Abstract
Background
The availability of complete genome sequences of versatile Pseudomonas occupying remarkably diverse ecological niches enabled to gain insights into their adaptative assets. The objective of this study was to analyze the complete genetic repertoires of metal transporters (metal transportomes) from four representative Pseudomonas species and to identify metal transporters with "Genomic Island" associated features.
Methods
A comparative metal transporter inventory was built for the following four Pseudomonas species: P.putida (Ppu) KT2440, P.aeruginosa (Pae) PA01, P.fluorescens (Pfl) Pf-5 and P.syringae (Psy)pv.tomato DC3000 using TIGR-CMR and Transport DB. Genomic analysis of essential and toxic metal ion transporters was accomplished from the above inventory. Metal transporters with "Genomic Island" associated features were identified using Islandpath analysis.
Results
Dataset cataloguing has been executed for 262 metal transporters from the four spp. Additional metal ion transporters belonging to NiCoT, Ca P-type ATPase, Cu P-type ATPases, ZIP and MgtC families were identified. In Psy DC3000, 48% of metal transporters showed strong GI features while it was 45% in Ppu KT2440. In Pfl Pf-5 and Pae PA01 only 26% of their metal transporters exhibited GI features.
Conclusion
Our comparative inventory of 262 metal transporters from four versatile Pseudomonas spp is the complete suite of metal transportomes analysed till date in a prokaryotic genus. This study identified differences in the basic composition of metal transportomes from Pseudomonas occupying diverse ecological niches and also elucidated their novel features. Based on this inventory we analysed the role of horizontal gene transfer in expansion and variability of metal transporter families.
Findings
Background
Metal ions play a variety of critical roles in the prokaryotic metabolism. Biological management of metal ions is accomplished by a complex interplay between metal ion transporters (transmembrane importers, transmembrane exporters) and their regulatory components [1]. In the post-genomic era, genetic landscapes of the organisms have unveiled the hidden facets of metal ion transporters and associated regulatory cascade thereby paving way for study of their metal transportomes.
The genus Pseudomonas is one of the most versatile and ecologically significant groups of bacteria on the planet that thrives well in metal rich biotopes. In recent studies, the unexpected capacity of Ppu KT2440 and Ppu CD2 genomes to endure exposure to heavy metals has been unraveled [2,3]. In Ppu KT2440 metal transporters acquired through HGT were also reported [4]. However, a systematic study of complete metal transportomes is unavailable for any of these Pseudomonas species. The availability of complete genome sequences for saprophytic P.putida (Ppu) KT2440, commensalistic P.fluorescens (Pfl) Pf-5, pathogenic P.aeruginosa (Pae) PA01 and P.syringae pv. tomato (Psy) DC3000 [5-8] provided an opportunity for analyzing their complete repertoire of metal transporters along with the role of HGT in their acquisition.
Methods
Compilation of metal transporter inventory
The genome sequence information of the four sequenced Pseudomonas spp Ppu KT2440, Pae PA01, Pfl Pf-5 and Psy DC3000 employed in this study were selected from TIGR-CMR database [9]. TransportDB, a relational database describing the predicted cytoplasmic membrane transport protein complement for organisms was used as a substrate for building the metal transportomes [10]. In order to search for homologs of a transporter family the best-annotated hit was used in a subsequent BLASTP search against the four genomes and retrieved the members. PFAM domain search was also carried out with the retrieved homologs, using CLC HMM algorithm [11] and PFAM database [12]. Transmembrane helix prediction for the membrane transporters was performed using TMHMM version 2.0 [13]. Genomic location and gene organization of transporters were identified from TIGR-CMR database.
Protein sequence analysis
CLC protein workbench has been employed to analyse the metal transporter proteins [11]. Genchek™ Graphical Viewer was used to represent GTPase signature motifs G1-G5 in the alignment generated for FeoBEco and FeoBPae.
Phylogenetic analysis
Phylogenetic analysis was performed with the characterised prototypes using CLC protein workbench [11]. For this analysis, multiple sequence alignments (MSA) were produced using progressive alignment algorithm. The generated pair wise alignments were used for finding the evolutionary distance between the pairs. Pair wise distances thus calculated was used to create a phylogenetic tree-employing neighbour joining (NJ) algorithm with 1000 bootstrap replicates.
Sequence Clusters
Metal transporters located in sequence clusters, were identified by cluster viewer from PEDANT database [14]. Cluster viewer aided us in identifying clusters of genes in the same organism that are paralogs. Sequence clusters thus obtained provided us with data regarding the metal transporters that have a BLAST similarity score greater than 45 bits in PSI-BLAST and also possess a similar Pfam domain with a significant E-value (typically 0.001). This tool helped us in determining paralogous genes of a family and aided in phylogenetic analyses.
Island Path Analysis
Island Path Analysis was used for the detection of metal transporters acquired through HGT [15]. After generating the complete inventory of metal transporters, we inspected the genomes of four Pseudomonas species with island path software (IPA version 1.0 tool) for the identification of those transporters located in GI's or exhibiting GI associated features like anomalous %G+C, dinucleotide bias above 1 STD DEV, presence of RNA genes (tRNA, rRNA genes) and mobility genes (transposons, insertion sequences). A putative GI can be identified with certainty by the presence of eight or more consecutive ORF's with dinucleotide bias alone or dinucleotide bias plus a mobility gene in proximity.
Results and discussion
From genome to metal transportome in Pseudomonas
Based on the global features of the four Pseudomonas genomes we could draw a comparison among the genome size, number of genes, transporter proteins and total number of metal transporters (Table 1). Among the four spp, Pfl has the largest metal transporter inventory (75 metal ion transporters) while Psy has the smallest (54 metal ion transporters). Other pathogenic species, Pae has more number of genes and transporter proteins but relatively less metal transporters (63 metal ion transporters) than Ppu (70 metal ion transporters).
