MCO5 were observed primarily in eggs and in the abdominal carcass of larvae. A phylogenetic analysis of insect MCO genes identified putative orthologs of MCO1 and MCO2 in all of the insect genomes tested, whereas MCO3, MCO4 and MCO5 were found only in the two mosquito species analyzed. MCO2 orthologs have especially high sequence similarity, suggesting that they are under strong purifying selection; the A isoforms are more conserved than the B isoforms. The mosquito specific group shares a common ancestor with MCO2. This initial study of mosquito MCOs suggests that MCO2 may be required for egg development or eggshell tanning in addition to cuticle tanning, while MCO1 and MCO3 may be involved in metal metabolism or immunity.
Keywords: laccase, multicopper oxidase, cuticle tanning, sclerotization, detoxification, immunity, metal metabolism
1. Introduction
The multicopper oxidase (MCO) family of enzymes includes laccases, ferroxidases, cuprous oxidases, ascorbate oxidases and bilirubin oxidases (Sakurai and Kataoka, 2007). Laccases have exceptionally broad substrate specificity whereas the other family members tend to oxidize a single substrate in vivo: iron, copper, ascorbic acid, and bilirubin, respectively. A partial list of laccase substrates includes diphenols, substituted monophenols, diamines, N-hydroxy compounds, and lignins (Thurston, 1994; Baldrian, 2006). Laccases have been discovered in bacteria, fungi, plants and insects. MCOs that function as metal oxidases have been identified from bacteria, fungi and plants but not insects; ascorbate oxidases have been found mainly in plants; and bilirubin oxidases are fungal enzymes (e.g., Huston et al., 2002; Quintanar et al., 2004; Larrondo et al., 2003; Hoopes and Dean, 2004; Sakurai and Kataoka, 2007). Some of the functions associated with multicopper oxidases include pigmentation, morphogenesis, detoxification, and lignin degradation (Hullo et al., 2001; Mayer and Staples, 2002).
Depsite their diverse functions, MCOs are evolutionarily and structurally related (Nakamura and Go, 2005; Hoegger et al., 2006). Most MCOs bind four copper atoms within two highly conserved copper centers (designated T1 and T2/T3). The substrate is bound near the T1 center, and dioxygen is bound near the T2/T3 center. Oxidation of a substrate ocurrs when an electron is transferred from the substrate to the T1 copper, which transfers the electron to the T2/T3 copper center where dioxygen is reduced to water (Solomon et al., 1996). The copper binding residues (10 histidines and one cysteine) in the T1 and T2/T3 centers can be used as signature residues to identify putative MCO genes (Nakamura and Go, 2005).
Of the insect genomes that have been investigated, each contains at least two putative MCO genes, and some genomes contain more (Dittmer et al, 2004). The genome of Anopheles gambiae, a species of mosquito, encodes five putative MCO genes (Dittmer et al., 2004). At present, the function of just one type of insect MCO is known: cuticle tanning catalyzed by laccase 2 (Lac2) orthologs. In Manduca sexta, MsLac2 is expressed in cuticle undergoing sclerotization (Dittmer et al., 2004), and it oxidizes substrates that have been implicated in cuticle sclerotization (Dittmer and Kanost, unpublished data). In Tribolium castaneum, RNAi mediated silencing of TcLac2 causes newly synthesized larval, pupal and adult cuticle to remain soft and pale instead of undergoing the normal hardening and darkening process (Arakane et al., 2005).
The functions of laccase 1 type enzymes and other insect MCOs are still unknown. In T. castaneum, silencing of TcLac1 had no effect on cuticle tanning or viability (Arakane et al., 2005). The putative Lac1 ortholog in Drosophila melanogaster (predicted gene CG3759) is expressed in Malpighian tubules (Wang, 2004) and is slightly upregulated in unspecified tissues in response to immune challenge suggesting a possible immune function (De Gregorio et al., 2001). MsLac1 is expressed in the midgut and Malpighian tubules of feeding stage larvae but not in later, non-feeding stage larvae or pupae. This expression pattern may indicate a role in detoxification of phenolic plant compounds or in metabolizing iron or copper in the larval diet (Dittmer et al., 2004).
Several additional functions of insect laccases have been proposed. One of the first suggested roles for an insect laccase was sclerotization of the egg case of a cockroach species, Periplaneta americana (Whitehead et al., 1960). A laccase-like activity in the saliva of a species of leafhopper, Nephotettix cincticeps, may detoxify monolignols found in plants by oxidizing the monolignols, which may then form nontoxic polymers prior to ingestion by the leafhopper (Hattori et al., 2005). It has been suggested that a laccase present in the midgut of Anopheles stephensi may play a role in defense against parasites (Sidjanski et al., 1997), that a laccase in parasitoid wasp venom may affect host responses to the parasitoid's eggs (Parkinson et al., 2003), and that laccases may be involved in hemolymph coagulation (Theopold et al., 2002). Finally, an uncharacterized phenoloxidase that could be a laccase has been implicated in mating plug formation in Drosophila nasuta (Asada and Kitagawa, 1988). It is important to note that these hypotheses of laccase function, except for catalyzing cuticle tanning, remain unproven.
The aim of this study was to characterize MCO genes in A. gambiae. Based on our understanding of the functions of non-insect MCOs and on the preliminary observations of insect MCOs described above, we predicted that mosquito MCOs may be involved in one or more of the following: cuticle sclerotization, pigmentation, eggshell tanning, mating plug formation, immune processes, detoxification of phenolics, or metal metabolism. Three approaches were used to identify potential functions of the A. gambiae MCOs. First, we analyzed the predicted amino acid sequences to identify similarities between the mosquito MCOs and MCOs from other organisms. Then, orthologs were identified by determining the phylogenetic relationships of MCO genes from seven insect species. Finally, mRNA expression profiles were analyzed by developmental stage, tissue specificity, response to blood feeding and response to immune challenge.
2. Materials and Methods
2.1 Mosquito culture
The G3 strain of Anopheles gambiae was obtained from the Malaria Research and Reference Reagent Resource Center. The mosquitoes were reared as described by Benedict (1997) with minor modifications. Briefly, larvae were reared at 27°C and were fed a mixture of baker’s yeast and ground fish food (Vitapro Plus Staple Power Flakes, Mike Reed Enterprises). Adults were maintained at 27°C with 85% relative humidity and were fed 10% sucrose. Adult females were fed heparinized equine blood with a membrane feeder (Hemotek).
