Abstract
The FAD prosthetic group of the ERV/ALR family of sulfhydryl oxidases is housed at the mouth of a 4-helix bundle and communicates with a pair of juxtaposed cysteine residues that form the proximal redox active disulfide. Most of these enzymes have one or more additional distal disulfide redox centers that facilitate the transfer of reducing equivalents from the dithiol substrates of these oxidases to the isoalloxazine ring where the reaction with molecular oxygen occurs. The present study examines yeast Erv2p and compares the redox behavior of this ER luminal protein with the augmenter of liver regeneration, a sulfhydryl oxidase of the mitochondrial intermembrane space, and a larger protein containing the ERV/ALR domain, quiescin-sulfhydryl oxidase (QSOX). Dithionite and photochemical reductions of Erv2p show full reduction of the flavin cofactor after the addition of 4-electrons with a mid-point potential of -200 mV at pH 7.5. A charge-transfer complex between a proximal thiolate and the oxidized flavin is not observed in Erv2p consistent with a distribution of reducing equivalents over the flavin and distal disulfide redox centers. Upon coordination with Zn2+, full reduction of Erv2p requires 6-electrons. Zn2+ also strongly inhibits Erv2p when assayed using tris(2-carboxyethyl)phosphine (TCEP) as the reducing substrate of the oxidase. In contrast to QSOX, Erv2p shows a comparatively low turnover with a range of small thiol substrates, with reduced Escherichia coli thioredoxin and with unfolded proteins. Rapid reaction studies confirm that reduction of the flavin center of Erv2p is rate-limiting during turnover with molecular oxygen. This comparison of the redox properties between members of the ERV/ALR family of sulfhydryl oxidases provides insights into their likely roles in oxidative protein folding.
Over the last decade a number of eukaryotic flavin-dependent sulfhydryl oxidases have been described that catalyze the net generation of disulfide bonds:
They include: yeast ERV1 (essential for respiration and vegetative growth) (1) and its mammalian counterpart augmenter of liver regeneration, ALR, (2–4); yeast Erv2p (5–7); an Arabidopsis homolog of these proteins (8, 9); yeast Ero1p (10–14) and its vertebrate orthologs Ero1α and Ero1β (15, 16); and finally larger multidomain sulfhydryl oxidases, containing an ERV/ALR module fused to a redox active thioredoxin domain, that are found principally in multicellular organisms (17–20).
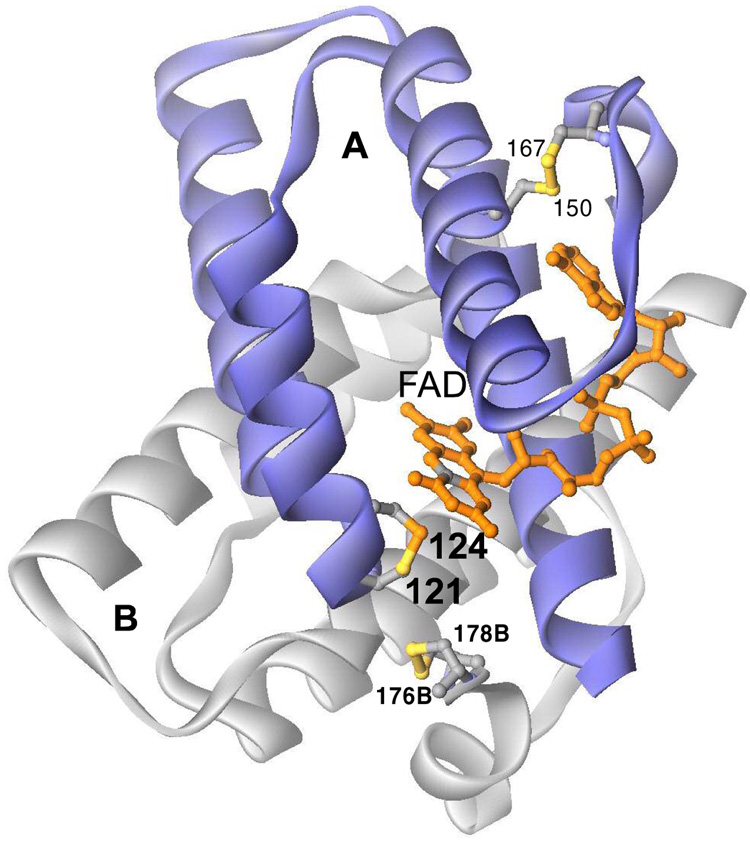
The ERV/ALR proteins and their larger cousins in the QSOX family all contain a diminutive helix-rich flavin binding domain first reported for yeast Erv2p by Fass and coworkers (6) and for rat ALR by Rose and colleagues (4). Subunit A of the Erv2p homodimer is shown in the foreground of Figure 1. The isoalloxazine ring is inserted at the end of a bundle of four helices in the A subunit with its C2/N3 region exposed to solvent (6). The proximal disulfide C121-124 is placed so that the sulfur of C124 is adjacent (3.4 Å) to the C4a position of the flavin. This locus is consistent with its expected role in formation of a C4a flavin adduct that serves as an intermediate in the transfer of reducing equivalents between (di)thiols and flavins (21–23). This proximal C121–C124 disulfide is not believed to be reduced by substrates directly in vivo, but rather receives reducing equivalents from a distal C176–C178 motif located on a conformationally mobile element at the C-terminus of the B subunit (6, 24).
Figure 1.
Structure of Saccharomyces cerevisae Erv2p (6). Three redox centers out of a total of 6 sites in dimeric Erv2p are shown towards the front of the Figure. The distal disulfide (176B–178B) of the flexible C terminus of the B (grey) subunit interacts with proximal disulfide C121–124 of the A (blue) subunit via disulfide exchange. Reducing equivalents are then transmitted to the flavin via a C124-flavin covalent adduct at the C4a position (grey). A structural disulfide is shown at the top right of the A subunit (C150–C167).
In vivo disulfide crosslinking has suggested that the immediate reductant of Erv2p is protein disulfide isomerase (PDI1p) (7). This has led to a scheme in which PDI serves as a general mediator between proteins undergoing oxidative folding in the yeast ER and the oxidase [where the arrows indicate the movement of electron pairs: (7, 24)]:
This model suggests that reduced PDI should be a better substrate of ERV2p than reduced client proteins undergoing oxidative folding. However, recent in vitro studies indicate that reduced PDI1p is actually a poor substrate of Erv2p (24). Kaiser and coworkers suggest that another ER protein may be the real physiological mediator for Erv2p, or a factor, which facilitates the redox interaction between PDI and Erv2p, may be missing from these in vitro experiments (24). Adding to the uncertainties concerning the catalytic specificity of Erv2p, we currently lack important basic information concerning the redox behavior of Erv2p to compare to the emerging data for human ALR (25), the plant mitochondrial AtErv1p (8, 9), and the avian QSOX1 (26–29). A comparison of the properties of these three rather diverse representatives of the ERV/ALR family of sulfhydryl oxidases will help clarify the basic features of flavin-linked catalysis in all of them.
MATERIALS AND METHODS
Materials
Dithiothreitol, reduced glutathione, cysteine, β-mercaptoethanol, DTNB, ultra-pure urea, superoxide dismutase, catalase, bovine pancreatic RNase, cytochrome c, and high molecular weight SDS protein standards were purchased from Sigma. Sodium dithionite was from the Virginia Smelting Company. Hydrogen peroxide was obtained as a 30% solution from Fisher Scientific. Triscarboxyethylphosphine (TCEP) was from Pierce. Thioredoxin and thioredoxin reductase were generously provided by Dr. Charles Williams. SDS-PAGE gels (4–15% and 18% Tris-HCl) were purchased from Bio-Rad. PD-10 desalting columns were purchased from Pharmacia Biotech.
General Methods
Unless otherwise stated, the buffer used here was 50 mM Tris-chloride, pH 7.5 (25 °C), containing 0.3 mM EDTA. Enzyme samples were concentrated and washed using Amicon Centricon 30 ultrafiltration devices. Visible and ultraviolet spectra were recorded on Hewlett-Packard 8452A or 8453 diode array spectrophotometers. Enzyme assays, using a Clarke-type oxygen electrode or discontinuous sampling using DTNB, were performed as described previously (26, 30, 31). Anaerobic manipulations and titrations were as described earlier (26, 32). Photoreductions were performed in semi-micro anaerobic cuvettes immersed in ice-water using a 150 W flood lamp, in Tris buffer containing 1.0 µM 5-deazaflavin and 1 mM EDTA. Where appropriate, TCEP solutions were adjusted with concentrated NaOH to the desired pH before assays. Stated stoichiometries are on a per flavin (per subunit) basis. Structures were visualized with DS ViewerPro (Accelerys).