Table 1.
Global features of four representative Pseudomonas genomes
| Topology | P.putida KT2440 | P.aeruginosa PA01 | P.fluorescens Pf5 | P.syringae DC3000 |
| Genome size (bp) | 6,181,862 | 6,264,403 | 7,074,893 | 6,538,260 |
| G+C content (%) | 61.6 | 66.5 | 63.3 | 58.3 |
| Total no: of genes | 5516 | 5565 | 6230 | 5843 |
| Total Transporter Proteins | 386 | 423 | 475 | 322 |
| Total Metal Transporters | 70 | 63 | 75 | 54 |
Based on the TransportDB [10] we compiled the metal transportomes for the entire complement of alkali/alkaline earth metals (Na+, K+, Ca2+, Mg2+), transition metals (Zn2+, Mn2+ Cu+, Ag+, Mo2+, Fe2+, Fe3+, Ni2+, Co2+) and heavy metals (Cr3+, As3+, Pb2+, Cd2+) in Pseudomonas (Additional files 1, 2, 3, 4). These transportomes furnish information for class/family/subfamily, transporter classification number (T.C.No), TIGR locus, protein name, genomic location, orientation, length of the protein, TMD's, substrate and the predicted role of the above metal transporters. Sequence cluster analysis helped in identifying paralogs for the transporter families (see Additional file 5). In this study we employed the Transporter Classification system-TC system for systematic classification of Pseudomonas metal transporters into the following four groups: ATP-dependent (ABC superfamily, P-type ATPase super family) Ion-channels, Secondary transporters and Unclassified. A comparative analysis depicting the basic differences in the composition of transporter classes and metal transporter families from the four Pseudomonas species is represented in Table 2 (This table also provides expanded names of the transporter families).
Table 2.
Comparative Analysis of Metal transporters from P.aeruginosa PA01, P.fluorescens Pf-5, P.putida KT2440 and P.syringae DC3000
| Metal Transporter Type/Family | Number of Transporters | |||
| P.aeruginosa PAO1 | P.fluorescens Pf5 | P.putida KT2440 | P.syringae Pv tomato DC 3000 | |
| I ATP Dependent | 17 (26.98%) | 18 (24. 32%) | 17 (24.28%) | 16 (29.62%) |
| The ATP-binding cassette (ABC) super family | 10 | 10 | 10 | 11 |
| The P-type ATPase (P-ATPase) super family | 7 | 8 | 7 | 5 |
| II. Ion Channels | 6 (9.52%) | 5 (6.66%) | 4 (5.71%) | 7(12.96%) |
| The CorA metal ion Transporter family (MIT family) | 3 | 3 | 3 | 4 |
| Small-conductance mechanosensitive ion channel (MSCS) family | 1 | 1 | - | 1 |
| The voltage-gated ion channel superfamily (VIC) | 2 | 1 | 1 | 2 |
| III Secondary Transporters | 36 (57.14%) | 47 (62.66%) | 45 (64.28%) | 28 (51.85%) |
| The Alanine/Glycine: Cation Symporter (AGCS) family | 3 | 1 | 1 | 1 |
| The Arsenite-Antimonite (ArsB) efflux family | 1 | 2 | 2 | 1 |
| The Bile acid: Na+ symporter (BASS) family | 3 | 2 | 2 | 1 |
| The Ca2+: Cation Antiporter (CaCA) family | - | 2 | 1 | 2 |
| The cation diffusion facilitator (CDF) family | 3 | 4 | 2 | 1 |
| The Chromate Ion Transporter (CHR) family | 1 | 1 | 1 | - |
| The Citrate Mg2+: H+ (CitM) Citrate-Ca2+: H+ (CitH) Symporter (CitMHS) family | - | 3 | 2 | 1 |
| The Monovalent cation: Proton Antiporter-1 (CPAI) family | 5 | 5 | 4 | 3 |
| The Monovalent cation: Proton Antiporter-2 (CPA2) family | 3 | 4 | 4 | 3 |
| The Monovalent cation: (K+or Na+) Proton Antiporter-3 (CPA3) family | 1 | 1 | 1 | 1 |
| The Dicarboxylate/AminoAcid: cation (Na+/H+) Symporter (DAACS) family | 1 | 3 | 2 | - |
| The Divalent Anion: Na+ symporter (DASS) family | 1 | 1 | 1 | - |
| The glutamate: Na+ symporter (ESS) family | 1 | 1 | 1 | |
| The K+ Uptake Permease (KUP) family | 1 | 1 | 1 | 1 |
| The Malonate: Na+symporter (MSS) family | 1 | 1 | - | - |
| The NhaA Na+:H+ Antiporter (NhaA) family | - | 1 | 2 | 2 |
| The NhaB Na+: H+ Antiporter (NhaB) family | 1 | 1 | 1 | - |
| The Ni2+-Co2+Transporter (NicoT) family | - | - | 1 | 1 |
| The Metal Ion (Mn2+- Fe2+) Transporter (Nramp) family | 2 | 1 | - | 3 |
| The Neurotransmitter sodium symporter (NSS) family | - | 1 | - | 1 |
| PNaS family: The Phosphate sodium symporter family | 2 | 2 | 1 | 1 |
| The Resistance-Nodulation Cell Division (RND) super family | 2 | 2 | 6 | - |
| The solute: sodium symporter (SSS) family | 2 | 4 | 5 | 3 |
| The K+ transporter (Trk) family | 1 | 1 | 2 | 1 |
| The Zinc (Zn2+)-Iron (Fe2+) Permease (ZIP) family | 1 | 2 | 2 | 1 |
| IV Unclassified | 4 (6.35%) | 5 (6.66%) | 4 (5.71%) | 3 (5.55%) |
| The Ferrous Iron uptake (FeoB) family | 1 | - | - | - |
| The Iron/Lead Transporter (ILT) super family | 1 | 2 | 1 | 2 |
| The Mg2+Transporter E (MgtE) family | 1 | 1 | 1 | 1 |
| The Mg2+Transporter C (MgtC) family | 1 | 2 | 2 | - |
| Total | 63 | 75 | 70 | 54 |
Salient Features of Pseudomonas metal transportomes
Apart from executing the dataset cataloguing, our analysis of Pseudomonas databases identified additional metal transporters belonging to Ca, ZIP, NiCoT, MgtC, ABC (copper), P-type ATPase (copper) families. Distinct features of transporters belonging to CPA3, CaCA, ZnuABC, MntABC, ZntA, Nramp, FeOB, OFeT, NikABC, Chr, PbrT, RND, CDF families and ABC super family were also reported.