2.2 Tissue preparation for RNA isolation
2.2.1 Developmental profile
Eggs from an overnight collection were combined with eggs that were between one and two days old. Two day old larvae were a mix of first and second instar larvae; six day old, third and fourth instar larvae; and nine day old, a mix of fourth instar larvae and pharate pupae. The pupae collected were tan or black (pharate adults). Female and male adults were two days old.
2.2.2 Tissue profiles
Insects were dissected in phosphate buffered saline (PBS, 10 mM sodium phosphate, 175 mM sodium chloride, pH 7.2), and tissues were placed in tubes on dry ice until all samples were collected. The samples were stored at −80°C until RNA isolation was done. Midguts and Malpighian tubules were removed from the abdomen of fourth instar larvae, and the remainder of the abdomen (“carcass”) was retained. Midguts, Malpighian tubules, ovaries, and testes plus accessory glands were dissected from 29 adult males and 17 adult females (nine days old).
2.2.3 Blood meal profiles
Adult females (4–6 days old) were fed heparinized equine blood with a membrane feeder. Mosquitoes that took a full blood meal were maintained at 27°C and 85% relative humidity, and they had access to sucrose solution but no dishes for egg laying. After 3, 6, 12, 24, 48, and 72 hours, mosquitoes were chilled on ice and dissected in PBS. The midguts (with the gut contents removed), Malpighian tubules, and ovaries were collected. Each sample represented 18 insects.
2.2.4 Immune challenge profile
Adult females 3 days old were chilled on ice to immobilize them, then pricked with a minuten pin dipped in a paste of freeze-dried Micrococcus luteus, a Gram positive species of bacteria. Control mosquitoes were not pricked. After 1, 3, 6, and 12 hours, mosquitoes were chilled on ice and dissected in PBS. The spermatheca, midgut, Malpighian tubules, ovaries and crop were removed from each mosquito, and the remaining abdominal tissue, which we refer to as the abdominal carcass, was retained for RNA isolation. Each sample represented six insects. The experiment was done twice.
2.3 RNA isolation and cDNA synthesis
Total RNA was isolated using the Ultraspec reagent method (Biotecx) or Tri reagent method (Sigma), and genomic DNA was removed with DNAseI. Pools of cDNA were synthesized with the SuperScript First-Strand Synthesis System for RT-PCR (Invitrogen) and oligo(dT) primers. One µl of cDNA was used for each PCR. Typically, 30 PCR cycles were done. For semiquantitative analysis of the effect of blood feeding or immune challenge on expression, the number of cycles required to produce faint but visible bands was emperically determined for each set of primers. The ribosomal protein S7 gene (Salazar et al., 1993) was used as a reference gene. At the start of this study, PCR products obtained from reactions using gene specific primers were cloned and sequenced to verify that the expected gene products had been amplified.
2.4 cDNA cloning
The cloning of a full length MCO1 cDNA was described previously (Dittmer et al., 2004). (Note that we have changed the name of this gene from laccase-1 to MCO1 because its predicted amino acid sequence suggests that it is a multicopper oxidase, but its substrate specificity is unkown and may not be laccase-like.) To obtain cDNA clones of MCO2A, MCO2B, MCO4 and MCO5, partial cDNA sequences were predicted from genomic DNA sequences (available in GenBank), primers for 5’ and 3’ Rapid Amplification of cDNA Ends (RACE) were designed based on the predicted sequences, and 5’ and 3’ regions were cloned with a RACE kit (GeneRacer, Invitrogen). The developmental profile (section 2.2.1) was used to identify life stages in which specific transcripts were abundantly expressed. Using primers encoding the start and stop codon regions or untranslated sequences, cDNAs encompassing the full coding regions of these genes were amplified from cDNA pools from pharate adults (MCO2A), day 2 larvae (MCO2B), day 9 larvae (MCO4), or day 6 larvae (MCO5). To obtain a cDNA clone of MCO3, primers based on an EST consensus sequence were used to amplify a full coding region cDNA from a pool of adult female cDNAs. Nucleotide sequences were confirmed by DNA sequencing of the full-length clones (GenBank accession numbers: MCO1, AY135184; MCO2A, AY943928; MCO2B, AY943929; MCO3, EF592176; MCO4, EU380796; MCO5, EU380797).
2.5 Analysis of gene structure
The gene structure of each of the A. gambiae MCO genes was determined by comparing cDNA sequences with genomic DNA sequences (available through Ensembl). The program Splign (Kapustin et al., 2004) was used to determine intron/exon boundaries. The MCO2 gene appeared to encode alternative exons (exons 6A-9A and 6B-9B). To determine which of the five potential combinations of exons are expressed, exon specific primers were designed, and PCR was used to look for each possibility in cDNA pools from eggs, day 6 larvae, pharate adults, and adult females.
2.6 Analysis of predicted amino acid sequences
Signal sequences and signal anchors were predicted by Signal P (Bendtsen et al., 2004).
2.7 Phylogenetic analysis
Phylogenetic analyses were performed using the MEGA version 3.1 software (Kumar et al., 2004). Sequences were aligned globally using the Clustal W program in MEGA and then adjusted manually by eye (see supplementary Figures S2 and S3 for the alignments). Trees were constructed by either the neighbor-joining method with a Poisson correction model, or by maximum parsimony using a heuristic approach with the close-neighbor-interchange method (with a search level of 1) and an initial tree generated by random addition with 10 repititions. For both methods gaps were treated as characters and statistical analysis was performed by the bootstrap method using 1,000 repetitions. The sequences (with GenBank accession number) used for the analyses were: Aedes aegypti AaLac1 (DQ011288), AaLac2 (DQ013890), AAEL007415 (XM_001658292), AAEL001632 (XM_001653679), AAEL001640 (XM_001653679), AAEL007802 (XM_001652867); Anopheles gambiae AgMCO1 (AY135184), AgMCO2A (AY943928), AgMCO2B (AY943929), AgMCO3 (EF592176), AgMCO4 (EU380796), AgMCO5 (EU380797); Apis mellifera LOC410365 (XM_393845), LOC552811 (XM_625186), LOC724890 (XM_001120790); Bombyx mori BmMCO1 (BK006377), BmLac2 (EU093074), BmLac2B (BK006378); Drosophila melanogaster CG3759 (NM_135443), CG30437-PA (NM_165431), CG5959 (NM_143184), CG32557 (NM_133021); Manduca sexta MsLac1 (AY135185), MsLac2 (AY135186); Pimpla hypochondriaca PhLac1 (AJ427356); Strongylocentrotus purpuratus LOC586543 (XP_786321); Tribolium castaneum TcLac1 (AY884065), TcLac2A (AY884061), TcLac2B (AY884062). The Drosophila laccase 2B sequence used is a composite of four overlapping EST sequences: CA807619 (reverse complement), CA806339 (reverse complement), EC213902, and CO313037, and is a correction of the predicted gene CG30437-PC (NM_136326).