Protein Expression and Purification
A truncated version of Erv2p, without the N-terminal 34 residues, was cloned into a pET24(a+) plasmid carrying kanamycin resistance. The construct encoded a hexahistidine tag at the C-terminus, and was 4 residues shorter than the Erv2-ΔN construct initially employed by Fass et al. for crystallography (6, 7). The plasmid was maintained and amplified in DH5α Escherichia coli strain. Glycerol stocks were made and kept at −80 °C for long term storage. The protein was expressed from the E. coli strain BL21(DE3) or BL21(DE3 star). Starter cultures were grown overnight at 37 °C in 5 ml LB containing 30 µg/mL of kanamycin, and used to inoculate each of four 500 mL of the same media in 2 L flasks. Cells were incubated at 200 rpm at 37 °C to an OD600 of 1.2–1.4 and then the media was supplemented with 1 mM isopropyl-L-thio-β-D-galactopyranoside and 10 µM riboflavin and shaking resumed at 25 °C. After overnight growth, cells were harvested at 4000 g for 20 min at 4 °C. Combined pellets (about 20 g) were resuspended in 25 mL of 50 mM potassium phosphate buffer, pH 7.5, without EDTA but containing 300 mM NaCl, 0.1 mg/mL lysozyme, the manufacturer’s recommended level of protease inhibitor (Sigma Protease Inhibitor Cocktail P8849) and 50 µM FAD. The suspension was disrupted using two passes through a French press at 10,000 psi, and then treated with five 5 sec pulses of sonication to shear DNA. The lysate was centrifuged at 6000 g for 30 min, and the supernatant was mixed for 1 h at 4 °C end-over-end with Invitrogen Probond resin (8 mL extract per mL of resin). The now yellow-brown resin was packed into a column and washed with three 4 mL aliquots of 50 mM phosphate buffer containing 300 mM NaCl and 10 mM imidazole at pH 7.5 followed by the same washes omitting NaCl. The column was developed with 5 mL aliquots of phosphate buffer, without added NaCl, but supplemented with 50, 200 and 500 mM imidazole respectively. Elution of the bright yellow ERV2p occurred in the 200 mM imidazole fraction. To ensure maximal FAD content, a two-fold excess of FAD over total concentration of subunits, was incubated overnight at 4 °C, or for 1 h at 25 °C, followed by gel filtration into Tris buffer over a PD10 column. Samples were concentrated by centrifuge ultrafiltration as needed. N-terminal sequencing, performed by Dr. Yu-Chu Huang on an Applied Biosystems gas-phase Procise Sequencer (Model 470A/120A/900A), confirmed the expected start of the protein sequence.
Determination of Extinction Coefficient of ERV2p
The extinction coefficient of FAD bound to ERV2p was determined by releasing the FAD cofactor upon the addition of 0.5 mM cetyltrimethylammonium bromide (CTAB; from a 50 mM solution made up in 50 mM phosphate buffer, pH 7.5, 1 mM EDTA). The spectrum of ERV2p was recorded before and after the addition of detergent, repeating the measurements with free FAD. Using an extinction coefficient of 11.3 mM−1 cm−1 for free FAD in the absence of detergent (33), and correcting for dilution prior to the addition of the detergent, yields a comparable value of 11.7 mM−1 cm−1 at 450 nm for Erv2p.
Reduced Protein Substrate Preparation
Reduced RNase (20 mg in 1 mL of Tris buffer containing 6 M guanidine hydrochloride adjusted to pH 8.0 with KOH) was incubated at 37 °C for 1 h under nitrogen with a 50-fold molar excess of DTT over RNase disulfides. The mixture was adjusted to pH 3.5 with glacial acetic acid and immediately gel-filtered on a PD10 column pre-equilibrated with deoxygenated 0.1% acetic acid containing 3 mM EDTA. Small aliquots of each fraction were tested with DTNB to allow reduced RNase to be collected without contamination from the following peak of DTT. The combined RNase fractions were deoxygenated by repeated evacuation and flushing with oxygen-free nitrogen and could be stored for weeks without significant loss of thiol titer.
Stopped-Flow Spectroscopy
Stopped-flow experiments were performed at 25 °C in a Hi-Tech Scientific SF-61 SX2 double-mixing stopped-flow system with the software provided by the vendor. For anaerobic experiments, the entire flow system, including the driving syringes and flow cells were soaked for hours with a solution of 10 mM dithionite. Photoreduced ERV2p was prepared in a tonometer as described earlier (32, 34).
RESULTS AND DISCUSSION
Expression, Purification, and Visible Spectrum of Erv2p
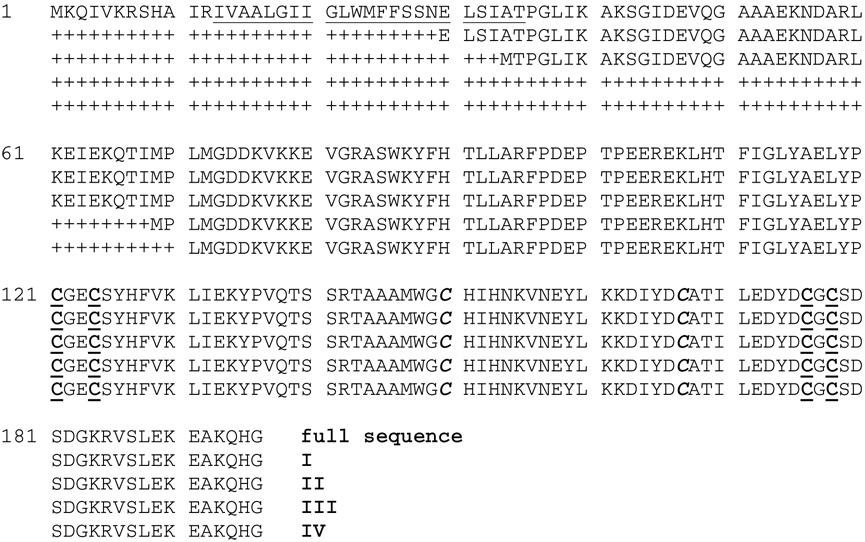
Figure 2 shows the sequence of full length Erv2p. It contains a putative N-terminal transmembrane span tethering Erv2p to the luminal surface of the ER (7). Because expression of the full-length protein results in either lysis of E. coli (5), or a much reduced yield of protein (7), truncated versions of Erv2p have been employed. The protein used here retains most of the sequence (Figure 1) between the transmembrane peptide and the ERV core domain (6) and is terminated by a hexahistidine tag. Bright yellow fractions containing ERV emerge from the Ni2+- NTA column and were freed of imidazole by gel filtration (see Methods). The protein was ~95% pure on SDS-PAGE and the N-terminal sequence, MTPGLIKAKS- was consistent with the construct, II in Figure 2 (see Methods). ERV2p could be stored at 4 °C or frozen at −20 °C in Tris buffer, pH 7.5, containing 1 mM EDTA for more than 1 year without noticeable decline in activity in the standard assay system (see Methods).
Figure 2.
Sequences of Erv2p constructs. The full length sequence of Saccharomyces cerevisiae Erv2p (51) is shown with a putative transmembrane sequence underlined. The constructs used are labeled I–IV at their C-terminii: I, (7); II, this work; III, (5); IV, Gross et al. (6). The redox-active cysteines residues are underlined in bold and those forming the structural disulfide are in bold italics.