Search for primary ATP-dependent calcium pumps in Pseudomonas using two characterized members (SP1551 and SP1623) of S.pneumoniae [16] identified a Ca P-type ATPase (PA1429) in Pae alone with no reciprocal BLAST hits from other three species.
ZIP family of transporters in Pseudomonas were retrieved using the experimentally characterized transporter ZupTEco [17]. Psy and Pae have single entities PA4467 and PSPTO_2053 while paralog expansion is seen for Ppu (PP_1836, PP_0947) and Pfl (PFL_4718, PFL_0910).
Using the experimentally characterized efflux member RcnAEco of NiCoT subfamily 2 [18] as a BLAST query against Pseudomonas databases, we identified two new efflux members PP_2968 (RcnAPpu) and PSPTO_4280 (RcnAPsy).
The experimentally characterized MgtC family transporter Pae (PA4635) [19] was used in the BLAST P analysis to identify the following MgtC homologs: PP_3244, PP_3541 (Ppu), PFL_4077, PFL_2871 (Pfl), PA4635 (Pae). No hits were identified for this family in Psy.
Pae uniquely harbours an ABC copper transporter PA3393 that is related to periplasmic copper binding protein NosD in Rhizobium meliloti whose function was presumed to insert copper into the exported reductase apoenzyme (NosZ) [20]. All the CuPATP1 transporters of Pseudomonas (PFL_0710, PP_0586, PA3920, PSPTO_0750) were found to be the best BLAST hits for experimentally characterized CopAEhi, while the other group of CuPATP2 transporters (PP_4261, PA1549, PFL_1915, PSPTO_1996) did not show significant BLAST identities with the characterized proteins of CopA, CopB.
CPA3 family members constituting multicomponent K+ efflux system are present in all the three species except in Pae where this component is involved in sodium ion homeostasis. Ppu (PP_2225 to PP_2230) and Pfl Pf-5 (PFL_2606 to PFL_2611) have six component K+ efflux systems while it is a five-component system in Psy.
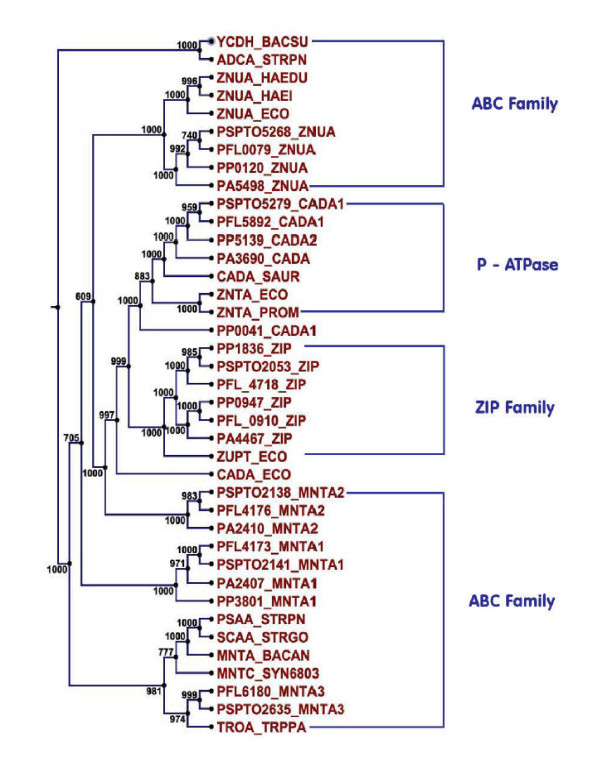
CaCA transporters i.e. secondary calcium transporters in Pfl and Psy (PFL_0722 and PSPTO_0764) are annotated as Ca2+/H+ (ChaA) antiporters that exhibit significant homology to ChaA of E.coli [21]. Proteins resembling mammalian Na2+/Ca2+exchangers (YrbG) [22] are present in Ppu (PP_0928), Pfl Pf-5 (PFL_0891) and Psy (PSPTO_4477). Pae PA01 lacks a distinct homologue for CaCA transporter. Representative Zinc transporters of Pseudomonas species are presented in table 3. ABC zinc transporters (ZnuABC) that recognize Zn2+ as their substrate and ABC manganese transporters (MntABC) that recognize Mn2+/Zn2+, were included in the phylogenetic profiling to study their overlapping specificities (Figure 1). Search for ZntA homologs in the Pseudomonas genomes using experimentally characterized ZntAEco [23] resulted in identification of CadA as the closest homologue with 37% identity from all the four species. However in Pseudomonas, only CadA proteins with five (PFL_5892, PSPTO_5279, PA3690) or six TMD's (PP_0041, PP_5139, PA2435) were identical to ZntA. The other CadA proteins with seven TMD's (PFL_6191, PSPTO_5532) did not show any significant identity in BLAST analysis (less than 24% identity) and appear to be unrelated to ZntA.
Table 3.