3. Results
3.1 Cloning and sequence analyses
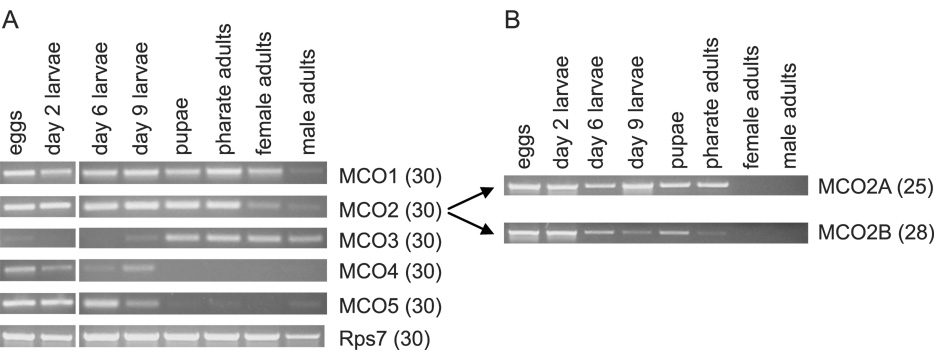
We identified five putative MCO genes in the A. gambiae genome and cloned cDNAs that encompass the full protein coding regions of these genes. An analysis of the MCO2 genomic DNA sequence suggested alternative exons for the carboxyl-terminal part of the protein. Using exon-specific PCR primers, we detected isoform A (encoded by exons 1-5, 6A-9A) and isoform B (encoded by exons 1-5, 6B-9B) but no other isoforms (supplementary Figure S1, Figure 4B). Eighty-one percent of the residues encoded by the alternatively spliced exons were identical. MCO3, 4 and 5 were found to be clustered on chromosome arm 3R and are highly similar (with amino acid identities ranging from 45–59%).
Figure 4. Developmental expression profiles of five putative MCO genes.
RT-PCR was used to determine whether a gene is expressed at a particular developmental stage. Pools of cDNAs were synthesized from 2.5 µg of total RNA. The number of PCR cycles is in parentheses. Eggs were mixed stage. Day 2 larvae = first and second instar, day 6 = third and fourth instar, day 9 = late fourth instar. A) Primers specific to each of the MCO genes were used. The MCO2 primers were designed to amplify both the A and B isoforms. Analysis of Rps7 (ribosomal protein gene 7) was included to verify the integrity of the cDNA samples. B) Primers specific to the MCO2A and MCO2B isoforms were used to detect differences in expression between the two isoforms. Note that MCO2A and 2B bands are not visible in the lanes corresponding to the adult stages, but both isoforms were detected in adults when more PCR cycles were used (data not shown, see Figure 6 for evidence of expression in ovaries).
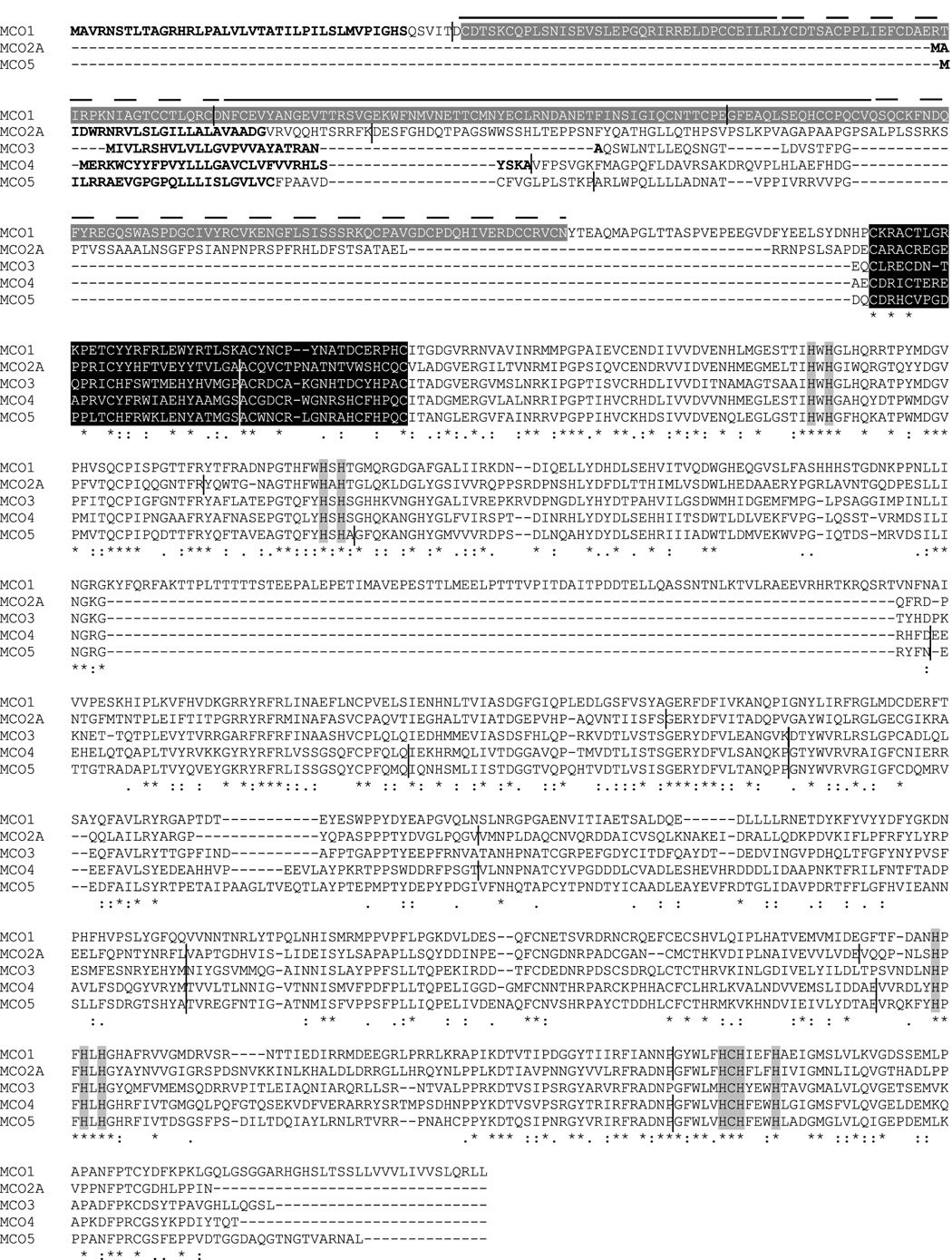
The predicted amino acid sequences were analyzed to identify key features (Figure 1). Most significantly, all of the expected putative copper binding residues (Nakamura and Go, 2005) were found in each of the MCOs. A cysteine rich domain unique to invertebrate MCOs (Dittmer et al., 2004) was also observed in each of the sequences. MCO1 was unique in having von Willebrand factor repeats at its amino terminus and an extra 88 residues in the middle section of the sequence. A signal anchor was predicted for MCO4 suggesting that it may be membrane bound. The other MCO sequences contain a putative signal peptide and thus are likely to be secreted proteins.