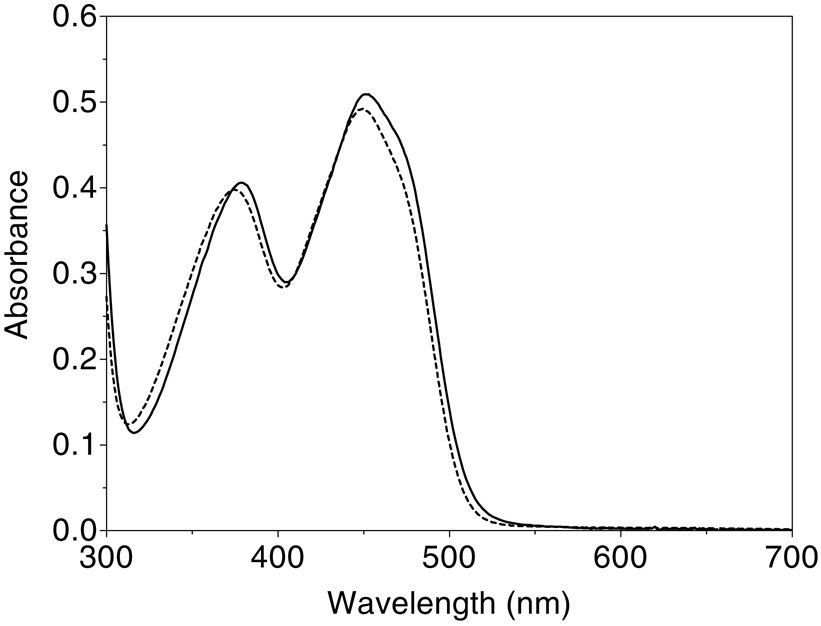
Prior constructs of Erv2p were reported to contain slightly sub-stoichiometric (0.7–0.8) levels of flavin per subunit (5, 7), and we therefore pretreated the purified protein with excess FAD (see Methods). After subsequent gel filtration, Erv2p shows a comparatively unresolved flavin spectrum with maxima at 450 nm and 380 nm, a comparatively low trough at 300 nm, and no significant turbidity at longer wavelengths (Figure 3). Since prior quantitation of Erv2p has used extinction coefficients for enzyme-bound flavin of either 10 mM−1cm−1 [an assumed value; (5)] or 12.5 mM−1cm−1 (7), we re-determined this parameter by release of the flavin using cetyltrimethylammonium bromide (see Methods). Two determinations gave identical values of 11.7 mM−1cm−1 at 450 nm (Figure 3). The protein to flavin (280/450 nm) absorbance ratio of the material used in Figure 2 is 4.6; significantly lower than the value of 6.4 observed earlier (7). A second gel filtration of this material, after storage for several months at 4 °C, yielded the same FAD/protein ratio indicating insignificant loss of flavin (not shown).
Figure 3.
Visible spectrum of Erv2p before and after denaturation. The visible spectrum of Erv2p, incubated with free FAD before gel filtration, was recorded in 50 mM Tris, pH 7.5, before (solid line) and after the addition of 0.5 mM CTAB (dashed line; see Methods).
Catalytic activity of ERV2p
Erv2p was first recognized as a sulfhydryl oxidase because it showed a turnover number of 4.5/min using a relatively low concentration of reduced lysozyme [7 µM lysozyme; 55 µM thiols; (5)]. Neither GSH nor DTT were reported to be significant substrates of this form of Erv2p (5). Using a longer version of Erv2p (Figure 2), Sevier et al. al so found that GSH is an insignificant substrate of the enzyme at pH 7.5 (7) and a poor one (about 2/min) at pH 8.0 (24). However, they observed significant activity with DTT (kcat 239/min, Km 6.9 mM at pH 7.5 (7)). In view of these differences, we first undertook a survey of substrate specificity using our preparations of Erv2p.
Table 1 records turnover numbers for potential substrates of Erv2p held at the fixed reductant concentrations indicated in the Table (see Methods). Our studies show that, at least in vitro, Erv2p is a modest catalyst of disulfide bond formation compared to QSOX. None of the monothiols (at 10 mM), including reduced glutathione, proved detectable substrates of this yeast oxidase at pH 7.5. In contrast, as observed by Sevier et al., (7, 24) dithiols are significant substrates. At 5 mM DTT we observe a turnover number of 63/min (with kcat and Km values of 206/min and 11.3 mM for DTT respectively). These latter values are in reasonable accord with the 239/min and 6.9 mM obtained previously (7). Two other dithiols were also effective substrates, 2,3-dimercapto-l-propanol (British Anti-Lewisite or BAL), and bis-(2-mercaptoethyl)sulfone (Table 1) together with trans-1,2-bis(2- mercaptoacetamide) recently reported by Vala et al. (24). A reduced dithiol peptide (VTWCGACKM-NH2), that was inspired by the CxxC motif of thioredoxin and previously found to exhibit rapid turnover with avian QSOX (35), showed a turnover number with Erv2p of <1/min (data not shown).
TABLE 1.
Substrates specificity of yeast Erv2p.
| monothiols | ||
| β-mercaptoethanol | (10 mM SH) | <0.1 /min |
| N-acetylcysteamine | (10 mM SH) | < 0.1/min |
| GSH | (10 mM SH) | <0.1/min |
| CoASH | (10 mM SH) | <0.1/min |
| dithiols | ||
| DTT | (5 mM) | 63/min |
| BAL: 2,3-dimercapto-1-propanol | (5 mM) | 58/min |
| Bis-(2-mercaptoethyl)sulfone | (5 mM) | 34/min |
| proteins | ||
| reduced lysozyme (5) | (50 µM –SH) | 2.3/min |
| RNasered | (200 µM -SH) | 3/min |
| Trx E. coli | (200 µM SH) | 2.5/min |
| phosphine | ||
| TCEP | (5 mM) | 96/min |
Two protein substrates were tested here. Reduced RNase, at 200 µM thiols (25 µM RNase protein; see Methods) showed a turnover number of 3 disulfides formed/min. Since reduced PDI1p has already been reported to be a "much slower" substrate than small molecular weight thiols (24), it was of interest to examine the much more reducing (36, 37) Escherichia coli thioredoxin as a potential substrate of Erv2p. Prior work has shown that E. coli thioredoxin is an effective substrate of QSOX1 (30); yeast Ero1p (13) and an Erv1-like sulfhydryl oxidase from plant mitochondria (8). Thioredoxin is only modestly reactive here (Table 1) confirming recent studies on Erv2p under slightly different buffer conditions (a turnover number of 0.8/min was reported by Vitu et al. (9)). Finally the sulfhydryl oxidases QSOX and Ero1p have been shown to catalyze the oxidation of phosphines, such as TCEP, its methyl esters, and trishydroxypropylphosphine, to their corresponding phosphine oxides (38). Of all the reductants selected in Table 1, TCEP (at 5 mM), is the best substrate. In terms of kcat/Km TCEP (kcat 60/s and Km 60mM; not shown) is a marginally better substrate than DTT (1000 M−1s−1 compared to 580 M−1s−1 respectively). These catalytic efficiencies are some 100-fold lower than the corresponding values recorded for QSOX (26, 38). Overall, highly active substrates for Erv2p have yet to emerge from in vitro studies (see later).
Finally, we investigated whether preparations of Erv2p which lack a full complement of FAD (with A280 /A450 ratios of 9.0 vs. 4.6) showed significant differences in steady state kinetic behavior toward DTT. In principle disulfide bridges in flavin-free “apo”-subunits could participate in redox-relays with active FAD-containing subunits.
Such an effect could occur at the level of dimeric forms of apo- and holo-ERV2p or between a mixed dimer of apo- and holo-subunits. However, when normalized for flavin content, there appeared no significant difference in steady-state kinetic parameters for DTT between fully-loaded Erv2p and preparations containing substantial amounts of apoenzyme (data not shown). This suggests that the apo form did not significantly contribute to the overall activity under these conditions.