Representatives of Zinc Transporters from P.putida KT2440, P.aeruginosa PA01, P.fluorescens Pf-5 and P.syringae DC3000
| Transporter protein | Functional family | Transport direction | Pseudomonas species | Number of Paralogs/Orthologs |
| ZnuABC | ABC ATPase | Uptake | Pputida | 2 |
| P.aeruginosa | 3 | |||
| P.fluorescens | 4 | |||
| P.syringae | 4 | |||
| ZupT | ZIP chemiosmotic (20 transporters) | Uptake | P.putida | 2 |
| P.aeruginosa | 1 | |||
| P.fluorescens | 2 | |||
| P.syringae | 1 | |||
| CorA | MIT family (Ion channels) | Uptake | P.putida | 1 |
| CadA | P-type ATPases | Efflux | P.putida | 2 |
| P.aeruginosa | 1 | |||
| P.fluorescens | 1 | |||
| P.syringae | 1 | |||
| CzcCBA | RND chemiosmotic (20 transporters) | Efflux | P.putida | 2 |
| P.aeruginosa | 1 | |||
| P.fluorescens | 1 | |||
| CzcD | CDF chemiosmotic (20 transporters) | Efflux | P.putida | 1 |
| P.fluorescens | 1 | |||
Figure 1.
Phylogenetic tree for Zinc/Manganese transporters using neighbour joining method with 1000 bootstrap replicates. It depicts the grouping of various orthologs of Pseudomonas species from three families (ABC, P-type ATPase and ZIP families). BACSU, Bacillus subtilis; STRPN, Streptococcus pneumoniae; HAEDU, Haemophilus ducreyi; HAEIN, Haemophilus influenzae; ECO, Escherichia coli; PROM, Proteus mirabilis; STRPN, Streptococcus pneumoniae; STRGO, Streptococcus gordonii; BACAN, Bacillus anthracis; SYN6803, Synechococystis sp strain PCC6803; TRPPA, Treponema pallidum.
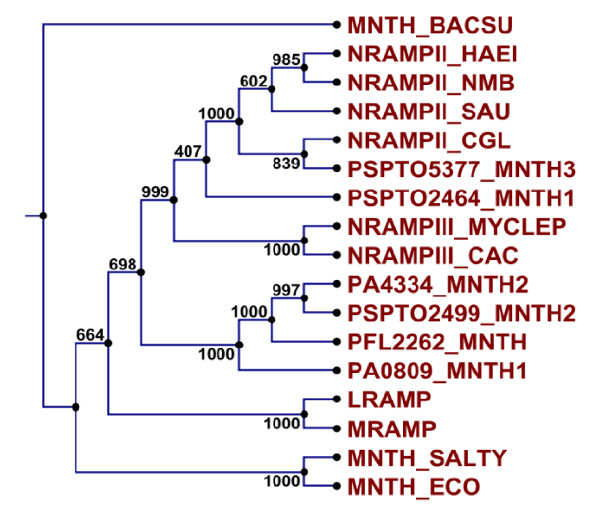
Only one Nramp member, MntH (PFL_2262) was observed for Pfl while Pae has two members MntH1 (PA0809), MntH2 (PA4334). Psy was found to possess three distinct members MntH1 (PSPTO_2464), MntH2 (PSPTO_2499) and MntH3 (PSPTO_5377) in this family. No conserved motifs characteristic for Nramp super family were observed for PSPTO_5377. However, phylogenetic analysis with Nramp family members confirmed its position in this family (Figure 2). Analysis of Lineage Specific Regions (greater than 2 Kb, enriched in mobile genetic elements and has genes specific to Psy DC3000) of Psy showed that PSPTO_5377 is the only metal transporter located in its LSR's (LSR no: 42).
Figure 2.
Phylogenetic tree for Nramp family of manganese transporters using neighbour joining method with 1000 bootstrap replicates. It depicts the grouping of various orthologs of Pseudomonas species within this family. BACSU, Bacillus subtilis; HAEI, Haemophilus influenzae; SAU, Staphylococcus aureus; CGL, Corynebacterium glutamicum; MYCLEP, Mycobacterium leprae; CAC, Corynebacterium acetylicum; SALTY, Salmonella typhimurium; ECO, Escherichia coli.
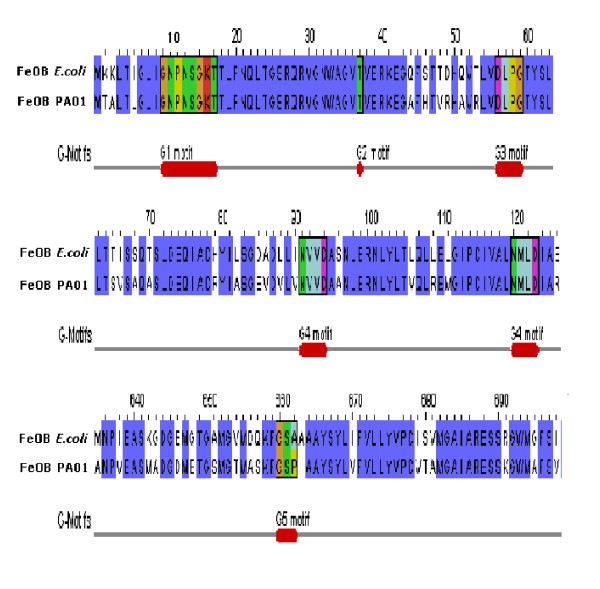
Three types of iron transporters (ABC, FeOB and OFeT) were found in the Pseudomonas species with the restriction of FeOB transporter (PA4358) to Pae and OFeT transporter (PFL_3255) to Pfl (Figure 3). Based on the previous reports of GTPase motifs in FeoBEco [24] highly conserved G1-G5 GTPase motifs were present in FeoBPae (Figure 3). In OFeTPfl two REXXE motifs (10REGIE14, 129REGLE133) were identified basing on the characterised prototype YcdNEco [25]. No iron transporters were reported for pathogenic Borrelia burgdorferi and Treponema pallidum while we reported substantial number of iron transporters for the pathogenic Pae and Psy (six and four respectively).