Figure 1. Alignment of the predicted amino acid sequences of MCO1-5.
Predicted signal peptides (or signal anchor for MCO4) are in bold type. Four von Willebrand factor domains in MCO1 are indicated by gray highlighting surrounding white type and plain or dashed lines. A class of conserved cysteine rich domains that have been found exclusively in invertebrate MCO sequences is shown in white type surrounded by black highlighting. The ten histidines and one cysteine that are predicted to bind to copper are indicated by gray highlighting. The positions of introns are indicated by short vertical bars.
We had hoped that comparisons between the A. gambiae MCO sequences and laccase, ascorbate oxidase and ferroxidase sequences would suggest possible functions for the AgMCOs; however, the AgMCO sequences were just about equally similar to each category of MCO, for example, MCO2A is 34% identical to a laccase from Trametes versicolor, 33% identical to an ascorbate oxidase from Cucurbita pepo, and 30% identical to a ferroxidase from Saccharomyces cerevisea (within the approximately 580 amino acid segment alignable from each sequence). In addition, we could not identify any signature sequences for laccases, ascorbate oxidases or ferroxidases that could be used to categorize the mosquito MCOs. Several residues critical for the ferroxidase activity of Fet enzymes are known (Stoj et al., 2006), and none of the AgMCOs contain all of these critical residues (data not shown). However, at least one non-Fet type ferroxidase lacks two of these important amino acids (Larrondo et al., 2003); therefore, they are not a completely reliable signature sequence. Because we could not predict the substrate specificity of the mosquito enzymes based on their primary amino acid sequences, we chose to name them MCOs rather than assign them to a more specific group of MCO.
3.2 Phylogenetic analyses
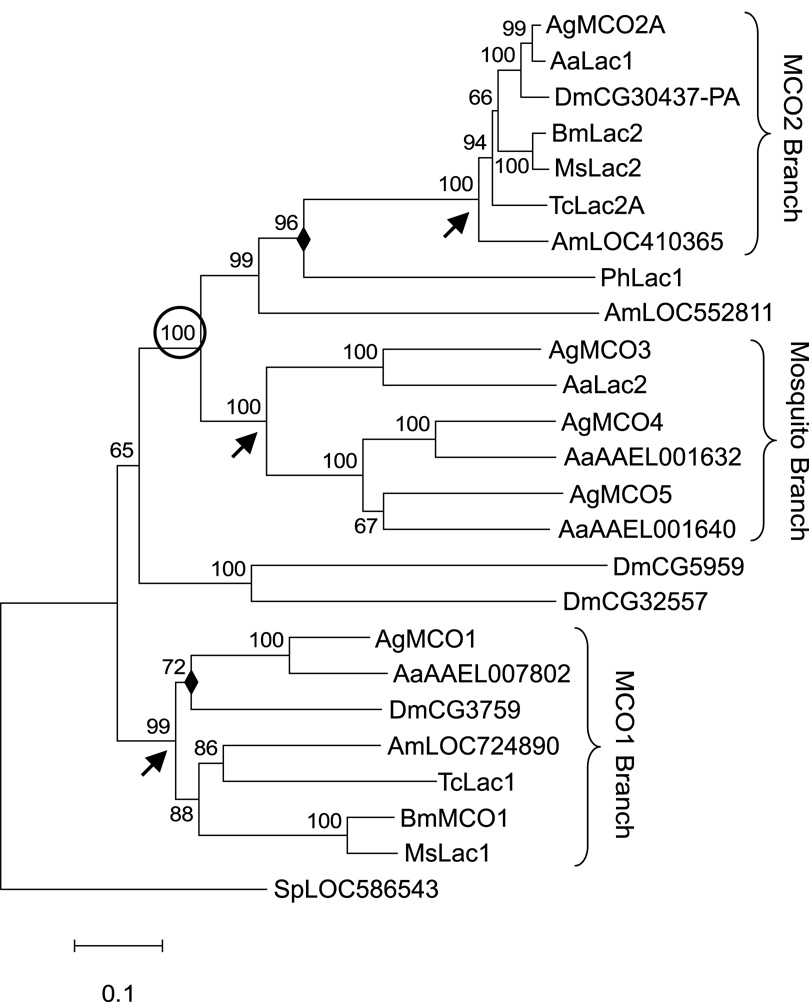
To learn the relationships among insect MCOs, we performed a phylogenetic analysis of genes from seven insect species representing four orders. In all of the insect genomes examined, we identified putative orthologs of MCO1 and MCO2. The average amino acid identity between pairs of genes in the MCO1 group was 54 ± 10% (mean ± standard deviation) whereas the identity between pairs in the MCO2 group was remarkably high at 88 ± 4% (see supplementary Table S1). The two main branches of the phylogenetic tree were strongly supported with a bootstrap value of 99 for the MCO1 group and 100 for the MCO2 group (Figure 2). MCO3, MCO4 and MCO5 were found only in A. gambiae and another species of mosquito, Aedes aegypti. Our analysis demonstrated that the genes in this mosquito specific cluster share a common ancestor with MCO2 (supported by a bootstrap of 100) (Figure 2).
Figure 2. Bootstrap consensus phylogenetic tree of insect MCO genes.