Reductive titrations of Erv2p
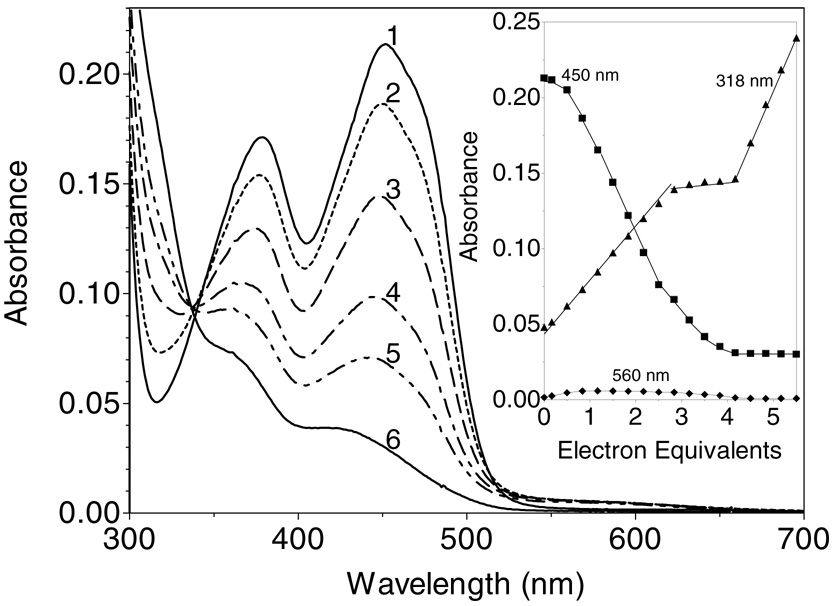
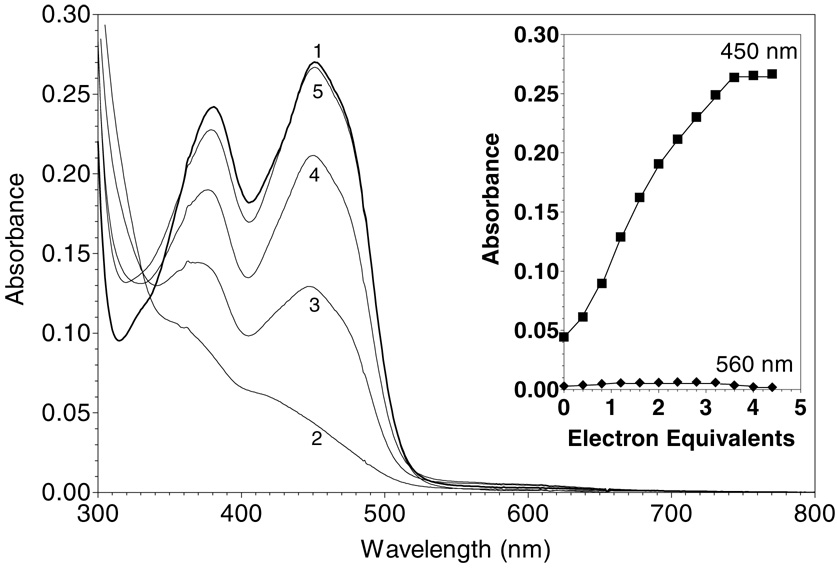
As shown in Figure 1, ERV2p has three potential redox centers: two disulfides (C121–124 and C176–178) and one FAD per subunit [together with a conserved structural disulfide, C150–167; (6)]. As expected, all of the 6 cysteine residues in Erv2p, as isolated from E. coli, appear to be present in disulfide bonds, and no reaction with DTNB is observed (not shown; see Methods). Figure 4 shows a dithionite titration of ERV2p at pH 7.5, 25 °C. The spectral changes were recorded 12 min after each addition of titrant, to allow completion of the spectral changes. Complete reduction of the flavin requires the addition of approximately 4-electrons per flavin center (see inset, consistent with the additional reduction of one disulfide equivalent). During this phase of the titration an isosbestic point is seen at 341 nm (Figure 4). Thereafter, excess dithionite accumulation is notably evident by following the absorbance at 318 nm (see inset). The non-linearity in the absorbance changes at 450 nm has been observed repeatedly, and suggests that the flavin ring in Erv2p is slightly more reducing than the disulfide redox center which also undergoes reduction under these conditions (see later). The observation that dithionite reduction does not reveal all 3 redox centers of Erv2p has precedent in experiments with native and truncated versions of avian QSOX (26, 29). Interestingly, Figure 4, and all our other static and kinetic experiments with Erv2p, show no evidence for the prominent thiolate to flavin charge-transfer spectrum seen upon reduction of QSOX (27, 31). While a small long wavelength feature is observed at the midpoint of the titration in Figure 4 (see inset), enlargement of this region of the spectrum beyond 500 nm shows the clear resolved signature of a blue semiquinone (see later).
Figure 4.
Dithionite titration of Erv2p. A solution of 18 µM ERV2p in Tris buffer, pH 7.5, containing 0.3 mM EDTA (curve 1) was titrated anaerobically with 0.83, 1.5, 2.2, 2.8, and 4.5 electron-equivalents of sodium dithionite/mol of flavin (curves 2-6, respectively). Intermediate spectra are omitted for clarity. The inset plots absorbance changes at 318 nm (triangles), 450 nm (squares) and 560 nm (diamonds) respectively.
Photoreduction of ERV2p with catalytic levels of 5-deazaflavin/EDTA (39) generates a similar progression of spectral changes to those seen upon dithionite titration (Figure 5) . Here, back-titration requires approximately 3.6 equivalents of ferricyanide for complete reoxidation of the flavin (inset, Figure 5). Unlike all of the other titrants used in these anaerobic titrations, ferricyanide reacts with Erv2p in the time required for mixing the reagents in the anaerobic cuvette (see later). The slightly sigmoidal regain of absorbance at 450 nm is comparable to that seen by retracing the dithionite titration shown in the inset to Figure 4. Thus, these data are again consistent with the delivery of reducing equivalents to both the flavin and to an additional disulfide redox center in each subunit of Erv2p. Again very small amounts of the blue semiquinone were observed at intermediate stages of the ferricyanide back-titration (Figure 5). Anaerobic reduction of Erv2p with up to 3 equivalents of DTT showed the same general spectral appearance observed with dithionite and photochemical reductions, including the accumulation of small levels of the blue semiquinone (data not shown). However redox equilibration with DTT was so sluggish that stoichiometries could not be reliably obtained from these titration attempts.
Figure 5.
Ferricyanide back titration of photochemically reduced ERV2p. Photoreduced ERV2p (23 µM in Tris buffer, pH 7.5; see Methods; curve 2) was titrated anaerobically with 1.2, 2.4, and 4.4 equivalents of ferricyanide/mol of flavin (curves 3–5 respectively). Curve 1 was ERV2p before photoreduction. Intermediate spectra are omitted for clarity. The inset plots absorbance changes at 450 nm (squares) and 560 nm (diamonds) respectively.
To place Erv2p in a thermodynamic context with respect to its potential thiol substrates (see later), it was important to assess the mid-point potential of this poorly understood catalyst of oxidative protein folding. Using either anthraquinone-2-sulfonate (E'° = − 226 mV anthraquinone-2-sulfonate) or anthraquinone-2,6-disulfonate (E'° = −184 mV anthraquinone-2,6-disulfonate) we obtained mid-point values for Erv2p of −204 mV and −197 mV at pH 7.5, 25 °C respectively.