Figure 3.
Multiple sequence alignment of FeoB transporters from Escherichia coli and Pseudomonas aeruginosa PA01 depicting conserved GTPase signature motifs (G1–G5).
In Ppu and Psy there are high affinity nickel uptake systems (Nik) of ABC family whose PBP's (PP_3342 and PSPTO_3088) belong to nickel/peptide/opine ABC transporter family. Two IMP's were identified for each of the two Nik systems, in NikPpu (PP_3343, PP_3344) and NikPsy (PSPTO_3089, PSPTO_3090). They displayed a characteristic transmembrane topology of five (PSPTO_3089, PP_3344) and six TMD's (PSPTO_3090, PP_3343).
Chromate transporters of Chr family mediating chromate resistance are present in all the three Pseudomonas species (PP_2556, PA4289, PFL_3149) except Psy. PbrT members conferring lead resistance are also present in all the four Pseudomonas species (PA5248, PFL_5990, PP_0180). As an exception Psy possess two PbrT systems, PbrT1 and PbrT2 (PSPTO_0141, PSPTO_3596). PbrT2Psy did not show the conserved motifs found in other lead transporters of this group. Uniquely there is one ABC family member for Pb in Ppu (PP_5165) with no existing sequence homology to PbrTPpu.
The best-characterized heavy metal resistant strains R.metallidurans have eight HME-RND proteins on chromosome. In comparison, Ppu has six RND chromosomal determinants (PP_0043, PP_2410, PP_3302, PP_5173, PP_1517, PP_5387), Pfl and Pae have two determinants each (PFL_2558, PFL_5218 PA0158, PA2520) while no RND determinant was identified in Psy. Pfl Pf-5 has a maximum of four CDF transporters (PFL_2508, PFL_5419, PFL_0604, PFL_5222) while they are three in Pae PA01 (PA0397, PA1297, PA3963) and two in Ppu KT2440 (PP_0026, PP_4774). However Psy DC3000 has only one distinct ortholog (PSPTO_4790) in this family.
Analysis of ABC super family members from the above inventory unravelled a distinct transmembrane topology for their integral membrane proteins (IMP's) that varied with the metal ion specificity of their associated PBP's. Accordingly, five/six TMD's were observed for nickel transporters, twelve for iron transporters, seven for zinc, eight for manganese and five for molybdenum
Multiplicity of paralogs in metal transporter families
Pseudomonas species has multiple paralogs for most of the metal ion transporters that are briefed out in Additional table 5. Some of the significant features of paralog distribution are highlighted in this section.
Sodium ion transporters constitute the bulk of secondary transporters and has maximum number of paralogs distributed in different families (Table 4).
Table 4.
Distribution of Sodium transporters in P.putida KT2440, P.aeruginosa PA01, P.fluorescens Pf-5 and P.syringae DC3000.
| Family | P.putida KT2440 | P.aeruginosa PA01 | P.fluorescens Pf-5 | P.syringae DC3000 |
| AGCS | 1 | 3 | 1 | 1 |
| BASS | 2 | 3 | 2 | 1 |
| CaCA | 1 | - | 1 | 1 |
| CPA1 | 4 | 5 | 5 | 3 |
| CPA2 | 1 | 1 | 2 | 1 |
| CPA3 | - | 1 | - | - |
| DAACS | 2 | 1 | 3 | - |
| DASS | 1 | 1 | 1 | - |
| ESS | 1 | 1 | 1 | - |
| MSS | - | 2 | 2 | - |
| NhaA | 2 | - | 1 | 2 |
| NhaB | 1 | 1 | 1 | - |
| NSS | - | - | 1 | 1 |
| PNAS | 1 | 2 | 2 | 1 |
| SSS | 5 | 2 | 4 | 3 |
| Total | 22 | 23 | 27 | 14 |
A multitude of K+ transporters are present in Pseudomonas species among which Kdp system (in Pfl), VIC super family (in Pae and Psy), CPA2 family (in Ppu, Pae, Pfl, Psy) and Trk family (in Ppu) possessed paralogs.
All the four Pseudomonas species were found to possess paralogs for PBP's of ABC Zn2+ and Mn2+ transporters. Psy alone has paralogs for PBP of ABC Mo2+ transporter.
Ppu has two paralogs for CitMHS family, which are presumed to facilitate transport of Citrate-Mn2+ complex while in Pfl this family has three paralogs with specificity for Mg2+/citrate.
All the four Pseudomonas species have paralogs for MIT family and P-type ATPases. Except for Psy all the three species possessed paralogs for RND and CDF families.
Psy and Pfl have paralogs for ILT family, mediating lead transport alone in the former while both ferrous iron and lead transport are mediated in the later.
In Ppu ArsB family has two paralogs that are part of separate arsRBCH operons while in Pfl out of the two paralogs (PFL_2185, PFL_3288) only former is located within an arsRBCH operon.
Horizontal Gene Transfer: a source of gene innovation in metal transportomes
In view of the mosaic nature of genomes of the Pseudomonas spp we identified metal transporters with GI associated features from our inventory (see Additional files 6, 7, 8, 9). In our analysis we identified three distinct categories of transporters: those with anomalous composition (%G+C) alone, anomalous composition plus other GI associated features, normal %G+C plus GI associated features (Additional files 6, 7, 8 and 9). We identified thirty-two transporters (45.71%) in Ppu possessing highly significant GI features while they were twenty-six (48.10%) in Psy, nineteen (26%) in Pfl and seventeen (26%) in Pae.
Families with strong GI features that were present uniquely in a species were: CPA3 family (sodium ion homeostasis), ABC family (Cu transporter), P-type ATPase (Cu transporter) in Pae, NhaB family, NiCoT family in Ppu, NSS, MScS and CitMHS families in Psy. ABC transporter for Molybdenum was found to have GI features in the three Pseudomonas except for Pfl. Strong GI features were observed only for certain paralogs of metal transporter families indicating that paralog expansion could have occurred through HGT (Additional 10). These results show how different metal transporter families vary in their propensity for HGT among the four spp.