The tree shown was constructed by the neighbor-joining method; analysis by maximum parsimony gave a tree of similar topology. Nodes not conserved by the two methods are marked with a filled diamond (◆). Numbers above branches are bootstrap values expressed as a percentage. Arrows denote nodes signifying the MCO1, MCO2, and mosquito-specific branches. The circled bootstrap value indicates strong support for a common ancestor of the MCO2 group and the mosquito-specific group. Abbreviations used are: Aa, Aedes aegypti; Ag, Anopheles gambiae; Am, Apis melifera; Bm, Bombyx mori; Dm, Drosophila melanogaster; Ms, Manduca sexta; Sp, Strongylocentrotus purpuratus; Tc, Tribolium castaneum. A sea urchin (S. purpuratus) MCO gene was used as the outgroup for the analysis. The scale bar indicates the number of substitutions per site.
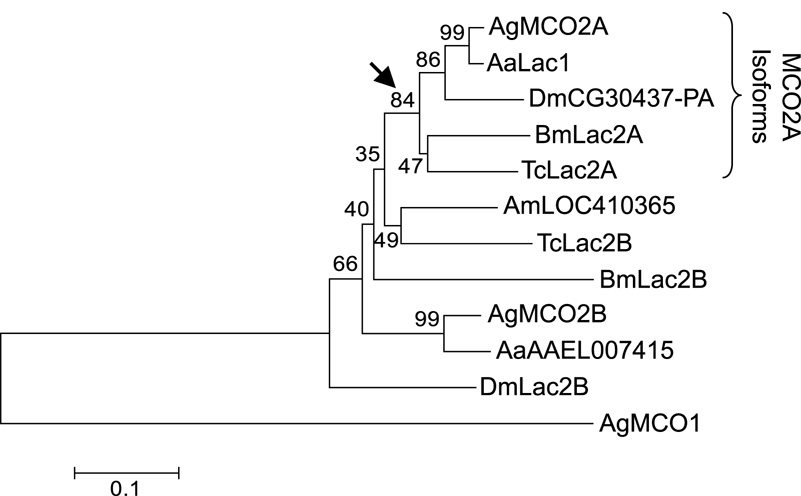
Several members of the MCO2 group have alternatively spliced isoforms. An analysis of the MCO2 group suggests that the A isoforms are more conserved than the B isoforms, and the A isoforms form a well supported clade with a bootstrap value of 84 (Figure 3).
Figure 3. Bootstrap consensus phylogenetic tree of MCO2 isoforms.
The tree was constructed by the neighbor-joining method. The arrow indicates the node signifying the “isoform A” branch. AgMCO1 was used as the outgroup for the analysis. Numbers, abbreviations, and scale bar are as described in the legend for Figure 2.
3.3 Expression profiles
To determine developmental expression profiles of the MCO genes, we isolated RNA from eggs, larvae, pupae, pharate adults and adults, and then performed RT-PCR. The expression patterns varied among the five genes (Figure 4A). MCO1 and MCO2 mRNAs were detected in all stages; MCO3 was observed in nine day old larvae, pupae and adults; and MCO4 and MCO5 were detected in the egg and larval stages. Barely detectable bands corresponding to MCO3 in eggs and larvae and MCO4 and MCO5 in post larval stages were also observed; therefore, these genes may be expressed at lower levels during these stages. The A and B isoforms of MCO2 had similar but non-identical patterns of expression (Figure 4B).
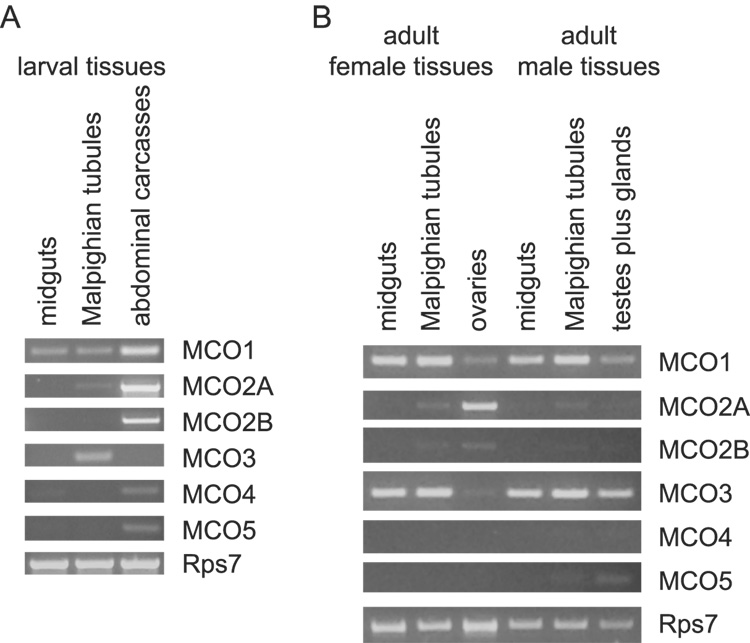
An initial survey of the tissues in which the MCO genes are expressed was achieved by isolating RNA from various larval and adult tissues followed by RT-PCR (Figure 5). We were particularly interested in whether any of the genes were expressed in cuticular epithelial cells (which synthesize enzymes required for cuticle sclerotization), Malpighian tubules (which express detoxifying enzymes), or male reproductive tissues (which are expected to produce enzymes involved in mating plug formation). RNAs corresponding to each of the MCO genes except MCO3 were identified in larval abdominal carcasses, which contain cuticular epithelial cells, fat body, muscles, and some additional tissues. MCO1, MCO2A and MCO3 mRNAs were detected in larval Malpighian tubules, and MCO1 mRNA was also detected in larval midguts. MCO1 and MCO3 mRNAs were present in adult midguts and Malpighian tubules, ovaries, and testes plus accessory glands. MCO2A and 2B mRNAs were present in ovaries, and faint bands were amplified from Malpighian tubule cDNAs. Very low expression of MCO5 in male Malpighian tubules and testes plus accessory glands was observed.
Figure 5. Expression of MCO genes in larval and adult tissues.
RT-PCR was used to determine whether MCO genes are expressed in various tissues. Tissues from fourth instar larvae (A) and adults (B) were analyzed. The amount of total RNA used for the cDNA reactions was 2 µg for larval tissues and 0.27 µg for all of the adult tissues except testes plus acessory glands, for which 0.11 µg was used. The number of PCR cycles used was 30 for the reactions in (A), 33 for the MCO gene reactions in (B) and 22 for the Rps7 reactions in (B).