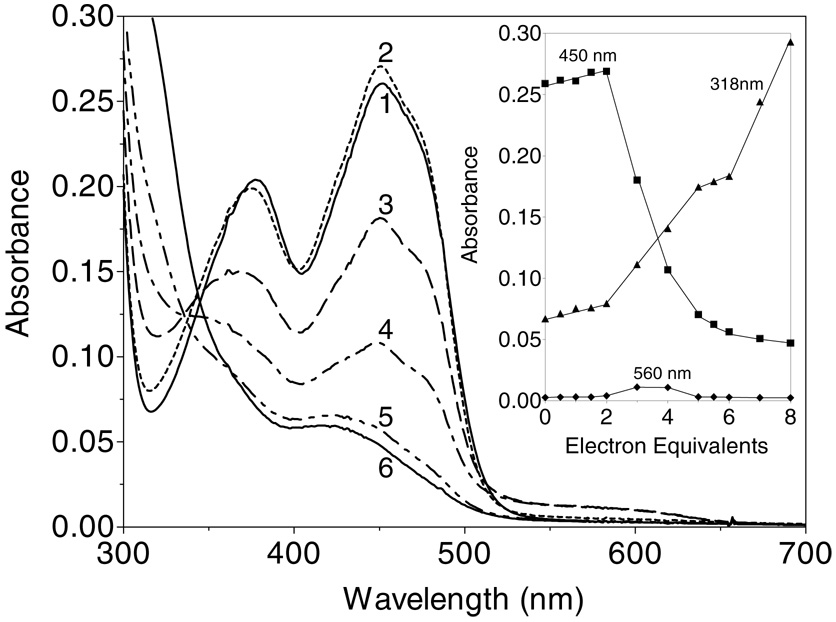
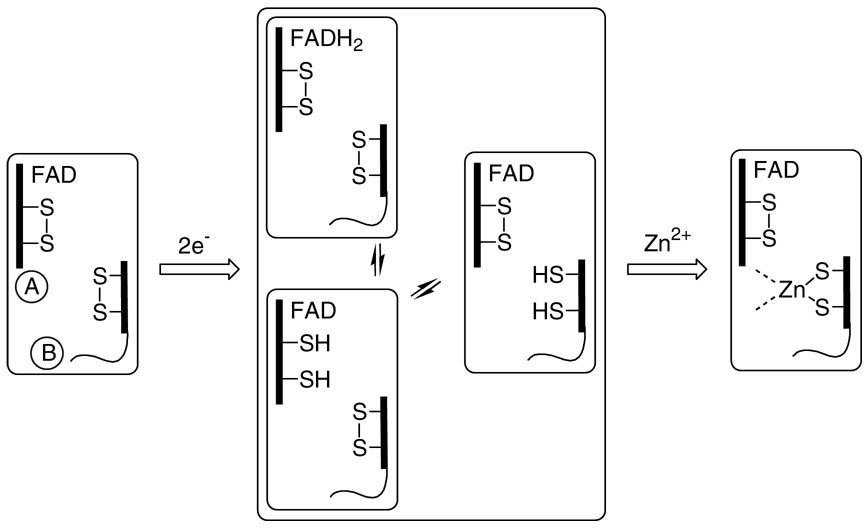
Zinc ions were recently shown to profoundly modulate the redox behavior of QSOX (28), and so we wished to examine whether the simpler Erv2p catalyst could be similarly perturbed by this divalent metal ion. Indeed, the course of the dithionite titration in Figure 4 is markedly altered by zinc: now, the addition of the first two electrons lead to a slight increase in absorbance and a slight blue shift in the main flavin absorbance envelope (curves 1 and 2; Figure 6). Such small blue shifts are suggestive of minor changes in the environment of the isoalloxazine ring (28). The second phase of the titration requires an additional 4 electrons and results in reduction of the flavin and an additional disulfide center. Thus a total of 6-electrons can be delivered to Erv2p in the presence of this thiophilic metal ion (Figure 6). It is plausible that zinc coordination involves the conformationally flexible C-terminus of Erv2p since it contains multiple potential ligands (e.g. in the sequence EDYDCGCSDSD containing the distal CGC (C176–178) motif. Scheme 1 is consistent with the data. Three of the possible 2-electron redox states of Erv2p are shown (omitting interchain disulfides between A and B subunits). The capture of one of them by zinc coordination (Scheme 1) would lead to the observed 2-electron equivalent lag in the reduction of the remaining two redox centers (flavin and the proximal disulfide: an additional 4-electrons). Further, Zn2+ proves to be a strong inhibitor of Erv2p when oxygen consumption is driven by the phosphine TCEP. Complete inhibition of the enzyme was achieved in 1 min at 10 µM Zn2+ in Tris buffer pH 7.5 without EDTA. The restoration of 1 mM EDTA effects rapid recovery of 80% of the original activity (data not shown; see Methods).
Figure 6.
Reduction of ERV2p in the presence of zinc ions. Erv2p (23 µM in Tris buffer, pH 7.5; containing 50 µM Zn2+; curve 1) was titrated anaerobically with 2, 3, 4, 6 and 8 electron-equivalents of dithionite/mol of flavin (curves 2–6 respectively). Intermediate spectra are omitted for clarity. The inset plots the absorbance changes at 318 nm (triangles), 450 nm (squares) and 560 nm (diamonds) respectively.
SCHEME 1.
Two electron reduction of Erv2p generates a Zn2+ binding site and modulates reductive titration of the protein. Only representative states for 2-electron reduced Erv2p are shown. A redox-active CxC disulfide at the C terminus of the B subunit reacts across the subunit interface of the homodimer with the proximal disulfide of the A subunit (see also Figure 1).
Enzyme monitored turnover of Erv2p
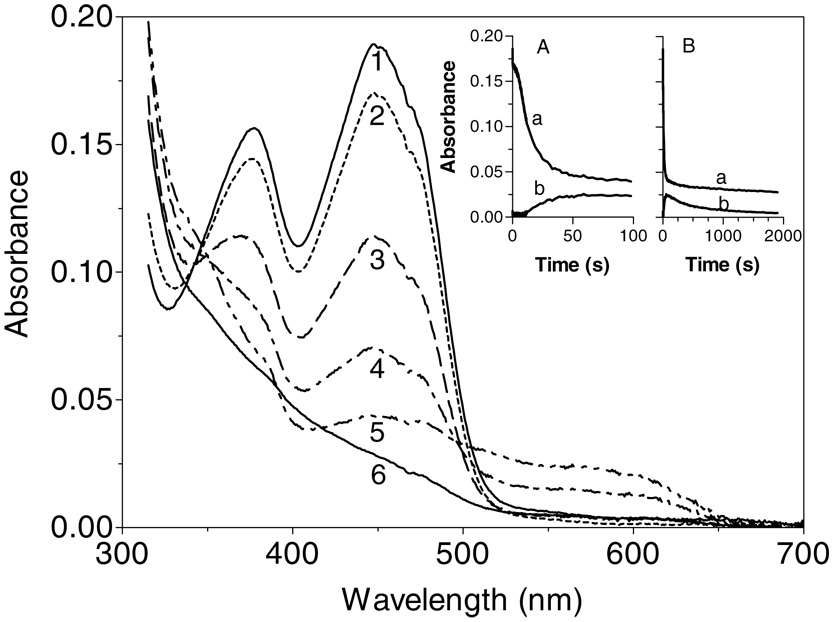
We next wanted to monitor the flavin chromophore of Erv2p during aerobic turnover in the presence of DTT (Figure 7). After mixing in the stopped-flow, to give a final concentration of 80 mM DTT, a small decrease (of about 10% of the original absorbance at 450 nm) is rapidly observed followed by a brief lag phase (clearly evident in inset A). These data show that the oxidized flavin component dominates in the steady state: the same conclusion was reached by Sevier et al. (7) monitoring absorbance values at 454 nm and using a 16-fold lower DTT concentration (Figure 7). One of the steps leading to reduction of the flavin prosthetic group clearly limits overall turnover with DTT (7). In Figure 7, further bleaching of the flavin (curves 2–5) accompanies the depletion of dissolved oxygen. Between curves 3 and 4 (at about 10 sec; inset A) there is the comparatively abrupt appearance of significant levels of the blue semiquinone. This feature resembles the blue semiquinone encountered on treatment of aerobic solutions of ALR with DTT but is approximately 3-fold less intense. The appearance of the blue radical species as reduction approaches completion is consistent with comproportionation via electron tunneling between FlH2 and Flox bound on adjacent subunits (25, 40). However, in contrast to ALR in which the blue semiquinone is very stable in the presence of excess DTT, the radical decays in Erv2p with a half-time of 6.5 min (Figure 7, inset panel B).
Figure 7.
Enzyme-monitored reduction of Erv2p by DTT under aerobic conditions. Erv2p and DTT (in 50 mM Tris buffer, pH 7.5, 25 °C, containing 240 µM dissolved oxygen) were mixed in the stopped-flow spectrophotometer to give final concentrations of 16 µM and 80 mM respectively. The main panel shows spectra taken at 0.02, 0.34, 11, 23, 63 and 1900 sec (curves 1–6 respectively). Insets A and B record absorbance traces at 450 and 560 nm (curves a and b respectively) over 100 and 2000 sec.