Conclusion
In this article we performed a dataset cataloging for 262 metal transporters from four representative Pseudomonas species. This is the first comprehensive genomic comparison of metal transporters, providing potentially important insights into the fundamental molecular aspects and novel facets of Pseudomonas metal transportomes.
Our comparative inventory identified and analyzed novel metal transporters belonging to the following families: NiCoT (PP_2968, PSPTO_4280), Ca P-type ATPase (PA1429), Cu P-type ATPases (PP_4261, PA1549), ZIP (PP_1836, PP_0947, PFL_4718, PFL_0910, and PSPTO_2053) and MgtC (PP_3244, PP_3541, PA4635, PFL_4077, PFL_2871).
Psy possessing least number of metal transporters showed maximum percentage (48%) of transporters with strong GI features. Our data is substantiated by the previous observations where majority of Psy ORF's shared features with horizontally transferred genes [26]. Ppu has 45% of its transporters possessing strong GI features. On the contrary, Pfl and Pae have only 26% of their metal transporters exhibiting GI features.
This comparative inventory can therefore provide a window for Pseudomonas community in mining large and heterogeneous data sets obtained from metagenome projects to identify new biologically relevant patterns of metal transporters resembling those in this study. A combinatorial approach of transcriptomics and functional genomics will aid in deducing the functions of these diverse metal transporters and assembling a complete picture of metal homeostasis.
Abbreviations
Ppu/PP/Ppu: P.putida KT2440; Pae/PA/Pae: P.aeruginosa PA01; Pfl/PFL/Pfl: P.fluorescens Pf-5; Psy/PSPTO/Psy: P.syringae DC3000; TMD's: Transmembrane domains; aa: Amino acid residues; bp: Base pairs; T.C.No: Transporter classification number; Me2+ transporter: Hypothetical metal transporter; MFS: Major facilitator super family; IMP's: Integral membrane proteins; HGT: Horizontal gene transfer.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AH performed the complete insilico analysis and wrote the draft of the manuscript. AR contributed to critical evaluation of the work and improvement of the manuscript organization. PMM conceived the study, guided the complete work and corrected the manuscript.
Supplementary Material
Catalogue of P.putida KT2440 metal transporters. Description: This catalogue was compiled for the entire complement of alkali/alkaline earth metal transporters (Na+, K+, Ca2+, Mg2+), transition metal transporters (Zn2+, Mn2+ Cu2+, Ag+, Mo2+, Fe2+, Fe3+, Ni2+, Co2+) and heavy metal transporters (Cr3+, As3+, Pb2+, Cd2+) from P.putida KT2440. Information provided in this catalogue includes the class/family/subfamily, Transporter classification number (T.C.No), TIGR locus, protein name, genomic location, orientation, length of the protein, Transmembrane domains (TMD’s), substrate and the predicted role of all the metal transporters.
Catalogue of P.aeruginosa PA01 metal transporters. Description: This catalogue was compiled for the entire complement of alkali/alkaline earth metal transporters (Na+, K+, Ca2+, Mg2+), transition metal transporters (Zn2+, Mn2+ Cu2+, Ag+, Mo2+, Fe2+, Fe3+, Ni2+, Co2+) and heavy metal transporters (Cr3+, As3+, Pb2+, Cd2+) from P.aeruginosa PA01. Information provided in this catalogue includes the class/family/subfamily, Transporter classification number (T.C.No), TIGR locus, protein name, genomic location, orientation, length of the protein, Transmembrane domains (TMD’s), substrate and the predicted role of all the metal transporters.
Catalogue of P.fluorescens Pf-5 metal transporters. Description: This catalogue was compiled for the entire complement of alkali/alkaline earth metal transporters (Na+, K+, Ca2+, Mg2+), transition metal transporters (Zn2+, Mn2+ Cu2+, Ag+, Mo2+, Fe2+, Fe3+, Ni2+, Co2+) and heavy metal transporters (Cr3+, As3+, Pb2+, Cd2+) from P.fluorescens Pf-5. Information provided in this catalogue includes the class/family/subfamily, Transporter classification number (T.C.No), TIGR locus, protein name, genomic location, orientation, length of the protein, Transmembrane domains (TMD’s), substrate and the predicted role of all the metal transporters.
Catalogue of P.syringae DC3000 metal transporters. Description: This catalogue was compiled for the entire complement of alkali/alkaline earth metal transporters (Na+, K+, Ca2+, Mg2+), transition metal transporters (Zn2+, Mn2+ Cu2+, Ag+, Mo2+, Fe2+, Fe3+, Ni2+, Co2+) and heavy metal transporters (Cr3+, As3+, Pb2+, Cd2+) from P.syringae DC3000. Information provided in this catalogue includes the class/family/subfamily, Transporter classification number (T.C.No), TIGR locus, protein name, genomic location, orientation, length of the protein, Transmembrane domains (TMD’s), substrate and the predicted role of all the metal transporters.
Sequence Cluster Analysis for detection of paralogs from four Pseudomonas spp. Description: This data provides evidence for the paralogous nature of metal transporters from various metal transporter families.
Metal transporters exhibiting GI associated features from P.putida KT2440. Description: This table lists out the metal transporters with GI associated features from P.putida KT2440 and provides evidence for the metal transporters with strong and weak GI features.
Metal transporters exhibiting GI associated features from P.aeruginosa PA01. Description: This table lists out the metal transporters with GI associated features from P.aeruginosa PA01 and provides evidence for the metal transporters with strong and weak GI features
Metal transporters exhibiting GI associated features from P.fluorescens Pf-5. Description: This table lists out the metal transporters with GI associated features from P.fluorescens Pf-5 and provides evidence for the metal transporters with strong and weak GI features.