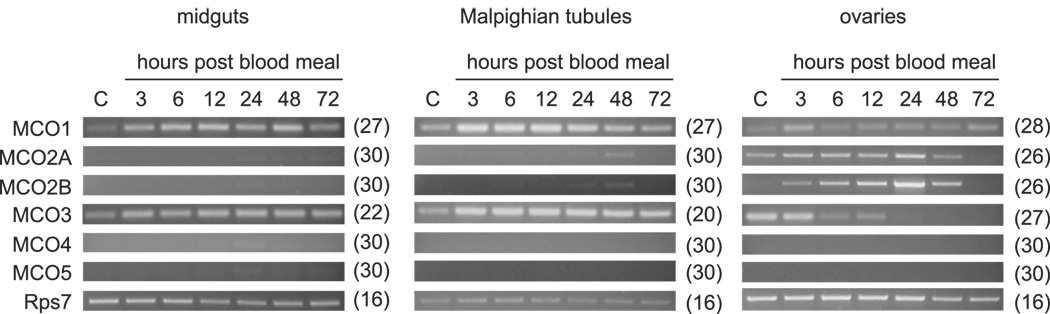
Genes that are involved in iron metabolism may be upregulated in midguts and/or Malpighian tubules in response to a blood meal, which is a source of iron. MCOs that catalyze eggshell tanning may be upregulated in ovaries following a blood meal because the ovaries synthesize chorion proteins in response to blood feeding. To identify such genes, we dissected tissues from mosquitoes 3, 6, 12, 24, 48 and 72 hours after they had taken a blood meal and from non-fed controls; we then isolated RNA and performed semiquantitative RT-PCR to look for changes in transcript abundance (Figure 6). MCO1 and MCO3 were upregulated in midguts and Malpighian tubules in response to blood feeding, whereas MCO2B transcript abundance in ovaries increased considerably following a blood meal.
Figure 6. Effect of blood feeding on MCO expression.
Semiquantitative RT-PCR was used to determine whether blood feeding has an effect on the expression of the MCO genes. Pools of cDNAs were synthesized from 2.5 µg (midguts or ovaries) or 1 µg (Malpighian tubules) of total RNA. The number of PCR cycles done is indicated in paretheses. Note: at 72 hours after blood feeding, eggs are fully developed.
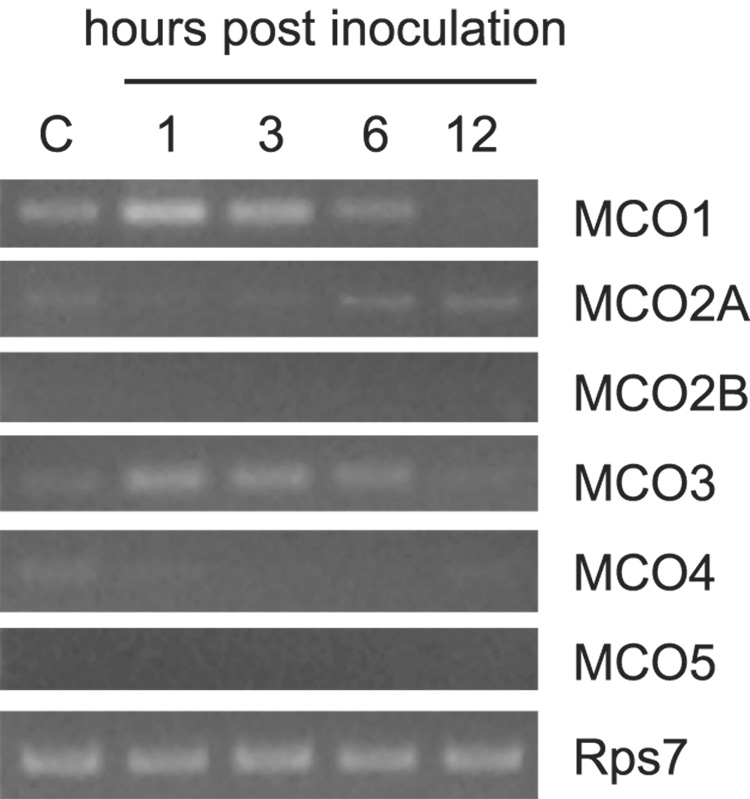
Genes with immune functions often are transcriptionally upregulated in the fat body of insects that have been inoculated with microbes. Fat body tissue is difficult to dissect from A. gambaie adults because the cells tend to disperse in the dissecting medium; therefore, for this experiment we used abdominal carcasses (with spermatheca, gut, Malpighian tubules, ovaries and crop removed) as a source of fat body cells. Mosquitoes were inoculated with Micrococcus luteus, and abdominal carcasses were collected after 1, 3, 6, and 12 hours. Naive mosquitoes were used as negative controls. RNA was isolated, and semiquantitative RT-PCR was performed (Figure 7). The transcript abundance of the MCO1 and MCO3 genes was higher at one and three hours post-inoculation.
Figure 7. Effect of immune challenge on MCO expression.
Semiquantitative RT-PCR was used to determine whether septic wounding has an effect on the expression of the MCO genes. Pools of cDNAs were synthesized from 0.5 µg of total RNA isolated from the abdominal carcasses of naive (c) mosquitoes or inoculated mosquitoes. The number of PCR cycles done was 33 for the MCO genes and 19 for Rps7. The results of one experiment are shown. A second experiment produced the same results.
4. Discussion
We identified two or more MCO genes in all of the insect genomes we examined. Orthologs of MCO1 and MCO2 were discovered in three dipteran species (A. gambiae, A. aegypti, D. melanogaster), two lepidopteran (B. mori, M. sexta), one coleopteran (T. castaneum), and one hymenopteran (A. mellifera). A preliminary analysis of the genomes of two hemimetabolous insects, the pea aphid (Acyrthosiphon pisum) and the human body louse (Pediculus humanus corporus) indicated orthologs for MCO1 and MCO2 in these species also (data not shown). Given the existence of MCO1 and MCO2 in a diverse group of holometabolous insects as well as hemimetabolous insects, we predict that orthologs of these two MCOs are present in nearly all insect species. In addition to these two genes, several insect species contain additional MCO genes with no obvious orthologs in other species, at least outside of the family level. In this group are the MCO3, 4 and 5 orthologs, which were identified only in the two mosquito species.