Reoxidation kinetics of photochemically reduced Erv2p
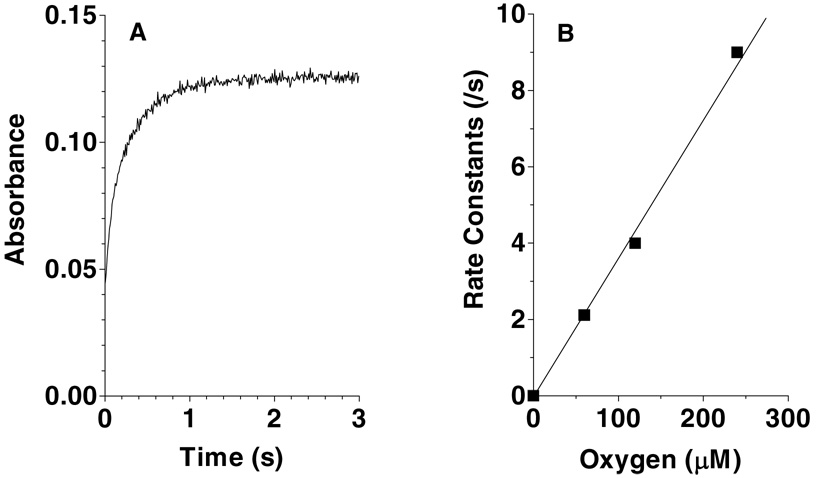
Figure 8 (panel A) shows the first-order absorbance changes at 450 nm when the reduced enzyme is mixed with air-saturated buffer (to give final concentrations of 11 µM enzyme and 120 µM oxygen). Panel B shows the apparent first-order rate constants at several oxygen concentrations, yielding a second-order rate constant of 3.5 × 104 M−1 s−1 at pH 7.5, 25 °C. The linear dependence on oxygen is typical for flavin oxidases (41–43). As expected for a flavoprotein oxidase (41–44), reoxidation of Erv2p is not accompanied by flavin radical intermediates (not shown). While Erv2p behaves normally in this respect, its reactivity in Figure 8 is more than two orders of magnitude slower than the corresponding second order rate constants for reduced flavin in avian QSOX (27). The molecular explanation for this relatively modest reactivity in such a small flavin-binding scaffold awaits further work (42), especially since there appears to be a solvent channel accessing the flavin ring in Erv2p (9). Mattevi has shown that solvent accessibility deduced from crystal structures does not necessarily correlate with the reactivity of the reduced flavoprotein towards molecular oxygen (42).
Figure 8.
Reoxidation of photochemically reduced Erv2p by oxygen. Panel A shows the absorbance increase at 450 nm on mixing reagents to give final concentrations of 11 µM reduced Erv2p and 120 µM dissolved oxygen. Panel B plots the pseudo first-order rate constants determined at a range of oxygen concentrations.
The ERV/ALR family of sulfhydryl oxidases: a comparison
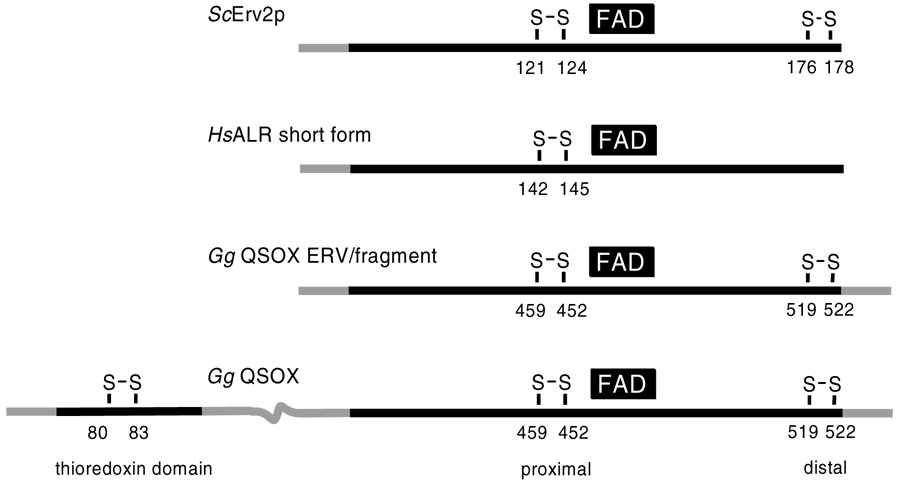
A major impetus for the present study was to collect basic information concerning the redox behavior of Erv2p: not only to get a better appreciation of this catalyst of oxidative protein folding in the yeast ER, but also for perspective on all enzymes which contain an Erv/ALR domain. There is now sufficient data on four proteins to allow meaningful comparisons (Table 2). The location of the redox centers in these proteins is depicted schematically in Figure 9.
Table 2.
Comparison of Selected Members of the ERV/ALR Family
| Protein a | # electrons to dihydroflavinb | Maximum # of electronsc | Flavin/Thiolate CT complexd | Blue Semiquinoned | TN/min with 5 mM DTT | TN/min with RNasered (200 µM –SH) |
|---|---|---|---|---|---|---|
| Erv2pe | 4 | 6 | No | Yes | 63 | 6 (200 uM SH) |
| ALR short formf | 2 | 4 | No | Yes | 46 | ~0 |
| QSOX-Erv domaing | 2 | 6 | No | Yes | 20 | ~0 |
| QSOX completeh | 4 | 8 | Yes | No | 1005 | 630 |
Proteins sources: Erv2p from S. cerevisiae, human ALR short form, QSOX Erv domain and complete protein were from avian egg white.
Denotes the number of electrons delivered during dithionite titrations when full reduction of the flavin is observed.
Electron equivalents derived from multiplying the total number of known redox centers by 2-electrons.
Thiolate to oxidized flavin charge-transfer complex or blue semiquinone observed during anaerobic reductive titrations of the proteins.
This work.
Farrell and Thorpe (25).
Raje and Thorpe (29).
Figure 9.
Domain organization of selected flavin-dependent sulfhydryl oxidases.
The critical common feature among all of these examples is a flavin with a closely interacting (proximal) disulfide. Evidently catalytic interactions between thiols and flavins have evolved independently multiple times. Prior experience with the pyridine nucleotide disulfide oxidoreductase family (23, 45) and with avian QSOX (26, 27), lead to the expectation that a thiolate-oxidized flavin charge-transfer complex would be observed on 2-electron reduction of Erv2p. Possible reasons why such absorbance bands have not been observed in native Erv2p are outlined below.
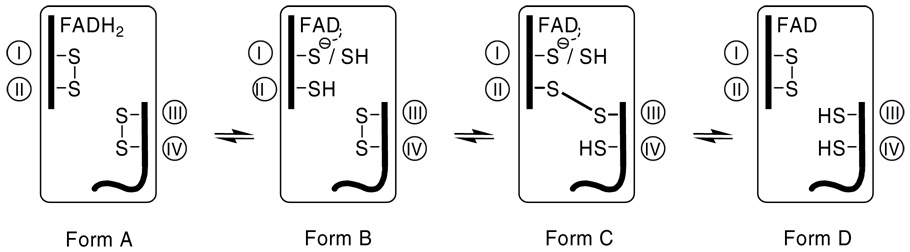
First it should be noted that the ALR-short form (25) and the ERV/ALR fragment of QSOX (29) do not form detectable thiolate to oxidized flavin charge-transfer complexes because the flavin is reduced stoichiometrically (that is before the disulfide redox centers; Table 2; Form A, Scheme 2).
SCHEME 2.
Some of the multiple forms of 2-electron reduced Erv2p. The forms B–D have an oxidized flavin ring and various redox states of cysteine I–IV. CYS I is the one closest to the flavin and could participate as a charge-transfer donor and C4a adduct formation. CYS II can form an interchain disulfide bridge with the distal disulfide (here depicted as involving CYS III). Charge-transfer complex would only occur with the deprotonated thiolate form of CYS I in either of states B and C.
In these cases, a thiolate to flavin charge-transfer complex would be impossible. However when the flavin and disulfide redox centers are similar in redox poise, as in Erv2p, then form A will be mixed with some combination of species carrying an oxidized flavin (such as B–D in Scheme 2). One potential reason for the absence of a thiolate to oxidized flavin complex is that conformational changes accompanying reduction of the proximal disulfide move CYS I out of charge-transfer range of the flavin. While a crystal structure of reduced Erv2p has yet to appear, the rigidity of this small 4-helix bundle make this possibility appear unlikely. Furthermore, mutation of CYS II to either SER or ALA in ERV1 (46) and in ALR (Farrell & Thorpe unpublished observation) shows the now-isolated CYS I can indeed form prominent charge-transfer absorbance at wavelengths > 530 nm. Another possible explanation is that the pK values of CYS I in forms B or C in Scheme 2 are too high to observe significant thiolate to flavin charge-transfer at pH 7.5. We therefore repeated the anaerobic dithionite reduction of Erv2p at pH 9.0 and also failed to generate noticeable charge-transfer absorbance (data not shown). In view of the above arguments, a more likely explanation for the lack of a charge-transfer feature is that the equilibrium between forms B–D in Scheme 2 favors form D. This would divert reducing equivalents to the distal redox center, where they cannot directly interact electronically with the flavin. Form C (Scheme 2) has already been suggested as a catalytic intermediate in Erv2p (6, 9, 24). Further a series of experiments using chimeras of the flexible C-terminus, containing the distal disulfide, and core Erv/ALR domains of AtErv1p and Erv2p are beginning to delineate the interactions between distal (III–IV) and proximal (1-II) disulfides and the functional consequences of these catalytic elements of the Erv/ALR family (9, 24). While formation of thiolate to flavin charge-transfer complexes have not been observed in Erv2p, variable levels of the blue flavosemiquinone are found (e.g. Figure 4 and Figure 7). However, there is currently no evidence that this radical form of Erv2p is of catalytic significance since it forms relatively slowly during aerobic turnover with DTT (Figure 7, panel A).