Metal transporters exhibiting GI associated features from P.syringae DC3000. Description: This table lists out the metal transporters with GI associated features from P.syringae DC3000 and provides evidence for the metal transporters with strong and weak GI features.
Role of HGT in paralogous expansion and acquisition of novel transporter families in Pseudomonas. Description: This table demarcates the role of HGT in paralogous expansion and acquisition of novel transporter families in each of the four Pseudomonas species.
Acknowledgments
Acknowledgements
AH was funded by grant from CSIR [9/132/663-03], New Delhi. This work was also supported by Indo-French Project grant no: 3709-1 [20(A)/PMM/IFP/BCD-07].
Contributor Information
Adhikarla Haritha, Email: adhikarla.haritha@gmail.com.
Agnes Rodrigue, Email: agnes.rodrigue@insa-lyon.fr.
Pamarthi Maruthi Mohan, Email: maruthipm@osmania.ac.in.
References
- Wood JM. Evolutionary aspects of metal ion transporters through cell membranes. In: Helmut Sigel, editor. Metal ions in biological systems. Vol. 18. New York: CRC Press; 1984. pp. 224–228. [Google Scholar]
- Canovas D, Cases I, de Lorenzo V. Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ Microbiol. 2003;5:1242–1256. doi: 10.1111/j.1462-2920.2003.00463.x. [DOI] [PubMed] [Google Scholar]
- Hu N, Zhao B. Key genes involved in heavy-metal resistance in Pseudomonas putida CD2. FEMS Microbiology Letters. 2007;267:17–22. doi: 10.1111/j.1574-6968.2006.00505.x. [DOI] [PubMed] [Google Scholar]
- Weinel C, Nelson KE, Tummler B. Global features of the Pseudomonas putida KT2440 genome sequence. Environ Microbiol. 2002;4:809–818. doi: 10.1046/j.1462-2920.2002.00331.x. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinel C, Paulsen IT, Dodson RJ, Hilbert H, Martins dos Santos VA, Fouts DE, Gill SR, Pop M, Hoolmes M, Brinkac L, Beanan M, DeBoy RT, Daugherty S, Kolonay J, Madupu R, Nelson W, White O, Peterson J, Khouri H, Hance I, Chris Lee P, Holtzapple E, Scanlan D, Tran K, Moazzez A, Utterback T, Rizzo M, Lee K, Kosack D, Moestl D, Wedler H, Lauber J, Stjepandic D, Hoheisel J, Straetz M, Heim S, Kiewitz C, Eisen JA, Timmis KN, Dusterhoft A, Tummler B, Fraser CM. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Press CM, Ravel J, Kobayashi DY, Myers GS, Mavrodi DV, DeBoy RT, Seshadri R, Ren Q, Madupu R, Dodson RJ, Durkin AS, Brinkac LM, Daugherty SC, Sullivan SA, Rosovitz MJ, Gwinn ML, Zhou L, Schneider DJ, Cartinhour SW, Nelson WC, Weidman J, Watkins K, Tran K, Khouri H, Pierson EA, Pierson LS, 3rd, Thomashow LS, Loper JE. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat Biotechnol. 2005;23:873–878. doi: 10.1038/nbt1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westhrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK-S, Wu Z, Paulsen IT, Relzer J, Saier MH, Hancock REW, Lory S, Olson MV. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Daugherty S, Brinkac L, Beanan MJ, Haft DH, Nelson WC, Davidsen T, Zafar N, Zhou L, Liu J, Yuan Q, Khouri H, Fedorova N, Tran B, Russell D, Berry K, Utterback T, Van Aken SE, Feldblyum TV, D'Ascenzo M, Wen-Ling Deng, Ramos AR, Alfano JR, Cartinhour S, Chatterjee AK, Delaney TP, Lazarowitz SG, Martin GB, Schneider DJ, Xiaoyan Tang, Bender CL, White O, Fraser CM, Collmer A. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv tomato DC3000. Proc Natl Acad Sci USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TIGR-CMR http://cmr.tigr.org/
- Ren Q, Kang KH, Paulsen IT. Transport DB: A Relational database of cellular membrane transport systems. Nucleic Acids Research. 2004;32:D284–D288. doi: 10.1093/nar/gkh016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLC bio http://www.clcbio.com/
- Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucleic Acids Res. 2004:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;3:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- PEDANT genome database http://pedant.gsf.de/
- Hsiao W, Wan I, Jones SJ, Brinkman FS. IslandPath: Aiding detection of genomic islands in prokaryotes. Bioinformatics. 2003;19:418–420. doi: 10.1093/bioinformatics/btg004. [DOI] [PubMed] [Google Scholar]
- Ambudkar SV, Lynn AR, Maloney PC, Rosen BP. Reconstitution of ATP dependent calcium transport from Streptococci. J Biol Chem. 1986;261:15596–15600. [PubMed] [Google Scholar]
- Grass G, Wong MD, Rosen BP, Smith RL, Rensing C. ZupT Is a Zn(II) uptake system in Escherichia coli. J Bacteriol. 2002;184:864–866. doi: 10.1128/JB.184.3.864-866.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigue A, Effantin G, Mandrand-Berthelot MA. Identification of rcnA (yohM), a Nickel and Cobalt Resistance Gene in Escherichia coli. J Bacteriol. 2005;187:2912–2916. doi: 10.1128/JB.187.8.2912-2916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cécile R, Eric A, Christine F, Annie H, Lena T, Potard B, Béatrice A. Dual role of the MgtC virulence factor in host and non-host environments. Molecular Microbiology. 2007;63:605–622. doi: 10.1111/j.1365-2958.2006.05542.x. [DOI] [PubMed] [Google Scholar]
- Holloway P, McCormick W, Watson RJ, Chan YK. Identification and analysis of the dissimilatory nitrous oxide reduction genes, nosRZDFY, of Rhizobium meliloti. J Bacteriol. 1996;178:1505–1514. doi: 10.1128/jb.178.6.1505-1514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey DM, Guffanti AA, Zemsky J, Pinner E, Karpel R, Padan E, Schuldiner S, Krulwich TA. Cloning and characterization of a putative Ca2+/H+ antiporter gene from Escherichia coli upon functional complementation of Na+/H+ antiporter-deficient strains by the over expressed gene. J Biol Chem. 1993;268:11296–11303. [PubMed] [Google Scholar]