Of the insect MCO genes, MCO2 orthologs are particularly well conserved. The short branch lengths and high degree of sequence similarity across orders indicate that relatively few amino acid changes have occurred and that MCO2 orthologs are likely to be under strong purifying selection to reduce the occurrence of non-synonomous substitutions. We propose, based on the significant sequence conservation of the MCO2 genes and the known function of two of the orthologs, TcLac2 and MsLac2 (Arakane et al, 2005; Dittmer et al, 2004; Dittmer and Kanost, unpublished data), that MCO2 enzymes are involved in cuticle sclerotization. Given the universal importance of cuticle sclerotization to all insects, it is not surprising that MCO2 orthologs are highly conserved. Our observations that AgMCO2 is expressed predominantly in developmental stages that undergo cuticle tanning and that AgMCO2 mRNA is abundant in larval abdominal carcasses suggest that this enzyme has a role in cuticle tanning. The discovery of AgMCO2 in cuticles cast off during molting is strong corroborating evidence for this function (He et al., 2007).
Alternative exons are present in the MCO2 genes of A. gambiae, A. aegypti, D. melanogaster, B. mori and T. castaneum, but not A. mellifera. The well supported clade of MCO2A isoforms suggests a conserved function for this isoform. In contrast, low sequence conservation among B isoforms from different species suggests that they have more diverse functions. The only functional study of MCO2 isoforms has been of the Lac2 gene in T. castaneum (Arakane et al., 2005). RNAi mediated gene silencing demonstrated a requirement for both isoforms during cuticle tanning in T. castaneum, although a lack of functional redundancy could be concluded from the mutant phenotypes: beetles lacking the A isoform had a more extreme phenotype than those lacking the B isoform, and those lacking both had the least amount of cuticle tanning. The AgMCO2 isoforms were both expressed in larval abdominal carcasses, consistent with a role in cuticle sclerotization; however, like the TcLac2 isoforms, we expect that the functions of the AgMCO2 isoforms are not entirely redundant because they are expressed in non-identical developmental patterns and have different transcriptional responses to a blood meal. The upregulation of AgMCO2B in ovaries in response to a blood meal suggests that the B isoform of MCO2 may play a role in eggshell tanning or egg development.
We were interested to know whether the alternative exons of the MCO2 gene were lost in the A. mellifera lineage or whether a duplication of carboxyl-terminal exons occurred after the emergence of the hymenopterans but before the divergence of coleopterans, dipterans, and lepidopterans. Unfortunately, because there is a disagreement about whether the hymenopterans or colepterans are the more basal order of the holometabolous insects (Savard et al., 2006), we cannot currently answer this question. When the assembled genome sequences of more insects are available, especially other hymenopterans or hemimetabolous species, it should be possible to determine when the duplication of the carboxyl-terminal exons occurred.
MCO1 orthologs were identified in all species tested, but their sequence similarity was much less than what we observed for the MCO2 orthologs. In addition, MCO1 orthologs have significant differences in their amino-terminal regions. The three dipteran species have amino-terminal von Willebrand factor domains (two in D. melanogaster, four in A. gambiae and A. aegypti) whereas the other MCO1 genes encode unrelated amino-terminal domains. The function of these von Willebrand factor domains is unknown, but they may be involved in multimer formation or other types of protein-protein interactions (discussed in Dittmer et al., 2004). Little is known about the function of MCO1 orthologs. A test of TcLac1 function was inconclusive because gene silencing resulted in no observable phenotype (Arakane et al., 2005). One notable characteristic of AgMCO1 is that it was expressed in all developmental stages and in all of the tissues that we analyzed. This apparently ubiquitous expression pattern is similar to that of MsLac1 and may suggest a basic physiological role for MCO1 orthologs. Two observations that suggest possible functions are the upregulation of MCO1 in Malpighian tubules and midgut in response to a blood meal and in abdominal carcasses in response to infection. Vertebrate blood contains a large amount of iron, and, given that some MCOs oxidize iron, we propose that AgMCO1 may be involved in iron metabolism. Expression in male mosquito tissues and in the Malpighian tubules of D. melanogaster and M. sexta (species that do not feed on blood) indicate that if MCO1 orthologs function in iron metabolism, the function is not as specific as simply detoxifying blood meal iron. Upregulation of AgMCO1 and its ortholog in D. melanogaster in response to injection of bacteria strongly suggests an immune function. Two possible mechanisms for antimicrobial action of an MCO are decreasing the amount of ferrous iron available to microbes and catalyzing reactions required for melanin synthesis. If MCO1 orthologs function in metal metabolism and/or immunity, we have a possible explanation for why TcLac1 silencing had no observable effect in insects that were not exposed to atypical amounts of iron or to microbes.
AgMCO3-5 have high sequence identity and are clustered on the right arm of chromosome 3. These properties and our phylogenetic analysis suggest that these genes arose by duplication and divergence events relatively recently but prior to the divergence of the two mosquito species (see below). MCO4 and 5 have similar intron/exon structures and similar expression patterns; in contrast, MCO3 lacks most of the introns that MCO4 and 5 share and has expression patterns unlike those of MCO4 and 5. MCO4 and 5 are expressed primarily in eggs and larvae, whereas MCO3 is expressed mainly in pupae and adults, and, of the three genes, only MCO3 is upregulated by infection and blood feeding, and only MCO3 transcripts are abundant in a sample containing testes and male acessory glands. Our current data do not provide a function for the MCO4 and 5 orthologs, but they offer a putative role for MCO3 in iron metabolism and immunity. Because MCO3 is expressed in male reproductive tissues, it is a candidate for oxidizing substrates during mating plug formation.
We were interested in whether the mosquito specific group of MCOs were derived from an MCO1 or MCO2 common ancestor. Our phylogenetic analysis grouped the MCO3-5 orthologs with the MCO2 orthologs. If having two MCO genes is the ancestral state for the holometabolous insects, these results demonstrate that the mosquito specific group is derived from a lineage specific duplication of an MCO2-like gene. It is interesting then that MCO3 has an adult expression pattern similar to MCO1 but not MCO2.
The presence of a laccase-like activity in insects was discovered over 50 years ago (discussed in Yamazaki, 1972), but a function for insect laccases was not proved until Arakane et al. (2005) demonstrated the requirement of TcLac2 in cuticle tanning. Additional functions for insect laccases and related MCOs have been proposed, but the diverse set of possible substrates for these enzymes has made predicting specific functions difficult. Our study of MCO genes in A. gambiae has enabled us to narrow our focus to a limited set of putative functions. In the future, we will test these hypotheses with the use of immunohistochemistry, substrate specificity studies, and gene silencing experiments, and, thus, advance our currently limited understanding of MCO function in insects.