A notable aspect of the data in Table 2 is the modest catalytic activities of Erv2p and ALR towards either DTT or reduced RNase when compared to full-length QSOX. Thus for the single domain oxidases kcat/Km for DTT are 200- to 400-fold lower than observed with QSOX. Further, QSOX is some 100-fold faster at generating disulfides in reduced RNase (200 µM thiols) than Erv2p or ALR. The enhanced reactivity of QSOX presumably reflects fusion of a redox-active thioredoxin (PDI-like) domain to an ERV/ALR fold (9, 19, 20, 29). The finding that the reduced thioredoxin portion of QSOX is a substrate of the flavin-containing fragment strengthens the notion that a critical aspect of efficient catalysis in this multidomain oxidase is the communication between these domains (19, 29).
In this regard, the ancient fusion of thioredoxin and Erv/ALR domains in QSOX may represent a Rosetta stone protein (47, 48): one that points the way to possible physiological substrates of Erv2p. This is currently an issue because the natural redox partner(s) of yeast Erv2p have yet to be identified. Although crosslinking experiments suggest that PDI1p interacts with Erv2p (7, 49), in vitro experiments with reduced PDI1p show that it is a poor substrate of this oxidase (24). This kinetic sluggishness has led to the suggestion that other ER redox proteins may mediate transfer of reducing equivalents from client unfolded proteins to Erv2p (24). The domain structure of QSOX hints that the physiological substrate(s) of Erv2p in the yeast ER do indeed contain the thioredoxin fold (even though it may not be those found in conventional PDIs). Another suggestion to address the weak reactivity between Erv2p and PDI1p is that essential factors required to accelerate the reaction may be absent in vitro (24). If PDI1p is a physiological reductant of Erv2p, the reaction is not thermodynamically favorable when considered merely in terms of the standard redox potentials for the partners: the two CxxC centers of yeast PDI are reported at −150 and −187 mV (50)whereas the redox poise of Erv2p determined here is more negative (about −200 mV; see above).
Finally it is worth considering whether a particularly facile oxidation of reduced PDI by Erv2p (or indeed Ero1p) would be desirable. If this were to happen, then oxidized PDI would rapidly accumulate and any activities that rely on reduced PDI (for example its redox-neutral isomerase activity) would be impaired. Secondly an accumulation of oxidized PDI would lead to subsequent reduction by endogenous GSH levels and this could, in principle, drive further hydrogen peroxide generation and depletion of cellular reducing power. It seems clear that if oxidized PDI is the immediate physiological oxidant in the lumen of the ER the pathways for its generation need to be carefully modulated.
ACKNOWLEDGMENT
We thank Dr. Yu-Chu Huang for gas phase sequencing and Drs. Karen Hoober and Charles Williams for gifts of avian sulfhydryl oxidase and Escherichia coli thioredoxin respectively.
Footnotes
This work was supported in part by NIH GM26643.
Abbreviations: ALR, augmenter of liver regeneration; CTAB, cetyltrimethylammonium bromide; DTT, dithiothreitol; ER, endoplasmic reticulum; ERV, protein essential for respiration and viability in yeast; GSH, reduced glutathione; QSOX, flavin-dependent sulfhydryl oxidases homologous to Quiescin Q6; and TCEP, tris(2-carboxyethyl)phosphine.
REFERENCES
- 1.Lee J, Hofhaus G, Lisowsky T. Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett. 2000;477(1–2):62–66. doi: 10.1016/s0014-5793(00)01767-1. [DOI] [PubMed] [Google Scholar]
- 2.Rose JP, Wu C-K, Dailey TA, Dailey HA, Wang BC. American Crystallographic Association. Minnesota: St. Paul; 2000. p. E0049. [Google Scholar]
- 3.Lisowsky T, Lee JE, Polimeno L, Francavilla A, Hofhaus G. Mammalian augmenter of liver regeneration protein is a sulfhydryl oxidase. Dig. Liver Dis. 2001;33:173–180. doi: 10.1016/s1590-8658(01)80074-8. [DOI] [PubMed] [Google Scholar]
- 4.Wu CK, Dailey TA, Dailey HA, Wang BC, Rose JP. The crystal structure of augmenter of liver regeneration: A mammalian FAD-dependent sulfhydryl oxidase. Protein Sci. 2003;12:1109–1118. doi: 10.1110/ps.0238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerber J, Muhlenhoff U, Hofhaus G, Lill R, Lisowsky T. Yeast ERV2p is the first microsomal FAD-linked sulfhydryl oxidase of the Erv1p/Alrp protein family. J. Biol. Chem. 2001;276:23486–23491. doi: 10.1074/jbc.M100134200. [DOI] [PubMed] [Google Scholar]
- 6.Gross E, Sevier CS, Vala A, Kaiser CA, Fass D. A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat. Struct. Biol. 2002;9:61–67. doi: 10.1038/nsb740. [DOI] [PubMed] [Google Scholar]
- 7.Sevier CS, Cuozzo JW, Vala A, Aslund F, Kaiser CA. A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat. Cell Biol. 2001;3:874–882. doi: 10.1038/ncb1001-874. [DOI] [PubMed] [Google Scholar]
- 8.Levitan A, Danon A, Lisowsky T. Unique features of plant mitochondrial sulfhydryl oxidase. J. Biol. Chem. 2004;279:20002–20008. doi: 10.1074/jbc.M312877200. [DOI] [PubMed] [Google Scholar]
- 9.Vitu E, Bentzur M, Lisowsky T, Kaiser CA, Fass D. Gain of function in an ERV/ALR sulfhydryl oxidase by molecular engineering of the shuttle disulfide. J. Mol. Biol. 2006;362:89–101. doi: 10.1016/j.jmb.2006.06.070. [DOI] [PubMed] [Google Scholar]
- 10.Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplamic reticulum. Mol. Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- 11.Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol. Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- 12.Tu BP, Ho-Schleyer SC, Travers KJ, Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–1574. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- 13.Gross E, Sevier CS, Heldman N, Vitu E, Bentzur M, Kaiser CA, Thorpe C, Fass D. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. U S A. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross E, Kastner DB, Kaiser CA, Fass D. Structure of Ero1p, source of disulfide bonds for oxidative protein folding in the cell. Cell. 2004;117:601–610. doi: 10.1016/s0092-8674(04)00418-0. [DOI] [PubMed] [Google Scholar]
- 15.Cabibbo A, Pagani M, Fabbri M, Rocchi M, Farmery MR, Bulleid NJ, Sitia R. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J. Biol. Chem. 2000;275:4827–4833. doi: 10.1074/jbc.275.7.4827. [DOI] [PubMed] [Google Scholar]
- 16.Pagani M, Fabbri M, Benedetti C, Fassio A, Pilati S, Bulleid NJ, Cabibbo A, Sitia R. Endoplasmic reticulum oxidoreductin 1-lbeta (ERO1-Lbeta), a human gene induced in the course of the unfolded protein response. J. Biol. Chem. 2000;275:23685–23692. doi: 10.1074/jbc.M003061200. [DOI] [PubMed] [Google Scholar]
- 17.Benayoun B, Esnard-Fève A, Castella S, Courty Y, Esnard F. Rat seminal vesicle FAD-dependent sulfhydryl oxidase:biochemical characterization and molecular cloning of a member of the new sulfhydryl oxidase/quiescin Q6 gene family. J. Biol. Chem. 2001;276:13830–13837. doi: 10.1074/jbc.M010933200. [DOI] [PubMed] [Google Scholar]