- Saaf A, Baars L, von Heijne G. The internal repeats in the Na+/Ca2+ exchanger-related Escherichia coli protein YrbG have opposite membrane topologies. J Biol Chem. 2001;276:18905–18907. doi: 10.1074/jbc.M101716200. [DOI] [PubMed] [Google Scholar]
- Rensing C, Mitra B, Rosen BP. The zntA gene of Escherichia coli encodes a Zn (II)-translocating P-type ATPase. Proc Natl Acad Sci USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. The membrane protein FeoB contains an intramolecular G protein essential for Fe (II) uptake in bacteria. Proc Natl Acad Sci USA. 2002;99:16243–16248. doi: 10.1073/pnas.242338299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse C, Scherer J, Koch D, Otto M, Taudte N, Grass G. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Molecular Microbiology. 2006;62:120–131. doi: 10.1111/j.1365-2958.2006.05326.x. [DOI] [PubMed] [Google Scholar]
- Joardar V, Lindeberg M, Schneider DJ, Collmer A, Buell CR. Lineage-specific regions in Pseudomonas syringae pv tomato DC3000. Mol Plant Pathol. 2005;6:53–64. doi: 10.1111/j.1364-3703.2004.00263.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Catalogue of P.putida KT2440 metal transporters. Description: This catalogue was compiled for the entire complement of alkali/alkaline earth metal transporters (Na+, K+, Ca2+, Mg2+), transition metal transporters (Zn2+, Mn2+ Cu2+, Ag+, Mo2+, Fe2+, Fe3+, Ni2+, Co2+) and heavy metal transporters (Cr3+, As3+, Pb2+, Cd2+) from P.putida KT2440. Information provided in this catalogue includes the class/family/subfamily, Transporter classification number (T.C.No), TIGR locus, protein name, genomic location, orientation, length of the protein, Transmembrane domains (TMD’s), substrate and the predicted role of all the metal transporters.
Catalogue of P.aeruginosa PA01 metal transporters. Description: This catalogue was compiled for the entire complement of alkali/alkaline earth metal transporters (Na+, K+, Ca2+, Mg2+), transition metal transporters (Zn2+, Mn2+ Cu2+, Ag+, Mo2+, Fe2+, Fe3+, Ni2+, Co2+) and heavy metal transporters (Cr3+, As3+, Pb2+, Cd2+) from P.aeruginosa PA01. Information provided in this catalogue includes the class/family/subfamily, Transporter classification number (T.C.No), TIGR locus, protein name, genomic location, orientation, length of the protein, Transmembrane domains (TMD’s), substrate and the predicted role of all the metal transporters.
Catalogue of P.fluorescens Pf-5 metal transporters. Description: This catalogue was compiled for the entire complement of alkali/alkaline earth metal transporters (Na+, K+, Ca2+, Mg2+), transition metal transporters (Zn2+, Mn2+ Cu2+, Ag+, Mo2+, Fe2+, Fe3+, Ni2+, Co2+) and heavy metal transporters (Cr3+, As3+, Pb2+, Cd2+) from P.fluorescens Pf-5. Information provided in this catalogue includes the class/family/subfamily, Transporter classification number (T.C.No), TIGR locus, protein name, genomic location, orientation, length of the protein, Transmembrane domains (TMD’s), substrate and the predicted role of all the metal transporters.
Catalogue of P.syringae DC3000 metal transporters. Description: This catalogue was compiled for the entire complement of alkali/alkaline earth metal transporters (Na+, K+, Ca2+, Mg2+), transition metal transporters (Zn2+, Mn2+ Cu2+, Ag+, Mo2+, Fe2+, Fe3+, Ni2+, Co2+) and heavy metal transporters (Cr3+, As3+, Pb2+, Cd2+) from P.syringae DC3000. Information provided in this catalogue includes the class/family/subfamily, Transporter classification number (T.C.No), TIGR locus, protein name, genomic location, orientation, length of the protein, Transmembrane domains (TMD’s), substrate and the predicted role of all the metal transporters.
Sequence Cluster Analysis for detection of paralogs from four Pseudomonas spp. Description: This data provides evidence for the paralogous nature of metal transporters from various metal transporter families.
Metal transporters exhibiting GI associated features from P.putida KT2440. Description: This table lists out the metal transporters with GI associated features from P.putida KT2440 and provides evidence for the metal transporters with strong and weak GI features.
Metal transporters exhibiting GI associated features from P.aeruginosa PA01. Description: This table lists out the metal transporters with GI associated features from P.aeruginosa PA01 and provides evidence for the metal transporters with strong and weak GI features
Metal transporters exhibiting GI associated features from P.fluorescens Pf-5. Description: This table lists out the metal transporters with GI associated features from P.fluorescens Pf-5 and provides evidence for the metal transporters with strong and weak GI features.
Metal transporters exhibiting GI associated features from P.syringae DC3000. Description: This table lists out the metal transporters with GI associated features from P.syringae DC3000 and provides evidence for the metal transporters with strong and weak GI features.
Role of HGT in paralogous expansion and acquisition of novel transporter families in Pseudomonas. Description: This table demarcates the role of HGT in paralogous expansion and acquisition of novel transporter families in each of the four Pseudomonas species.