Supplementary Material
Acknowledgments
We thank Dr. Karl Kramer for information about the biochemistry of multicopper oxidases and Celeste Yang and Brian Ransom for cloning an MCO3 cDNA. This is contribution 08-266-J from the Kansas Agricultural Experiment Station. The project described was supported by Grant Numbers R03AI057816 and R01AI070864 from the National Institutes of Allegery and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proc. Natl. Acad. Sci. USA. 2005;102:11337–11342. doi: 10.1073/pnas.0504982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrian P. Fungal laccases - occurence and properties. FEMS Microbiol. Rev. 2006;30:215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Benedict MQ. Care and maintenance of anopheline mosquito colonies. In: Crampton JM, Beard CB, Louis C, editors. Molecular Biology of Insect Disease Vectors: A Methods Manual. CITY: Chapman & Hall; 1997. pp. 3–12. [Google Scholar]
- De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA. 2001;98:12590–12595. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer NT, Suderman RJ, Jiang H, Zhu Y-C, Gorman MJ, Kramer KJ, Kanost MR. Characterization of cDNAs encoding putative laccase-like multicopper oxidases and developmental expression in the tobacco hornworm, Manduca sexta, and the malaria mosquito, Anopheles gambiae. Insect Biochem. Mol. Biol. 2004;34:29–41. doi: 10.1016/j.ibmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Hattori M, Konishi H, Tamura Y, Konno K, Sogawa K. Laccase-type phenoloxidase in salivary glands and watery saliva of the green rice leafhopper, Nephotettix cincticeps. J. Insect. Physiol. 2005;51:1359–1365. doi: 10.1016/j.jinsphys.2005.08.010. [DOI] [PubMed] [Google Scholar]
- He N, Botelho JMC, McNall RJ, Belozerov V, Dunn WA, Mize T, Orlando R, Willis JH. Proteomic analysis of cast cuticles from Anopheles gambiae by tandem mass spectrometry. Insect Biochem. Mol. Biol. 2007;37:135–146. doi: 10.1016/j.ibmb.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Hoegger PJ, Kilaru S, James TY, Thacker JR, Kues U. Phylogenetic comparison and classification of laccase and related multicopper oxidase protein sequences. FEBS J. 2006;273:2308–2326. doi: 10.1111/j.1742-4658.2006.05247.x. [DOI] [PubMed] [Google Scholar]
- Hoopes JT, Dean JFD. Ferroxidase activity in a laccase-like multicopper oxidase from Liriodendron tulipifera. Plant Physiol. Biochem. 2004;42:27–33. doi: 10.1016/j.plaphy.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Hullo M-F, Moszer I, Danchin A, Martin-Verstraete I. CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 2001;183:5426–5430. doi: 10.1128/JB.183.18.5426-5430.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston WM, Jennings MP, McEwan AG. The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol. Microbiol. 2002;45:1741–1750. doi: 10.1046/j.1365-2958.2002.03132.x. [DOI] [PubMed] [Google Scholar]
- Kapustin Y, Souvorov A, Tatusova T. Splign – a hybrid approach to spliced alignments. In: Gramada A, Bourne P, editors. Eighth Annual International Conference on RECOMB 2004 – Currents in Computational Molecular Biology. 2004. p. 741. [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Larrondo LF, Salas L, Melo F, Vicuna R, Cullen D. A novel extracellular multicopper oxidase from Phanerochaete chrysosporium with ferroxidase activity. Appl. Environ. Microbiol. 2003;69:6257–6263. doi: 10.1128/AEM.69.10.6257-6263.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer AM, Staples RC. Laccase: new functions for an old enzyme. Phytochemistry. 2002;60:551–565. doi: 10.1016/s0031-9422(02)00171-1. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Go N. Function and molecular evolution of multicopper blue proteins. Cell. Mol. Life Sci. 2005;62:2050–2066. doi: 10.1007/s00018-004-5076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanar L, Gebhard M, Wang T-P, Kosman DJ, Solomon EI. Ferrous binding to the multicopper oxidases Saccharomyces cerevisiae Fet3p and human ceruloplasmin: contributions to ferroxidase activity. J. Am. Chem. Soc. 2004;126:6579–6589. doi: 10.1021/ja049220t. [DOI] [PubMed] [Google Scholar]
- Sakurai T, Kataoka K. Basic and applied features of multicopper oxidases, CueO, bilirubin oxidase, and laccase. Chem. Rec. 2007;7:220–229. doi: 10.1002/tcr.20125. [DOI] [PubMed] [Google Scholar]
- Salazar CE, Mills-Hamm D, Kumar V, Collins FH. Sequence of a cDNA from the mosquito Anopheles gambiae encoding a homologue of human ribosomal protein S7. Nucleic Acids Res. 1993;21:4147. doi: 10.1093/nar/21.17.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard J, Tautz D, Richards S, Weinstock GM, Gibbs RA, Werren JH, Tettelin H, Lercher MJ. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Research. 2006;16:1334–1338. doi: 10.1101/gr.5204306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidjanski S, Mathews GV, Vanderberg JP. Electrophoretic separation and identification of phenoloxidases in hemolymph and midgut of adult Anopheles stephensi mosquitoes. J. Parasitol. 1997;83:686–691. [PubMed] [Google Scholar]
- Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases. Chem. Rev. 1996;96:2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- Stoj CS, Augustine AJ, Zeigler L, Solomon EI, Kosman DJ. Structural basis of the ferrous iron specificity of the yeast ferroxidase, Fet3p. Biochemistry. 2006;45:12741–12749. doi: 10.1021/bi061543+. [DOI] [PubMed] [Google Scholar]
- Theopold U, Li D, Fabbri M, Scherfer C, Schmidt O. The coagulation of insect hemolymph. Cell. Mol. Life Sci. 2002;59:363–372. doi: 10.1007/s00018-002-8428-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston CF. The structure and function of fungal laccases. Microbiology. 1994;140:19–26. [Google Scholar]
- Wang J, Kean L, Yang J, Allan AK, Davies SA, Herzyk P, Dow JAT. Function-informed transriptome analysis of Drosophila renal tubule. Genome Biol. 2004;5:R69. doi: 10.1186/gb-2004-5-9-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead DL, Brunet PCJ, Kent PW. Specificity in vitro of a phenoloxidase system from Periplaneta americana (L.) Nature. 1960;185:610. doi: 10.1038/185610a0. [DOI] [PubMed] [Google Scholar]
- Yamazaki HI. Cuticular phenoloxidase from the silkworm, Bombyx mori: properties, solubilization, and purification. Insect Biochem. 1972;2:431–444. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.