- 18.Hoober KL, Glynn NM, Burnside J, Coppock DL, Thorpe C. Homology between egg white sulfhydryl oxidase and quiescin Q6 defines a new class of flavin-linked sulfhydryl oxidases. J. Biol. Chem. 1999;274:31759–31762. doi: 10.1074/jbc.274.45.31759. [DOI] [PubMed] [Google Scholar]
- 19.Coppock DL, Thorpe C. Multidomain flavin-dependent sulfhydryl oxidases. Antioxid. Redox Signal. 2006;8:300–311. doi: 10.1089/ars.2006.8.300. [DOI] [PubMed] [Google Scholar]
- 20.Thorpe C, Hoober K, Raje S, Glynn N, Burnside J, Turi G, Coppock D. Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch. Biochem. Biophys. 2002;405:1–12. doi: 10.1016/s0003-9861(02)00337-5. [DOI] [PubMed] [Google Scholar]
- 21.O'Donnell ME, Williams CH., Jr Reconstitution of Escherichia coli Thioredoxin Reductase with 1-Deaza-FAD: Evidence for 1-Deaza-FAD-C4a Adduct Linked to the Ionization of an Active Site Base. J. Biol. Chem. 1984;259:2243–2251. [PubMed] [Google Scholar]
- 22.Thorpe C, Williams CH. Spectral evidence for a flavin adduct in a monoalkylated derivative of pig heart lipoamide dehydrogenase. J. Biol. Chem. 1976;251:7726–7728. [PubMed] [Google Scholar]
- 23.Williams CH., Jr . In: Chemistry and Biochemistry of Flavoenzymes. Müller F, editor. CRC Press, Chemistry and Biochemistry of Flavoenzymes; 1992. pp. 121–211. [Google Scholar]
- 24.Vala A, Sevier CS, Kaiser CA. Structural determinants of substrate access to the disulfide oxidase Erv2p. J. Mol. Biol. 2005;354:952–966. doi: 10.1016/j.jmb.2005.09.076. [DOI] [PubMed] [Google Scholar]
- 25.Farrell SR, Thorpe C. Augmenter of liver regeneration: a flavin dependent sulfhydryl oxidase with cytochrome C reductase activity. Biochemistry. 2005;44:1532–1541. doi: 10.1021/bi0479555. [DOI] [PubMed] [Google Scholar]
- 26.Hoober KL, Joneja B, White HB, III, Thorpe C. A Sulfhydryl Oxidase from Chicken Egg White. J. Biol. Chem. 1996;271:30510–30516. doi: 10.1074/jbc.271.48.30510. [DOI] [PubMed] [Google Scholar]
- 27.Hoober KL, Thorpe C. Egg white sulfhydryl oxidase: Kinetic mechanism of the catalysis of disulfide bond formation. Biochemistry. 1999;38:3211–3217. doi: 10.1021/bi9820816. [DOI] [PubMed] [Google Scholar]
- 28.Brohawn SG, Rudik I, Thorpe C. Avian sulfhydryl oxidase is not a metalloenzyme: adventitious binding of divalent metal ions to the enzyme. Biochemistry. 2003;42:11074–11082. doi: 10.1021/bi0301385. [DOI] [PubMed] [Google Scholar]
- 29.Raje S, Thorpe C. Inter-domain redox communication in flavoenzymes of the quiescin/sulfhydryl oxidase family: role of a thioredoxin domain in disulfide bond formation. Biochemistry. 2003;42:4560–4568. doi: 10.1021/bi030003z. [DOI] [PubMed] [Google Scholar]
- 30.Hoober KL, Thorpe C. Flavin-dependent sulfhydryl oxidases in protein disulfide bond formation. Methods Enzymol. 2002;348:30–34. doi: 10.1016/s0076-6879(02)48622-3. [DOI] [PubMed] [Google Scholar]
- 31.Hoober KL, Sheasley SS, Gilbert HF, Thorpe C. Sulfhydryl oxidase from egg white: a facile catalyst for disulfide bond formation in proteins and peptides. J. Biol. Chem. 1999;274:22147–22150. doi: 10.1074/jbc.274.32.22147. [DOI] [PubMed] [Google Scholar]
- 32.Gorelick R, Schopfer LM, Ballou DP, Massey V, Thorpe C. Interflavin oxidation-reduction reactions between pig kidney general acyl-CoA dehydrogenase and electron-transferring flavoprotein. Biochemistry. 1985;24:6830–6839. doi: 10.1021/bi00345a015. [DOI] [PubMed] [Google Scholar]
- 33.Beinert H. In: The Enzymes. Boyer PD, Lardy HA, Myrback K, editors. New York: Academic Press; 1960. pp. 339–416. [Google Scholar]
- 34.Williams CH, Arscott LD, Matthews RG, Thorpe C, Wilkinson KD. Methodology employed for anaerobic spectrophotometric titrations and for computer-assisted data analysis. Methods Enzymol. 1979:185–198. doi: 10.1016/0076-6879(79)62217-6. [DOI] [PubMed] [Google Scholar]
- 35.Cline DJ, Thorpe C, Schneider JP. Structure Based Design of a Fluorimetric Redox Active Peptide Probe. Anal. Biochem. 2003;325:144–150. doi: 10.1016/j.ab.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Moore EC, Reichard P, Thelander L. Enzymatic Synthesis of Deoxyribonucleotides.V. Purification and Properties of Thioredoxin Reductase from Escherichia Coli B. J. Biol. Chem. 1964;239:3445–3452. [PubMed] [Google Scholar]
- 37.Chivers PT, Prehoda KE, Raines RT. The CXXC motif: a rheostat in the active site. Biochemistry. 1997;36:4061–4066. doi: 10.1021/bi9628580. [DOI] [PubMed] [Google Scholar]
- 38.Cline DJ, Redding SE, Brohawn SG, Psathas JN, Schneider JP, Thorpe C. New water-soluble phosphines as reductants of peptide and protein disulfide bonds: reactivity and membrane permeability. Biochemistry. 2004;43:15195–15203. doi: 10.1021/bi048329a. [DOI] [PubMed] [Google Scholar]
- 39.Massey V, Hemmerich P. Photoreduction of flavoproteins and other biological compounds catalyzed by deazaflavins. Biochemisty. 1978;17:9–16. doi: 10.1021/bi00594a002. [DOI] [PubMed] [Google Scholar]
- 40.Kay CWM, Elsasser C, Bittl R, Farrell SR, Thorpe C. Determination of the distance between the two neutral flavin radicals in augmenter of liver regeneration by pulsed ELDOR. J. Amer. Chem. Soc. 2006;128:76–77. doi: 10.1021/ja057308g. [DOI] [PubMed] [Google Scholar]
- 41.Massey V. Activation of molecular oxygen by flavins and flavoproteins. J. Biol. Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 42.Mattevi A. To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem. Sci. 2006;31:276–283. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Massey V. Flavins and Flavoproteins, International Congress Series. Elsevier Science; 2002. pp. 3–11. [Google Scholar]
- 44.Massey V, Hemmerich P. Active-site probes of flavoproteins. Biochem. Soc. Trans. 1980;8:246–257. doi: 10.1042/bst0080246. [DOI] [PubMed] [Google Scholar]
- 45.Argyrou A, Blanchard JS. Flavoprotein disulfide reductases: advances in chemistry and function. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:89–142. doi: 10.1016/S0079-6603(04)78003-4. [DOI] [PubMed] [Google Scholar]
- 46.Hofhaus G, Lee JE, Tews I, Rosenberg B, Lisowsky T. The N-terminal cysteine pair of yeast sulfhydryl oxidase Erv1p is essential for in vivo activity and interacts with the primary redox centre. Eur. J. Biochem. 2003;270:1528–1535. doi: 10.1046/j.1432-1033.2003.03519.x. [DOI] [PubMed] [Google Scholar]
- 47.Marcotte EM, Pellegrini M, Ng HL, Rice DW, Yeates TO, Eisenberg D. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 48.Bowers PM, Pellegrini M, Thompson MJ, Fierro J, Yeates TO, Eisenberg D. Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol. 2004;5:R35. doi: 10.1186/gb-2004-5-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson B, Xiao R, Gilbert HF. A structural disulfide of yeast protein-disulfide isomerase destabilizes the active site disulfide of the N-terminal thioredoxin domain. J. Biol. Chem. 2005;280:11483–11487. doi: 10.1074/jbc.M414203200. [DOI] [PubMed] [Google Scholar]
- 51.Stein G, Lisowsky T. Functional comparison of the yeast scERV1 and scERV2 genes. Yeast. 1998;14:171–180. doi: 10.1002/(SICI)1097-0061(19980130)14:2<171::AID-YEA209>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]